Abstract
Flow cytometry is currently underutilized for bacterial phenotyping and standard microbiological techniques do not provide phenotypic information about the state of the bacterial disease. Pseudomonas aeruginosa is a human pathogen of increased importance in public health due to both the ability to cause chronic diseases and the prevalence of functionally different subsets that can be difficult to treat and diagnose. In the present study, we used flow cytometry to analyze the growth phase of P. aeruginosa. A simple method for single cell quantitative detection of bacterial biofilm and planktonic cells was established with a combination of membrane permeable (SYTO 60) and impermeable (TOTO-1) dyes plus the addition of polystyrene counting beads. The specificity of the dye combination for biofilm detection was determined by comparison with impaired biofilm forming strains of P. aeruginosa LasI/RhlI−/− and ΔPfPhage. Results suggest that flow cytometric bacterial phenotyping serves as an expandable platform that may be useful for enumeration of population level variation in P. aeruginosa studies.
INTRODUCTION
Biofilm diseases are a growing public health problem with limited resolution using traditional antibacterial therapies and diagnostics (Costerton et al., 1995; 1999). The World Health Organization priority pathogen list encompasses biofilm forming pathogens, and it is estimated that the large majority of infectious bacteria are capable of biofilm formation (WHO, 2017; Lebeaux et al., 2014; Romling and Balsalobre, 2012). As a biofilm-forming pathogen, Pseudomonas aeruginosa forms multicellular communities consisting of planktonic cells that contribute to the spread of infection and cells within a matrix-protected biofilm including unflagellated cells, persister cells, and viable but non-culturable cells (VNBCs) (Ayrapetyan et al., 2018; Magana et al., 2018). High percentages of persister cells and VNBCs can contribute to underestimation of colony forming units (CFU) and both sensitivity limits and culture conditions for these cell types can occasionally lead to culture-negative results for patients (Ayrapetyan et al., 2018; Magana et al., 2018). Assessment of these cell types is limited to research laboratories, where it is known that the phenotype can impact host response (Secor et al., 2011; Yamada et al., 2018). Inability to rapidly identify emerging infections can contribute to increasing hospital-acquired infection rates and prolong hospitalization. Inefficient antibiotic exposure can compound difficulties for success rates of clinical treatment (Magana et al., 2018).
Bacterial growth phase assessment of whether a cell is in a planktonic or biofilm state can be enumerated by sample staining coupled with microscopy or spectroscopy but these techniques are limited in their ability to retrieve additional population level information and staining protocols decay the viability of the sample. Crystal violet staining is commonly used for biofilm quantification, though it does not reveal the proportion of planktonic cells or provide any information on living versus dead cells in a biofilm matrix (Magana et al., 2018). Recently, a combination of membrane permeable and impermeable DNA-binding dyes has been reported for use in fluorescent microscopy (Okshevsky and Meyer, 2014b). This approach characterizes the biofilm based on identification of both extracellular DNA (eDNA) from the biofilm matrix and living cells using TOTO-1 and SYTO 60, respectively (Allesen-Holm, 2006; Das et al., 2010; Hall-Stoodley et al., 2008; Jennings et al., 2015; Mann et al., 2009; Okshevsky and Meyer, 2014a; Seper et al., 2011; Qin et al., 2007; Whitchurch et al., 2002). While shown to be accurate for biofilm identification, fluorescence microscopy preparation decays the sample and is time consuming for resolution on the population-level (Magana et al., 2018).
Flow cytometry (FCM) tools are being developed for bacteria and fluorescence-activated cell sorting (FACS) to maintain the isolation of viable cells post-sort (Brown et al., 2019; Fontana et al., 2017; Steen 2000; Trevathan-Tackett et al., 2014). Indeed, several indicators of viability, as well as phenotypic and functional attributes such as phosphatase activity and membrane fluidity have now been assessed via FCM (Duhamel et a., 2008; Marielle et al., 2000; Nescerechka et al., 2016; Stocks 2004). The FCM approach to quantify biofilm uses TO-PRO-3 iodide as a single-stain viability dye. The benefit of this method is that TO-PRO-3 iodide stains the DNA of dead cells, as well as the cell wall of viable cells. The difference in fluorescence intensity enables the identification of both viable and dead cells. Accordingly, TO-PRO-3 has been proposed to discriminate and quantify viable and dead cells with a single stain. However, recent evidence from fluorescent stains for visualizing eDNA in biofilms revealed that TO-PRO-3 lacked specificity for eDNA and overestimated biofilm formation because it penetrated both viable and dead cells (Okshevsky and Meyer, 2014b). This finding suggests that the erroneous results due to the passive diffusion of TO-PRO-3 inside the viable cells will be significant if the cells are not analyzed quickly after staining. To address this problem, we established a dual-stain procedure based on cell permeable SYTO 60 and the eDNA stain TOTO-1, which display excellent sensitivity for visualization of eDNA versus TO-PRO-3. In the present paper, we describe how this technique could be applied for growth phase determination. We demonstrate that the combination of TOTO-1 with SYTO 60 and FCM allow for an accurate identification and quantitation of biofilm cells.
