Abstract
Yeast display is an efficient technology for selection of antibodies and other proteins with high affinity and thermal stability. Here, we describe a method for affinity maturation of engineered antibody domains (eAds) using yeast display. EAd yeast libraries of relatively large size (~109) were generated and subjected to alternating rounds of magnetic-activated cell sorting (MACS), fluorescent-activated cell sorting (FACS), and random mutagenesis. The highest affinity clones from the final round of maturation were identified and analyzed. We discuss extensively each key step, and provide detailed protocols and helpful notes.
Keywords: Yeast display, Antibody domain, MACS, FACS
1. Introduction
Yeast display is emerging as an effective technology for isolating and engineering antibodies or proteins for therapeutics development and a variety of biomedical applications. In the yeast display system, the antibody or protein is displayed on the yeast surface by fusing to the yeast agglutinin protein Aga2p, which attaches to Aga1p through two disulfide bonds. Expression of the antibody/protein-Aga2p and Aga1p are under the control of galactose-inducible GAL1 promoter (1). One of the main advantages this technology offers is its eukaryotic system providing very sophisticated protein folding and chaperones machinery, which allows efficient and consistent display of variety of proteins. Recently, yeast surface display has been successfully used to engineer not only antibodies (2–5), but also a wide variety of proteins like fibronectin (6, 7), T cell receptors (TCRs) (8), natural killer cell receptors (9), and proteins of the major histocompatibility complex (MHC) (10) with dramatic improvements in stability and affinity. More importantly, a high-efficacy yeast electroporation protocol has been described (11) recently, which enables construction of yeast display library with large size up to 1010. This makes yeast display comparable to phage display system in terms of library size and thus further simplifies the initial antibody or protein isolation process.
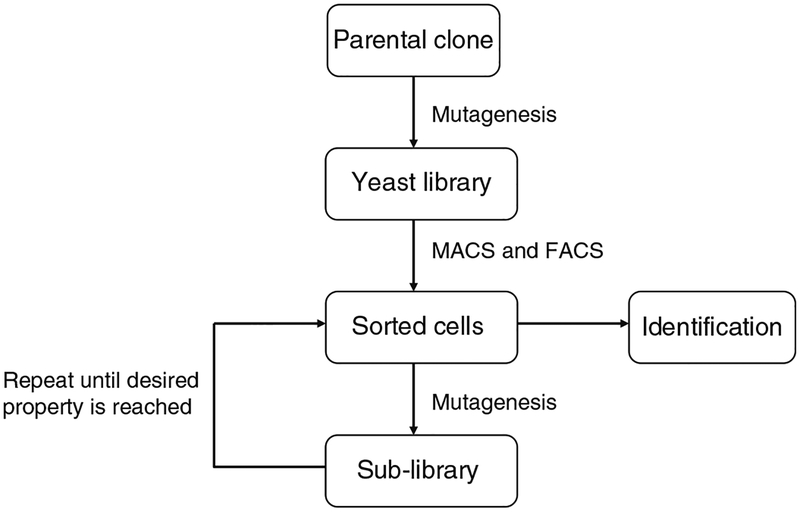
Isolated or engineered human immunoglobulin constant γ1 CH2 domains were proposed as novel scaffolds for construction of libraries containing diverse binders of small size but still conferring some effector functions (12–15). Here, we describe a protocol for displaying, engineering, and characterization of CH2 domains against antigens of interest using yeast display (Fig. 1). The yeast-displayed CH2 mutant library with size at 109 was generated by transformation of yeast with linearized vector and mutant DNA inserts derived from CH2 domain through error-prone PCR. One round of magnetic-activated cell sorting (MACS) against the biotinylated antigen was performed first to downsize the initial library. The library was then sorted several times by fluorescent-activated cell sorting (FACS) to enrich for specific binders. The enriched CH2 domain mutants were further mutated by error-prone PCR of the entire gene pool to yield a new sub-library. The process of sorting and mutagenesis was then cyclically repeated until the cell population reached the desired property. Clones with the highest affinity from the final round of maturation were identified and their sequences were analyzed.
