Abstract
Free calcium ions are common second messengers in plant cells. The calcineurin B-like protein (CBL) is a special calcium sensor that plays an important role in plant growth and stress response. In this study, we obtained three CBL genes (GenBank accession nos. KX013374, KX013375, and KX013376) from sugarcane variety ROC22. The open reading frames of ScCBL genes ranged from 642 to 678 base pairs in length and encoded polypeptides from 213 to 225 amino acids in length. ScCBL2-1, ScCBL3-1, and ScCBL4 were all located in the plasma membrane and cytoplasm. ScCBL2-1 and ScCBL3-1 expression was up-regulated by treatment with salicylic acid (SA), methyl jasmonate (MeJA), hydrogen peroxide (H2O2), polyethylene glycol (PEG), sodium chloride (NaCl), or copper chloride (CuCl2). ScCBL4 expression was down-regulated in response to all of these stresses (abscisic acid (ABA), SA, MeJA, and NaCl) except for H2O2, calcium chloride (CaCl2), PEG, and CuCl2. Expression in Escherichia coli BL21 cells showed that ScCBLs can enhance tolerance to NaCl or copper stress. Overexpression of ScCBLs in Nicotiana benthamiana leaves promoted their resistance to infection with the tobacco pathogen Ralstonia solanacearum. The results from the present study facilitate further research regarding ScCBL genes, and in particular, their roles in the response to various stresses in sugarcane.
Subject terms: Abiotic, Biotic
Introduction
Calcium (Ca2+) is an important inorganic nutritive element and a ubiquitous second messenger1. Ca2+ not only plays a vital role in maintaining the stability of the cell wall, cell membrane, and membrane binding proteins, but also is widely involved in the regulation and control of plant growth and development, as well as response to external environmental stimuli2. In plants, intracellular Ca2+ sensors accurately recognize specific Ca2+ signatures that are generated in response to different external stimuli3. Under adverse conditions, cell signal transduction receptors on the plant cell membrane recognize the stimulus signal, and then activate the Ca2+ channel protein through phosphorylation, thereby leading to an instantaneous increased in the Ca2+ concentration in the cytoplasm, which produces the “Ca2+ signal”4. Ca2+ sensors in plants detect this stress-induced Ca2+ signal and deliver it to downstream effectors, activating a signal cascade reaction that regulates resistance and tolerance5. Ca2+ sensors can be divided into two main types according to their structural features. One type is the sensor responders, including calcium-dependent protein kinases and calmodulins, which have all the functions of Ca2+ sensor relay proteins, as well as the kinase activity6. The other type is the sensor relays, including calmodulin-like proteins and CBL, which do not have kinase activity. Sensor relays can specifically target downstream proteins to transfer the perceived Ca2+ signals in response to various environmental stimuli6.
CBL, a Ca2+ sensor relay protein that is expressed in a wide range of plants, can interact with a family of serine-threonine protein kinases known as CBL-interacting protein kinases (CIPKs)7. CBLs were initially identified in Arabidopsis thaliana and are closely related to both the regulatory B subunit of calcineurin and neuronal calcium sensors in animals8. CBLs have been found in many terrestrial plants, such as mosses, ferns, gymnosperms, monocots, and dicots9. CBLs are a multigene family. The A. thaliana, Oryza sativa, and Populus trichocarpa genomes are each predicted to contain approximately 10 CBL family members10–12. Eight CBL genes have been identified in Sorghum biocolor13,14. In addition, CBL genes have been investigated in Brassica napus15, Solanum melongena16, and other plant species17,18. CBL proteins contain a classical EF-hand helix-loop-helix motif with a 12-residue loop19. In EF-hand motifs, the Ca2+-binding sites are located at residues 1 (X), 3 (Y), 5 (Z), 7 (Y), 9 (X), and 12 (Z)10,19. Different CBL proteins have different degrees of variation in the EF-hand structure, but the number of EF-type regions and the distance between them is the same in all known CBL proteins11.
The function of CBL genes has been studied in A. thaliana, O. sativa, and other plant species. In A. thaliana, AtCBLs play a role in the response to multiple abiotic stresses20–22. For instance, AtCBL1 functions as a positive regulator in response to salt and drought but as a negative regulator in response to cold20. Abscisic acid (ABA) is a signaling molecule that plays a role in the plant response to aging and stress23. AtCBL9 is a common element in the ABA signaling and stress-induced ABA biosynthesis pathways21. Ten OsCBL genes in rice are expressed in various organs at the adult stage and have also been found to respond to different stress conditions [sodium chloride (NaCl), polyethylene glycol (PEG), and cold]24. In addition, OsCBL8 overexpressing transgenic rice seedlings showed more tolerance to salt stress than non-transgenic seedlings24. S. bicolor CBL genes are thought to regulate sodium carbonate stress-specific cellular adaptation responses and influence the plant growth and developmental patterns14. Analysis of CBL transcripts in Populus euphratica under abiotic stress suggested that seven CBL (PeCBL1, 2, 3, 4, 5, 9, and 10) members may play important roles in responding to specific external stimuli12.
Sugarcane (Saccharum spp.) is an economically attractive polyploid C4 grass that is used not only to produce approximately 60% of the world’s sugar but also to produce ethanol, a low-carbon-emission fuel25. To date, there have been few reports on CBL genes in sugarcane26–28. Zhang (2013) cloned five CBL genes (GenBank accession Nos. KC800815, KC800816, KC800817, KC800818, and KC800819) from Saccharum hybrid variety GT28 and found that CBL5 and CBL6 may play key roles in adaptation to low temperatures28. Using real-time quantitative polymerase chain reaction (qRT-PCR) analyses, Ling et al. (2018) found that SsCBL1 and SsCBL6 play important regulatory roles in response to a variety of stresses (low potassium, drought, and salt)26. Yeast two-hybrid assays showed that ScCIPK8 interacts with ScCBL1, ScCBL3, and ScCBL627. In the past 15 years, ROC22 is the most widely cultivated sugarcane in China due to its high yield and high sugar and good ratoon properties. Previous research found that ROC22 can well resist infection by Pokkah boeng disease29,30. Lan et al. (2014) found that ROC22 has better drought tolerance compared to five sugarcane varieties31. However, a systematic analysis of CBL genes in sugarcane variety ROC22 especially on the view of function differentiation, however, has not yet been reported.
In this study, we successfully cloned three sugarcane ScCBL genes by reverse transcription-PCR (RT-PCR) and subjected the cloned sequences to bioinformatics analysis. The expression patterns of these three ScCBL genes in different sugarcane tissues and under various exogenous stresses were investigated by qRT-PCR. In addition, we assessed the subcellular localization of these ScCBL proteins and analyzed their function by expression in Escherichia coli BL21 and transient expression in N. benthamiana. This study aims to provide useful information about the sequence characteristics of these three ScCBL proteins as well as their expression patterns in response to phytohormones and various stresses. This increased knowledge of ScCBL genes could be applied by sugarcane breeder to develop resistant variety.
Materials and Methods
Plant materials and treatments
The sugarcane variety ROC22 were provided by the Key Laboratory of Sugarcane Biology and Genetic Breeding, Ministry of Agriculture (Fuzhou, China).
We sampled sugarcane tissues as described by Wang32. Selecting three healthy and mature ROC22 sugarcane stalks with uniform growth rates from the field. The bud, stem pith, stem skin, meristem, and the youngest fully expanded viz + 1 leaf with a visible dewlap (the collar between the leaf blade and sheath) were sampled. These tissues were wrapped, frozen in liquid nitrogen, and stored at −80 °C until total RNA extraction. We performed the sampling after abiotic stress treatment as follows33: uniform four-month-old cultured ROC22 plantlets were transferred to water for one week and then treated with eight exogenous treatments, including 100 μM ABA, 5 mM salicylic acid (SA), 100 μM methyl jasmonate (MeJA), 50 μM calcium chloride (CaCl2), 10 μM hydrogen peroxide (H2O2), 25% PEG 8000, 250 mM NaCl, or 100 mM copper chloride (CuCl2), by root dipping at 28 °C with 16 h light and 8 h darkness34. Whole plantlets treated with CaCl2, SA, MeJA, or ABA were collected at 0 h, 3 h, 6 h, and 12 h. Plantlets treated with H2O2, PEG, or NaCl were collected at 0 h, 6 h, 12 h, and 24 h. Plantlets subjected to Cu stress were harvested at 0 h, 12 h, 24 h, and 48 h. Each treatment group contained three biological replicates, and plantlets were stored at −80 °C until extraction of total RNA.