A pair of P. aeruginosa strains known to produce less robust biofilms were chosen to test the specificity of the panel for planktonic and biofilm cells: ΔPfPhage and LasI/RhlI−/− P. aeruginosa. The LasI/RhlI−/− strain lacks regulators of quorum sensing important for early and late biofilm stage development (Siehnel et al., 2010; Kalia et al., 2018; Nelson et al., 2009). Pf bacteriophage is a viral infection of bacteria that has been observed to change the biophysical properties of biofilm including crystal assembly and mucoid birefringence and the ΔPfPhage strain lacks this (Secor et al., 2015; Sweere et al., 2019). These strains were compared to the wildtype parent strain P. aeruginosa PA01.
METHODS
Reagents and instruments.
An LSRII equipped with Blue, Violet, Green, Red Lasers was used for analysis. SSC was used as a threshold; it was set using a buffer only control and adjusted until minimal noise was seen (Rudy et al., 2014).
Bacterial strains and culture methods.
P. aeruginosa wildtype strain PA01 and a strain reduced in biofilm growth P. aeruginosa ΔPfPhage, derived from PA01 ATCC 47085 (via the Rice Lab of Nanyang Technological University) were donated from the laboratory of Prof. Paul Bollyky, M.D./Ph.D. (Stanford University Medical Center) (Secor et al., 2015; Sweere et al., 2019). Another strain with reduced biofilm growth, P. aeruginosa LasI/RhlI−/− were supplied by the Singh Lab (University of Washington) via the Secor Lab (University of Montana) (Siehnel et al., 2010). P. aeruginosa PA14 was donated from the Hancock lab (University of British Columbia) (Rahme et al., 1995; Pletzer et al., 2017).
Logarithmic growth phase was obtained by shaking at 200 RPM at 37°C for 3 hours. For biofilm preparation, overnight cultures were diluted to an OD 0.1 at 600nm in Luria-Bertani broth. Bacterial suspensions were aliquoted into 96-well microtiter plates (Cole-Palmer) and incubated under static growth at 37°C for 24 or 48 hours. Suspension was then removed and plates were washed twice with equal volume of sterile PBS. Wells were scraped with a syringe and moved to polystyrene tubes for staining. Samples were homogenized and sonicated at 120W (CO-Z) for 5 minutes before dilution in equivalent volume of PBS. Samples were vortexed and centrifuged prior to staining as below.
To confirm staining signal is coming from live bacteria, some samples received a dose of 10% Triton-X 100 for 30 minutes after culture, before staining.
Bacterial growth phase staining.
The staining protocol was adapted from Okshevsky and Meyer (2014b). The fluorescent stains proposed are generally assayed via microscopy: TOTO-1 (T3600, Invitrogen, Carlsbad, CA USA) and SYTO 60 (S11342, Invitrogen, Carlsbad, CA USA) (Table 1). TOTO-1 is an excellent stain for visualizing eDNA biofilm structures because it is a very sensitive and stable DNA-binding stain, and membrane-impermeable for live cells. SYTO 60 is cell permeable and recommended to use as a counterstain with TOTO-1 to produce high quality images of eDNA in an array of Gram-positive and Gram-negative bacterial species. This stain is permeable in both live and dead cells allowing for identification of bacteria without a fluorescent reporter construct.
Table 1.
Absorption, fluorescence maxima, and dye concentration used for staining for the DNA of the dyes tested in this study.
| Dye | Absorbance(nm) | Emission(nm) | Dye Concentration |
|---|---|---|---|
| SYTO 60 | 652 | 678 | 10μM |
| TOTO-1 | 514 | 533 | 2 μM |
1mM of Toto-1 (2 mM stock) and 5 mM of SYTO 60 (10 mM stock) were added to 300 μl of sterile PBS and mixed by vortexing (Okshevsky and Meyer, 2014b). Samples were stained protected from light for 10 minutes at room temperature. Dye was then washed off by centrifugation and samples were suspended in equal volumes of FACS Buffer (PBS and 1% BSA) for analysis.