Fig. 1.
Sketch of the protocol for selection and maturation of binders.
2. Materials
2.1. Error-Prone PCR
Bio-Rad MyCycler™ thermal cycler (Bio-Rad).
GeneMorph® II Random Mutagenesis Kit (Agilent).
8-Oxo-deoxyguanosine triphosphate and 2′-deoxy-p-nucleoside-5′-triphosphate.
Gel extraction kit (Qiagen).
Primers shown in Table 1.
Table 1.
PCR primers used
| ERRORF | 5′ CTTCAGTTTTGGCCCAGGCGGCC 3′ |
| ERRORR | 5′ ACCACTAGTTGGGCCGGCCTG 3′ |
| YDRDF | 5′ CTTCGCTGTTTTTCAATATTTTCTGTTATTGCTTCAGTTTTGGCCCAGGCGGCC 3′ |
| YDRDR | 5′ GAGCCGCCACCCTCAGAACCGCCACCCTCAGAGCCACCACTAGTTGGGCCGGCCTG 3′ |
2.2. Preparation of DNA Inserts and Vector for Library Construction
2.3. Preparation of Electroporation Competent Cells and Transformation of the Yeast Library
0.2-cm Gene Pulser/MicroPulser Cuvettes (Bio-Rad).
Gene Pulser (Bio-Rad).
Yeast strain EBY100.
YPD medium: dissolve 20 g dextrose, 20 g peptone, and 10 g yeast extract in deionized H2O to a volume of 1 L and sterilize by filtration. This medium can be stored for 2 months at 4°C.
Lithium acetate (LiAc) and dithiothreitol (DTT) solution: dissolve 6.6 g LiAc and 1.54 g DTT in 1 L H2O.
1 M sorbitol solution: dissolve 182 g sorbitol in 1 L H2O.
1 M CaCl2 solution: dissolve 111 g in 1 L H2O.
Electroporation buffer: add 1 ml of 1 M CaCl2 solution into 1 L of 1 M sorbital solution.
SDCAA medium: dissolve 20 g dextrose, 6.7 g Difco yeast nitrogen base w/o amino acid, 5 g Bacto casamino acids, 5.4 g Na2 HPO4, and 8.56 g NaH2PO4·H2O in H2O to a volume of 1 L and sterilize by filtration.
SDCAA plate: dissolve 5.4 g Na2HPO4 and 8.56 g NaH2PO4·H2O, 182 g sorbitol and 15 g agar in H2O to 0.9 L and autoclave. Dissolve 20 g dextrose, 6.7 g Difco yeast nitrogen base, and 5 g Bacto casamino acids in 100 ml H2O and sterilize by filtration. Cool until 50°C, add filter-sterilized solution, and pour plates.
2.4. Growth and Induction of Yeast Cells
SG/RCAA medium: dissolve 20 g galactose, 20 g raffinose, 1 g dextrose, 6.7 g Difco yeast nitrogen base w/o amino acid, 5 g Bacto casamino acids, 5.4 g Na2HPO4, and 8.56 g NaH2PO4·H2O in H2O to a volume of 1 L and sterilize by filtration (see Note 2).
2.5. MACS Selection
AutoMACS equipment (Miltenyi).
Streptavidin-conjugated microbeads (Miltenyi).
PBSA buffer: dissolve 8 g NaCl, 0.2 g KCl, 1.44 g Na2HPO4, 0.24 g KH2PO4, 1 g bovine serum albumin in 1 L of H2O, adjust the pH to 7.4 with HCl, and sterilize by filtration.
2.6. FACS Selection
FACSAria sorter (BD Biosciences).
Mouse anti-c-myc IgG (AbD Serotec).
Alexa Fluor 488 goat anti-mouse IgG (Invitrogen).
R-phycoerythrin-conjugated streptavidin (Invitrogen).
