Abstract
Salt-inducible kinase 2 (SIK2) has been established as a regulator of diverse biological processes including cell metabolism. A recent study has reported that SIK2 is required for adipocyte-induced ovarian cancer (OC) survival through facilitating fatty acid oxidation. However, whether SIK2 also plays a role in the lipid synthesis in OC cells remains elusive. Here, we showed that SIK2 significantly promoted the lipid synthesis in OC cells. On the one hand, SIK2 enhanced fatty acid synthesis through upregulating the expression of sterol regulatory element binding protein 1c (SREBP1c) and thus the transcription of major lipogenic enzyme FASN. On the other hand, SIK2 promoted cholesterol synthesis through upregulating the expression of sterol regulatory element binding protein 2 (SREBP2) and thus the transcription of major cholesterol synthesis enzymes HMGCR. Moreover, PI3K/Akt signaling pathway was found to be involved in the upregulation of SREBP1c and SREBP2 in OC cells. Moreover, in vitro and in vivo assays indicated that the SIK2-regulated fatty acid and cholesterol synthesis played a critical role in the growth of OC cells. Our findings demonstrate that SIK2 is a critical regulator of lipid synthesis in OC cells and thus promotes OC growth, which provides a strong line of evidence for this molecule to be used as a therapeutic target in the treatment of this malignancy.
Subject terms: Cancer metabolism, Cancer therapy, Oncogenes
Introduction
Dysregulation of fatty acid metabolism has been increasingly recognized as a component of malignant transformation in many different cancers, including ovarian cancer1,2. Elevated de novo fatty acid synthesis provides cancer cells with building blocks, signaling molecules and post-translational modifications to promote rapid cell proliferation. To date, many enzymes which are involved in de novo fatty acid biosynthesis, such as ATP citrate lyase (ACLY), acetyl-CoA carboxylase (ACC), fatty acid synthase (FASN) and stearoyl-CoA desaturase1 (SCD1), are overexpressed and contributed to poor clinical outcomes in many different types of cancers3,4. Compared with most current studies focusing on de novo fatty acid synthesis, the functional roles of cholesterol in cancer development has received less attention5,6. Hypercholesterolemia has been considered as an important risk factor for cancers7,8. Except for serum cholesterol, intracellular cholesterol also has been shown to play a crucial role in the regulation of tumor progression. Elevation of intracellular cholesterol level has been observed in tumor tissues9,10, which promoted the proliferation, migration and invasion of cancer cells. Besides, several recent studies also have demonstrated that the increased expression of cholesterol synthesis genes is associated with the decreased patient survival. Chushi Li et al. have reported that 3-hydroxy-3-methyl-glutaryl-coenzyme A reductase (HMGCR), the rate-limiting enzyme for cholesterol synthesis is up-regulated in gastric cancer and positively regulates the growth and migration of cancer cells11. The oncogenic roles of HMGCR have also been revealed in glioblastoma and esophageal squamous cell carcinoma12,13. Recently, there has been a revival of enthusiasm amongst investigators to study how lipid metabolism pathways are reprogrammed in cancer cells. However, mechanisms underlying the increased de novo fatty acid and cholesterol synthesis in cancer cells are still not completely understood and further study is still required.
Salt-inducible kinase 2 (SIK2) is an AMP-activated protein kinase (AMPK)-related protein kinase that plays important roles in the regulation of cellular metabolism. Besides, several studies have reported that SIK2 activates a series of signaling pathways, such as PI3K/Akt, Hippo-YAP and LKB1-HDAC, which are associated with diverse cellular processes14. Recent studies have unraveled the role of SIK2 in cancer development and progression. It has shown that SIK2 is required for the proliferation of both prostate cancer and ovarian cancer cells15,16. Moreover, a recent study has reported that SIK2 is highly expressed in adipocyte-rich metastases and required for adipocyte-induced proliferation of metastatic ovarian cancer through facilitating fatty acid oxidation17, implying that SIK2 may play a critical role in the regulation of fatty acid metabolism. However, the role of SIK2 in the regulation of lipid synthesis in cancer cells, especially in ovarian cancer (OC) cells, is still unclear.
In this study, we explored the functional role and the underlying molecular mechanisms of SIK2 in the regulation of lipid synthesis, including fatty acid and cholesterol synthesis, in OC cells.
Materials and methods
Antibodies and reagents
The primary antibodies used in this study and their working concentration were listed in Supplementary Table S1. The PI3K inhibitor LY294002 (Cat. no. HY-10108), SIK inhibitor HG-9-91-01 (Cat. no. HY-15776), FASN inhibitor C75 (Cat. no. HY-12364) and HMGCR inhibitor Mevastatin (Cat. no. HY-17408) were purchased from MedChemExpress (New Jersey, USA).
Cell lines and tissue samples
Human OC cell lines A2780, HEY, SKOV3 and ES2 were obtained from the American Type Culture Collection (ATCC) and cultured in recommended Dulbecco’s Modified Eagle Medium (DMEM) and RPMI-1640 medium supplemented with 10% FBS, respectively. In addition, tumor tissues and matched peritumor tissues were obtained from 121 OC patients from the Department of Gynaecology and Obstetrics at Xijing Hospital between 2011 and 2015. The study has been approved by the Ethics Committee of Xijing Hospital and all patients provided written informed consent.
Knockdown and overexpression of target genes
Small interference RNA (siRNA) targeting SIK2, SREBP1c, SREBP2 and control siRNA (Genepharma, Shanghai, China) were transfected into OC cell lines using the transfection reagent lipofectamine 2000 (Thermo Fisher Scientific, MA, USA) according to the manufacturer’s protocols. Sequences of siRNA used in the study were listed in Supplementary Table S2.
SIK2 overexpression vector and empty vector were kind gifts from Dr. Ahmed Ashour Ahmed (University of Oxford, UK). SREBP1c knockdown (shSREBP1c) and overexpression vectors have been constructed in our previous study18. SREBP2 overexpression vector were purchased from (Genepharma, Shanghai, China).
Quantitative real-time PCR analysis
Total RNA was extracted from OC cells using RNA extraction Kit (TaKaRa, 9767, Japan) following the manufacturer’s instructions. Reverse transcription for complementary DNA (cDNA) synthesis was performed using Transcriptor Reverse Transcriptase (TaKaRa, RR036A, Japan). Quantitative real-time PCR (qRT-PCR) was used to determine the gene expression level using 2 × SYBR Green qPCR Master Mix (S2014, Everbright USA) with the primers listed in Supplementary Table S2. β-actin was used as an internal control.
Western blot analysis
Proteins extracted from lysed OC cells were run on SDS-PAGE for separation and then transferred onto PVDF membranes followed by incubation with primary and HRP-conjugated second antibody antibodies as recommended by the manufacturer. Antibodies and their dilutions were listed in Supplementary Table S1 and β-actin served as an internal control. Quantification of the blots was done using Image J software.
Quantification of free fatty acid, triglyceride, phospholipids and cholesterol
Lipids were extracted from OC cell homogenates. OC cells homogenates were prepared for lipids extraction using chloroform/methanol (2:1) and then quantitatively estimated for the levels of free fatty acid, triglycerideand, phospholipids and cholesterol with EnzyChromTM free fatty acid, triglyceride, phospholipidassay and cholesterol kits (Bioassay Systems, Hayward, CA, USA), following their manufacturers’ protocols.