Identification and quantification by flow cytometry.
The P. aeruginosa gate was determined by stopping the specimen sample to determine noise threshold with the use of a buffer only control. Quantification utilized the Bacterial Counting Kit for flow cytometry (Invitrogen, Carlsbad, CA USA) adapted from manufacturer’s protocol as follows. Microspheres (6 μm, B7277, Invitrogen, Carlsbad, CA USA) were sonicated in a water bath for 5 minutes. Within a biosafety cabinet, 10 μL of the microsphere suspension was added to equivalent dilutions of bacterial specimens. All experiments utilized control specimens of buffer only (no beads), beads only, and single color stains. For quantitation with this kit, specimens were collected based on time. In accordance with the manufacturer’s protocol, the bacteria frame was divided by microsphere frame to determine number of bacteria per 10−6 mL of the sample, otherwise known as the count of bacterial per 0.001 μL of solution. A total of 100,000 events were acquired. All samples were tested with 2 to 3 biological replicates and repeated a minimum of twice.
Data analysis.
Data analysis was performed using FlowJo (BD Biosciences, New Jersey, USA0. Analysis was performed using GraphPad Prism 7 to test statistical significance. P values <0.05 were considered significant.
RESULTS
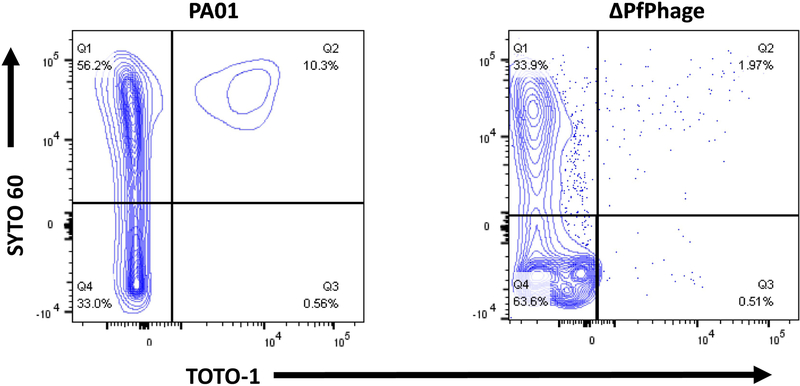
The specificity of the panel for planktonic and biofilm cells was tested on P. aeruginosa strains wildtype (WT) PA01, ΔPfPhage and LasI/RhlI-/-. Wildtype PA01, LasI/RhlI−/−, and ΔPfPhage were compared for dual positive staining efficiency. After 10h incubation, the majority of cells for the ΔPfPhage mutant strain were negative for TOTO-1 (Fig. 1). Substantial dual positive staining was evident in our older WT PA01 planktonic cultures, which maintained a 5% increase in dual-staining versus ΔPfPhage.
Figure 1.
Biofilm Analysis of Biofilm-Impaired Strain by Flow Cytometry.
Two bacteria strains, WT PA01 and ΔPfphage PA, were cultured by shaking to generate old stationary phase (10h). Bacteria were stained with SYTO 60 and TOTO-1 and processed for flow cytometry. The panels feature WT PA01 (L) and ΔPfphage PA (R) dual stained with SYTO 60 and TOTO-1. All samples were gated on SSC/FSC and threshold level set to SSC 200. The experiment was performed in duplicate and independently repeated. Representative figures are shown.
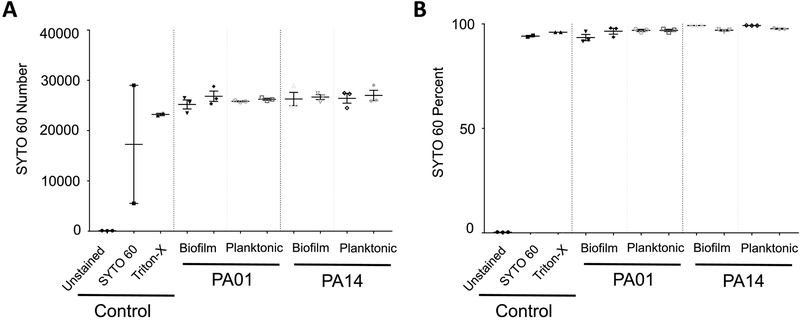
We hypothesized that, versus stationary phase, more of the cells from biofilm cultures would have an extracellular matrix detectable by TOTO-1 staining. Static biofilm cultures were compared to levels of planktonic cells from early logarithmic phase cultures. Initially, we determined that both biofilm and planktonic cultures had positive signal for SYTO 60, indicative of live cells (Fig. 2). These values did not significantly differ for PA01 or PA14 cells due to culture condition. The cell concentration of biofilm cultures was enumerated as number of bacteria per equivalent number of recorded events.
