Abstract
Smaller recombinant antibody fragments are now emerging as alternatives of conventional antibodies. Especially, immunoglobulin (Ig) constant CH2 domain and engineered CH2 with improved stability are promising as scaffolds for selection of specific binders to various antigens. We constructed a yeast display library based on an engineered human IgG1 CH2 scaffold with diversified loop regions. A group of CH2 binders were isolated from this yeast display library by panning against nucleolin, which is a tumor- associated antigen involved in cell proliferation, tumor cell growth and angiogenesis. Out of 20 mutants, we selected 3 clones exhibiting relatively high affinities to nucleolin on yeasts. However, recombinant CH2 mutants aggregated when they were expressed. To find the mechanism of the aggregation, we employed computational prediction approaches through structural homology models of CH2 binders. The analysis of potential aggregation prone regions (APRs) and solvent accessible surface areas (ASAs) indicated two hydrophobic residues, Val264 and Leu309, in the b-sheet, in which replacement of both charged residues led to significant decrease of the protein aggregation. The newly identified CH2 binders could be improved to use as candidate therapeutics or research reagents in the future.
Keywords: Antibody domain, CH2, Nucleolin, Yeast display, Monoclonal antibody, Aggregation prone region
1. Introduction
Monoclonal antibodies (mAbs) have been highly successful as targeted therapeutic agents for diagnosing and treating diseases [1,2]. Despite their widespread application, it is widely accepted that full-size antibodies exhibit poor penetration into solid diseased tissues (such as solid tumor tissues) as well as inability to enter occluded regions of protein molecules due to their relatively large size (150 kDa) [3—5]. Protein scaffolds with relatively small size have distinct advantages because of greater and more rapid tissue accumulation and the ability to potentially target epitopes not accessible by full-size antibodies. Antibody-based small scaffolds include Fabs (60 kDa), single chain Fv fragments (scFvs) (20—30 kDa) and domain antibodies (dAbs) (12—15 kDa). Meanwhile, non-antibody-based scaffolds, such as fibronectin type III subunit, tenascin type III subunit, and ankyrin repeat proteins, are under development as novel therapeutics [6—8]. Through protein engineering technologies, these small scaffolds are being optimized for enhanced affinity, stability and expression levels [9]. Redesigned antibody-like scaffolds offer a novel class of therapeutics with unique properties that are not possessed by full-size antibodies. They are promising as imaging reagents and candidate therapeutics.
Human Ig constant γ1 CH2 domain is an independent folding domain that comprises part of the Fc portion of Ig. Previous studies have proved that CH2 can be expressed and refolded as a soluble, monomeric domain at a high level [10]. CH2 domains can be engineered so as to retain some of the effectors functions that are not possessed by other smaller antibody-like fragments. For example, isolated CH2 domains can confer full or partial functions of Fc receptor-binding and relatively long half-lives [11]. It was also proved that engineered CH2 exhibits remarkable thermostability with almost 20 °C higher Tm than wild-type CH2 [10,12]. Importantly, CH2-based reagents are likely to be well tolerated in concentrations needed for achieving the long half-life, according to the fact that CH2 is a portion of all Ig subtypes, which exist in human whole blood with high concentrations [11]. Engineered CH2 domains are under development as scaffolds for construction of libraries containing diverse binders that provide novel candidate therapeutics [10,13]. In recent years, several binders against the HIV-1 Env gp41 and gp120 have been successfully isolated from CH2-based phage libraries with diversified loop regions [14,15].
Yeast display is a powerful technology for isolating and engineering antibodies or proteins to increase their affinity, specificity and stability. Over past years, yeast display has emerged as an effective alternative to phage display technology and has particular advantages in the selection and affinity maturation of mAbs [16—20]. In some cases, because the protein folding and secretory of yeast is similar to mammalian cells, it finds out more or different mAb or protein candidates rather than phage display [21,22]. Here, we constructed a large-size yeast display library based on a stabilized CH2 scaffold with CDR-grafting or mutagenesis of loops. Some binders were isolated from this yeast library by panning against a cancer-specific antigen nucleolin that is not selected using CH2 phage display libraries. In order to solve the aggregation issues, through potential aggregation prone region (APR) analysis, we demonstrated that two facial hydrophobic residues (V264 and L309) located in the β-sheet likely play an important role in the stability and aggregation resistance. Replacement of both residues to charged residue remarkably reduced the aggregation forming of nucleolin binders. Our results could be applicable to design of candidate CH2 therapeutics.
