Abstract
JC virus (JCV) Agnoprotein (Agno) plays critical roles in successful completion of the viral replication cycle. Understanding its regulatory roles requires a complete map of JCV-host protein interactions. Here, we report the first Agno interactome with host cellular targets utilizing “Two-Strep-Tag” affinity purification system coupled with mass spectroscopy (AP/MS). Proteomics data revealed that Agno primarily targets 501 cellular proteins, most of which contain “coiled-coil” motifs. Agno-host interactions occur in several cellular networks including those involved in protein synthesis and degradation; and cellular transport; and in organelles, including mitochondria, nucleus and ER-Golgi network. Among the Agno interactions, Rab11B, Importin and Crm-1 were first validated biochemically and further characterization was done for Crm-1, using a HIV-1 Rev-M10-like Agno mutant (L33D + E34L), revealing the critical roles of L33 and E34 residues in Crm-1 interaction. This comprehensive proteomics data provides new foundations to unravel the critical regulatory roles of Agno during the JCV life cycle.
Keywords: JCV, Mitochondria, Golgi, Interactome, Coiled-coil, Leucine zipper, α-helix, Progressive multifocal leukoencephalopathy, Polyomavirus, Nucleus, Endosome trafficking, SV40, BK virus, Merkel cell carcinoma virus, Crm-1, Importin, Rab11B and HIV-1 Rev M10 mutant
1. Introduction
All viruses are obligatory intracellular parasites and encode a limited number of regulatory and structural proteins, which are, by all means, inadequate to propagate their progeny without the host cell environment. In order to overcome this deficiency, they developed virus-specific strategies to modify the host cell environment by targeting specific host proteins, protein complexes, networks and organelles to create a more conducive environment to achieve a successful propagation cycle (Brito and Pinney, 2017; Franzosa and Xia, 2011). JC virus (JCV) is a human polyomavirus with a small double-stranded DNA genome and is the etiological agent of a fatal demyelinating disease of the central nervous system (CNS), called progressive multifocal leukoencephalopathy (PML), which affects a subpopulation of immunocompromised patients including those with the AIDS, multiple sclerosis and cancer (Berger et al., 1987; Berger, 2000; Ferenczy et al., 2012; Kleinschmidt-DeMasters and Tyler, 2005; Langer-Gould et al., 2005; Padgett et al., 1971; Van Assche et al., 2005). Recent studies indicate that JCV is also associated with another brain pathology termed “granule cell neuronopathy” (GCN) (Dang et al., 2012; Du Pasquier et al., 2003; Soleimani-Meigooni et al., 2017; Wuthrich et al., 2009, 2016). The initial infection by this virus is asymptomatic and the virus remains latent in several tissues and organs, including, kidneys, the hematopoietic progenitor cells, peripheral blood B lymphocytes, and tonsillar stromal cells and even it may reside latent in the brain (Monaco et al., 1998; White et al., 1992). Reactivated JCV sets up a primary infection in oligodendrocytes (the myelin producing cells of CNS) and astrocytes; and causes PML (Ferenczy et al., 2012; Saribas et al., 2019).
JCV, like any other virus, encodes a limited number of regulatory and structural proteins from its coding regions including large T antigen, small t antigen, T′-proteins, Agno, ORF1, ORF2, VP1, VP2 and VP3 (Frisque et al., 1984; Saribas et al., 2019). Among those, Agno, one of the important regulatory proteins of JCV, plays critical roles during the viral life cycle (Sariyer et al., 2011) and was structurally and functionally studied the most (Saribas et al., 2016). It is a small and highly basic phospho-protein (71 aa long) and primarily localizes to the cytoplasmic compartment of the infected cells with high concentrations accumulating at the perinuclear area (Ellis and Koralnik, 2015; Gerits and Moens, 2012; Nomura et al., 1983; Rinaldo and Hirsch, 2007; Saribas et al., 2012, 2016) However, a small amount of the protein is also consistently detected in the nucleus of the infected cells (Okada et al., 2001; Saribas et al., 2012), suggesting that it plays regulatory roles in both compartments during the viral life cycle.