Figure 2.
SYTO 60 Analysis by Flow Cytometry.
Two bacteria strains, WT PA01 and PA14, were cultured by shaking to generate planktonic cultures (3h) or under static conditions to generation biofilm cultures (1d). Bacteria were stained with SYTO 60 and TOTO-1 and processed for flow cytometry. Cumulative data includes, from L to R: unstained, SYTO 60 only, and Triton-X control for WT PA01. Panel A is direct counts and Panel B is percent of cells in bacteria gate. All samples were gated on SSC/FSC and threshold level set to SSC 200. The experiment was performed in triplicate and independently repeated.
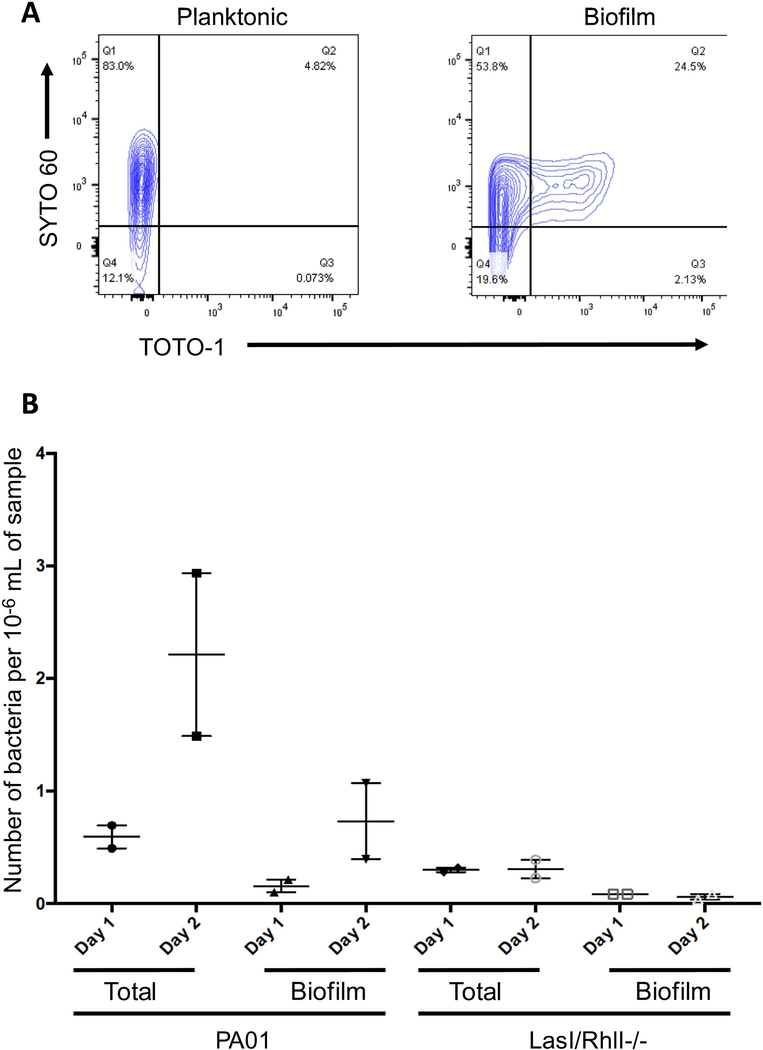
Biofilm has been shown to begin to form in shaking stationary phase cultures after extended time (Schleheck et al., 2009). In agreement with previous reports, we found that WT PA01 had a 2.4-fold increase in dual positive signal for SYTO 60 and TOTO-1 in 10h shaking stationary phase cultures compared to the early logarithmic phase (Fig. 1A & Fig. 3A). Biofilm culture conditions (without shaking) further increase dual positive staining ~5-fold versus early logarithmic planktonic culture conditions (Fig. 3A). After 2d in static incubation, dual SYTO 60 and TOTO-1 staining increased in WT PA01 but the LasI/RhlI−/− mutant eliminated 90% of the cells positive for these properties (Fig. 3B).
Figure 3.
Biofilm Analysis of Early Planktonic Cultures and Biofilm by Flow Cytometry. Bacteria were stained with SYTO 60 and TOTO-1 and processed for flow cytometry.