2. Materials and methods
2.1. Construction of CH2 yeast-displayed library
The sequences of CH2 variants were amplified by two primers RDlinker1F (5’ GATATATCCATGGCCCAGGCGGCC 3’) and ERRORR (5’ ACCACTAGTTGGGCCGGCCTG 3’) by PCR. Reaction products were purified and concentrated by an ultrafilter (Millipore) in pure water. The pYD7 vector was linearized with restriction zyme Sfi I (NEB). Multiple aliquots of DNA inserts and digested vector DNA were mixed in water. The mixture was then transformed into yeast cells EBY100 using electroporation transformation method as previously described [18,23].
2.2. Yeast library selection
Selection of yeast library was performed as described previously with some modification [18,23]. Briefly, yeast cells (5 × 1010) were incubated with biotinylated nucleolin protein (the final concentration 100 nM) for 1 h at room temperature in 20 ml PBSM buffer (2 mM EDTA and 0.5% BSA in PBS), followed 10 min on ice. After washing once with 100 ml PBSM buffer, the yeast cell pellet was resuspended in 10 ml PBSM buffer. Approximate 0.5 ml of Strep- tavidin microbeads (Miltenyi) were added to the yeast and incubated for 10 min at 4 °C with rotation. The yeast cells were made up to 100 ml with PBSM buffer and loaded onto auto magnetic- activated cell sorting machine (Miltenyi) in 25-mL aliquots. The yeasts recovered from magnetic beads are then sorted several times by fluorescent-activated cell sorting (FACS). Approximately 1 —5 × 108 yeast cells were incubated with biotinylated nucleolin and a 1:100 dilution of mouse anti-c-myc antibody (Life Technologies). After incubation, both 1:100 dilution of PE-streptavidin (Life Technologies) and AlexaFluor 488-labelled goat anti-mouse antibody (Life Technologies) were added for FACS selection. Four rounds of selections were performed.
2.3. Affinity measurement of CH2 domains on yeast
The equilibrium dissociation constant for a clone was determined using the method as previously described [18,24].
2.4. Flow cytometric analysis
Individual yeast clones were grown, induced and analyzed by flow cytometry (BD Calibur). The assays were performed as described previously [23].
2.5. ELISA
Purified CH2 domain proteins in the concentration of 2 mg/ml were coated on 96-well ELISA plates overnight at 4 ° C. Serially concentrations of biotinylated proteins with triplicate samples were added into wells and incubated for 1 h. Bound proteins were detected with HRP conjugated streptavidin (1:1000; Sigma). The o- Phenylenediamine substrate (Sigma) was added and the reaction was read at 450 nm.
2.6. Computational aggregation predication
Homology based molecular models of CH2 variants were built using the crystal structure of the CH2 domain (PDB entry: 3DJ9) in MOE-model [25]. The resulting models for three CH2 variants were aligned. Potential APRs were identified in the amino acid sequences of CH2 variants by using a combination of prediction tools, Tango [26], AGGRESCAN [27], and AMYLPRED2 [28]. Distribution of surface charged and hydrophobic residues were also analyzed by inhouse programs. The structural models were used to compute solvent accessible surface area (ASA) for all the amino acids. The ASA values were used to identify potential residues for disruption of aggregation prone regions. Structural modeling and analysis were made with PyMOL software.
2.7. Site-directed mutagenesis
PCR was performed using Stratagene’s QuikChange® Site- Directed Mutagenesis Kit according to the instructions of the manufacturer.
2.8. Expression and purification of CH2 domains
Proteins were expressed and purified as described previously [29]. Escherichia coli strain HB2151 was used for protein expression. Purified proteins was dialyzed against PBS and filtered through a 0.2 mm low protein binding filter (Pall).
2.9. Size exclusion chromatography
The purified CH2 proteins were loaded into the Superdex 75 HR 10/300 column (GE Healthcare) running on AKTA chromatography system (GE Healthcare) to assess possible oligomer formation.
3. Results
3.1. CH2 library construction and selection
A semi-synthetic CH2-based library has been made by grafting CDRs or introducing mutagenesis onto loop regions of engineered CH2 scaffold and cloned into the phage vector by Gong et al. [5,10].