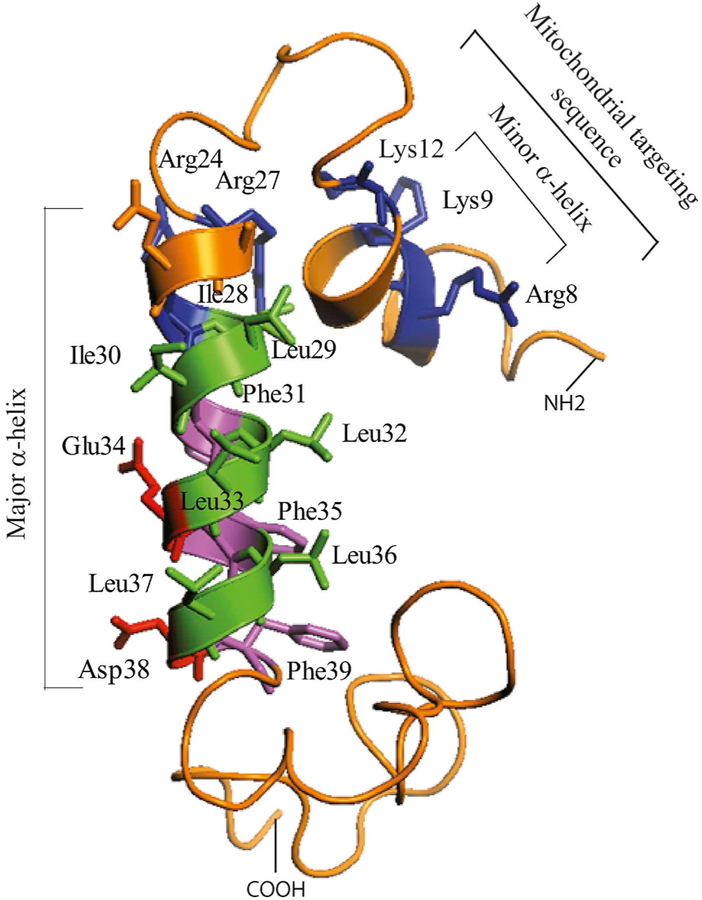
One of the remarkable structural features of Agno is to form highly stable homodimers and oligomers through its major α-helix domain which spans amino acids 24 through 39 (Saribas et al., 2013; Sariyer et al., 2011). We recently resolved the three dimensional (3D) structure of Agno using a highly pure synthetic Agno peptide (Coric et al., 2014, 2017), which revealed that the protein harbors two α-helical domains designated as minor and major α-helices, which span Leu6-Lys13 and Arg24-Phe39 respectively and the rest of the protein adopts an intrinsically disordered conformation (Coric et al., 2017). In other words, amino acids 1–5, 14–23 and 40–71 do not have any defined tertiary structure (Fig. 2). Such disordered regions are suggested to provide considerable flexibility to any protein including Agno, to diversify its binding partners and thereby amplify its functions in cells (Dyson and Wright, 2005; Fink, 2005; Ou et al., 2012).
Fig. 2.
A flow chart of the experimental designs to determine the Agno-associated proteins by AP/MS.
Another interesting feature of Agno is its release from the Agno positive cells (Otlu et al., 2014; Saribas et al., 2018). This phenomenon was first reported by Otlu et al. (2014), but later through the structure-based follow up mechanistic studies by Saribas et al., it was determined that the major α-helix domain of Agno plays a critical role in its release (Saribas et al., 2018). In particular, the negatively (Glu34 and Asp38) and positively (Lys22 and Lys23) charged residues which are located on the hydrophilic face of the major α-helix domain of Agno are the major contributing residues to its release (Fig. 7B) (Saribas et al., 2018). In addition, an aromatic residue, Phe31, located adjacent to the hydrophilic face was also shown to play a significant role in the release process (Saribas et al., 2018). The function of the released Agno from infected cells are currently unknown, however, initial characterization studies showed that it strongly interacts with unidentified cell surface components (Saribas et al., 2018) and perhaps subsequently enters the cells (Craigie et al., 2018), suggesting that it exerts its activity either through the cell surface or by entering the cells.