Panel A features early logarithmic planktonic culture (L) and biofilm culture (R) dual stained with SYTO 60 and TOTO-1. Bacteria (WT PA01) were cultured in two ways, either shaking to generate early logarithmic planktonic culture (3h) or static to generate biofilm (24h). Panel B features dual stained WT PA01 and LasI/RhlI-/−. Bacteria were cultured in static conditions to generate biofilm (24h or 48h). Microsphere gate was divided by bacteria gate to generate counts per unit volume of 1/106 mL. All samples were gated on SSC/FSC and threshold level set to SSC 200. The experiment was performed in duplicate and independently repeated.
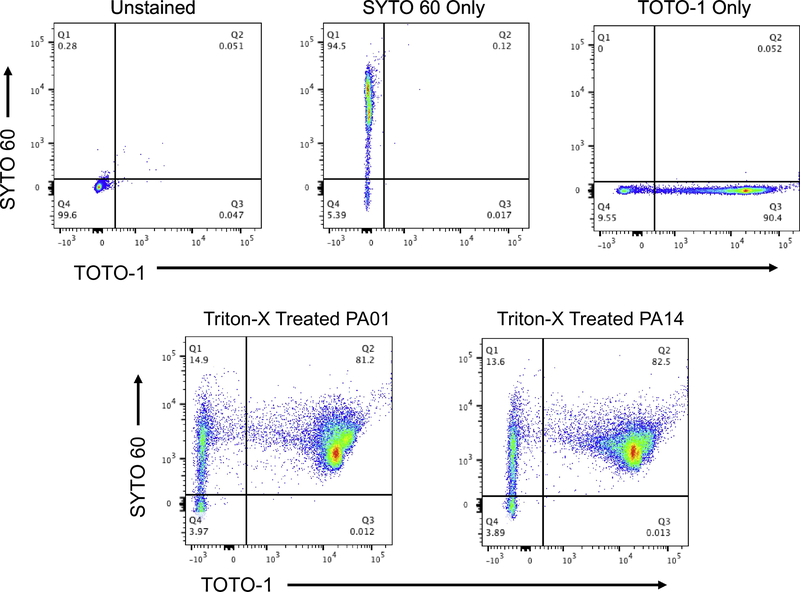
As a positive control for this staining panel, Triton-X treatment was used to reduce membrane discrepancy for TOTO-1. This pretreatment increases the permeability of viable cells, which enhance the diffusion of cell impermeable dye TOTO-1. Complete staining of over ~80% double positive was observed with pre-addition of dilute Triton-X 100, but total cell death equivalent to detergent addition was not observed in any other tested populations (Fig. 4). Cultured recovery confirmed death of Triton-X treated cells. As expected, controls had null values for dual-staining (upper panels, Fig. 4).
Figure 4.
Experimental Controls including Detergent Treated Cells assayed by Flow Cytometry. Bacteria were cultured by shaking to generate planktonic cells (3h).
Bacteria were stained with SYTO 60 and TOTO-1 and processed for flow cytometry. The top panels feature specificity controls, from L to R: unstained, SYTO 60 only, and TOTO-1 only. The bottom panels: Triton-X 100 pretreated (30 min, 10% Triton-X 100 prior to staining) planktonic culture WT PA01 (L); and planktonic culture PA14 (R). All samples were gated on SSC/FSC and threshold level set to SSC 200. Representative images for three independent experiments are shown.
Altogether, these results demonstrate that flow cytometry is a robust system for enumeration of population level variation in P. aeruginosa research.
DISCUSSION
Flow cytometry has many benefits including an adaptable panel design and high throughput single cell screening. Additionally, this technology is more sensitive than other platforms, allowing for quantitative measurements with high resolution and minimal downtime. This work aimed to evaluate the flow cytometric assessment of bacterial growth phase as a quantifiable phenotypic parameter. We found that WT PA01 generated significantly higher dual positive cells in biofilm culture versus logarithmic phase; and that strains reduced in biofilm growth were largely absent for dual positive staining. Although bacteria such as Pseudomonas are known for paralog evolution and gene redundancy, both of the mutants used in this paper are reduced in biofilm-forming capacity compared to wildtype PA01 (Gevers et al., 2004; Ghosh and O’Connor, 2017). We attributed the residual dual-staining of ΔPfPhage as assay noise due to the overlap of this staining panel with viable cell identification.
Flow cytometry panels designed for bacteria are an area of growing need for both clinical diagnostics and research purposes. Biofilm diseases and VNBCs convolute traditional microbiological culture techniques. It will be of interest to add additional markers to the panel and learn whether increased differentiation of matrice-associated molecules can distinguish other subsets in biofilms, such as VNBCs and persister cells, and to ensure viability of each cell subset for downstream analysis.