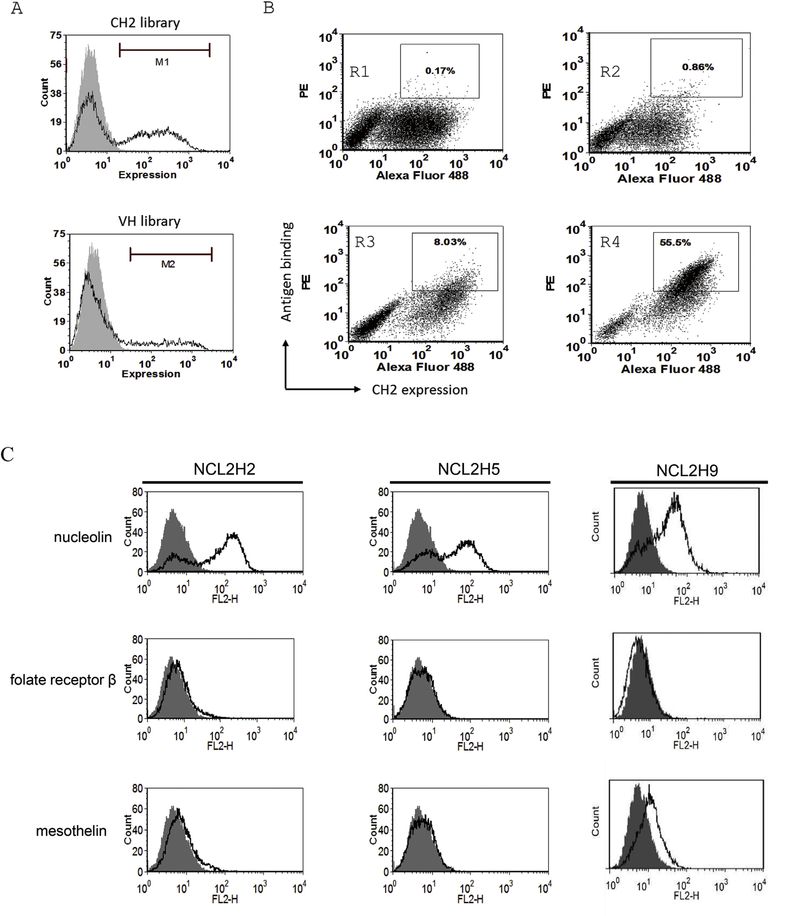
The CH2 phage library was estimated to have size about 109 sequences. In this study, the CH2 library was cloned into a pYD7 vector and displayed on the surface of yeast cells. After calculation, the yeast library contains 1 × 1010 transformants over 10-fold larger than that of the original library size. Thus, flow cytometric analysis verified more than 40% of the transformed cells displaying CH2 inserts (Fig. 1a). In contrast, we cloned a previously reported recombinant VH domain library on yeasts [4], which only showed 20% expression of library inserts on yeasts (Fig. 1a). After sequencing, most of clones showed the correct grafting or mutagenesis in loop 1 (V264-K274), loop2 (Q295-R301) and loop3 (K322-P331) regions (Supplementary Fig. S1).
Fig. 1.
Expression and selection of the yeast display CH2 domain library. (A) The grafted CH2 yeast library indicated 43% of expressed cell. In contrast, the grafted VH yeast library indicated ~20% of expressed cell. (B) The sorting rounds of CH2 yeast library against nucleolin. The double-positive yeast cells were selected from sort gates. Four figures shows sorting of yeast library against nucleolin at rounds 1,2,3, and 4. (C) Binding of yeast-expressed CH2 binders to antigens. Three antigens nucleolin, folate receptor b, and mesothelin, were used to detect CH2 binders.
MACS step is used to deplete yeast cells that do not express CH2 inserts or bind weakly to nucleolin antigen from the library. Sequentially, the library was subjected to 4 rounds of FACS sorting for nucleolin. After sorting, a population of clones was enriched to bind nucleolin (Fig. 1b). After sequencing and antigen-binding analysis, we identified three clones (NCL2H2, NCL2H5, NCL2H9) exhibiting relatively high-binding affinities to nucleolin. The antigen specificity of three clones was further confirmed against several tumor-associated antigens by flow cytometry (Fig. 1c). As the result, fluorescence intensity shifts were observed to nucleolin in all three clones, but not folate receptor beta and mesothelin.
3.2. Binding affinities of CH2 binders to nucleolin
Yeast-displayed method enables to quantitatively measure binding affinity or dissociation kinetics of antibodies or proteins by antigen titration analysis of flow cytometry [30]. Binding parameters determined on yeast are typically close to that measured using soluble proteins in Surface Plasmon Resonance (SPR) assays [18]. After analysis (Supplementary Fig. S2), affinity titrations indicated that all three clones had equilibrium dissociation constants (Kd) for antigen nucleolin: 84 nM (clone NCL2H2), 116 nM (clone NCL2H5), and 141 nM (clone NCLH9).