Quantification of neutral lipid
The content of neutral lipids in OC cells was monitored with a fluorescence dye BODIPY 493/503 (Invitrogen) following the manufacturer’s instruction. Briefly, after fixed with 4% paraformaldehyde, OC cells were then stained with BODIPY 493/503 (1 μg/mL) for 30 min at 37 °C. DAPI (20 μL/mL) was used for nuclei counterstain for 15 min. Confocal images were acquired with an Olympus FV-1000 confocal microscope (Olympus, Tokyo, Japan). Quantification of LDs was done using Image J software (Average numbers of LDs per cell, percentage of cellular area occupied by LDs).
Immunohistochemical staining
Immunohistochemical (IHC) analysis was performed as previously described19. Briefly, 3-µm thick tissue sections were deparaffinized and hydrated. Hot citrate buffer (pH = 6.0) was used for antigen retrieval. Antigen was retrieved by treating with under pressure. The sections were then incubated with primary antibodies overnight at 4 °C. Color was developed using DAB and sections were then counterstained with hematoxylin. The immunostaining results were independently scored by two pathologists according to the intensity of staining as previously described20. The primary antibodies used in the study were listed in Supplementary Table S1.
MTS cell viability and colony formation assay
Cell proliferation was measured using the CellTiter 96 Aqueous One Solution cell proliferation assay (Promega Corp, Madison, WI) following the manufacturer’s instructions. Briefly, OC cells were seeded into 96-well plates and incubated for 5 days. Cell proliferation was detected daily based on the absorbance at 490 nm using a microplate reader. For colony formation assay, 1000 OC cells were seeded into 6-well plate and cultured about 2 weeks. The colonies were stained with crystal violet and counted.
EDU assay
A 5-ethynyl-2′-deoxyuridine (EdU)-incorporation assay kit (Ribobio, Guangdong, China) was used to measure the proliferation ability of OC cells following the manufacturer’s protocol.
Wound-healing and transwell invasion assays
Wound-healing assay was performed to assess the migration of OC cells following the procedures as described previously18. For determination of OC cell invasion, a 6.5-mm Transwell chamber (Corning Costar, China) was used. Briefly, OC cells were collected and added into the top chamber of 24-well transwell plates. Simultaneously, cell culture medium containing 20% FBS were added into the bottom of transwell chamber. After culture for 48 h, invaded cells were stained with crystal violet and counted under a microscopy. The average number of inserted cells in five randomly selected fields was calculated.
In vivo tumor growth assay
Six-week-old athymic female BALB/c nude mice were randomly divided into groups (six mice/group). A total of 1 × 107 OC cells were injected subcutaneously into nude mice and the tumor size was assessed each week. The mice were sacrificed 40 days after injection and tumors were harvested and tumor size was compared between groups. All animal experiments were approved by the Institutional Animal Experiment Committee of Xijing Hospital.
Statistical analysis
All values are presented as mean ± S.E.M from three independent experiments. Statistical significance was determined by one-way ANOVA or unpaired two-tailed Student’s t-test. P-value <0.05 was considered statistically significant. Correlations between measured variables were analyzed by Spearman rank correlation test.
Results
SIK2 significantly increased lipid contents in ovarian cancer cells
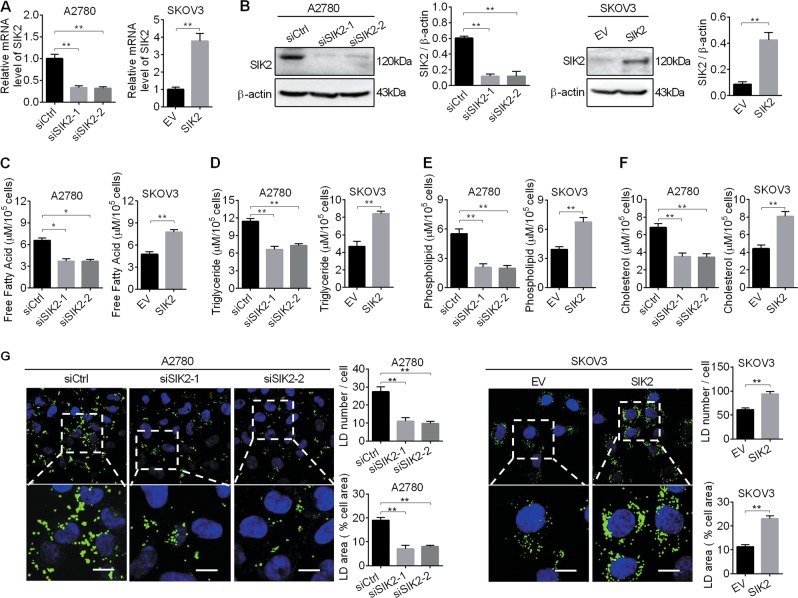
To investigate the potential role of SIK2 in the regulation of aberrant lipid metabolism in OC cells, the effect of SIK2 knockdown and overexpression on lipid content in A2780 cells with relative high SIK2 expression at both mRNA and protein levels and SKOV3 cells with relative low SIK2 expression at both mRNA and protein levels (Supplementary Fig. S1A, B) were firstly evaluated. The successful knockdown and overexpression of SIK2 was verified by qRT-PCR and Western blot analysis (Fig. 1a, b). Our data showed that A2780 cells with SIK2 knockdown had significantly decreased levels of intracellular free fatty acid, triglyceride, phospholipids and cholesterol when compared with control cells (Fig. 1c–f). In contrast, overexpression of SIK2 in SKOV3 cells increased the levels of intracellular free fatty acid, triglyceride, phospholipids and cholesterol. Keeping with the findings, cellular staining with the lipophilic dye BODIPY 493/503 provided further support, showing that SIK2 increased the intracellular contents of neutral lipids in OC cells (Fig. 1g). Collectively, our findings indicate that SIK2 promotes the reprogramming of lipid metabolism in OC cells.
Fig. 1. SIK2 significantly increased lipid contents in ovarian cancer (OC) cells.
a, b Quantitative real-time PCR (a) and Western blot (b) analyses of SIK2 expression in ovarian cancer A2780 and SKOV3 cells with treatment as indicated. siSIK2–1 and siSIK2–2, siRNAs against SIK2; siCtrl, control siRNA; SIK2, expression vector encoding SIK2; EV, empty vector. c–f Cellular content of free fatty acid (c), triglyceride (d), phospholipids (e) and cholesterol (f) were detected in A2780 and SKOV3 cells with treatment as indicated. g The content of neutral lipids were detected by BODIPY 493/503 dye and counterstained with DAPI in A2780 and SKOV3 cells with treatment as indicated. Scale bars, 50 μm. Average numbers of LDs per cell (upper panel), percentage of cellular area occupied by LDs (lower panel). Data are shown as mean ± S.E.M from three independent experiments. *p < 0.05; **p < 0.01.