Phenotypic quantification presented herein remedies issues related to the underestimation of bacterial counts due to the presence of viable but non-culturable cells; as well as the underestimation of CFUs from selective culture conditions and/or the inability to control selective culture conditions with multi-species competition. Density determination reliant on growth is also lengthy, requiring a minimum of 1d turnaround. Another standard way to assess biofilms, electron microscopy, is largely qualitative, leaving a need to quickly perform a quantitative determination of biofilm phenotype (reviewed in Magana et al., 2018).
Despite trial of five different bacterial fixatives, this dye combination did not retain equivalent discrimination parameters under fixation. This is a limitation of this work that could potentially be overcome by substitution of extracellular biofilm matrix stains that don’t overlap with cell death or fixatives that can exhibit their function without altering membrane permeability.
Viability stains and aptamer-based species identification coupled with genetic reporters can create bacteria panels that can provide as much information about infectious bacterial diseases as modern immune panels do for clinical diagnostics. Coupled with automated sample handling and high throughput adaptors, unindentified bacterial samples can be derisked and run in real-time. Additional R&D can advance the versatility of bacterial flow cytometric technology for pressing unmet clinical solutions. Future exploratory research for capturing resistance and tolerance by flow cytometry could lead to an interesting era of ex vivo diagnostic platforms for bacterial infections.
CONCLUSION
Using P. aeruginosa, we observed a time- and culture condition- dependent dual positive percentage in SYTO 60 and TOTO-1 staining by flow cytometry. In accordance with published fluorescent microscopy techniques, these data support the formation of biofilm dependent strongly on time in culture and RPM during incubation (Okshevsky and Meyer, 2014b). We propose that flow cytometry for bacterial phenotyping can be used to identify and count biofilm cells and that this assay has substantial potential to significantly impact future clinical diagnostics. From the data produced, we have validated two fluorescent dyes, TOTO-1 and SYTO 60, that are effective stains to quantify biofilm bacteria at the single cell level.
Highlights:
A combination of the DNA-binding dyes SYTO 60 and TOTO-1 were used to stain for biofilm extracellular matrix.
Bacterial single cell suspension was prepared for flow cytometry from P. aeruginosa wildtype PA01, LasI/RhlI−/− and ΔPfPhage.
Normal logarithmic growth phase was largely negative for TOTO-1 staining.
Biofilm growth phase was identified as double positive for SYTO 60 and TOTO-1.
Quantitation is realized by the input of bacterial counting beads prior to analysis.
Acknowledgments.
We thank the Santa Maria lab for helpful discussions and critical reading of the manuscript. Special thanks to Dr. Lisa Nichols at Stanford Shared FACS Facility for supporting flow cytometry analysis and to Stanford Otolaryngology Intern Gisela Gomez for sample preparation.
Funding. We would like to thank the Department of Otolaryngology for funding this study through startup funds for PSM. Our thanks for core support from the Stanford Initiative to Cure Hearing Loss through generous gifts from the Bill and Susan Oberndorf Foundation. This work was supported by grants NIH LRP from NCATS to KMK and Stanford MCHRI through SPARK to KMK, LAB, and PSM. Data was collected with analyzers at the Stanford Shared FACS Facility (SSFF) obtained using NIH S10 Shared Instrument Grant (S10RR027431-01).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing interests. LAB and PSM are inventors on a patent application, novel drugs against P. aeruginosa.
References.