2.7. Affinity Determination
FACScalibur Flow cytometer (BD Biosciences).
3. Methods
3.1. Error-Prone PCR
Randomly mutate the entire CH2 DNA fragment using Stratagene GeneMorph® II Random Mutagenesis Kit. Oligonucleotides used for error-prone PCR are described in Table 1.
Perform PCR in a 50-μl reaction containing 1 × Mutazyme II reaction buffer, 0.5 μM each of primers ERRORF and ERRORR, 0.2 mM (each) dNTPs, 1 ng of plasmid template, 2 μM 8-oxo-deoxyguanosine triphosphate, 2 μM 2′-deoxy-p-nucleoside-5′-triphosphate, and 2.5 U of Mutazyme II DNA polymerase (see Note 3).
Error-prone PCR was performed using the following conditions: denature at 95°C for 2 min, 35 cycles of 95°C for 1 min, 60°C for 1 min, and 72°C for 1 min, followed by extension at 72°C for 10 min.
Purify the amplified products from agarose gel with the QIAquick Gel Extraction Kit and determine the DNA concentration through spectrophotometer measurement.
3.2. Preparation of DNA Inserts and Vector for Library Construction (see Note 4)
Oligonucleotides used for PCR amplification are described in Table 1.
Perform PCR in four 100-μl PCR reactions containing 1 × Accuprime PCR reaction mix, 1 μM of primers YDRDF and YDRDR, 120 ng of error-prone PCR product, and 2.5 U of Accuprime pfx DNA polymerase.
Denature at 95°C for 2 min, cycled 30 times at 95°C for 30 s, 60°C for 30 s, and 72°C for 30 s, and finally extended at 72°C for 2 min.
Purify all amplified products from agarose gel with the QIAquick Gel Extraction Kit and determine the DNA concentration through spectrophotometer measurement.
Digest the pYD7 vector with SfiI restriction enzyme and purify it with gel extraction kit following the instructions from manufacturer.
Combine 12 μg of mutagenized insert DNA and 4 μg of digested vector and concentrate the volume to 20 μl using a vacuum concentrator.
3.3. Preparation of Electroporation Competent Cells and Transformation of the Yeast Library
Inoculate EBY100 yeast colony into 5 ml YPD medium and grow overnight at 30°C.
Inoculate the overnight culture into 50 ml fresh YPD medium and grow cells at 30°C until OD at 600 nm reach 1.6 (about 4–5 h).
Once cells have reached an absorbance of about 1.6 at 600 nm, pellet cells at 2,500 × g for 3 min at 4°C and wash with 25 ml cold water twice. Wash cell once with 25 ml cold electroporation buffer.
Resuspend cells in 10 ml of LiAc/DTT solution and incubate at 30°C with shaking at 250 rpm for 30 min.
Pellet cells at 2,500 × g for 3 min at 4°C and washed once with 25 ml of electroporation buffer.
Resuspend cells in electroporation buffer to reach 0.4 ml volume. Mix concentrated DNA with resuspended cells and keep cells on ice.
Aliquot 0.4 ml of resuspended cell-DNA mixture per prechilled electroporation cuvette. Keep electroporation cuvettes on ice until pulsed.
Load cuvette into gene pulser and electroporate at 2.5 kV and 25 μF. Immediately add 1 ml of the warm (30°C) mixture of 1 M sorbitol: YPD medium (1:1) to the cuvette. Typical time constants for electroporation range from about 3 ms to 4 ms without greatly affecting transformation efficiency.
Transfer cells from pulsed cuvettes to a 50-ml Falcon tube. Wash each cuvette with an additional 1 ml of the mixture of 1 M sorbitol: YPD medium to recover the remaining cells from the cuvettes.
Make up the volume of cells to 10 ml by the mixture of 1 M sorbitol: YPD medium and incubate it at 30°C with shaking at 250 rpm for 1 h.