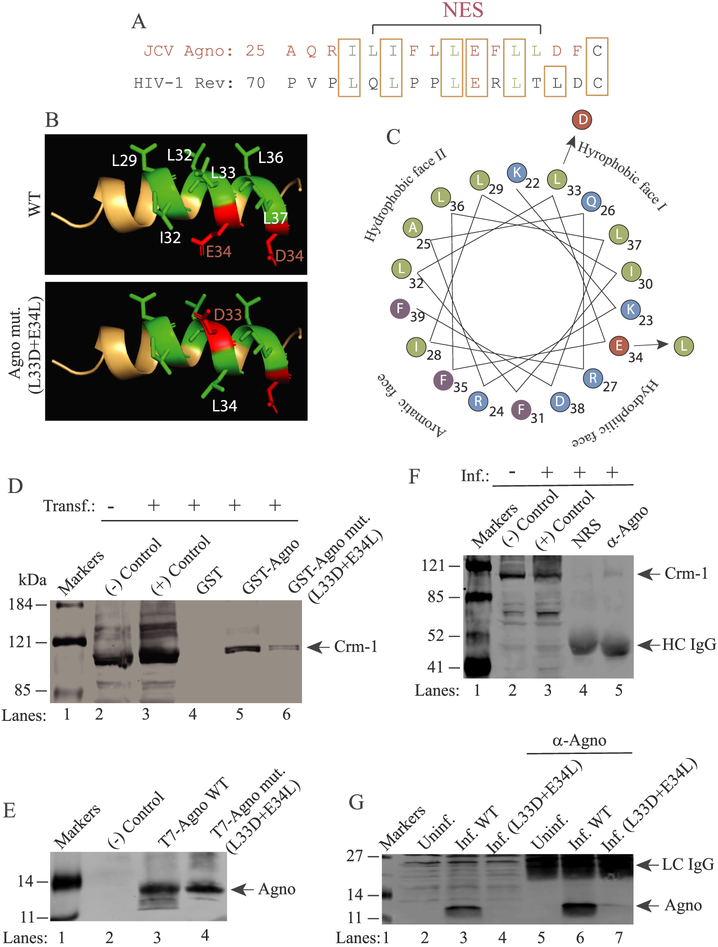
Fig. 7. Validation of the Agno interaction with Crm-1 by co-immunoprecipitation in infected cells.
(A) Alignment of JCV Agno NES with that of HIV-1 Rev. (B) 3D NMR structure of Agno major α-helix domain (NES domain) shown in wild-type form and HIV-1 Rev M10-like mutant form (L33D + E34L). The PyMOL Molecular Graphics System, Version 2.0 Schrödinger was used to obtain this 3D presentation. (C) Critical amino acids are designated. Helical-wheel representation of the major α-helix of Agno. The four different faces of the helix and amino acid substitutions are indicated. (D) GST pull down assay demonstrates the attenuated interaction of Rev M10-like mutant of Agno (L33D + E34L) with Crm-1. Pull down assays were performed as described in the materials and methods using extracts (0.5 mg) prepared from HEK293 cells transfected with T7–2xStrep-tagged Crm-1. In lanes 1 and 2, 40 μg extract were loaded as negative and positive controls from the untransfected and transfected cells respectively. (E) Analysis of the expression levels of Agno WT and its L33D + E34L mutant by Western blotting. (−) Control: Extracts from untransfected cells. (F) Agno-Crm-1 interaction analyzed by Co-IP in infected cells. Whole-cell extracts prepared from JCV-infected SVG-A cells were immunoprecipitated either with normal rabbit serum (NRS) (10 μl) or α-Agno (10 μl) polyclonal antibody. Immunocomplexes were then resolved on a SDS-8% PAGE and analyzed by Western blotting using an α-Crm-1 antibody as described in materials and methods. In lanes 1 and 2, whole-cell extracts (40 μg) prepared from uninfected (lane 1) and infected (lane 2) were loaded as negative (−) and positive (+) controls respectively. (G) Western blot analysis of the expression levels of WT Agno and its L33D + E34L mutant in JCV-infected cells (6th day post-infection). Both proteins were analyzed by a direct Western blotting (lanes 3 and 4 respectively, 40 μg/lane) and IP/Western (lanes 6 and 7 respectively, 200 μg/lane) as described in materials and methods. In lane 2 and 3, 40 μg extract prepared from uninfected and infected cells were loaded as negative and positive controls respectively. Inf.: Infection, Uninf.: Uninfected., HC IgG: Antibody heavy chain. LC IgG: Antibody light chain.