Total RNA extraction and the first-strand cDNA synthesis
Total RNA were extracted from all samples with Trizol® Reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s specifications. RNA samples with an OD260/OD280 of between 1.8 and 2.0 were selected and treated with DNase I (Promega, Madison, WI, USA) to remove DNA contamination. First-strand cDNA was synthesized using a Prime-ScriptTM RT Reagent Kit (TaKaRa, Dalian, China) according to the manufacturer’s protocol, and then checked by 1% agarose gel electrophoresis.
Isolation of sugarcane ScCBL genes and gateway entry vector construction
The Z. mays sequence (GenBank Acc No. NM_001155706) which derived from ZmCBL3 (GenBank Acc No.EU962348.1) was used as a probe and the NCBI BlastN tool was applied to retrieve homologous EST sequences in the sugarcane genome. The BioEdit Contig Assembly Program (CAP) was employed to assemble one sugarcane CBL sequence (ScCBL2-1). The other two sugarcane CBL sequences (ScCBL3-1 and ScCBL4) were selected from our previous transcriptome data of sugarcane infected with Sugarcane Mosaic virus35. The specific primers were designed using Primer 5.0 and the NCBI primer designing tool (http://www.ncbi.nlm.nih.gov/tools/primer-blast/) (Table S1).
The RT-PCR reactions (Table S2) were designed according to the specifications for Ex Taq (TaKaRa, Dalian, China). Subsequently, the PCR products were detected by 1% agarose gel electrophoresis and purified, ligated into the pMD-19-T vector, transformed into E. coli DH5α and then sequenced (Shenggong, Shanghai, China).
The ScCBL ORFs were amplified from pMD19-T-ScCBLs with Gateway entry adapters attB1 and attB2 using the primers shown in Table S1. The PCR conditions are shown in Table S2. The PCR amplification products were gel-purified and transformed into the Gateway@ donor vector pDONR221 (Invitrogen) following the manufacturer’s instructions and using Gateway® BP ClonaseTM II Enzyme Mix (Invitrogen). The BP reaction mixtures were transformed into competent E. coli DH5α cells and sequenced (Sangon, Shanghai, China). Then the verified pDONR221-ScCBL plasmids were used to construct the expression vectors.
Sequence analysis of ScCBL genes
The ORF Finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html) was used to translate and analyze the three ScCBL genes. Molecular weight (MW), isoelectric points (pIs), and grand average of hydropathicity (GRAVY) of each protein sequence were analyzed by ExPASY (http://web.expasy.org/protparam/). The DNAMAN software was applied to conduct multiple alignment. The phylogenetic tree was constructed by the neighbor-joining bootstrap method (1,000 replicates) using the MEGA X program36.
qRT-PCR analysis
The 7500 qRT-PCR system (Applied Biosystems, San Francisco, CA, USA) was applied to detect and analyze the expression of ScCBL genes in different sugarcane tissues and under various exogenous stresses. The qRT-PCR primers (Table S1) were designed using Beacon Designer 8.12 software. Cullin (CUL)37 and clathrin adaptor complex (CAC)37 were employed as the internal controls (Table S1). The 20 μL qRT-PCR reaction contained 10 μL SYBR Green Master Mix, 0.8 μL each of the 10 μM primers, 1.0 μL cDNA templates (20 × diluted cDNA), and 7.4 μL double distilled water. Each qRT-PCR reaction was repeated three times, and the conditions were as follows: 50 °C for 2 min, 95 °C for 10 min, 40 cycles of 95 °C for 15 s, and 60 °C for 1 min. The 2-ΔΔCt method was used to analyze the qRT-PCR data38 and the statistical analysis was conducted by using Data Processing System v9.50 software (China). Data were expressed as the mean ± standard error (SE). Significance (p < 0.05) was calculated using one-way analysis of variance (ANOVA), followed by Duncan’s new multiple range test.
Subcellular localization assay
The pFAST-R05-ScCBL2–1-GFP, pFAST-R05-ScCBL3-1-GFP, and pFAST-R05-ScCBL4-GFP vectors were constructed by LR reaction using the LR ClonaseTM II Enzyme Mix (Invitrogen) according to the manufacturer’s instructions. The three recombinant ScCBL-GFP vectors were transformed into competent Agrobacterium tumefaciens GV3101 cells. The Agrobacterium-mediated transient expression in N. benthamiana leaves was performed according to the method described by Su et al.39. After infiltration for 48 h, the subcellular localization of the fusion protein were visualized by laser scanning confocal microscopy (Leica TCS SP5, Wetzlar, Germany).
Expression in E. coli BL21 (DE3) cells
Prokaryotic expression vectors were constructed based on pEZYHb from the pDONR221-ScCBL plasmids by LR reaction. The recombinant pEZYHb-ScCBL plasmids and the empty vector pEZYHb (control) were transformed into competent E. coli BL21 (DE3) cells for the prokaryotic expression experiments.
A spot assay was conducted to characterize the expression of ScCBLs in competent E. coli BL21 cells in response to NaCl and CuCl2 stresses. When the OD600 of E. coli BL21 cells containing pEZYHb-ScCBLs or pEZYHb (control) in LB medium (containing 80 μg/mL ampicillin) reached 0.6, 1.0 mM Isopropyl β-D-thiogalactoside (IPTG) was added, and the cells were cultured at 37 °C for another 12 h. The concentration of the cultures was adjusted to OD600 = 0.6, and the samples were then diluted 10−3 and 10−4 in LB medium40. Next, 10 μL from each of the 10−3 and 10−4 dilutions was spotted on LB agar plates. For the salt tolerance assay, we prepared LB media with 250 mM, 500 mM, or 750 mM of NaCl. For the heavy metal tolerance assay, we added 250 μM, 500 μM, or 750 μM of CuCl2 to the LB media36. All of the plates were incubated at 37 °C overnight and then photographed.
Transient assay for ScCBL genes in N. benthamiana leaves
To understand how ScCBL expression changes in response to pathogen infection and whether the plant hypersensitive reaction is also activated, pEarleyGate 203-ScCBLs overexpressing vectors were constructed using the Gateway cloning technique. Competent Agrobacterium GV3101 cells were transformed with recombinant pEarleyGate 203-ScCBL plasmids. The empty pEarleyGate 203 vector was transformed into Agrobacterium GV3101 cells for use as a control. The cells were then cultured in LB liquid medium (supplemented with 50 μg/mL kanamycin and 35 μg/mL rifampicin) overnight at 28 °C. After incubation, the cells were centrifuged and resuspended in MS liquid medium (containing 200 μM acetosyringone) at an OD600 of 0.8. After infiltration into the N. benthamiana leaves, the plants were cultured at 24 °C for 24 h (16 h light/8 h darkness)39. RT-PCR was exploited to detect whether ScCBL genes have been overexpressed in N. benthamiana, with the RNA of treated leaves and ScCBL genes specific primers (gScCBL2-1, gScCBL3-1, and gScCBL4, Table S1), and the NtEF1-α was treated as control. The treated N. benthamiana leaves were used for the transcriptional analysis of the eight tobacco immunity-associated marker genes (Table S1)41.
To analyze the inhibitory effect of ScCBL genes on pathogen infection, Ralstonia solanacearum was cultured to an OD600 of 0.8 in potato dextrose water (PDW) liquid medium at 28 °C. Then, N. benthamiana leaves that had been infiltrated with pEarleyGate 203-ScCBLs or pEarleyGate 203 for 24 h were infected with R. solanacearum. All of the treated plant materials were cultured at 24 °C (16 h light/8 h darkness) for 7 days and then photographed. DAB and trypan blue staining were utilized to analyze the Agrobacterium-infiltrated leaves as described by Liu et al.33.