SIK2 upregulated the expressions of lipogenic enzymes in OC cells
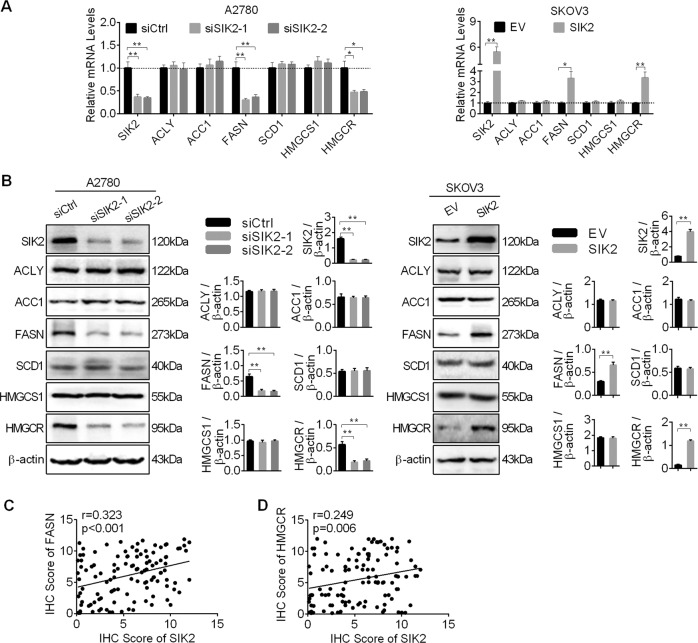
To study the role of SIK2 in the regulation of de novo fatty acid synthesis and cholesterol synthesis, we analyzed the expression of key enzymes involved in both fatty acid (ACLY, ACC1, FASN, SCD1) and cholesterol synthesis (HMGCS1, HMGCR). Our results showed that the expression levels of FASN and HMGCR were significantly decreased at both mRNA and protein levels in A2780 cells with SIK2 knockdown when compared with control cells, while SIK2 overexpression increased the expressions of FASN and HMGCR in SKOV3 cells (Fig. 2a, b). However, no significant changes of the expressions of ACLY, ACC1, SCD1 and HMGCS1 were observed when SIK2 was knockdown or overexpressed in A2780 and SKOV3 cells. To provide further support, the expression of SIK2 and FASN were determined by IHC analysis in tumor tissue samples from 121 OC patients. Spearman rank correlation analysis indicated a significant positive correlation of IHC scores between SIK2 and FASN (r = 0.323, p < 0.001) (Fig. 2c and Supplementary Fig. S2A), as well as between SIK2 and HMGCR (r = 0.249, p = 0.006) (Fig. 2d and Supplementary Fig. S2A).
Fig. 2. SIK2 upregulated the expressions of lipogenic enzymes in OC cells.
a Quantitative RT-PCR analysis for mRNA expression levels of lipogenic enzymes ACLY, ACC1, FASN, SCD1, HMGCS1 and HMGCR in A2780 and SKOV3 cells with treatment as indicated. b Western blot analysis for protein expression levels of lipogenic enzymes of ACLY, ACC1, FASN, SCD1, HMGCS1 and HMGCR in A2780 and SKOV3 cells with treatment as indicated. c, d Spearman correlation analysis of the relationship between the protein expression levels of SIK2 and FASN, as well as between SIK2 and HMGCR based on IHC staining results. Data are shown as mean ± S.E.M from three independent experiments. *p < 0.05; **p < 0.01.
SIK2 upregulated FASN expression to promote fatty acid synthesis via SREBP1c
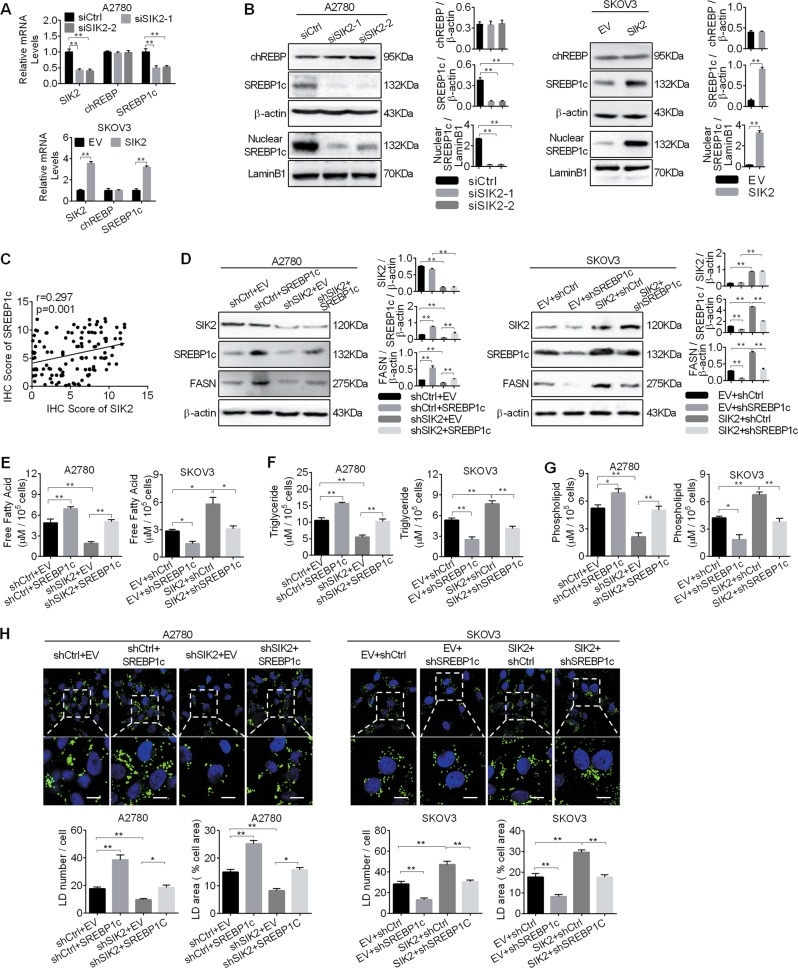
Carbohydrate-responsive element-binding protein (chREBP) and sterol regulatory elementary binding protein1c (SREBP1c) are important transcription factors in the regulation of fatty acid synthesis through transcriptional activation of lipogenic enzymes21. To explore the molecular mechanism of SIK2-mediated upregulation of lipogenic enzymes, the expression levels of chREBP and SREBP1c (the dominant isoform of SREBP1 in the ovary tissue22) in A2780 and SKOV3 cells were determined by qRT-PCR and Western blot analyses (Fig. 3a, b). Our results showed that only SREBP1c was significantly decreased at both mRNA and protein levels when SIK2 was knocked-down in A2780 cells, while no significant change was seen on the expression of chREBP. In contrast, SIK2 overexpression in SKOV3 cells increased SREBP1c expression. In addition, the level of nuclear SREBP1c, which represents the transcriptional activity of this molecule, showed a similar expression pattern to that of total SREBP1c, implying that SIK2 promoted the transcriptional activity of SREBP1c in OC cells. To provide further supports, the levels of SREBP1c was examined in tumor tissues from 121 OC patients. Spearman rank correlation analysis showed a positive correlation of expression levels between SIK2 and SREBP1c (r = 0.297, p = 0.001) (Fig. 3c and Supplementary Fig. S2A).