3.3. Prediction of aggregation prone regions
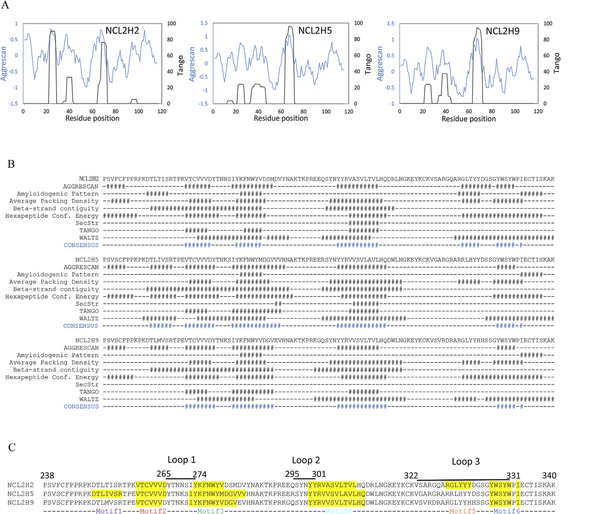
A combination of computational methods, including at least eight different protein aggregation prediction tools, was used to avoid potential biases. This led to the generation of relative objective and accurate results by comparison of the different algorithms. Firstly, we performed TANGO and AGGRESCAN analysis on all the three anti-nucleolin CH2 variants. The TANGO and AGGRESCAN profiles of NCL2H2, NCL2H5 and NCL2H9 are shown in Fig. 2a. Three major APRs (motif 1 —3) obtained from this analysis were well conserved in all three sequences. Relatively high percent of hydrophobic amino acids was presented in all of the three motifs. Among them, the second motif (259-VTCVVV-264) in NCL2H2 and the third motif (302-VV/ASVLT/AVL-309) in all three sequences showed the relatively high aggregation propensities. The APR prediction was further analyzed by a consensus algorithm for the prediction of ‘aggregation-prone’ peptides on the basis of a strong agreement among eight different existing methods. The consensus of APRs obtained from the calculation are highlighted in the alignment of eight algorithms (Fig. 2b). Three aggregation-prone motifs were consistent with the combination analysis of TANGO and AGGRESCAN. The consensus method predicted an additional APR (101-GYWSYW-106) in all the three sequences. Besides, NCL2H2 contained an APR (GLYYYD) as well as NCL2H5 contained another predicted APR (TLIVSR) in loop regions. These two APRs were not found in the other two sequences. All motifs were mapped on to the sequence alignment of three CH2 variants in blue, green, and red in Fig. 2c.
Fig. 2.
Prediction of potential aggregation prone regions (APRs) in three CH2 variants. (A) Overlap of TANGO and AGGRESCAN profiles for three CH2 binders. X-axis shows sequence number. Left y-axis and blue curve are for AGGRESCAN score. Right y-axis and black curve are for TANGO aggregation percentage. (B) The consensus histograms of APRs analyzed by eight different predicting methods. (C) Indications of potential APR motifs in sequence alignment of three CH2 variants.
3.4. Identification of the aggregation ‘hot spots’ on APRs
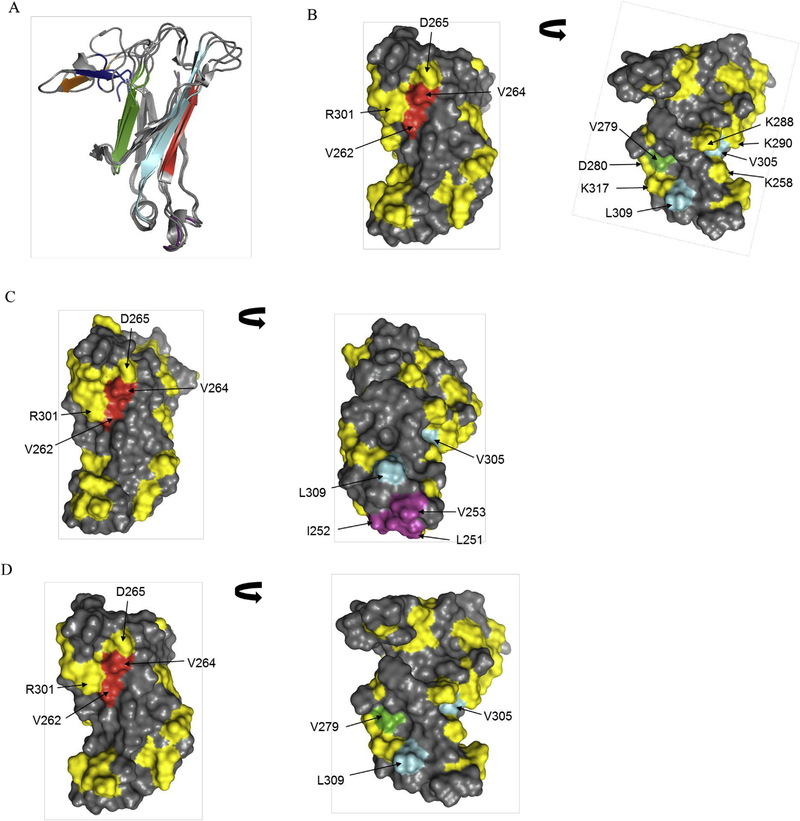
Molecular models of three CH2 variants were built based on the available crystal structure of human CH2. The structural alignment models of 3 molecules yielded RMSD value of 3.19 A. The sequence identity varied between 83 and 88%. Fig. 3a shows structural alignment of three molecules along with APRs marked in difference color. We observed that motif 1 of NCL2H5 located in the helix region (purple color). In the three APR motifs (2—4), certain hydrophobic residues point out at the surface of strands B, C, and E as shown in red, green and cyan, respectively. Two additional APR motifs (5 in orange, 6 in blue) locate at the loop 3 regions of CH2s that are mainly involved in antigen binding regions. Hydrophobic residues, especially b-branched aliphatic residues could contribute to potential risk of misfolding and aggregation [31].