Results
Identification of CBL genes in sugarcane
Three ScCBL genes were successfully amplified by RT-PCR from sugarcane variety ROC22. According to the homology with AtCBLs (Figure S1), three ScCBL genes were designated as ScCBL2-1, ScCBL3-1, and ScCBL4, respectively. Basic information about these three genes is shown in Table 1. The length of the ScCBLs open reading frames (ORFs) ranged from 642 to 678 base pairs, and they encoded polypeptides from 213 to 225 amino acids in length. The isoelectric points (pIs) of the polypeptides ranged from 4.77 to 4.82, and the grand average of hydropathicity (GRAVY) of each ScCBL was negative. The molecular weight (MW) of these ScCBLs ranged from 24.31 to 25.85 kDa.
Table 1.
Features of CBL genes in sugarcane.
| Gene | GenBank Acc No. | ORF Length (bp) | Peptide Length | MW (kDa) | PI | GRAVY |
|---|---|---|---|---|---|---|
| ScCBL2-1 | KX013374 | 678 | 225 | 25.79 | 4.82 | −0.208 |
| ScCBL3-1 | KX013375 | 678 | 225 | 25.85 | 4.8 | −0.234 |
| ScCBL4 | KX013376 | 642 | 213 | 24.31 | 4.77 | −0.275 |
Notes: MW, molecular weight; PI, isoelectric point; GRAVY, grand average of hydropathicity.
Phylogenetic analysis of ScCBLs
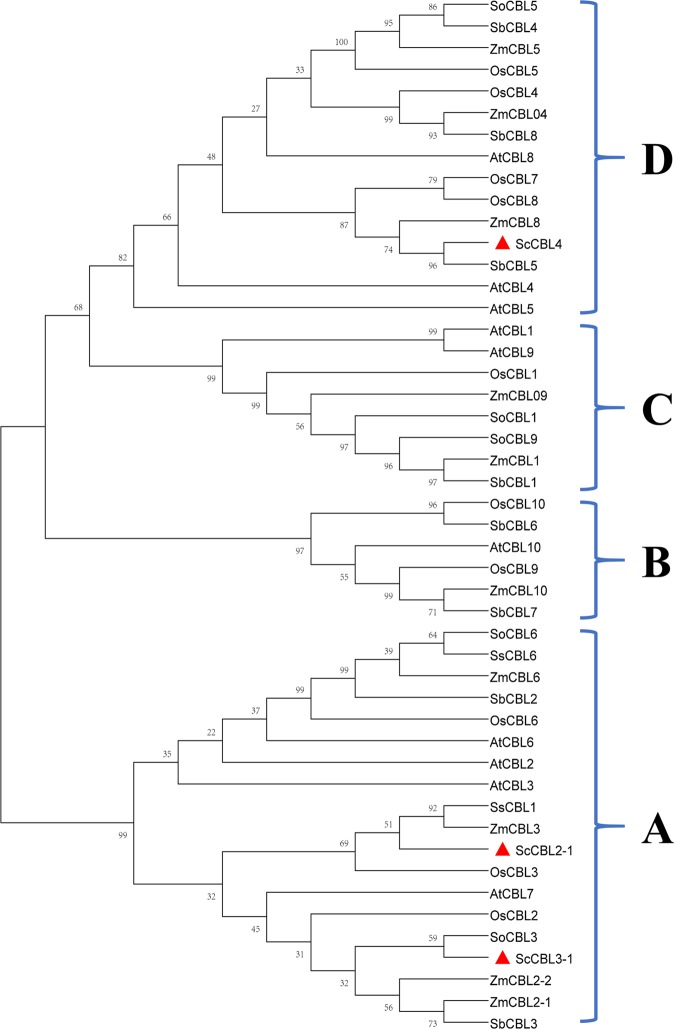
Sequences for CBL proteins identified in A. thaliana11,42, O. sativa11,24, Zea mays43, and S. bicolor44 were obtained from GenBank (https://www.ncbi.nlm.nih.gov/). A phylogenetic tree analysis was performed and the CBL proteins were grouped into four clades (A–D; see Fig. 1). The three sugarcane CBL proteins fell into two different groups: ScCBL2-1 and ScCBL3-1 were in group A, and ScCBL4 was in group D.
Figure 1.
Phylogenetic analysis of the predicted amino acid sequence of CBL proteins from sugarcane and other plant species. The phylogenetic tree was generated using MEGA X software, and the sequence phylogram was constructed by the neighbor-joining bootstrap method (1,000 replicates). At, Arabidopsis thaliana; Os, Oryza sativa; Zm, Zea mays; Sb, Sorghum bicolor; So, Saccharum spp. (GT28); Ss, Saccharum spp. (ROC22); Sc, Saccharum spp. (ROC22). ScCBLs in this study were indicated in red triangles. All the corresponding GenBank Accession numbers were listed in Table S3.
Sequence analysis of the ScCBL proteins
A multiple alignment analysis was performed using the amino acid sequences of the three ScCBLs and ten AtCBLs11. As shown in Figure 2, all of these CBL proteins contained more than two EF-hand domains, which are essential for CBL to bind Ca2+19. The C-terminal region of all three ScCBL proteins contained an FPSF motif. Note that only ScCBL4 contained an N-terminal MGCVSSK sequence, which is a unique CBL protein domain referred to as the myristoylation domain45,46. ScCBL2-1 and ScCBL3-1 had N-terminal tonoplast targeting sequences (TTSs), which may mediate their subcellular localization47. The size of the linker regions between the EF-hand loops was absolutely invariant in all three of the proteins: 23 amino acids separated EF1 and EF2, whereas 25 amino acids separated EF2 and EF3, and 32 amino acids separated EF3 and EF4.
Figure 2.
Sequence homology analysis of sugarcane and A. thaliana CBLs. The black box indicated the ScCBL4 myristoylation site. The yellow box indicated the ScCBL2-1 and ScCBL3-1 tonoplast targeting sequences. The red box indicated the FPSF motif. The EF notations indicated the EF-hand domain. Sequences highlighted in dark blue, red, and light blue indicated homology 100%, ≥ 75%, and ≥ 50% homology, respectively. aa, amino acids. All the corresponding GenBank Accession were listed in Table S3.
Tissue-specific expression of the ScCBL genes
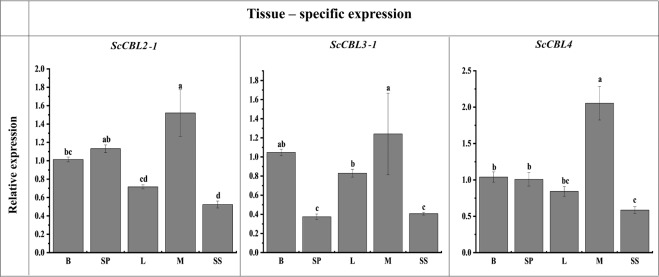
ScCBL expression in different sugarcane tissues (bud, stem pith, leaf, meristem, and stem skin) was detected by qRT-PCR. Figure 3 shows that the three ScCBLs were expressed in all of the tissues tested. ScCBL2-1, ScCBL3-1, and ScCBL4 were expressed at the highest levels in the meristem. ScCBL2-1 and ScCBL4 were expressed at the lowest levels in the stem skin, while ScCBL3-1 had the lowest transcription in the stem pith.
Figure 3.
Expression of ScCBL genes in different sugarcane tissues, as assessed by qRT-PCR. Data were normalized to cullin (CUL) and clathrin adaptor complex (CAC) expression levels. All data points were means ± SE (n = 3). Different lowercase letters indicated a significant difference, as determined by the least-significant difference test (p-value < 0.05). B, bud; SP, stem pith; L, leaf; M, meristem; SS, stem skin.