Although much progress has been made in revealing the structural and functional features of Agno, understanding its complete functions and targets during the viral life cycle still remains elusive. Previous efforts helped to identify a number of viral and cellular partners in cells by protein-protein interaction assays including large T-antigen (LT-Ag) (Safak et al., 2001), small t-antigen (Sm t-Ag) (Sariyer et al., 2008), JCV capsid protein VP1 (Suzuki et al., 2012), YB-1 (Safak et al., 2002), tubulin (Endo et al., 2003), FEZ1 and HP1-α (Okada et al., 2005; Suzuki et al., 2005), p53 (Darbinyan et al., 2002), and adaptor protein complex 3 (AP-3) (Suzuki et al., 2013). Additionally, Agno was implicated to be involved in various aspects of the JCV life cycle, including viral transcription and replication (Safak et al., 2001), virion formation (Sariyer et al., 2006; Suzuki et al., 2012), functioning as a viroporin (Suzuki et al., 2010, 2013) and deregulation of cell cycle progression (Darbinyan et al., 2002).
In order to further unravel the molecular targets of Agno in cells, we have employed an elegant affinity purification (AP system) –“Strep Tag”- (Schmidt and Skerra, 2007) to capture Agno-associated proteins followed by identification of those by Mass Spectroscopy (MS), (AP/MS). The proteomics data was then analyzed by using “FunRich Software” – a functional enrichment analysis tool - (http://www.funrich.org), to establish protein-protein interaction networks developed by Pathan et al., 2015, 2017, and by using the “STRING database”- (Search Tool for the Retrieval of Interacting Genes/Proteins), a comprehensive protein-protein interaction network building web resource (https://string-db.org). Detailed analysis of the proteomics data revealed that Agno primarily interacts with cellular proteins that contain “coiled-coil” motifs. Agno was also shown to target numerous protein networks such as ubiquitin-proteasome complexes, tRNA synthases, and eukaryotic translation initiation factors and organelles including mitochondria, nucleus, endoplasmic reticulum and Golgi apparatus. Among Agno targets, interaction of Agno with Rab11B, Importin and Exportin/Crm-1 was validated by protein-protein interaction studies and further characterized for Agno-Crm-1 interaction using a HIV-1 Rev-M10 like mutant of Agno, (L33D + E34L) in the viral background. Results showed that these residues are critical for interaction of Agno with Crm-1. This is the first comprehensive proteomics data analysis of Agno targets in cells, which lays down a new foundation to further unravel the critical regulatory roles of Agno during the JCV life cycle. Our findings will also help to understand the progression of JCV-caused diseases in the infected individuals.
2. Materials and methods
2.1. Construction of a T7–2xStrep affinity purification system
Creation of a Twin Strep Tag and use of Strep-Tactin affinity purification system were previously reported (Twin-Strep Tag: GG-SA-WNHPQFEK-GGGSGSGG-SA-WSHPQFEK-GS) (Schmidt et al., 2013). This cassette was further modified to enhance the expression of JCV Agno in cells by fusion of a T7 tag to the N-terminus of the Twin Strep tag, (MASRMASMTGGQQM-GG-SA-WNHPQFEK-GGGSGSGG-SA-WSHPQFEK-GS) and named “T7–2xStrep Tag”. The T7 tag was successfully used earlier for high-levels of expression of JCV Agno in mammalian cells (Otlu et al., 2014; Saribas et al., 2018). The nucleotide sequence of the corresponding tag was commercially synthesized by the IDT technologies (https://www.idtdna.com) as follows: Forward primer: 5′AGCTTCGTATGGCTTCTAGGATGGCATCGATGACAGGTGGCCAACAG ATGGGTGGTAGCGCGTGGAGCCATCCGCAGTTTGAAAAAGGCGGCGG CAGCGGCGGCGGCAGCGGCGGCAGCGCGTGGAGCCATCCGCAGTTTG AAAAAG-3, where “AGCTT” represents a partial HindIII restriction site.
Reverse primer: 5′-AGCTTCGTATGGCTTCTAGGATGGCATCGATGA CAGGTGGCCAACAGATGGGTGGTAGCGCGTGGAGCCATCCGCAGTTT GAAAAAGGCGGCGGCAGCGGCGGCGGCAGCGGCGGCAGCGCGTGGA GCCATCCGCAGTTTGAAAAAG-3’, where “AGCTT” represents a partial BamHI restriction site. These two DNA sequences were annealed and subcloned into the pcDNA3.1 (+) vector at HindIII/BamHI restriction sites. Subsequently, JCV full-length Agno gene was inserted into the same vector at BamH1/XhoI restriction sites in-frame with T7–2xStrep-tag. The resulting plasmid is designated as pcDNA3.1 (+)-T7–2xStrep-JCVAgno. For a control vector, a stop codon was introduced at BamH1 site of the pcDNA3.1(+)-T7–2xStrep-JCVAgno plasmid using Quik Change® mutagenesis kit (Agilent, catalog no. 200523) and specific mutagenic oligonucleotides to block Agno expression, which still allows the expression of the T7–2xStrep tag only and designated as pcDNA3.1(+)-T7–2xStrep-Stop-JCVAgno. The plasmids were then sequenced to confirm the integrity of the constructs.