Fig. 3.
Mapping of potential APR residues on three-dimensional structures of three CH2 variants. (A) Structural alignment of three CH2 variants (NCL2H2, NCL2H5 and NCL2H9). The APR motifs are shown in purple, red, green, cyan, orange and blue. (BeD) Surface diagrams for structures of NCL2H2 (B), NCL2H5 (C), and NCL2H9 (D) illustrating hydrophobic APR residues in red, green, cyan and purple. The distribution of charged residues is shown in yellow.
The predicted APRs were further analyzed in structural context by computing hydrophobicity, amphipathicity, pi and K-D hydrophobic moment. The values are shown in supplementary Table S1. Overall values computed from sequences alone appeared to be quite similar. Structural models of the three candidates were used to compute ASA values for each amino acid in the protein. The ASA values of the amino acid residues located within predicted APRs were examined to locate potential surface exposed residues suitable for mutagenesis. The goal of this exercise was identify potential sites for APR disruption. Table 1 lists the hydrophobic and aromatic residues with ASA >50% (that is these residues are practically floating in water) that are part of the APRs. We found a hydrophobic residue Leu309 with high scores indicated in all three CH2 variants.
Table 1.
Hydrophobic or Aromatic residues with ASA >50% in the APRs found in the three sequences
| Peptide sequences | NCL2H2 | NCL2H5 | NCL2H5 |
|---|---|---|---|
| 259-VTCVVV-264 | - | - | - |
| 274-KFNWYV-279 | - | I272, V282, V284 | - |
| 272-IYKFNWYMDGVVV-284 | |||
| 302-VASVLTVL-309 | L309 | L309 | L309 |
| 302-VASVLAVL-309 | |||
| 302-VVSVLTVL-309 | |||
| 8-GLYYYD-14 (loop 3)a | G8, L9, Y10, Y11, Y12 | - | - |
| 18-GYWSYW-6 (loop 3)a | Y19, W20, W23 | Y19, Y22, W23 | Y19, Y22, W23 |
Numerals indicate residue positions in loop 3.
The hydrophobic residues in APR motifs, not including motifs of loops, were mapped on the structures of CH2 domains. As shown in Fig. 3b—d, in all three variants, two hydrophobic residues (V262 and V264) of motif 2 are partially exposed on the surface of the b-sheet, and are completely protected by positively charged residues, R265 and R301. The hydrophobic residue (V279) of motif 3 is surrounded by charged residues D280 and K317. In motif 4, one hydrophobic residue V305 is buried by K/E258, K288 and K290. However, it is clearly shown that another hydrophobic residue L309 in the strand E is protruded on the surface of all three CH2s. In addition, we found a few hydrophobic residues L251, I252 and V253 of motif 1 are exposed at the helical regions of NCL2H5.
In conclusion, two hydrophobic residues, V264 and L309, are likely to contribute to the aggregation according to the ASA and structure analysis.
3.5. Improving aggregation by targeted mutagenesis
Mutagenesis of hydrophobic residues at APRs was used to directly evaluate the involvement of these residues on aggregation. Two predicted residues, V264 and L309, were replaced with charged amino acid lysine in NCL2H2 and NCL2H9. Purified soluble proteins were analyzed by a size exclusion chromatography column. Ratios of monomer at total proteins were summarized in Table 2. The introduction of V264K and L309K onto NCL2H2 had largely improved the level of aggregation detected by size exclusion. The monomer portion was produced at 40% of H2VLmut compared to 7.5% of NCL2H2. We did not find the obvious improvement of aggregation in another clone NCL2H9. Our studies proved that the hydrophobic residues identified as the aggregation hot spots likely contributed to the aggregation prone of anti-nucleolin CH2 binders.