Expression of the ScCBL genes in response to phytohormones and various abiotic stresses
qRT-PCR analysis showed that the three ScCBL genes exhibited different expression patterns in response to ABA, SA, MeJA, H2O2, CaCl2, PEG, NaCl, or CuCl2 stress (Fig. 4). When subjected to ABA stress, ScCBL2-1, ScCBL3-1 and ScCBL4 transcription were all inhibited. Under SA, ScCBL2-1 and ScCBL3-1 were expression-induced, but ScCBL4 was down-regulated. As for MeJA, the three genes have the similar expression pattern with that under SA. Handling with H2O2, the expression of all ScCBLs peaked at 12 h. ScCBL2-1 and ScCBL3-1 expression was inhibited in response to treatment with CaCl2. We did not find significant difference, however, in the expression of ScCBL4 between treatment and control. PEG stress did not induce a significant expression level difference of ScCBL4 in compared with the control, but ScCBL2-1 and ScCBL3-1 expression was up-regulated (ScCBL2-1 peaked at 24 h at a value 17.1 times higher than that of the control, and ScCBL3-1 peaked at 6 h at a value 3.0 times higher than that of the control). Treatment with NaCl inhibited ScCBL4 expression but induced ScCBL2-1 and ScCBL3-1 expression, with the highest expression levels (16.7 times and 2.5 times higher than that of the control, respectively) occurring at 12 h. In response to CuCl2 treatment, ScCBL2-1, ScCBL3-1, and ScCBL4 were up-regulated (ScCBL2-1 expression sharply increased at 48 h to a value 9.3 times higher than that of the control, and ScCBL3-1, and ScCBL4 expression peaked at 12 h at values that were 6.1 and 2.9 times higher, respectively, than that of the control).
Figure 4.
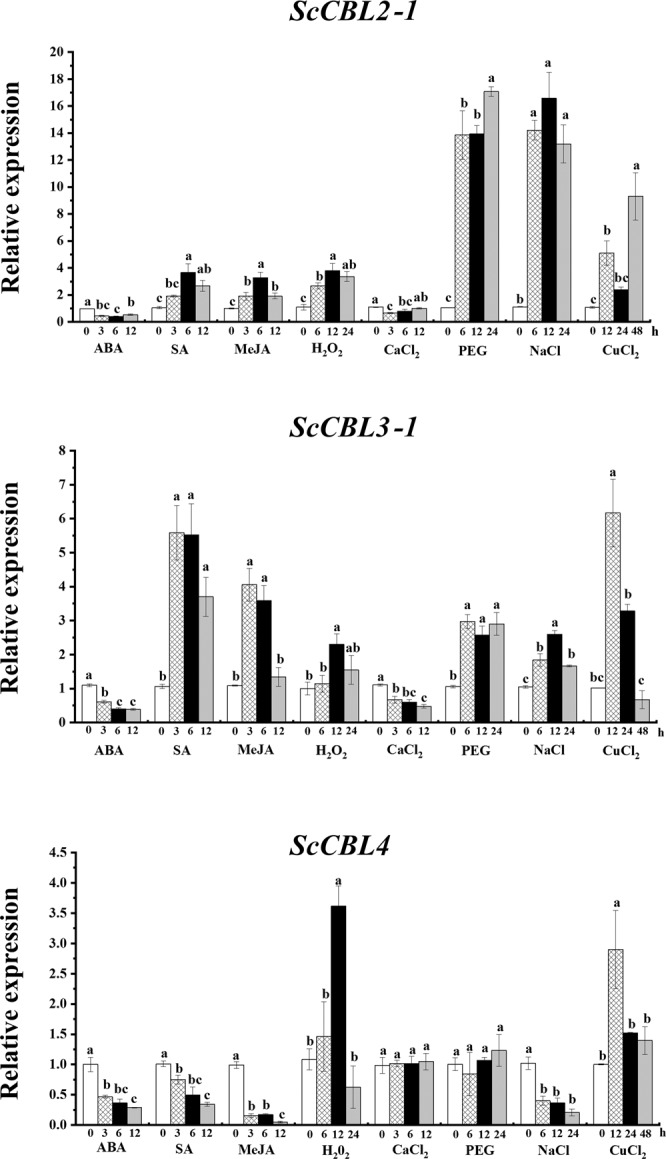
Expression of ScCBL genes in ROC22 plantlets after treatment with 100 μM ABA, 5 mM SA, 100 μM MeJA, 10 μM H2O2, 50 μM CaCl2, 25% PEG, 250 mM NaCl, or 100 mM CuCl2, as determined by qRT-PCR. Data were normalized to the cullin (CUL) and clathrin adaptor complex (CAC) expression levels. All data points were means ± SE (n = 3). Different lowercase letters indicate a significant difference, as determined by the least-significant difference test (p-value < 0.05). ABA, abscisic acid; SA, salicylic acid; MeJA, methyl jasmonate; H2O2, hydrogen peroxide; CaCl2, calcium chloride; PEG, polyethylene glycol; NaCl, sodium chloride; CuCl2, copper chloride.
Subcellular localization of the ScCBL proteins
To understand the subcellular localization of ScCBLs, ScCBL::GFP fusion proteins were expressed transiently in N. benthamiana leaf cells (Fig. 5). At 48 h post-infiltration, ScCBL2-1::GFP, ScCBL3-1::GFP, and ScCBL4::GFP were observed to locate in the plasma membrane and cytoplasm.
Figure 5.
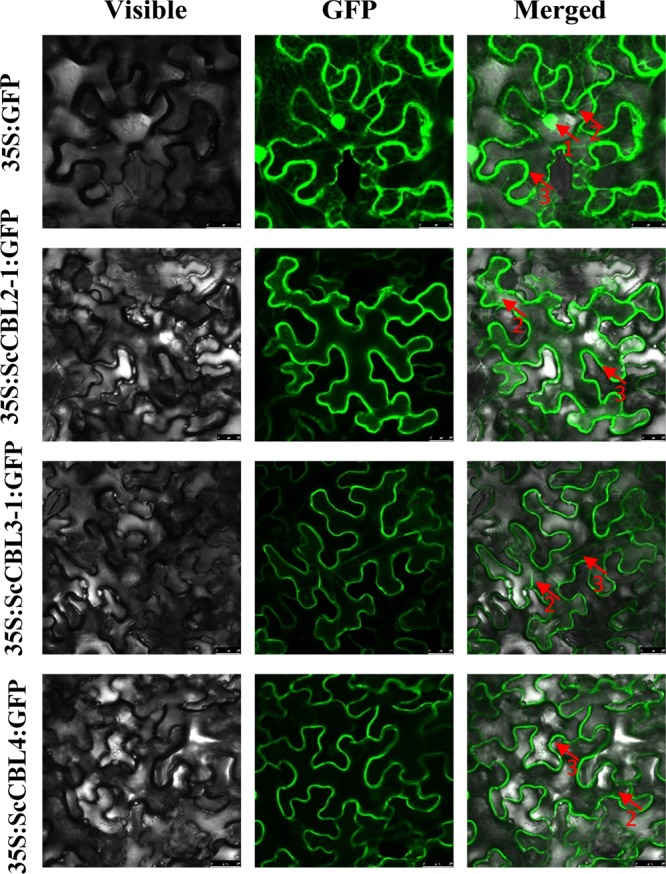
Subcellular localization of ScCBLs in Nicotiana benthamiana leaves 48 h after infiltration. Epidermal cells were imaged using visible light, green fluorescence, and merged light. Red arrows labeled 1, 2, and 3 indicated the nucleus, cytoplasm, and plasma membrane, respectively.
ScCBL genes expression in E. coli BL21 (DE3) strain
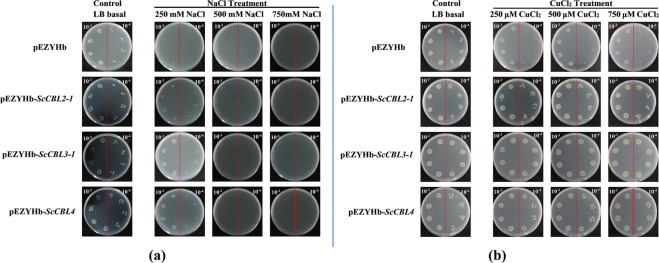
Bacterial cells overexpressing pEZYHb-ScCBLs had similar growth to control cells on solid LB medium (control), whereas the cells grown on media containing different concentrations of salt or CuCl2 showed marked differences in growth (Fig. 6). None of the cells grew on LB plates supplemented with 500 or 750 mM NaCl (Fig. 6(a)). Cells transformed with pEZYHb-ScCBLs, however, exhibited better survival on LB plates supplemented with 250 mM NaCl compared with untransformed cells (Fig. 6(a)). These results indicated that bacterial cells overexpressing pEZYHb-ScCBLs had better tolerance to high salt stress, which halted the growth of control cells. We monitored the role of pEZYHb-ScCBLs in metal stress by supplementing the growth media with various concentrations of CuCl2. Cells containing pEZYHb-ScCBLs grew more abundantly on LB plates supplemented with CuCl2 compared with control cells (Fig. 6(b)), suggesting that bacteria harboring pEZYHb-ScCBLs had significant tolerance to CuCl2.