Fig. 3. SIK2 upregulated FASN expression to promote fatty acid synthesis via SREBP1c.
a, b Quantitative RT-PCR (a) and Western blot (b) analyses for expressions of chREBP and SREBP1c in A2780 and SKOV3 cells with treatment as indicated. c Spearman correlation analysis of the relationships between the protein expression levels of SIK2 and SREBP1c based on the IHC staining results. d Western blot analysis of SREBP1c and FASN in A2780 and SKOV3 cells with treatment as indicated (shSIK2, shRNA against SIK2; shCtrl, control shRNA; SREBP1c, expression vector encoding SREBP1c; EV, empty vector). e–g Cellular content of free fatty acid (e), triglyceride (f) and phospholipids (g) were detected in A2780 and SKOV3 cells with treatment as indicated. h The content of neutral lipids were detected by BODIPY 493/503 dye and counterstained with DAPI in A2780 and SKOV3 cells with treatment as indicated. Scale bars, 50 μm. Average numbers of LDs per cell (left), percentage of cellular area occupied by LDs (right). Data are shown as mean ± S.E.M from three independent experiments. *p < 0.05; **p < 0.01.
We further explored whether SREBP1c was involved in SIK2-mediated upregulation of FASN in OC cells. Our results showed that SIK2 knockdown significantly reduced the expressions of FASN, the contents of intracellular free fatty acid, triglyceride and phospholipids, as well as the intensity of BODIPY staining in A2780 cells, whereas overexpression of SREBP1c significantly restored the reduction. In contrast, the expression of FASN, content of intracellular free fatty acid, triglyceride and phospholipids, and intensity of BODIPY staining were clearly increased when SIK2 was overexpressed in SKOV3 cells (Fig. 3d–h). Expectedly, silencing of SREBP1c remarkably suppressed the lipogenisis-promoting effect of SIK2 overexpression.
SIK2 upregulated HMGCR expression to promote cholesterol synthesis via SREBP2
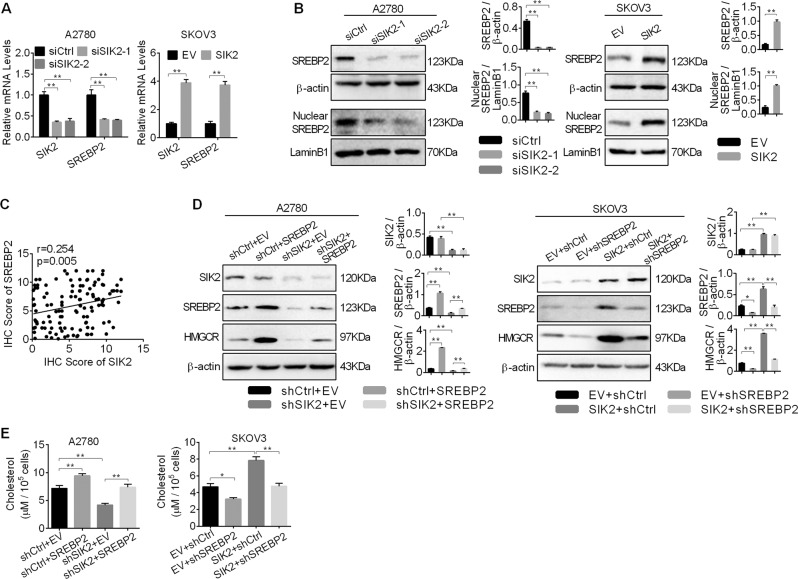
Next, we explored the molecular mechanism of SIK2-mediated upregulation of cholesterol synthesis enzyme HMGCR. Considering that SREBP2 is well-known as a key regulator of cholesterol synthesis through transactivation of genes involved in cholesterol biosynthesis, the effect of SIK2 knockdown on the expression of SREBP2 was examined by qRT-PCR and Western blot analyses. As shown in Fig. 4a, b, SREBP2 was significantly decreased at both mRNA and protein levels when SIK2 was knocked-down in A2780 cells, whereas SIK2 overexpression significantly increased SREBP2 expression in SKOV3 cells. In addition, the nuclear expression of SREBP2, which represents the transcriptional activity of this molecule, showed a similar expression pattern to that of total SREBP2 (Fig. 4b), implying that SIK2 promotes the transcriptional activity of SREBP2 in OC cells. Moreover, spearman rank correlation analysis also showed a positive correlation of expression levels between SIK2 and SREBP2 in tumor tissues from 121 OC patients (r = 0.254, p = 0.005) (Fig. 4c and Supplementary Fig. S2A).
Fig. 4. SIK2 upregulated HMGCR to promote cholesterol synthesis via SREBP2.
a, b Quantitative RT-PCR (a) and Western blot (b) analyses for expressions of SREBP2 in A2780 and SKOV3 cells with treatment as indicated. c Spearman correlation analysis of the relationships between the protein expression levels of SIK2 and SREBP2 based on the IHC staining results. d Western blot analysis of SREBP2 and HMGCR in A2780 and SKOV3 cells with treatment as indicated (shSIK2, shRNA against SIK2; shCtrl, control shRNA; SREBP2, expression vector encoding SREBP2; EV, empty vector). e The intracellular levels of cholesterol was detected in A2780 and SKOV3 cells with treatment as indicated. Data are shown as mean ± S.E.M from three independent experiments. *p < 0.05; **p < 0.01.
We further explored whether SREBP2 was involved in SIK2-upregulated HMGCR and cholesterol synthesis in OC cells. Our results showed that SIK2 knockdown significantly reduced the expression of HMGCR and the intracellular levels of cholesterol, whereas overexpression of SREBP2 significantly restored the reduction (Fig. 4d, e). In contrast, the expression of HMGCR and the intracellular levels of cholesterol was clearly increased when SIK2 was overexpressed in SKOV3 cells, whereas silencing of SREBP2 remarkably suppressed the intracellular levels of cholesterol of SIK2 overexpression (Fig. 4d, e).