Pellet cells at 2,500 × g for 3 min and remove supernatant. Resuspend cells in 500 ml SDCAA medium. Incubate at 30°C with 250 rpm shaking for 24–48 h. Plate serial dilutions of the transformed cells on SDCAA plates to determine transformation efficiency (see Note 5).
Passage the library at least once before use to reduce the number of untransformed cells. Store the library at 4°C or −80°C (see Note 6).
3.4. Growth and Induction of Yeast Cells
3.4.1. Libraries
Thaw frozen aliquots of yeast library at room temperature and grow overnight (about 20 h) in 0.5–1 L of SDCAA medium in an incubator shaker set to 30°C with 250 rpm (see Note 7).
Pellet at least 5 × 109 cells from a freshly passaged library culture at 2,500 × g for 3 min and resuspend cells in SG/RCAA medium to an absorbance of about 0.5–1 at 600 nm.
Induce the library in SG/RCAA medium at 20°C with shaking at 250 rpm for 16–18 h.
3.4.2. Individual Clones
Inoculate single colony into 1–2 ml of SDCAA medium and grow at 30°C with shaking at 250 rpm overnight.
Resuspend cell pellet in S/GRCAA to an absorbance of about 0.5–1 at 600 nm and incubate at 20°C with 250 rpm for 16–18 h.
Wash cells once in wash buffer before staining for flow cytometry, or store the cells by placing the tubes at 4°C for up to 1 month.
3.5. MACS Selection (see Note 8)
Pellet 5 × 109 freshly induced yeast cells at 2,500 × g for 5 min and aspirate the supernatant. To wash, resuspend cells in 50 ml PBSA buffer, repellet cells, and discard supernatant.
To label yeast, resuspend cells in 10 ml PBSA buffer. Add biotinylated antigen to a final concentration of 100 nM and mix by gentle inversion (see Note 9).
Incubate cell suspension at room temperature with gentle rotation on a tube rotator for 1 h, followed by 10 min on ice.
Pellet cells at 2,500 × g for 3 min and wash once with 25 ml PBSA buffer.
Resuspend pellet in 5 ml PBSA buffer, add 100 μl streptavidin-conjugated microbeads to the suspension and mix by gentle inversion.
Incubate on ice for 10 min with gentle mixing by inversion every 2 min.
Add 20 ml PBSA buffer to the suspension and gently mix by inversion.
Turn on autoMACS and run the “Clean” program. For detailed instructions on using the autoMACS system, refer to the user’s manual.
Under the “pos2” port of the instrument, place a 15-ml tube filled with 2 ml SDCAA medium to collect eluted cells. Under the “neg1” port, place an empty 50-ml tube to collect the flow-through. Place the cell suspension under the intake port and choose separation protocol “Possel_s” to begin the separation.
Pellet eluted cells at 2,500 × g for 3 min to get rid of the sodium azide in the mixture and resuspend cells in fresh 10 ml SDCAA medium (see Note 10).
Propagate eluted cells overnight at 30°C in a shaker with speed set at 250 rpm, followed by induction in SG/RCAA medium at 20°C before subsequent sorting by FACS.
3.6. FACS Selection
Induce 5 × 107 cells from MACS sorted pool at 20°C with shaking speed set at 250 rpm for 16–18 h.
Pellet 5 × 107 induced cells at 2,500 × g for 3 min.
Label yeast cells with mouse anti-c-myc IgG (2 μg/ml) and an appropriate concentration of biotinylated antigen in an appropriate final volume of PBSA buffer. Antigen concentrations are chosen based on the expected dissociation constant (KD) of the parental binder. The antigen incubation volume must be large enough to allow at least tenfold excess of antigen over binders displayed on yeasts (see Note 11).
Incubate at room temperature for 3 h or 4°C overnight with rotation (see Note 12).
After incubation, wash cells three times with PBSA buffer and then resuspend cells in 1 ml PBSA buffer.
Stain cells with both 1:100 dilution of R-phycoerythrin-conjugated streptavidin and Alexa Fluor 488 conjugated goat anti-mouse IgG antibody at 4°C for 30 min.