Figure 6.
Growth of BL21/pEZYHb and BL21/pEZYHb-ScCBLs in E. coli BL21 cells. IPTG (1.0 mM) was added to the cultures to induce expression of the recombinant protein. The cultures were diluted 1,000-fold (left side of the red line on the plate) and 10,000-fold (right side of the red line on the plate), and 10 μL of the dilutions were spotted onto LB plates. LB plates without any supplements were used as the control. To explore the impact of BL21/pEZYHb and BL21/pEZYHb-ScCBLs on the response to salt and metal stresses, LB plates were supplemented with NaCl (250 mM, 500 mM, and 750 mM) or CuCl2 (250 μM, 500 μM, and 750 μM).
Transient overexpression of ScCBL genes in N. benthamiana leaves
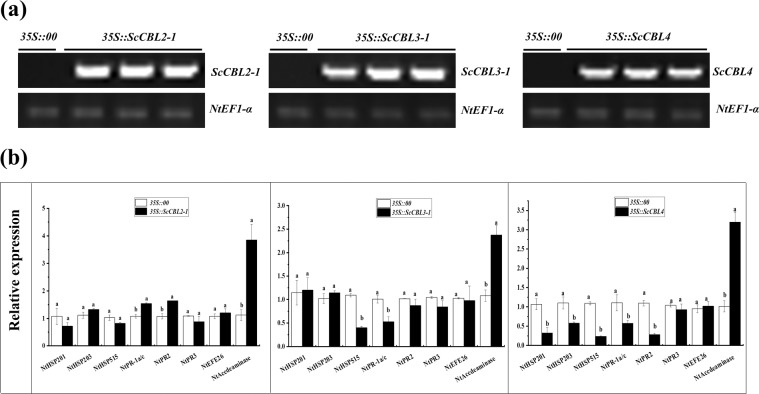
After transient overexpression of ScCBL genes in N. benthamiana leaves at 24 h post-infiltration, ScCBL genes were detected by RT-PCR (Fig. 7(a)). The expression levels of eight immunity-associated marker genes in N. benthamiana were detected by qRT-PCR, including the hypersensitivity response (HR) marker genes NtHSR201, NtHSR203, and NtHSR515; the salicylic acid–related gene NtPR−1a/c; the jasmonic acid pathway–associated genes NtNPR2 and NtNPR3; and the ethylene synthesis–dependent genes NtEFE26 and NtAccdeaminase (Fig. 7(b)). After 24 h of infiltration, the expression levels of NtHSR201, NtHSR203, NtHSR515, NtNPR3, and NtEFE26 remained unchanged, while NtPR-1a/c, NtNPR2, and NtAccdeaminase were up-regulated in 35 S::ScCBL2-1 leaves. In 35 S::ScCBL3-1 leaves, NtHSR515 and NtPR-1a/c were down-regulated, but NtAccdeaminase was up-regulated. Except for NtAccdeaminase was up-regulated, the expression of the other five HR marker genes (NtHSR201, NtHSR203, NtHSR515, NtPR-1a/c, and NtPR2) was down-regulated in 35 S::ScCBL4 leaves.
Figure 7.
Transient overexpression of ScCBLs in Nicotiana benthamiana leaves. (a) RT-PCR analysis of ScCBLs in the N. benthamiana leaves at 24 h post-infiltration (The gels were selected from the same gel with the same exposure, and the unmodified figure was shown in Figure S2). (b) Relative expression of immunity-associated marker genes in the N. benthamiana leaves at 24 h post-infiltration. The transcript levels were normalized to NtEF1-α. Mock, the Agrobacterium strain carrying 35 S::00. All data points were means ± SE (n = 3). Different lowercase letters indicated a significant difference, as determined by the Duncan’s new multiple range test (p-value < 0.05).
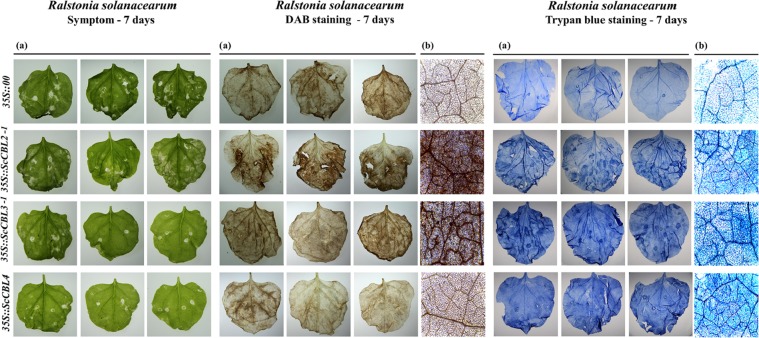
The results of pathogen infection of tobacco plants transiently expressing the ScCBLs are shown in Fig. 8. At 7 days post-inoculation with R. solanacearum, we observed disease symptoms in both the 35 S::ScCBLs and the control (35 S::00) leaves, but the yellow necrotic spots on the control leaves were larger than those observed on 35 S::ScCBLs leaves. In addition, darker 3, 3′-diaminobenzidine (DAB) staining was detected in 35 S::ScCBL2-1 and 35 S::ScCBL3-1 leaves than in the control leaves. And there were more intensely trypan blue–stained cells in 35 S::ScCBLs leaves than in the control leaves.
Figure 8.
Resistance effect of transient overexpression of ScCBL genes in Nicotiana benthamiana leaves. Images were captured using a (a) stereoscope and (b) a light microscope. Disease symptoms, DAB staining and trypan blue staining results from N. benthamiana leaves infected with R. solanacearum after infiltration with 35 S::00 (control) or 35 S::ScCBLs-containing Agrobacterium. Disease symptoms of infected leaves were observed at 7 days post-inoculation.
Discussion
Studies show that plant signal transduction processes under stress are accompanied by changes in cellular calcium concentration48,49. As a unique Ca2+ sensor in plants, CBL plays an important role in signal pathways of plant development and response to various stresses50. In this study, we characterized and discussed the possible functions of ScCBL2-1, ScCBL3-1, and ScCBL4 from sugarcane based on the results of bioinformatic analysis and experiments.
Sequences and phylogenetic analysis of ScCBLs
In the present study, three CBL genes were isolated from sugarcane. The ScCBL proteins appear to be rather conserved in size and structure. These ScCBL genes were predicted to encode polypeptides ranging from 24.31 to 25.85 kDa. The results are similar to those in A. thaliana and O. sativa, in which most AtCBLs and OsCBLs ranged from 23.5 to 25.9 kDa in size11. The sequence comparison results showed that the three ScCBL proteins all contained a C-terminal FPSF motif, and the serine residue in these FPSF motifs could be phosphorylated by the CIPK protein kinase51,52. A TTS motif, which mediates subcellular localization47, was clearly seen in the N-terminus of ScCBL2-1 and ScCBL3-1. The N-terminus of ScCBL4 contained a MGCVSSK sequence, which is a unique CBL protein domain known as the myristoylation domain45,46. This domain has been hypothesized to be the ancestral localization domain for CBLs53. The TTS motif of ScCBL2-1 and ScCBL3-1 had the same consensus motif as that found in AtCBL2 and AtCBL3, which spanned 19 amino acids47. The TTS in AtCBL2 or AtCBL3 was necessary and sufficient for targeting GFP fusion proteins to the tonoplast in A. thaliana mesophyll cells47. We thus speculate that ScCBL2-1 and ScCBL3-1 may have the same function to AtCBL2 or AtCBL347. However, we found that ScCBL2-1 and ScCBL3-1 not only located in the plasma membrane, but also in the cytoplasm. Phylogenetic analysis placed the three ScCBL proteins into two clades. The ScCBL members investigated here clustered closely with SoCBL, SsCBL, ZmCBL, SbCBL, and OsCBL orthologs, which indicated that the closer evolutionary relationship between the four species from the gramineae family (Saccharum spp., S. bicolor, Z. may, and O. sativa) compared to those of A. thaliana. In addition, interestingly, we found that CBL family members in A. thaliana (AtCBL1, 4, 5, and 9) and O. sativa (OsCBL1, 4, 5, 7, and 8) that harbor an N-myristoylation motif were distributed into two neighboring subgroups of the phylogenetic tree (C and D), and ScCBL4 fell into subgroup D. This site diversity may have enabled the evolutionary separation of CBL-type membrane-associated and membrane-independent calcium signaling pathways11.