- Allesen-Holm M, Barken KB, Yang L, Klausen M, Webb JS, et al. , 2006. A characterization of DNA release in Pseudomonas aeruginosa cultures and biofilms. Mol. Microbiol 59, 1114–1128. [DOI] [PubMed] [Google Scholar]
- Ayrapetyan M, Williams T, & Oliver JD, 2018. Relationship between the viable but nonculturable state and antibiotic persister cells. J. Bacteriol 200(20), e00249–18. doi: 10.1128/JB.00249-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MR, Hands CL, Coello-Garcia T, Sani BS, Davenport RJ, 2019. A flow cytometry method for bacterial quantification and biomass estimates in activated sludge. J. Microbiol. Methods 160, 73–83. [DOI] [PubMed] [Google Scholar]
- Costerton JW, Lewandowski Z, Caldwell DE, Korber DR, Lappin-Scott HM, 1995. Microbial biofilms. Annu. Rev. Microbiol 49, 711–45. [DOI] [PubMed] [Google Scholar]
- Costerton JW, Stewart PS, Greenberg EP, 1999. Bacterial biofilms: A common cause of persistent infections. Science. 284(5418), 1318–1322. [DOI] [PubMed] [Google Scholar]
- Das T, Sharma PK, Busscher HJ, van der Mei HC, Krom BP, 2010. Role of extracellular DNA in initial bacterial adhesion and surface aggregation. Appl. Environ. Microbiol 76, 3405–3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duhamel S, Gregori G, Wambeke FV, Mauriac R, Nedoma J, 2008. A method for analysing phosphatase activity in aquatic bacteria at the single cell level using flow cytometry. J. Microbiol. Methods 75(2), 269–278. [DOI] [PubMed] [Google Scholar]
- Fontana C, Crussard S, Simon-Dufay N, Pialot D, Reyes J, 2017. Use of flow cytometry for rapid and accurate enumeration of live pathogenic Leptospira strains. J. Microbiol. Methods 132, 34–40. [DOI] [PubMed] [Google Scholar]
- Ghosh S, and O’Connor TJ, 2017. Beyond paralogs: The multiple layers of redundancy in bacterial pathogenesis. Front. Cell. Infect. Microbiol 7:467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gevers D, Vandepoele K, Simillion C, Van de Peer Y, 2004. Gene duplication and biased functional retention of paralogs in bacterial genomes. 12(4), 148–154. [DOI] [PubMed] [Google Scholar]
- Hall-Stoodley L, Nistico L, Sambanthamoorthy K, Dice B, Nguyen D, et al. , 2008. Characterization of biofilm matrix, degradation by DNase treatment and evidence of capsule downregulation in Streptococcus pneumoniae clinical isolates. BMC Microbiol. 8, 173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings LK, Storek KM, Ledvina HE, Coulon C, Marmont LS, 2015. Pel is a cationic exopolysaccharide that cross-links extracellular DNA in the Pseudomonas aeruginosa biofilm matrix. PNAS, 112(36), 11353–8. PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalia M, Yadav VK, Singh PK, Sharma D, Narvi SS, Agarwal V, 2018. Exploring the impact of parthenolide as anti-quorum sensing and anti-biofilm agent against Pseudomonas aeruginosa. Life Sci. 199, 96–103. [DOI] [PubMed] [Google Scholar]
- Kerstens M, Boulet G, Van Kerckhoven M, Clais S, Lanckacker E, Delputte P, Maes L, Cos P, 2015. A flow cytometric approach to quantify biofilms. Folia Microbiol (Praha). 60(4), 335–42. [DOI] [PubMed] [Google Scholar]
- Lebeaux D, Ghigo JM, Beloin C, 2014. Biofilm-related infections: Bridging the gap between clinical management and fundamental aspects of recalcitrance toward antibiotics. Microbiol. Mol. Biol. Rev 78(3): 510–543. PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magana M, Sereti C, Ioannidis A, Mitchell CA, Ball AR, et al. , 2018. Options and limitations in clinical investigation of bacterial biofilms. Clin Microbiol Rev 31(3):e00084–16. doi: 10.1128/CMR.00084-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann EE, Rice KC, Boles BR, Endres JL, Ranjit D, et al. , 2009. Modulation of eDNA release and degradation affects Staphylococcus aureus biofilm maturation. PLoS One 4, e5822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marielle B, Sarrah G, 2000. Assessment of bacterial membrane fluidity by flow cytometry. J. Microbiol. Methods 143, 50–57. [DOI] [PubMed] [Google Scholar]
- Nelson LK, D’Amours GH, Sproule-Willoughby KM, Morck DW, Ceri H, 2009. Pseudomonas aeruginosa las and rhl quorum-sensing systems are important for infection and inflammation in a rat prostatitis model. Microbiology 155, 2612–9. doi: 10.1099/mic.0.028464-0. Epub 2009 May 21. [DOI] [PubMed] [Google Scholar]