SIK2 promoted the upregulation of SREBP1c and SREBP2 by activating the PI3K/Akt signaling pathway
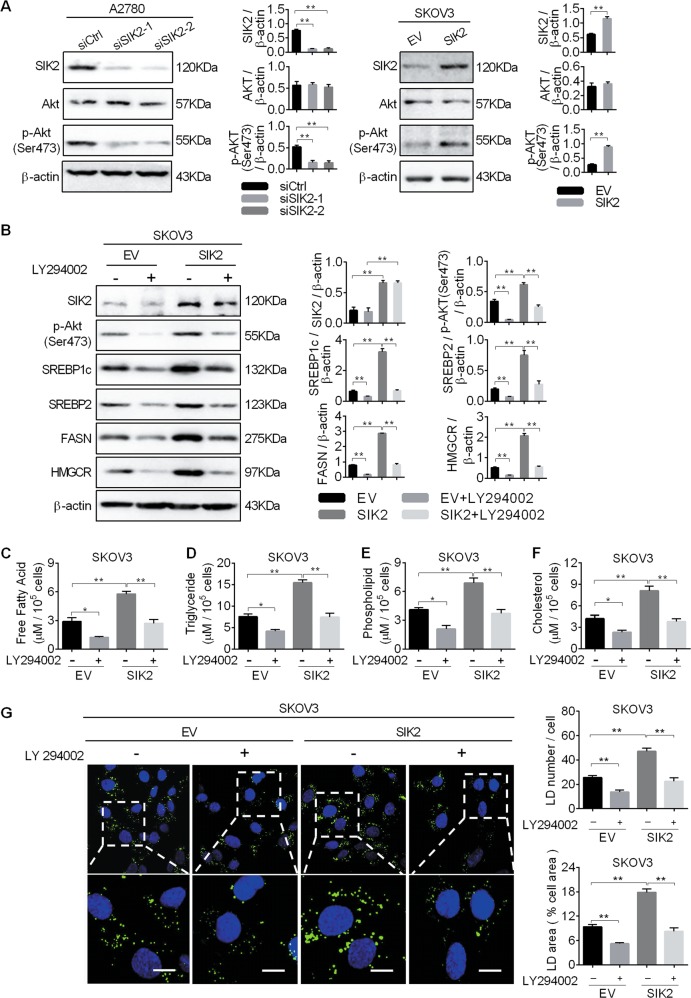
Several previous studies have demonstrated that activation of the oncogenic PI3K/Akt pathway plays a critical role in the lipid synthesis23,24. Because PI3K/Akt has been reported to be activated by SIK2 in OC cells17, we hypothesized that SIK2 activated PI3K/Akt pathway would promote the SREBP1c- and SREBP2-mediated upregulation of fatty acid and cholesterol synthetic enzymes. As shown in Fig. 5a, the phosphorylation level of Akt (Ser473) was significantly decreased in A2780 cells when SIK2 was knocked down, whereas SIK2 overexpression exhibited an opposite effect on the phosphorylation level of Akt (Ser473) in SKOV3 cells. To further study the involvement of PI3K/Akt, we treated OC cells with LY294002, a highly selective PI3K inhibitor. Our results showed that LY294002 treatment markedly decreased the phosphorylation level of Akt (Ser473) in SKOV3 cells with or without SIK2 over-expression. Meanwhile, the expression levels of SREBP1c, SREBP2, FASN and HMGCR were remarkably decreased by LY294002 treatment (Fig. 5b). Expectedly, the decreased levels of intracellular free fatty acid, triglyceride, phospholipids, cholesterol and BODIPY staining of neutral lipids were observed upon treatment with LY294002 (Fig. 5c–g).
Fig. 5. SIK2 promoted the upregulation of SREBP1c and SREBP2 by activating the PI3K/Akt signaling pathway.
a Western blot analysis for total Akt and phosphorylated Akt (Ser473) in A2780 and SKOV3 cells with treatment as indicated. b Western blot analysis for expression of p-Akt (Ser473), SREBP1c, SREBP2, FASN and HMGCR in SKOV3 cells treated with an Akt inhibitor LY294002 (LY294002, 5 μM PI3K/Akt signaling inhibitor for 12 h). c–f Cellular content of free fatty acid (c), triglyceride (d), phospholipids (e) and cholesterol (f) were detected in SKOV3 treated with Akt inhibitor LY294002. g The content of neutral lipids were detected by BODIPY 493/503 dye and counterstained with DAPI in SKOV3 cells treated with Akt inhibitor LY294002. Scale bars, 50μm. Average numbers of LDs per cell (upper panel), percentage of cellular area occupied by LDs (lower panel). Data are shown as mean ± S.E.M from three independent experiments. *p < 0.05; **p < 0.01.
SIK2 promoted the growth of OC cells by enhancing fatty acid and cholesterol synthesis
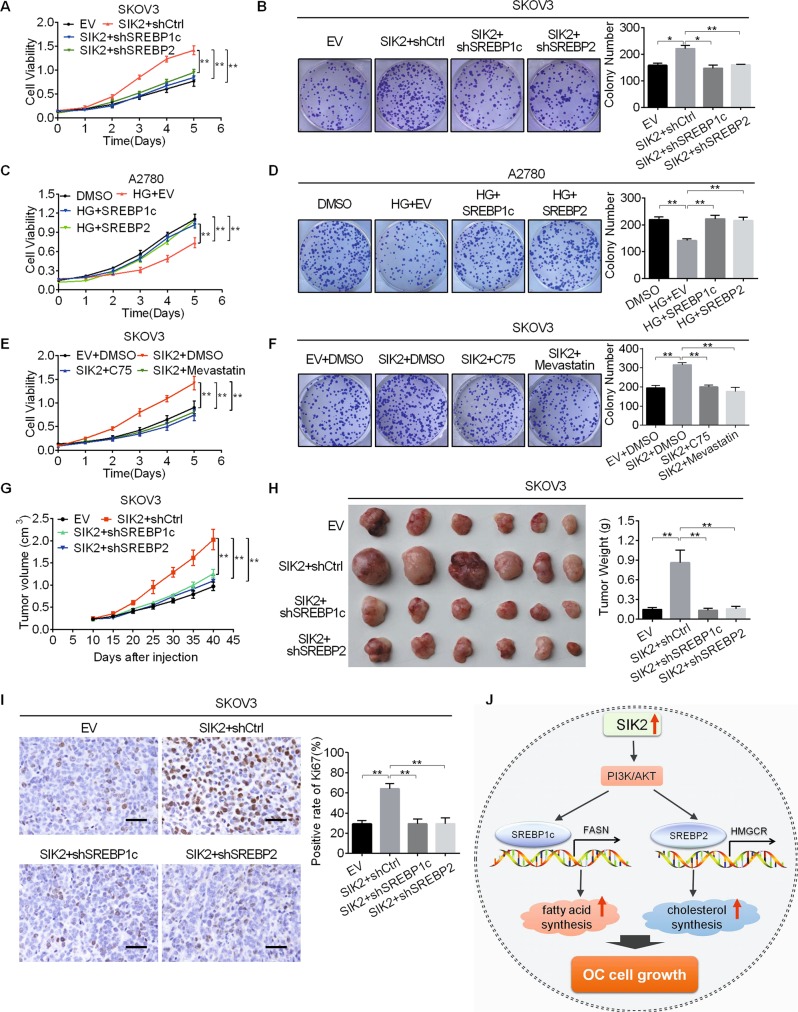
Considering the important roles of fatty acid and cholesterol synthesis in tumor growth, we hypothesized that SIK2 might promote OC growth through enhancing fatty acid and cholesterol synthesis. To test this hypothesis, pSilencerTM3.1-shSREBP1c and pSilencerTM3.1-shSREBP2 plasmids were respectively transfected into SKOV3 cells (Supplementary Fig. S3A, B) to construct stable cell lines. We found that SIK2 overexpression significantly promoted the proliferation (Fig. 6a and Supplementary Fig. S3C) and colony formation of SKOV3 cells (Fig. 6b), which were attenuated by either knockdown of SREBP1c or SREBP2. Whereas, overexpression of SREBP1c or SREBP2 restored the inhibitory effects of HG-9-91-01 (an inhibitor of SIK) treatment in A2780 cells (Supplementary Fig. S3D-G and Fig. 6c, d). To provide further supports, fatty acid and cholesterol synthesis were suppressed using C75 (an inhibitor of FASN) and Mevastatin (an inhibitor of HMGCR). As shown in Fig. 6E, F and Supplementary Fig. S3H, SIK2 overexpression significantly promoted the proliferation and colony formation of SKOV3 cells, which were significantly attenuated by treatment with C7525,26 at a concentration of 25 μM for 24 h or Mevastatin27 at a concentration of 1 μM for 24 h. To evaluate the potential role of enhanced fatty acid and cholesterol synthesis in SIK2-mediated OC growth in vivo, stable cell lines were established and injected subcutaneously into the nude mice. The SIK2 overexpression group exhibited a much faster tumor growth than the control group, whereas the tumor growth-promoting effect of SIK2 was attenuated by knockdown of SREBP1c or SREBP2 (Fig. 6g, h). Immunohistochemical staining analysis indicated a significantly high percentage of Ki-67 positive cells in SIK2-overexpressed xenografts when compared with control, whereas knockdown of SREBP1c or SREBP2 exhibited an opposite effect (Fig. 6i).