Table 2.
Portions of CH2 monomer in the total proteins.
| Clone name | Monomer (%) | Clone name | Monomer (%) | Mutation position |
|---|---|---|---|---|
| NCL2H2 | 7.5 | NCL2H9 | 60.5 | - |
| H2Vmut | 1.9 | H9Vmut | 1.2 | V264 |
| H2Lmut | 10 | H9Lmut | 48.4 | L309 |
| H2VLmut | 40.3 | H9VLmut | 16.6 | V264, L309 |
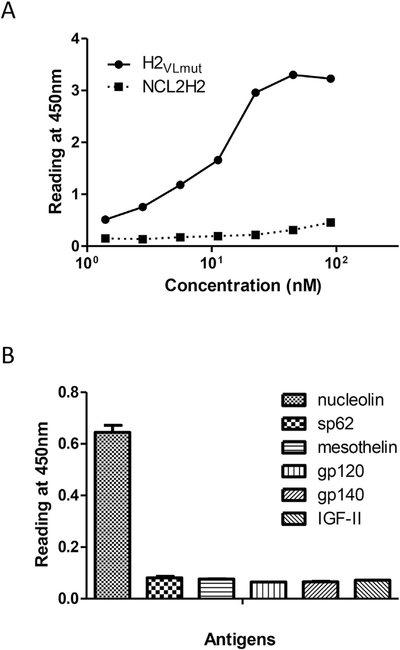
We analyzed antigen binding abilities of the improved mutants, H2VLmut and NCL2H2. These purified CH2 binders bound to nucle- olin as measured by ELISA. As improvement, H2VLmut showed stronger binding than the parental NCL2H2 in soluble protein formats (Fig. 4a). Further, the specificity of H2VLmut was also measured to bind to six irrelevant antigens in which only nucleolin was specifically bound (Fig. 4b).
Fig. 4.
Binding of NCL2H2 mutants to nucleolin with specificity. (A) Comparison of NCL2H2 and H2VLmut in ELISA. (B) Binding specificity of H2VLmut to nucleolin, meso- thelin, gp120, gp140, MPER peptide sp62, and IGF-II.
4. Discussion
Recently, human Ig CH2 domains have been extensively studied as novel scaffolds for the development of novel candidate therapeutics [5]. It has been shown that engineered human CH2 domain significantly increases its stability without affecting some other properties [10,13]. Previously from a semi-synthetic CH2-based phage library (size ~109), selection of binders against specific HIV-1 gp41 suggested that CH2 could be a promising scaffold for constructions of binder libraries [15].
The most frequent use of yeast display techniques facilitates the development of antibodies or proteins as potential therapeutic agents. Importantly, yeast display allows not only the isolation of many candidates that are not identified using phage display [22], but also the determination of binding affinity or dissociation kinetics [30]. In this work we explored the feasibility of engineered human CH2 scaffold for the development of binders against tumor- associated antigens. We cloned a semi-synthetic CH2-based phage library (comprises of 109 sequences) and displayed on the surface of yeast cells. The size of the yeast library was more than 5—10 fold than that of the original library size, which guaranteed to extensively cover the all diversity in original CH2 library. Indeed, FACS analysis proved that more than 40% of CH2s were displayed in the yeast library. The sufficient diversity enables to isolate CH2-based binders against certain antigens such as nucleolin, which was not selected from the phage CH2 library. The affinities of isolated CH2 binders specific for nucleolin range from 84 nM to 150 nM. The affinities of selected binders are modest but typical of those selected from CH2 scaffolds.
Protein aggregation is widely seen and often driven by the intrinsic instabilities [31]. Previous studies have shown that engineered CH2 domains have relatively high propensity coded in their sequences for aggregation [12]. We found that identified CH2 binders tend to aggregation, resulting in the exacerbation of solubility and binding. The findings of potential APR motifs are essential to minimize the aggregation propensity of proteins by removing the “hot spots” in APRs. Computational tools have been adopted to predict the potential propensity for protein aggregation formation. We analyzed potential aggregation regions by comparison of the different existing algorithms, which led to the generation of relative objective and accurate results. Most of the aggregation-prone motifs are especially rich in b-branched aliphatic residues, mainly including isoleucine, valine and leucine residues on the outer surfaces. Two of six APRs (259-VTCVVV-264 and 302-VV/ASVLT/AVL- 309) are suggested to be representative motifs with high frequency in commercial mAbs [31]. The further ASA analysis is used to identify potential sites for disruption of aggregation prone regions. The Leu309 in the motif (302-VV/ASVLT/AVL-309) of all three CH2 binders is ranked the top scores of hydrophobicity. The locations of APR motifs on three-dimensional structure were also addressed. Mapping of motifs reveals that charged residues largely protect some hydrophobic residues in APRs by covering over them. In addition, two hydrophobic residues, V264 and L309, are partly exposed to the surface of b-sheets. Altering of two hydrophobic residues in at least one of three CH2 binders led to the improved aggregation and the retained antigen binding. It is still an interesting challenge to further investigate the potential aggregation sites in other CH2 binders.