- Nescerecka A, Hammes F, Juhna T, 2016. A pipeline for developing and testing staining protocols for flow cytometry, demonstrated with SYBR Green I and propidium iodide viability staining. J. Microbiol. Methods 131, 172–180. [DOI] [PubMed] [Google Scholar]
- Okshevsky M, Meyer RL, 2014. The role of extracellular DNA in the establishment, maintenance and perpetuation of bacterial biofilms. Crit. Rev. Microbiol 10.3109/1040841X.2013.841639. [DOI] [PubMed] [Google Scholar]
- Okshevsky M, Meyer RL, 2014. Evaluation of fluorescent stains for visualizing extracellular DNA in biofilms. J. Microbiol. Methods 105, 102–104. PMID: 25017901 [DOI] [PubMed] [Google Scholar]
- Pletzer D, Mansour SC, Wuerth K, Rahanjam N, & Hancock RE, 2017. New Mouse Model for Chronic Infections by Gram-Negative Bacteria Enabling the Study of Anti-Infective Efficacy and Host-Microbe Interactions. mBio, 8(1), e00140–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahme LG, Stevens EJ, Wolfort SF, Shao J, Tompkins RG, Ausubel FM, 1995. Common virulence factors for bacterial pathogenicity in plants and animals. Science. 268,1899–1902. [DOI] [PubMed] [Google Scholar]
- Rudy CK, Etter E, Dryden M, Yeager KA, Klibanov M, A L, & Lannigan J., 2014. Imaging flow cytometry elucidates limitations of microparticle analysis by conventional flow cytometry. Cytometry Part A, 85(9), 756–770. [DOI] [PubMed] [Google Scholar]
- Römling U, Balsalobre C, 2012. Biofilm infections, their resilience to therapy and innovative treatment strategies. J. Intern. Med 272(6):541–61. [DOI] [PubMed] [Google Scholar]
- Schleheck D, Barraud N, Klebensberger J, Webb JS, McDougald D, Rice SA, & Kjelleberg S, 2009. Pseudomonas aeruginosa PAO1 preferentially grows as aggregates in liquid batch cultures and disperses upon starvation. PloS one, 4(5), e5513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Secor PR, Sweere JM, Michaels LA, Malkovskiy AV, Lazzareschi D, 2015. Filamentous Bacteriophage Promote Biofilm Assembly and Function. Cell Host Microbe. 11;18(5), 549–59. PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Secor PR, James GA, Fleckman P, Olerud JE, McInnerney K, Stewart PS, 2011. Staphylococcus aureus biofilm and planktonic cultures differentially impact gene expression, mapk phosphorylation, and cytokine production in human keratinocytes. BMC Microbiol. 11:143 PMID:21693040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seper A, Fengler VHI, Roier S, Wolinski H, Kohlwein SD, et al. , 2011. Extracellular nucleases and extracellular DNA play important roles in Vibrio cholerae biofilm formation. Mol. Microbiol 82, 1015–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siehnel R, Traxler B, An DD, Parsek MR, Schaefer AL, & Singh PK, 2010. A unique regulator controls the activation threshold of quorum-regulated genes in Pseudomonas aeruginosa. PNAS, 107(17), 7916–7921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steen HB, 2000. Flow cytometry of bacteria: glimpses from the past with a view to the future. J. Microbiol. Methods 42(1), 65–74. [DOI] [PubMed] [Google Scholar]
- Stocks S, 2004. Mechanism and use of the commercially available viability stain, BacLight. Cytometry Part A 61, 189–195. [DOI] [PubMed] [Google Scholar]
- Sweere JM, Van Belleghem JD, Ishak H, Bach MS, Popescu M, 2019. Bacteriophage trigger antiviral immunity and prevent clearance of bacterial infection. Science 363, 6434 PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevathan-Tackett S, Macreadie P, Ralph P, Seymour J, 2014. Detachment and flow cytometric quantification of seagrass-associated bacteria. J. Microbiol. Methods 102, 23–25. [DOI] [PubMed] [Google Scholar]
- Qin Z, Ou Y, Yang L, Zhu Y, Tolker-Nielsen T, et al. , 2007. Role of autolysin-mediated DNA release in biofilm formation of Staphylococcus epidermidis. Microbiol. 153, 2083–2092. [DOI] [PubMed] [Google Scholar]
- Whitchurch CB, Tolker-Nielsen T, Ragas PC, Mattick JS, 2002. Extracellular DNA required for bacterial biofilm formation. Science 295, 1487. [DOI] [PubMed] [Google Scholar]
- World Health Organization, 2017. Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. https://www.who.int/medicines/publications/WHO-PPL-Short_Summary_25Feb-ET_NM_WHO.pdf [Google Scholar]
- Yamada KJ, Heim CE, Aldrich AL, Gries CM, Staudacher AG, & Kielian T, 2018. Arginase-1 Expression in Myeloid Cells Regulates Staphylococcus aureus Planktonic but Not Biofilm Infection. Infection and immunity, 86(7), e00206–18. doi: 10.1128/IAI.00206-18 [DOI] [PMC free article] [PubMed] [Google Scholar]