Wash the cells three times with 5 ml PBSA buffer again and then resuspend in PBSA buffer for flow cytometric sorting. Sorting gates are determined to select only the population with higher antigen-binding signals (see Note 13).
Grow collected cells overnight in SDCAA medium at 30°C and induce in 10 ml SG/RCAA at 20°C for 18 h for the next round of sorting.
Repeat steps 2–8 for additional rounds. Approximately 1 × 107 yeast cells were used for staining at a concentration of 0.1 × to 0.01 × the concentration at KD. The addition of anti-c-myc and the staining with secondary reagents is as described above (see Note 14).
Isolate yeast plasmids using Zymoprep yeast plasmid kit according to the manufacturer’s instructions and use them as templates of sub-library construction (see Note 15).
Spread cells from the final round on SDCAA plates to identify single clones; affinities of monoclonal yeast display antibody can be measured and compared by FACS-based analysis.
3.7. Affinity Determination
Inoculate a 2-ml SDCAA culture with a clone of interest and grow overnight at 30°C.
Inoculate a 2-ml SG/RCAA culture with 5 × 106 cells and induce at 20°C for at least 18 h.
Set up nine tubes containing 1 × 105 induced cells. Pellet at 2,500 × g for 3 min and wash once using 200 μl PBSA buffer.
Choose nine different concentrations of biotinylated antigens around the equilibrium dissociation constant. Incubation volumes and number of yeast stained are chosen to keep the number of antigen molecules in tenfold excess above the number of binder (see Note 16). An example experimental setup is in Table 2.
To each tube, add the appropriate volumes of buffer, cells, and antigen.
Place tubes for 3 h at room temperature. This allows ample time for the binding reaction to reach equilibrium (see Note 17).
Pellet cells at 2,500 × g for 3 min, aspirate supernatant, and wash cells with 500 μl PBSA buffer.
Add 50 μl PBSA buffer with streptavidin-phycoerythrin (1:100 dilution) to each tube. Resuspend the cells and mix by pipetting.
Incubate on ice for 30 min, shielding from light.
Pellet cells at 2,500 × g for 3 min, aspirate supernatant, and wash cells with 500 μl PBSA buffer.
Analyze cells of each tube using a flow cytometry.
Plot the total mean fluorescence intensity (MFI) from the phycoerythrin channel against the concentration of antigen and use a nonlinear least squares to fit the curve (such as found in GraphPad) and determine the KD using the following equation: ϒ = Bmax × X/(KD + X) + Bmin, where ϒ = MFI at given antigen concentration, X = antigen concentration, Bmax = MFI at saturation, and Bmin = MFI of no antigen control.
Table 2.
Example setup for KD determination
| Tube | Concentration of Ag | Molecules of Ag | Molecules of Aba | Volume |
|---|---|---|---|---|
| 1 | 2 μM | 6 × 1013 | 5 × 109 | 50 μl |
| 2 | 1 μM | 3 × 1013 | 5 × 109 | 50 μl |
| 3 | 500 nM | 3 × 1013 | 5 × 109 | 100 μl |
| 4 | 250 nM | 1.5 × 1013 | 5 × 109 | 100 μl |
| 5 | 100 nM | 6 × 1012 | 5 × 109 | 100 μl |
| 6 | 50 nM | 3 × 1012 | 5 × 109 | 100 μl |
| 7 | 25 nM | 1.5 × 1012 | 5 × 109 | 100 μl |
| 8 | 12.5 nM | 7.5 × 1011 | 5 × 109 | 500 μl |
| 9 | 0 | 0 | 5 × 109 | 100 μl |
Ag antigen, Ab antibody
1 × 105 cells are used. Assuming 5 × 104 Abs per cell
4. Notes
pYD7 was modified from vector pCTCON2 (6). Two SfiI restriction sites before and after the insert match the cloning sites of phage display vector pComb3X were introduced into pYD7, which allows identified antibody genes shuttling among different vector for display on yeast and soluble expression in bacteria. In addition, protein of interest for display was put at the N-terminus of Aga2p instead of the C-terminus as it is in pCTCON2.