Differential responses of ScCBLs to phytohormones and abiotic stresses
Several studies have shown that CBL genes play an important role in the plant stress response12,21,43,54. ABA, SA and MeJA, which are phytohormones, play an important role in the response of plants to adverse environmental conditions55. H2O2 is a ROS molecule that mediates signaling functions56. Through the interaction with CIPK, CBL protein regulates the production of H2O2 in the presence of NADPH oxidase, so as to maintain the positive feedback mechanism of stress tolerance57. In this study, when subjected to ABA stress, the three ScCBLs genes were all down-regulated. In Arabidopsis, AtCBL2 and AtCBL3 were also not obviously altered by ABA47. Under SA and MeJA, ScCBL2-1 and ScCBL3-1 were up-regulated while ScCBL4 was inhibited. PsCBL which is orthologues to AtCBL2, was up-regulated in response to SA18,58. And the expression of ScCBLs was induced under H2O2. We guess that the up-regulated ScCBLs were in response to the regulation of exogenous H2O2. Besides, CaCl2 stress did not induce any significant change in ScCBL4 expression, whereas ScCBL2-1 and ScCBL3-1 were down-regulated. Recent biophysical evidence has indicated that Ca2+ does not stimulate the interaction between CBL2 and CIPK14, even though Ca2+ is required for kinase activation through CBL59,60. Different ScCBL genes showed various expression patterns in response to CaCl2 stress, so the interaction between these ScCBL genes and Ca2+ needs to be further investigated. We also found that ScCBL2-1 and ScCBL3-1 were up-regulated in response to PEG, NaCl, and CuCl2, while ScCBL4 was induced by CuCl2, and inhibited by NaCl. Studies have shown that ZmCBL4 can significantly improve the salt tolerance of transgenic Arabidopsis43. AtCBL2 and AtCBL3 were marginally induced by dehydration47. From all the above, we deduced that ScCBL genes have different expression patterns in response to various stresses. We thus speculate that the same or different expression patterns among family genes may be caused by the functional divergence during evolution, which is accordance with the previous research that a homologous pattern resulted from genome duplication, and it caused the gain or loss of function as part of fine-tuning cellular function due to new functionalization in the course of genome evolution61.
Prokaryotic expression of ScCBLs under NaCl and CuCl2
Previous studies showed that CBL1, CBL4, CBL9, and CBL10 play important roles in the response to high salt stress46, for example, ZmCBL4 can significantly improve the salt tolerance of transgenic A. thaliana43. AtCBL10 is mainly induced by salt62. At lower concentration of salt (250 mM NaCl), however, ScCBL-transformed bacterial cultures showed better survival compared with the untransformed cells. These results suggested that ScCBL genes can enhance cell tolerance to low concentrations of salt. Studies have shown that excessive Cu2+ can cause oxidative stress, leading to lipid peroxidation, which destroys cell membrane structure63. Ca2+ can connect phosphates, phospholipids, and protein carboxyl groups on cell membranes, increase the hydrophobicity of cell membranes, and at the same time, reduce membrane permeability and enhance membrane stability64. In this study, ScCBLs-overexpressing and control bacterial cells had similar growth on solid LB medium (control). Besides, interestingly, in our studies, under metal stress conditions (CuCl2), recombinant ScCBLs cells exhibited dramatically better survival compared with nonrecombinant cells. These results suggested that ScCBL Ca2+ sensors can enhance tolerance to CuCl2.
Transient expression of ScCBLs response to R. solanacearum
Ethylene is thought to act as an internal signal regulator during plant growth and development, and can respond to external adverse conditions including biotic and abiotic stresses65. In addition, Ca2+ signaling plays a critical role in the response to biotic and abiotic stimuli66. Fagerstedt et al. found that an increase in the concentration of Ca2+ ions can activate the CBL-CIPK system and cause ethylene-responsive gene activation67. In the present study, we found that ethylene synthesis–dependent immunity-associated marker gene (NtAccdeaminase) was up-regulated when transiently overexpressed ScCBLs in N. benthamiana leaves (Fig. 7). Moreover, since CBL proteins can function as Ca2+ sensor relays7, we can hypothesize that ScCBL genes may take part in the ethylene synthesis pathway and play a role in the response to external stressors67. Reactive oxygen species (ROS) act as signaling molecules to regulate development and stress responses68. As a relatively stable active oxygen, H2O2, plays different roles in plant responses to external stresses69. A previous study showed that, in plants, attempted infection by microbial pathogens is often accompanied by rapid cell death in and around the initial infection site and that this response is associated with restricted pathogen growth and represents a form of PCD70. In this study, to investigate changes in ScCBLs expression in response to pathogen infection, we injected R. solanacearum into N. benthamiana containing 35 S::ScCBLs and a control construct. Then, we used DAB staining and trypan blue staining to detect hydrogen peroxide (H2O2) accumulation and cell necrosis in the leaves. We observed darker DAB staining compared with the control leaves after overexpression of ScCBL in N. benthamiana leaves and inoculation with R. solanacearum (Fig. 8). Besides, we also observed more intense trypan blue staining of cells in N. benthamiana leaves overexpressing ScCBL genes after inoculation with R. solanacearum compared with control leaves. This result suggested that overexpression of ScCBL genes can effectively promote resistance to infections in tobacco plants.
Conclusion
Three CBL genes (ScCBL2-1, ScCBL3-1, and ScCBL4) in sugarcane that encode proteins harboring EF-hand motifs were cloned and identified. These ScCBL genes were constitutively expressed in the sugarcane bud, stem pith, leaf, meristem, and stem skin. And they showed different expression patterns in response to stimulation with phytohormones and various abiotic stresses. Overexpression of ScCBL genes enhanced E. coli BL21 cell growth under conditions of NaCl or CuCl2 stress. Additionally, transient overexpression of ScCBL genes in N. benthamiana leaves resulted in different expression levels of tobacco immunity-associated marker genes, as well as increased resistance to infection with R. solanacearum. The findings from this study of ScCBLs may serve as a basis for the elucidation of the mechanisms underlying sugarcane immunity.
Supplementary information
Acknowledgements
This research was funded by the National Key R&D Program of China (2018YFD1000503 and 2019YFD1000500), the National Natural Science Foundation of China (31871688, 31671752, 31101196 and 31340060), the Research Funds for Distinguished Young Scientists in Fujian Provincial Department of Education (SYC-2017) and the China Agriculture Research System (CARS-17).
Author contributions
W.S., L.H. and Y.Q. conceived, designed and initiated the project. N.H., H.L. and H.M. prepared materials. W.S., L.H., Y.R. and D.W. performed experiments and contributed to data analysis and validation. W.S. drafted the manuscript. L.X., K.M., Y.S. and Y.Q. revised the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-019-57058-7.