In summary, our studies shows the efficient yeast display selection and the aggregation resistance of antibody CH2 domains as novel candidate therapeutics. Our observations indicate that the prediction of sequence/structural motifs in engineered CH2 candidates is important to understand the mechanism of the aggregation. The protein aggregation improvement is useful and helpful for the development of CH2-based therapeutics.
Supplementary Material
Acknowledgements
We thank Dr. Sandeep Kumar for the assistance of protein prediction and analysis and Dr. Dane Wittrup for providing yeast display vector. This work was supported by the Guangdong Science and Technology Program (2016A050502034), Natural Science Foundation of Guangdong (Grand No. 2015A030313741), Shenzhen Peacock Innovation Plan Fund (Grand No. KQCX20140520154115029), Shenzhen Knowledge Innovation Program (Grand No. JCYJ20160422155108542, JCYJ20140901003939026), Macau Science and Technology Development Fund (FDCT 066/2015/A2), Intramural Research Program of University of Macau, Faculty of Health Sciences, and the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Footnotes
Appendix A. Supplementary data
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.bbrc.2017.02.058.
Transparency document
Transparency document related to this article can be found online at http://dx.doi.org/10.1016Zj.bbrc.2017.02.058.
References
- [1].Dimitrov DS, Therapeutic antibodies, vaccines and antibodyomes, MAbs 2 (2010) 347–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Chen Z, Wang L, Xu T, Wang Q, Kang L, Zhao Q, Generation of bispecific antibodies by Fc heterodimerization and their application, Curr. Pharm. Bio- technol. 17 (2016) 1324–1332. [DOI] [PubMed] [Google Scholar]
- [3].Jain RK, Physiological barriers to delivery of monoclonal antibodies and other macromolecules in tumors, Cancer Res. 50 (1990) 814s–819s. [PubMed] [Google Scholar]
- [4].Chen W, Zhu Z, Feng Y, Dimitrov DS, Human domain antibodies to conserved sterically restricted regions on gp120 as exceptionally potent cross-reactive HIV-1 neutralizers, Proc. Natl. Acad. Sci. U. S. A. 105 (2008) 17121–17126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Dimitrov DS, Engineered CH2 domains (nanoantibodies), MAbs 1 (2009) 26–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].De Laporte L, Rice JJ, Tortelli F, Hubbell JA, Tenascin C promiscuously binds growth factors via its fifth fibronectin type III-like domain, PLoS One 8 (2013) e62076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Bloom L, Calabro V, FN3: a new protein scaffold reaches the clinic, Drug Discov. Today 14 (2009) 949–955. [DOI] [PubMed] [Google Scholar]
- [8].Pluckthun A, Designed ankyrin repeat proteins (DARPins): binding proteins for research, diagnostics, and therapy, Annu. Rev. Pharmacol. Toxicol. 55 (2015) 489–511. [DOI] [PubMed] [Google Scholar]
- [9].Holliger P, Hudson PJ, Engineered antibody fragments and the rise of single domains, Nat. Biotechnol. 23 (2005) 1126–1136. [DOI] [PubMed] [Google Scholar]
- [10].Gong R, Vu BK, Feng Y, Prieto DA, Dyba MA, Walsh JD, Prabakaran P, Veenstra TD, Tarasov SG, Ishima R, Dimitrov DS, Engineered human antibody constant domains with increased stability, J. Biol. Chem. 284 (2009) 14203–14210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Gehlsen K, Gong R, Bramhill D, Wiersma D, Kirkpatrick S, Wang Y, Feng Y, Dimitrov DS, Pharmacokinetics of engineered human monomeric and dimeric CH2 domains, MAbs 4 (2012) 466–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Gong R, Wang Y, Ying T, Feng Y, Streaker E, Prabakaran P, Dimitrov DS, N- terminal truncation of an isolated human IgG1 CH2 domain significantly in¬creases its stability and aggregation resistance, Mol. Pharm. 10 (2013) 2642–2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Gong R, Wang Y, Feng Y, Zhao Q, Dimitrov DS, Shortened engineered hu¬man antibody CH2 domains: increased stability and binding to the human neonatal Fc receptor, J. Biol. Chem. 286 (2011) 27288–27293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Xiao X, Feng Y, Vu BK, Ishima R, Dimitrov DS, A large library based on a novel (CH2) scaffold: identification of HIV-1 inhibitors, Biochem. Biophys. Res. Commun. 