Fig. 6. SIK2 promoted the growth of OC cells by enhancing fatty acid and cholesterol synthesis.
a Cell proliferation ability was evaluated using the MTS assay in SKOV3 cells treated as indicated (EV, empty vector; shCtrl, control shRNA; SIK2, expression vector encoding SIK2; shSREBP1c, shRNAs against SREBP1c; shSREBP2, shRNAs against SREBP2). b Colony formation assay in SKOV3 cells treated as indicated. c Cell proliferation ability was evaluated using the MTS assay in A2780 cells treated as indicated (HG (SIK inhibitor HG-9-91-01), 4 μM for 8 h; SREBP1c, expression vector encoding SREBP1c; SREBP2, expression vector encoding SREBP2). d Colony formation assay in A2780 cells treated as indicated. e Cell proliferation ability was evaluated using the MTS assay in SKOV3 cells treated as indicated (EV, empty vector; SIK2, expression vector encoding SIK2; C75, FASN inhibitor, 25 μM for 24 h; Mevastatin, HMGCR inhibitor, 1 μM for 24 h). f Colony formation assay in SKOV3 cells treated as indicated. g Tumor growth curves of subcutaneous xenograft model for SKOV3 cells with treatment as indicated. h The dissected tumors from sacrificed mice and their weight were shown. i The expression of Ki-67 in subcutaneous xenografts from different groups were confirmed by immunohistochemistry (IHC) analysis. j Schematic depicting the regulation of fatty acid metabolism by SIK2 in OC cells. Scale bars, 50 μm. Data are shown as mean ± S.E.M from three independent experiments. *p < 0.05; **p < 0.01.
Take into account that SIK2 also promotes the metastasis of OC cells17, we explored whether SIK2 promoted OC metastasis through enhancing fatty acid and cholesterol synthesis on metastasis of OC cells. As shown in Supplementary Fig. S4A, B, SIK2 overexpression significantly promoted the migration and invasion abilities of OC cells, which were attenuated by either knockdown of SREBP1c or SREBP2. Whereas, overexpression of SREBP1c or SREBP2 restored the effects of HG-9-91-01 treatment in A2780 cells (Supplementary Fig. S4C, D). Expectedly, inhibition of fatty acid and cholesterol synthesis using C75 (inhibitor of FASN) and Mevastatin (inhibitor of HMGCR) showed similar results to the effects of SREBP1c or SREBP2 knockdown (Supplementary Fig. S4E, F). These findings indicate that SIK2 promotes both the growth and metastasis of OC cells through enhancing fatty acid and cholesterol synthesis.
Discussion
Reprogrammed cellular metabolic pathways of fatty acid and cholesterol synthesis have been recognized as common metabolic hallmarks in many different tumor types, including ovarian cancer2,28,29. However, the molecular mechanisms underlying the reprogramming process of lipid metabolism remains largely unknown. In this study, we demonstrated that the SIK2, a member of the AMPK family of kinases, enhanced the fatty acid and cholesterol synthesis in OC cells, which is important for the progression of OC. In addition, our findings indicated that SIK2 enhanced fatty acid and cholesterol synthesis through upregulating the expression of SREBP1c/FASN and SREBP2/HMGCR through activating PI3K/Akt signaling pathway. In contrast, several previous studies in nonmalignant cells such as adipocytes and preadipocytes have demonstrated that SIK2 negatively regulated the lipogenesis30,31. These findings suggest that SIK2 may play distinct and even opposite roles in the regulation of lipogenesis in tumor and non-tumor cells, which still needs further confirmation.
The increased de novo fatty acid synthesis is caused by multiple mechanisms, most of which includes the increased expression of lipogenic enzymes. It has been reported that the expression of FASN is increased in high grade serous carcinomas and its overexpression is correlated with poor outcome for women with OC32,33. In OC cell lines, overexpression of FASN has also been observed to promote tumor cell growth and its inhibition serves as a promising therapeutic strategy34,35. Consistently, our results demonstrated that SIK2 promoted fatty acid synthesis of OC cells through up-regulation of FASN, which further support FASN as a critical oncogene in cancer progression. Compared with most current studies focusing on de novo fatty acid synthesis, cholesterol metabolism in cancer development has received less attention. Currently, in several types of cancer cells, it has been demonstrated that tumor cells display the altered cholesterol metabolism which is characterized by the elevated intracellular cholesterol level. Chushi Li et al. have reported that the rate-limiting enzyme HMGCR for cholesterol synthesis is significantly up-regulated in gastric cancer, which promotes the malignant biological behavior of the cancer cells11. Besides, the oncogenic effect of HMGCR has also been demonstrated in glioblastoma and prostate cancer cells12,36. In OC cells, HMGCR has been reported to play a role in cisplatin resistance37 and its inhibition exhibits anti-metastatic and anti-tumorigenic effects38. In accordance, our results demonstrated that SIK2 promoted cholesterol biosynthesis and thus OC cell growth through the up-regulation of HMGCR.
Sterol regulatory element binding proteins (SREBPs) are the most important transcription factors that regulate lipid homeostasis. In mammalian cells, three SREBP isoforms including SREBP1a, SREBP1c, and SREBP2, have been identified. Among the three SREBP isoforms, SREBP1c preferentially activates enzymes involved in fatty acid and triglyceride metabolism, while SREBP2 preferentially activates enzymes involved in cholesterol metabolism39. Currently, the activation of both SREBP1c and SREBP2 have been observed in many different types of cancer18,40–43, including OC37,44. In accordance, our results showed that SREBP1c and SREBP2 expressions and activities were elevated by SIK2 in ovarian cancers. Cumulative evidences have indicated that the expression of SREBPs are tightly regulated by several well-known oncogenic signaling pathways, particularly the PI3K/Akt pathway23,45,46, which is frequently activated in human cancer cells47. A previous study has demonstrated that PI3K/Akt pathway could be activated by SIK2 in OC cells17. Our present study further showed that the activation of the PI3K/Akt signaling was involved in the upregulation of SREBP1c and SREBP2 by SIK2 in OC cells. In addition to SREBPs, ChREBP is another important transcriptional regulator involved in the regulation of lipogenesis. A previous study has demonstrated that SIK2 inhibited the activity of ChREBP transactivation through p300-mediated acetylation in mouse hepatocytes48. Accordingly, although no significant effect of SIK2 on the expression of ChREBP were observed in our present study, we still cannot rule out the possibility that ChREBP may also be activated by SIK2 in OC cells and thus play a role in SIK2-regulated lipogenesis and tumor progression.