References
- 1.Xiong LM, Schumaker KS, Zhu JK. Cell signaling during cold, drought, and salt stress. Plant Cell. 2002;14:165–183. doi: 10.1105/tpc.000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hepler PK. Calcium: a central regulator of plant growth and development. Plant Cell. 2005;17:2142–2155. doi: 10.1105/tpc.105.032508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allen GJ, et al. Alteration of stimulus-specific guard cell calcium oscillations and stomatal closing in Arabidopsis det3 mutant. Science. 2000;289:2338–2342. doi: 10.1126/science.289.5488.2338. [DOI] [PubMed] [Google Scholar]
- 4.Chinnusamy V, Schumaker K, Zhu JK. Molecular genetic perspectives on cross-talk and specificity in abiotic stress signalling in plants. J. Exp. Bot. 2004;55:225–236. doi: 10.1093/jxb/erh005. [DOI] [PubMed] [Google Scholar]
- 5.Reddy VS, Reddy AS. Proteomics of calcium-signaling components in plants. Phytochemistry. 2004;65:1745–1776. doi: 10.1016/j.phytochem.2004.04.033. [DOI] [PubMed] [Google Scholar]
- 6.Xi Y, Liu JY, Chao D, Cheng ZM. The CBL and CIPK gene family in Grapevine (Vitis vinifera): genome-wide analysis and expression profiles in response to various abiotic stresses. Front. Plant Sci. 2017;8:978. doi: 10.3389/fpls.2017.00978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shi JR, et al. Novel protein kinases associated with calcineurin B–like calcium sensors in Arabidopsis. Plant Cell. 1999;11:2393–2405. doi: 10.1105/tpc.11.12.2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kudla J, Xu Q, Harter K, Gruissem W, Luan S. Genes for calcineurin B-like proteins in Arabidopsis are differentially regulated by stress signals. Proc. Natl. Acad. Sci. USA. 1999;96:4718–4723. doi: 10.1073/pnas.96.8.4718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thoday-Kennedy EL, Jacobs AK, Roy SJ. The role of the CBL–CIPK calcium signalling network in regulating ion transport in response to abiotic stress. Plant Growth Regul. 2015;76:3–12. doi: 10.1007/s10725-015-0034-1. [DOI] [Google Scholar]
- 10.Batistič O, Kudla J. Plant calcineurin B-like proteins and their interacting protein kinases. Biochim. Biophys. Acta. 2009;1793:985–992. doi: 10.1016/j.bbamcr.2008.10.006. [DOI] [PubMed] [Google Scholar]
- 11.Kolukisaoglu U, Weinl S, Blazevic D, Batistic O, Kudla J. Calcium sensors and their interacting protein kinases: genomics of the Arabidopsis and rice CBL-CIPK signaling networks. Plant Physiol. 2004;134:43–58. doi: 10.1104/pp.103.033068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang HC, Yin WL, Xia XL. Calcineurin B-Like family in Populus: comparative genome analysis and expression pattern under cold, drought and salt stress treatment. Plant Growth Regul. 2008;56:129–140. doi: 10.1007/s10725-008-9293-4. [DOI] [Google Scholar]
- 13.Li LB, et al. Identification and bioinformatics analysis of SnRK2 and CIPK family genes in sorghum. Agr. Sci. China. 2010;9:19–30. doi: 10.1016/S1671-2927(09)60063-8. [DOI] [Google Scholar]
- 14.Zhang CX, Bian MD, Yu H, Liu Q, Yang ZM. Identification of alkaline stress-responsive genes of CBL family in sweet sorghum (Sorghum bicolor L.) Plant Physiol. Biochem. 2011;49:1306–1312. doi: 10.1016/j.plaphy.2011.08.010. [DOI] [PubMed] [Google Scholar]
- 15.Zhang HF, et al. Identification and characterization of CBL and CIPK gene families in canola (Brassica napus L.) BMC Plant Biol. 2014;14:8. doi: 10.1186/1471-2229-14-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li J, Jiang MM, Ren L, Liu Y, Chen HY. Identification and characterization of CBL and CIPK gene families in eggplant (Solanum melongena L.) Mol. Genet. Genomics. 2016;291:1769–1781. doi: 10.1007/s00438-016-1218-8. [DOI] [PubMed] [Google Scholar]
- 17.Kabir MH, Wang MH. Response of calcineurin B-like protein kinase gene in tomato to various abiotic stresses. J. Korean Soc. Appl. Bi. 2010;53:15–21. doi: 10.3839/jksabc.2010.003. [DOI] [Google Scholar]
- 18.Mahajan S, Sopory SK, Tuteja N. Cloning and characterization of CBL-CIPK signalling components from a legume (Pisum sativum) FEBS J. 2010;273:907–925. doi: 10.1111/j.1742-4658.2006.05111.x. [DOI] [PubMed] [Google Scholar]
- 19.Lewit-Bentley A, Réty S. EF-hand calcium-binding proteins. Curr. Opin. Struct. Biol. 2000;10:637–643. doi: 10.1016/S0959-440x(00)00142-1. [DOI] [PubMed] [Google Scholar]
- 20.Yong HC, et al. CBL1, a calcium sensor that differentially regulates salt, drought, and cold responses in Arabidopsis. Plant Cell. 2003;15:1833–1845. doi: 10.1105/tpc.012393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pandey GK, et al. The calcium sensor calcineurin B-like 9 modulates abscisic acid sensitivity and biosynthesis in Arabidopsis. Plant Cell. 2004;16:1912–1924. doi: 10.1105/tpc.021311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yong HC, et al. Constitutive overexpression of the calcium sensor CBL5 confers osmotic or drought stress tolerance in. Arabidopsis. Mol. Cells. 2010;29:159–165. doi: 10.1007/s10059-010-0025-z. [DOI] [PubMed] [Google Scholar]
- 23.Shen JQ, Zhang ZZ, Pan WH, Pan JW. Functions and action mechanisms of CBL-CIPK signaling system in plants. Plant Physiol. J. 2014;50:641–650. doi: 10.13592/j.cnki.ppj.2014.0052. [DOI] [Google Scholar]
- 24.Gu ZM, et al. Expression analysis of the calcineurin B-like gene family in rice (Oryza sativa L.) under environmental stresses. Gene. 2008;415:1–12. doi: 10.1016/j.gene.2008.02.011. [DOI] [PubMed] [Google Scholar]
- 25.D’Hont, A. et al. In Genomics of tropical crop plants Vol. 1 Sugarcane: a major source of sweetness, alcohol, and bio-energy (eds Paul H. Moore & Ray. Ming) Ch. 21, 483–513 (Springer, (2008).
- 26.Ling QP, et al. Expression analysis of CBL1 and CBL6 genes in sugarcane under abiotic stress. Mol. Plant Breed. 2018;16:377–385. doi: 10.13271/j.mpb.016.000377. [DOI] [Google Scholar]
- 27.Farani TF, Gentile A, Tavares RG, Ribeiro C, Menossi M. Characterization of a protein-protein interaction network of the CBL-interacting protein kinase 8 from sugarcane. Genet. Mol. Res. 2015;14:483–491. doi: 10.4238/2015.January.26.1. [DOI] [PubMed] [Google Scholar]
- 28.Zhang, B. Q. Study on the physiological and biochemical characteristics and differentially expressed proteome during late growth stage of sugarcane under cold stress Doctor thesis, Guangxi University, (2013).
- 29.Wang ZP, et al. Preliminary evaluation of resistance on Pokkah Boeng disease of major sugarcane varieties in the field of Guangxi. Chin. J. Tropi. Crop. 2016;37:107–112. doi: 10.3969/j.issn.1000-2561.2016.05.016. [DOI] [Google Scholar]
- 30.Xian W, et al. Detection of epicuticular wax on sugarcane leaves and its correlation with agronomic characters and stress resistance. Journal of southern agriculture. 2018;49:36–45. doi: 10.3969/j.issn.2095-1191.2018.09.05.. [DOI] [Google Scholar]
- 31.Lan J, et al. Comprehensive evaluation on drought resistance difference of six sugarcane varieties. Southwest China j. Agr. Sci. 2014;27:1374–1381. doi: 10.16213/j.cnki.scjas.2014.04.044. [DOI] [Google Scholar]
- 32.Wang, S. S. Cloning and identification of chitinase family genes from sugarcane Master thesis, Fujian Agriculture and Forestry University, (2014).