387 (2009) 387–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Gong R, Wang Y, Ying T, Dimitrov DS, Bispecific engineered antibody domains (nanoantibodies) that interact noncompetitively with an HIV-1 neutralizing epitope and FcRn, PLoS One 7 (2012) e42288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Zhao Q, Ahmed M, Guo HF, Cheung IY, Cheung NK, Alteration of electrostatic surface potential enhances affinity and tumor killing properties of anti-ganglioside GD2 monoclonal antibody hu3F8, J. Biol. Chem. 290 (2015) 13017–13027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Zhao Q, Ahmed M, Tassev DV, Hasan A, Kuo TY, Guo HF, O’Reilly RJ, Cheung NK, Affinity maturation of T-cell receptor-like antibodies for Wilms tumor 1 peptide greatly enhances therapeutic potential, Leukemia 29 (2015) 2238–2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Zhao Q, Feng Y, Zhu Z, Dimitrov DS, Human monoclonal antibodyfragments binding to insulin-like growth factors I and II with picomolar affinity, Mol. Cancer Ther. 10 (2011) 1677–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Ahmed M, Cheng M, Zhao Q, Goldgur Y, Cheal SM, Guo HF, Larson SM, Cheung NK, Humanized affinity-matured monoclonal antibody 8H9 has potent antitumor activity and binds to FG loop of tumor antigen B7-H3, J. Biol. Chem. 290 (2015) 30018–30029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Li D, Liu J, Zhang L, Xu T, Chen J, Wang L, Zhao Q, N-terminal residues of an HIV-1 gp41 membrane-proximal external region antigen influence broadly neutralizing 2F5-like antibodies, Virol. Sin. 30 (2015) 449–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Boder ET, Wittrup KD, Yeast surface display for screening combinatorial polypeptide libraries, Nat. Biotechnol. 15 (1997) 553–557. [DOI] [PubMed] [Google Scholar]
- [22].Bowley DR, Labrijn AF, Zwick MB, Burton DR, Antigen selection from an HIV-1 immune antibody library displayed on yeast yields many novel antibodies compared to selection from the same library displayed on phage, Protein Eng. Des. Sel. 20 (2007) 81–90. [DOI] [PubMed] [Google Scholar]
- [23].Zhao Q, Zhu Z, Dimitrov DS, Yeast display of engineered antibody domains, Methods Mol. Biol. 899 (2012) 73–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Chao G, Lau WL, Hackel BJ, Sazinsky SL, Lippow SM, Wittrup KD, Isolating and engineering human antibodies using yeast surface display, Nat. Protoc. 1 (2006) 755–768. [DOI] [PubMed] [Google Scholar]
- [25].Almagro JC, Beavers MP, Hernandez-Guzman F, Maier J, Shaulsky J, Butenhof K, Labute P, Thorsteinson N, Kelly K, Teplyakov A, Luo J, Sweet R, Gilliland GL, Antibody modeling assessment, Proteins 79 (2011) 3050–3066. [DOI] [PubMed] [Google Scholar]
- [26].Fernandez-Escamilla AM, Rousseau F, Schymkowitz J, Serrano L, Prediction of sequence-dependent and mutational effects on the aggregation of peptides and proteins, Nat. Biotechnol. 22 (2004) 1302–1306. [DOI] [PubMed] [Google Scholar]
- [27].de Groot NS, Castillo V, Grana-Montes R, Ventura S, AGGRESCAN: method, application, and perspectives for drug design, Methods Mol. Biol. 819 (2012) 199–220. [DOI] [PubMed] [Google Scholar]
- [28].Tsolis AC, Papandreou NC, Iconomidou VA, Hamodrakas SJ, A consensus method for the prediction of ‘aggregation-prone’ peptides in globular proteins, PLoS One 8 (2013) e54175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Zhao Q, Chan YW, Lee SS, Cheung WT, One-step expression and purification of single-chain variable antibody fragment using an improved hex- ahistidine tag phagemid vector, Protein Expr. Purif. 68 (2009) 190–195. [DOI] [PubMed] [Google Scholar]
- [30].Boder ET, Raeeszadeh-Sarmazdeh M, Price JV, Engineering antibodies by yeast display, Arch. Biochem. Biophys. 526 (2012) 99–106. [DOI] [PubMed] [Google Scholar]
- [31].Wang X, Das TK, Singh SK, Kumar S, Potential aggregation prone regions in biotherapeutics: a survey of commercial monoclonal antibodies,, MAbs 1 (2009) 254–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.