Raffinose and low concentration of dextrose in the SGCAA medium increase the efficiency of protein display on yeast cell surface.
Two nucleotide analogues (8-oxo-deoxyguanosine triphosphate and 2′-deoxy-p-nucleoside-5′-triphosphate) were used in the error-prone PCR reaction mixture to further broaden the mutation diversity during the error-prone PCR process. The amount of the template and the number of cycles used in the PCR can be adjusted according to desired mutation rate, decreasing the template concentration or increasing the PCR cycle number will increase the mutation rate. For determining the mutation frequency, the mutagenized genes can be ligated into sequencing vector, e.g., TA cloning vector, for sequencing.
In order to obtain large amounts of DNA insert and increase the efficacy of gap-repairing process in vivo, mutated DNA repertoire derived from error-prone PCR must be re-amplified using primers YDRDF and YDRDR under normal PCR conditions.
Typically, 1–4 × 108 transformants could be obtained from each transformation. The number of electroporation reaction is determined based on the desired size of the yeast library.
Grown library culture can be stored at 4°C for about 1 month. Frozen aliquots with 10% glycerol can be made for long-term storage at −80°C freezer or liquid nitrogen. Cells should be freshly passaged before induction.
The number of cells thawed from the frozen stock should be at least tenfold of the initial library size to reduce the probability of losing the diversity of the library.
The sorting capacity of MACS can reach to 5 × 109 yeast cells per sorting, which allows elimination of yeast cells that do not express antibodies, and quickly downsizes the initial library size and makes it compatible for subsequent FACS-based sorting. All of these steps except antigen incubation should use ice-cold buffers.
Antigen concentrations for initial sorting with MACS vary with the antigen of interest. 100 nM is generally sufficient to enrich antibody binders.
Remove buffer from sorted cells due to the presence of sodium azide in MACS running buffer before amplifying the cells in SDCAA medium. To avoid bacterial contamination after magnetic sorting, grow yeast cultures in SDCAA medium with pen–strep antibiotics.
Selections are generally performed after allowing the reaction to reach equilibrium. The volume for incubation of yeast is chosen to keep antigens at least a tenfold excess of the number of antibodies. For example, 5 × 107 cells are incubated in 1 ml labeling volume. Assuming 5 × 104 Ab per yeast cell, the Ab concentration in the sample is calculated as following: (5 × 104 Ab per cell) × (5 × 107 cells in 1 ml) = 4.15 nM. Therefore, the lowest antigen concentration is 41.5 nM.
Incubation times are chosen to approach equilibrium of the reaction. The time constant is defined as: t = −ln (1 – θ)/(kon × C + koff), where t is time, θ is equilibrium, kon is association rate constant, koff is dissociation rate constant, and C is antigen concentration (2). In general, 3 h at room temperature is sufficient for antigen concentrations that are in nanomolar range. Alternatively, incubate the antigen with cells overnight at 4°C.
Typically, sorting gate should be set to sort out the brightest0.1–0.3% of antigen-binding and c-myc positive clones.
The sorting at 1/10 antigen concentration at KD of the parental clone can differentiate affinity improved mutants from the parental clone and avoid the presence of the parental clone in the sort gate window.
Copies of plasmids are very low in yeast cells. Amount of the yeast cell culture used for the miniprep need to be optimized to ensure good-quality plasmid when used as template for subsequent error-prone PCR, using too many cells may result in even lower yield and quality.
If the KD is unknown, use several concentrations across a wide range and then focus the concentration range around KD in a subsequent experiment.
For the KD in nanomolar range, 3 h is usually sufficient. For the higher affinity binders, longer incubation times are required, see Note 12.