Increased fatty acid and cholesterol synthesis have been linked to tumorigenesis and cancer progression. Several previous pre-clinical studies have demonstrated that the decrease of cholesterol synthesis inhibits cell proliferation, invasion and metastasis in OC38,49. Previous studies have demonstrated that SIK2 plays an important role in the cell proliferation and survival in OC15,50. Considering that fatty acid and cholesterol function as the major building blocks and signaling molecules which are required in cancer cells, we propose that the SIK2-regulated fatty acid and cholesterol synthesis may be involved in the OC growth. As expected, our study demonstrated that either over-expression of SREBP1c or SREBP2 robustly reversed the inhibition of growth of OC cells both in vivo and in vitro by SIK2-silencing or inhibition.
In summary, we demonstrate a key regulatory role of SIK2 in lipid metabolism of OC cells. Our study provides novel insights to understand the molecular mechanisms underlying the reprogrammed lipid metabolism in cancer cells, as well as a strong line of evidence for this molecule to be used a therapeutic target in OC treatment.
Supplementary information
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Program No. 81972427 and 81672828), State Key Laboratory of Cancer Biology Project (Program No. CBSKL2017Z01), Natural Science Basic Research Plan in Shaanxi Province of China (Program No. 2018JM7018), National Natural Science Foundation of China (Program No. 81772618).
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Edited by J.E. Chipuk
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Jing Zhao, Xiaohong Zhang, Tian Gao
Contributor Information
Jibin Li, Email: lijibin2006@163.com.
Shujuan Liu, Email: hanliu@fmmu.edu.cn.
Supplementary information
Supplementary Information accompanies this paper at (10.1038/s41419-019-2221-x).
References
- 1.Rohrig F, Schulze A. The multifaceted roles of fatty acid synthesis in cancer. Nat. Rev. Cancer. 2016;16:732–749. doi: 10.1038/nrc.2016.89. [DOI] [PubMed] [Google Scholar]
- 2.Corbet C, Feron O. Emerging roles of lipid metabolism in cancer progression. Curr. Opin. Clin. Nutr. Metab. Care. 2017;20:254–260. doi: 10.1097/MCO.0000000000000381. [DOI] [PubMed] [Google Scholar]
- 3.Currie E, Schulze A, Zechner R, Walther TC, Farese RV., Jr. Cellular fatty acid metabolism and cancer. Cell Metab. 2013;18:153–161. doi: 10.1016/j.cmet.2013.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cha JY, Lee HJ. Targeting lipid metabolic reprogramming as anticancer therapeutics. J. Cancer Prev. 2016;21:209–215. doi: 10.15430/JCP.2016.21.4.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Murai T. Cholesterol lowering: role in cancer prevention and treatment. Biol. Chem. 2015;396:1–11. doi: 10.1515/hsz-2014-0194. [DOI] [PubMed] [Google Scholar]
- 6.Wang HH, Garruti G, Liu M, Portincasa P, Wang DQ. Cholesterol and lipoprotein metabolism and atherosclerosis: recent advances in reverse cholesterol transport. Ann. Hepatol. 2017;16:s27–s42. doi: 10.5604/01.3001.0010.5495. [DOI] [PubMed] [Google Scholar]
- 7.Wang J, Wang WJ, Zhai L, Zhang DF. Association of cholesterol with risk of pancreatic cancer: a meta-analysis. World J. Gastroenterol. 2015;21:3711–3719. doi: 10.3748/wjg.v21.i12.3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nelson ER, Chang CY, McDonnell DP. Cholesterol and breast cancer pathophysiology. Trends Endocrinol. Metab. 2014;25:649–655. doi: 10.1016/j.tem.2014.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuzu OF, Noory MA, Robertson GP. The role of cholesterol in cancer. Cancer Res. 2016;76:2063–2070. doi: 10.1158/0008-5472.CAN-15-2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Llaverias G, et al. Role of cholesterol in the development and progression of breast cancer. Am. J. Pathol. 2011;178:402–412. doi: 10.1016/j.ajpath.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chushi L, et al. HMGCR is up-regulated in gastric cancer and promotes the growth and migration of the cancer cells. Gene. 2016;587:42–47. doi: 10.1016/j.gene.2016.04.029. [DOI] [PubMed] [Google Scholar]
- 12.Qiu Z, et al. HMGCR positively regulated the growth and migration of glioblastoma cells. Gene. 2016;576:22–27. doi: 10.1016/j.gene.2015.09.067. [DOI] [PubMed] [Google Scholar]
- 13.Zhong C, et al. HMGCR is necessary for the tumorigenecity of esophageal squamous cell carcinoma and is regulated by Myc. Tumour Biol. 2014;35:4123–4129. doi: 10.1007/s13277-013-1539-8. [DOI] [PubMed] [Google Scholar]
- 14.Chen F, Chen L, Qin Q, Sun X. Salt-inducible kinase 2: an oncogenic signal transmitter and potential target for cancer therapy. Front Oncol. 2019;9:18. doi: 10.3389/fonc.2019.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ahmed AA, et al. SIK2 is a centrosome kinase required for bipolar mitotic spindle formation that provides a potential target for therapy in ovarian cancer. Cancer Cell. 2010;18:109–121. doi: 10.1016/j.ccr.2010.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bon H, et al. Salt-inducible kinase 2 regulates mitotic progression and transcription in prostate cancer. Mol. Cancer Res. 2015;13:620–635. doi: 10.1158/1541-7786.MCR-13-0182-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miranda F, et al. Salt-inducible kinase 2 couples ovarian cancer cell metabolism with survival at the adipocyte-rich metastatic niche. Cancer Cell. 2016;30:273–289. doi: 10.1016/j.ccell.2016.06.020. [DOI] [PubMed] [Google Scholar]
- 18.Li J, et al. CD147 reprograms fatty acid metabolism in hepatocellular carcinoma cells through Akt/mTOR/SREBP1c and P38/PPARalpha pathways. J. Hepatol. 2015;63:1378–1389. doi: 10.1016/j.jhep.2015.07.039. [DOI] [PubMed] [Google Scholar]
- 19.Li J, et al. Mitochondrial elongation-mediated glucose metabolism reprogramming is essential for tumour cell survival during energy stress. Oncogene. 2017;36:4901–4912. doi: 10.1038/onc.2017.98. [DOI] [PubMed] [Google Scholar]
- 20.Huang Q, et al. CD147 promotes reprogramming of glucose metabolism and cell proliferation in HCC cells by inhibiting the p53-dependent signaling pathway. J. Hepatol. 2014;61:859–866. doi: 10.1016/j.jhep.2014.04.035. [DOI] [PubMed] [Google Scholar]
- 21.Wang Y, Viscarra J, Kim SJ, Sul HS. Transcriptional regulation of hepatic lipogenesis. Nat. Rev. Mol. Cell Biol. 2015;16:678–689. doi: 10.1038/nrm4074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shimomura I, Shimano H, Horton JD, Goldstein JL, Brown MS. Differential expression of exons 1a and 1c in mRNAs for sterol regulatory element binding protein-1 in human and mouse organs and cultured cells. J. Clin. Invest. 1997;99:838–845. doi: 10.1172/JCI119247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krycer JR, Sharpe LJ, Luu W, Brown AJ. The Akt-SREBP nexus: cell signaling meets lipid metabolism. Trends Endocrinol. Metab. 2010;21:268–276. doi: 10.1016/j.tem.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 24.Liao X, et al. LAMP3 regulates hepatic lipid metabolism through activating PI3K/Akt pathway. Mol. Cell Endocrinol. 2018;470:160–167. doi: 10.1016/j.mce.2017.10.010. [DOI] [PubMed] [Google Scholar]