- 33.Liu F, et al. Plant jasmonate ZIM domain genes: shedding light on structure and expression patterns of JAZ gene family in sugarcane. BMC Genomics. 2017;18:771. doi: 10.1186/s12864-017-4142-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Su Y, et al. Isolation of a novel peroxisomal catalase gene from sugarcane, which is responsive to biotic and abiotic stresses. PLos One. 2014;9:e84426. doi: 10.1371/journal.pone.0084426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ling H, et al. Transcriptional insights into the sugarcane-sorghum mosaic virus interaction. Trop. Plant Biol. 2018;11:163–176. doi: 10.1007/s12042-018-9210-6. [DOI] [Google Scholar]
- 36.Su YC, et al. Molecular cloning and characterization of two pathogenesis-related β-1,3-glucanase genes ScGluA1 and ScGluD1 from sugarcane infected by Sporisorium scitamineum. Plant Cell Rep. 2013;32:1503–1519. doi: 10.1007/s00299-013-1463-9. [DOI] [PubMed] [Google Scholar]
- 37.Ling H, Wu QB, Guo JL, Xu LP, Que YX. Comprehensive selection of reference genes for gene expression normalization in sugarcane by real time quantitative RT-PCR. PLos One. 2014;9:e97469. doi: 10.1371/journal.pone.0097469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2 method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 39.Su YC, et al. ScChi, encoding an acidic class III chitinase of sugarcane, confers positive responses to biotic and abiotic stresses in sugarcane. Int. J. Mol. Sci. 2014;15:2738–2760. doi: 10.3390/ijms15022738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guo JL, et al. A novel dirigent protein gene with highly stem-specific expression from sugarcane, response to drought, salt and oxidative stresses. Plant Cell Rep. 2012;31:1801–1812. doi: 10.1007/s00299-012-1293-1. [DOI] [PubMed] [Google Scholar]
- 41.Su YC, et al. Isolation and characterization of ScGluD2, a new sugarcane beta-1,3-Glucanase D family gene induced by Sporisorium scitamineum, ABA, H O, NaCl, and CdCl stresses. Front. Plant Sci. 2016;7:1348. doi: 10.3389/fpls.2016.01348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim KN, Cheong YH, Gupta R, Luan S. Interaction specificity of Arabidopsis calcineurin B-like calcium sensors and their target kinases. Plant Physiol. 2000;124:1844–1853. doi: 10.1104/pp.124.4.1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang MY, et al. Overexpression of a putative maize calcineurin B-like protein in Arabidopsis confers salt tolerance. Plant Mol. Biol. 2007;65:733–746. doi: 10.1007/s11103-007-9238-8. [DOI] [PubMed] [Google Scholar]
- 44.Weinl S, Kudla J. The CBL–CIPK Ca -decoding signaling network: function and perspectives. New Phytol. 2009;184:517–528. doi: 10.1111/j.1469-8137.2009.02938.x. [DOI] [PubMed] [Google Scholar]
- 45.Ishitani M, Zhu JK. SOS3 function in plant salt tolerance requires N-myristoylation and calcium binding. Plant Cell. 2000;12:1667–1678. doi: 10.1105/tpc.12.9.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li RF, et al. Functions and mechanisms of the CBL–CIPK signaling system in plant response to abiotic stress. Progr. Nat. Sci. 2009;19:667–676. doi: 10.1016/j.pnsc.2008.06.030. [DOI] [Google Scholar]
- 47.Tang RJ, et al. Tonoplast calcium sensors CBL2 and CBL3 control plant growth and ion homeostasis through regulating V-ATPase activity in. Arabidopsis. Cell Res. 2012;22:1650–1665. doi: 10.1038/cr.2012.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sanders D, Brownlee C, Harper JF. Communicating with Calcium. Plant Cell. 1999;11:691. doi: 10.1105/tpc.11.4.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gilroy S, Trewavas A. Signal processing and transduction in plant cells: the end of the beginning? Nat. Rev. Mol. Cell Biol. 2001;2:307–314. doi: 10.1038/35067109. [DOI] [PubMed] [Google Scholar]
- 50.Li ZY, et al. Overexpression of soybean GmCBL1 enhances abiotic stress tolerance and promotes hypocotyl elongation in Arabidopsis. Biochem. Bioph. Res. Co. 2012;427:731–736. doi: 10.1016/j.bbrc.2012.09.128. [DOI] [PubMed] [Google Scholar]
- 51.Du W, et al. Phosphorylation of SOS3-like calcium-binding proteins by their interacting SOS2-like protein kinases is a common regulatory mechanism in. Arabidopsis. Plant Physiol. 2011;156:2235–2243. doi: 10.1104/pp.111.173377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hashimoto K, et al. Phosphorylation of calcineurin B-like (CBL) calcium sensor proteins by their CBL-interacting protein kinases (CIPKs) is required for full activity of CBL-CIPK complexes toward their target proteins. J. Biol. Chem. 2012;287:7956–7968. doi: 10.1074/jbc.M111.279331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kleist TJ, Spencley AL, Luan S. Comparative phylogenomics of the CBL-CIPK calcium-decoding network in the moss Physcomitrella, Arabidopsis, and other green lineages. Front. Plant Sci. 2014;5:187. doi: 10.3389/fpls.2014.00187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Albrecht V, et al. The calcium sensor CBL1 integrates plant responses to abiotic stresses. Plant J. 2003;36:457–470. doi: 10.1046/j.1365-313x.2003.01892.x. [DOI] [PubMed] [Google Scholar]
- 55.Verma V, Ravindran P, Kumar PP. Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 2016;16:86. doi: 10.1186/s12870-016-0771-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ilya G, et al. Transcriptomic footprints disclose specificity of reactive oxygen species signaling in Arabidopsis. Plant Physiol. 2006;141:436–445. doi: 10.1104/pp.106.078717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mohanta TK, et al. Early events in plant abiotic stress signaling: interplay between calcium, reactive oxygen species and phytohormones. J. Plant Growth Regul. 2018;37:1033–1049. doi: 10.1007/s00344-018-9833-8. [DOI] [Google Scholar]
- 58.Tuteja N, Mahajan S. Further characterization of calcineurin B-like protein and its interacting partner CBL-interacting protein kinase from Pisum sativum. Plant Signal Behav. 2007;2:358–361. doi: 10.4161/psb.2.5.4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tominaga M, Harada A, Kinoshita T, Shimazaki K. Biochemical characterization of calcineurin B-like-interacting protein kinase in Vicia guard cells. Plant Cell Physiol. 2010;51:408–421. doi: 10.1093/pcp/pcq006. [DOI] [PubMed] [Google Scholar]
- 60.Akaboshi M, et al. The crystal structure of plant-specific calcium-binding protein AtCBL2 in complex with the regulatory domain of AtCIPK14. J. Mol. Biol. 2008;377:246–257. doi: 10.1016/j.jmb.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 61.Ray S, Agarwal P, Arora R, Kapoor S, Tyagi AK. Expression analysis of calcium-dependent protein kinase gene family during reproductive development and abiotic stress conditions in rice (Oryza sativa L. ssp. indica) Mol. Genet. Genomics. 2007;278:493–505. doi: 10.1007/s00438-007-0267-4. [DOI] [PubMed] [Google Scholar]
- 62.Quan R, et al. SCABP8/CBL10, a putative calcium sensor, interacts with the protein kinase SOS2 to protect Arabidopsis shoots from salt stress. Plant Cell. 2007;19:1415–1431. doi: 10.1105/tpc.106.042291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Oren S, Rubin B, Rabinowitch HD, Tel-Or E. Loading beans with sublethal levels of copper enhances conditioning to oxidative stress. J. Plant Physiol. 2001;158:1415–1421. doi: 10.1078/0176-1617-00626. [DOI] [Google Scholar]
- 64.Jones RGW, Lunt OR. The function of calcium in plants. Bot. Rev. 1967;33:407–426. doi: 10.1007/BF02858743. [DOI] [Google Scholar]
- 65.Wang KL-C, Li H, Ecker JR. Ethylene biosynthesis and signaling networks. Plant Cell. 2002;14:S131–S151. doi: 10.1105/tpc.001768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Defalco TA, Bender KW, Snedden WA. Breaking the code: Ca sensors in plant signalling. Biochem. J. 2010;425:27–40. doi: 10.1042/BJ20091147. [DOI] [PubMed] [Google Scholar]
- 67.Mancuso, S. & Shabala, S. In Waterlogging signalling and tolerance in plants Oxygen deprivation, metabolic adaptations and oxidative stress (eds Olga. Blokhina & Kurt V. Fagerstedt) 119–147 (Springer, 2010).
- 68.Apel K, Hirt H. Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 2004;55:373–399. doi: 10.1146/annurev.arplant.55.031903.141701. [DOI] [PubMed] [Google Scholar]
- 69.Liu YH, Offler CE, Ruan YL. A simple, rapid, and reliable protocol to localize hydrogen peroxide in large plant organs by DAB-mediated tissue printing. Front. Plant Sci. 2014;5:745. doi: 10.3389/fpls.2014.00745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lam E, Kato N, Lawton M. Programmed cell death, mitochondria and the plant hypersensitive response. Nature. 2001;411:848–853. doi: 10.1038/35081184. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.