Acknowledgments
We thank Professor Dane Wittrup for providing the yeast display vector pCTCON2 and yeast strain EBY100 and members of our group for helpful discussions. This project was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research and by the Gates Foundation (DSD).
References
- 1.Chao G, Lau WL, Hackel BJ, Sazinsky SL, Lippow SM, Wittrup KD (2006) Isolating and engineering human antibodies using yeast surface display. Nat Protoc 1:755–768 [DOI] [PubMed] [Google Scholar]
- 2.Garcia-Rodriguez C, Levy R, Arndt JW, Forsyth CM, Razai A, Lou J, Geren I, Stevens RC, Marks JD (2007) Molecular evolution of antibody cross-reactivity for two subtypes of type A botulinum neurotoxin. Nat Biotechnol 25:107–116 [DOI] [PubMed] [Google Scholar]
- 3.Walker LM, Bowley DR, Burton DR (2009) Efficient recovery of high-affinity antibodies from a single-chain Fab yeast display library. J Mol Biol 389:365–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Z, Kim GB, Woo JH, Liu YY, Mathias A, Stavrou S, Neville DM Jr (2007) Improvement of a recombinant anti-monkey anti-CD3 diphtheria toxin based immunotoxin by yeast display affinity maturation of the scFv. Bioconjug Chem 18:947–955 [DOI] [PubMed] [Google Scholar]
- 5.Zhao Q, Feng Y, Zhu Z, Dimitrov DS (2011) Human monoclonal antibody fragments binding to insulin-like growth factors 1 and 2 with picomolar affinity. Mol Cancer Ther 10:1677–1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hackel BJ, Kapila A, Wittrup KD (2008) Picomolar affinity fibronectin domains engineered utilizing loop length diversity, recursive mutagenesis, and loop shuffling. J Mol Biol 381:1238–1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lipovsek D (2011) Adnectins: engineered target-binding protein therapeutics. Protein Eng Des Sel 24:3–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Richman SA, Healan SJ, Weber KS, Donermeyer DL, Dossett ML, Greenberg PD, Allen PM, Kranz DM (2006) Development of a novel strategy for engineering high-affinity proteins by yeast display. Protein Eng Des Sel 19:255–264 [DOI] [PubMed] [Google Scholar]
- 9.Dam J, Guan R, Natarajan K, Dimasi N, Chlewicki LK, Kranz DM, Schuck P, Margulies DH, Mariuzza RA (2003) Variable MHC class I engagement by Ly49 natural killer cell receptors demonstrated by the crystal structure of Ly49C bound to H-2K(b). Nat Immunol 4:1213–1222 [DOI] [PubMed] [Google Scholar]
- 10.Esteban O, Zhao H (2004) Directed evolution of soluble single-chain human class II MHC molecules. J Mol Biol 340:81–95 [DOI] [PubMed] [Google Scholar]
- 11.Benatuil L, Perez JM, Belk J, Hsieh CM (2010) An improved yeast transformation method for the generation of very large human antibody libraries. Protein Eng Des Sel 23:155–159 [DOI] [PubMed] [Google Scholar]
- 12.Dimitrov DS (2009) Engineered CH2 domains (nanoantibodies). MAbs 1:26–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiao X, Feng Y, Vu BK, Ishima R, Dimitrov DS (2009) A large library based on a novel (CH2) scaffold: identi fi cation of HIV-1 inhibitors. Biochem Biophys Res Commun 387:387–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gong R, Vu BK, Feng Y, Prieto DA, Dyba MA, Walsh JD, Prabakaran P, Veenstra TD, Tarasov SG, Ishima R, Dimitrov DS (2009) Engineered human antibody constant domains with increased stability. J Biol Chem 284:14203–14210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gong R, Wang Y, Feng Y, Zhao Q, Dimitrov DS (2011) Shortened engineered human antibody CH2 domains: increased stability and binding to the human neonatal receptor. J Biol Chem 286:27288–27293 [DOI] [PMC free article] [PubMed] [Google Scholar]