- 25.Uddin S, et al. Overexpression of fatty acid synthase in Middle Eastern epithelial ovarian carcinoma activates AKT and Its inhibition potentiates cisplatin-induced apoptosis. Mol. Med. 2011;17:635–645. doi: 10.2119/molmed.2011.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen HW, Chang YF, Chuang HY, Tai WT, Hwang JJ. Targeted therapy with fatty acid synthase inhibitors in a human prostate carcinoma LNCaP/tk-luc-bearing animal model. Prostate Cancer Prostatic Dis. 2012;15:260–264. doi: 10.1038/pcan.2012.15. [DOI] [PubMed] [Google Scholar]
- 27.Hsu CK, Lin CC, Hsiao LD, Yang CM. Mevastatin ameliorates sphingosine 1-phosphate-induced COX-2/PGE2-dependent cell migration via FoxO1 and CREB phosphorylation and translocation. Br. J. Pharm. 2015;172:5360–5376. doi: 10.1111/bph.13326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu Q, Luo Q, Halim A, Song G. Targeting lipid metabolism of cancer cells: a promising therapeutic strategy for cancer. Cancer Lett. 2017;401:39–45. doi: 10.1016/j.canlet.2017.05.002. [DOI] [PubMed] [Google Scholar]
- 29.Kouba S, et al. Lipid metabolism and Calcium signaling in epithelial ovarian cancer. Cell Calcium. 2019;81:38–50. doi: 10.1016/j.ceca.2019.06.002. [DOI] [PubMed] [Google Scholar]
- 30.Park J, et al. SIK2 is critical in the regulation of lipid homeostasis and adipogenesis in vivo. Diabetes. 2014;63:3659–3673. doi: 10.2337/db13-1423. [DOI] [PubMed] [Google Scholar]
- 31.Du J, Chen Q, Takemori H, Xu H. SIK2 can be activated by deprivation of nutrition and it inhibits expression of lipogenic genes in adipocytes. Obesity (Silver Spring) 2008;16:531–538. doi: 10.1038/oby.2007.98. [DOI] [PubMed] [Google Scholar]
- 32.Cai Y, et al. Expressions of fatty acid synthase and HER2 are correlated with poor prognosis of ovarian cancer. Med. Oncol. 2015;32:391. doi: 10.1007/s12032-014-0391-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ueda SM, et al. Expression of fatty acid synthase depends on NAC1 and is associated with recurrent ovarian serous carcinomas. J. Oncol. 2010;2010:285191. doi: 10.1155/2010/285191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wagner R, et al. Multi-level suppression of receptor-PI3K-mTORC1 by fatty acid synthase inhibitors is crucial for their efficacy against ovarian cancer cells. Oncotarget. 2017;8:11600–11613. doi: 10.18632/oncotarget.14591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rahman MT, et al. Fatty acid synthase expression associated with NAC1 is a potential therapeutic target in ovarian clear cell carcinomas. Br. J. Cancer. 2012;107:300–307. doi: 10.1038/bjc.2012.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ashida S, Kawada C, Inoue K. Stromal regulation of prostate cancer cell growth by mevalonate pathway enzymes HMGCS1 and HMGCR. Oncol. Lett. 2017;14:6533–6542. doi: 10.3892/ol.2017.7025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zheng L, Li L, Lu Y, Jiang F, Yang XA. SREBP2 contributes to cisplatin resistance in ovarian cancer cells. Exp. Biol. Med. 2018;243:655–662. doi: 10.1177/1535370218760283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stine JE, et al. The HMG-CoA reductase inhibitor, simvastatin, exhibits anti-metastatic and anti-tumorigenic effects in ovarian cancer. Oncotarget. 2016;7:946–960. doi: 10.18632/oncotarget.5834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eberle D, Hegarty B, Bossard P, Ferre P, Foufelle F. SREBP transcription factors: master regulators of lipid homeostasis. Biochimie. 2004;86:839–848. doi: 10.1016/j.biochi.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 40.Chen A, et al. FTO promotes SREBP1c maturation and enhances CIDEC transcription during lipid accumulation in HepG2 cells. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2018;1863:538–548. doi: 10.1016/j.bbalip.2018.02.003. [DOI] [PubMed] [Google Scholar]
- 41.Wen YA, et al. Downregulation of SREBP inhibits tumor growth and initiation by altering cellular metabolism in colon cancer. Cell Death Dis. 2018;9:265. doi: 10.1038/s41419-018-0330-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sethi, G., Shanmugam, M. K. & Kumar, A. P. SREBP-1c as a molecular bridge between lipogenesis and cell cycle progression of clear cell renal carcinoma. Biosci. Rep.37, 10.1042/BSR20171270 (2017). [DOI] [PMC free article] [PubMed]
- 43.Zhong, C., Fan, L., Li, Z., Yao, F. & Zhao, H. SREBP2 is upregulated in esophageal squamous cell carcinoma and cooperates with cMyc to regulate HMGCR expression. Mol. Med. Rep., 10.3892/mmr.2019.10577 (2019). [DOI] [PMC free article] [PubMed]
- 44.Do MT, Hwang YP, Kim HG, Na M, Jeong HG. Mollugin inhibits proliferation and induces apoptosis by suppressing fatty acid synthase in HER2-overexpressing cancer cells. J. Cell. Physiol. 2013;228:1087–1097. doi: 10.1002/jcp.24258. [DOI] [PubMed] [Google Scholar]
- 45.Shi Q, Hoffman B, Liu Q. PI3K-Akt signaling pathway upregulates hepatitis C virus RNA translation through the activation of SREBPs. Virology. 2016;490:99–108. doi: 10.1016/j.virol.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 46.Huang J, Chen S, Cai D, Bian D, Wang F. Long noncoding RNA lncARSR promotes hepatic cholesterol biosynthesis via modulating Akt/SREBP-2/HMGCR pathway. Life Sci. 2018;203:48–53. doi: 10.1016/j.lfs.2018.04.028. [DOI] [PubMed] [Google Scholar]
- 47.Mundi PS, Sachdev J, McCourt C, Kalinsky K. AKT in cancer: new molecular insights and advances in drug development. Br. J. Clin. Pharmacol. 2016;82:943–956. doi: 10.1111/bcp.13021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bricambert J, et al. Salt-inducible kinase 2 links transcriptional coactivator p300 phosphorylation to the prevention of ChREBP-dependent hepatic steatosis in mice. J. Clin. Invest. 2010;120:4316–4331. doi: 10.1172/JCI41624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martirosyan A, Clendening JW, Goard CA, Penn LZ. Lovastatin induces apoptosis of ovarian cancer cells and synergizes with doxorubicin: potential therapeutic relevance. BMC Cancer. 2010;10:103. doi: 10.1186/1471-2407-10-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.SIK2 Promotes Ovarian Cancer Spread. Cancer Discov. 2016;6:OF1. doi: 10.1158/2159-8290.CD-NB2016-109. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.