2.2. Peptide synthesis
“T7–2xStrep peptide” (MASRMASMTGGQQM-GG-SA-WNHPQFEK--GGGSGSGG-SA-WSHPQFEK-GS) was commercially synthesized by Biosynthesis company (Lewisville, TX, https://www.biosyn.com).
2.3. Expression of the T7–2xStrep-Agno
HEK293T cells (ATCC, catalog no. CRL-3216) were grown into ~80% confluency in DMEM, supplemented with 10% fetal bovine serum (FBS) and antibiotics (penicillin/streptomycin) (ThermoFisher, catalog no. 15070063) in tissue culture plates (100 mm in diameter, Becton Dickinson, catalog no. 353002) treated with Poly-l-lysine (Sigma, catalog no. p8920–100 ml). Cells were maintained at 37 °C in a humidified atmosphere and supplemented with 7% CO2. The cells were then separately transfected with pcDNA3.1(+)-T7–2xStrep-JCVAgno (experimental) and pcDNA3.1(+)-T7–2xStrep-Stop-JCVAgno (control) expression plasmids (30 μg each) using calcium-phosphate precipitation method (Graham and van der Eb, 1973). Then, the expression of the T7–2xStrep tagged Agno was evaluated by Western blotting.
2.4. Two independent rounds of affinity purification of the T7–2xStrep-Agno-associated proteins
The first round of the affinity purification:
At 24 h posttransfection, whole cell extracts were prepared from the transfected cells (HEK293T) as follows: Cells (grown at ~80% confluency and transfected in 100 mm dishes) were washed with 1xPBS twice, lysed in 2 ml of TNN buffer [50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1 mM EDTA and 1.0% NP-40 in the presence of protease inhibitors (Sigma, catalog no. P8340)], collected and incubated at 4 °C on a rocking platform for 30 min. The cell lysates were then cleared by centrifugation at 17,000×g for 10 min at 4 °C. The NP-40 concentration in the whole-cell extracts were adjusted to 0.3% and stored at - 80 °C until use. Thirty milligrams of whole-cell extracts (WCEs) (control and experimental) were incubated with 150 μl of MagStrep “Type 3” XT magnetic beads (IBA Lifesciences, catalog no. 2-4090-002) at 4 °C for 16 h on a racking platform to capture T7–2xStrep-tagged Agno and Agno-bound proteins. Note that 1 μg of T7–2xStrep peptide was also incubated with “the control extract” during the incubation and purification. Agno-interacting protein complexes were then washed in TNN buffer containing 0.3% NP40 by using a bead-capturing magnet system (“DynaMag”, ThermoFisher, catalog no. 12321D) and eluted in the same buffer containing 50 mM biotin.
The second round of the affinity purification:
The experimental conditions for the second round of the affinity purification of T7–2xStrep-Agno-associated proteins were the same as the one as described for the first round of purification steps except that the TNN buffer contained relatively higher concentration of NaCl (250 mM).
2.5. Western blotting, silver and colloidal blue staining
Thirty milligrams of WCE prepared from either transfected (experimental) or untransfected (control) cells were subjected to affinity purification using 150 μl of MagStrep type 3 XT magnetic beads for 16 h at 4 °C on a racking platform. The bead-protein complexes were then washed with washing buffer [50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1 mM EDTA and 0.3% NP-40] and split into three equal fractions. One fraction was resolved on a 15% SDS-PAGE and analyzed by Western blotting using a polyclonal anti-Agno antibody (Del Valle et al., 2002). The protein complexes from the other two fractions were eluted with biotin and separated on a “NUPAGE 4–12% Bis-Tris protein gel” (Invitrogen, catalog no. NP0337Box) using MES-SDS buffer (Invitrogen, catalog no. NP0002) followed by visualizing the samples either with “silver staining” (ThermoFisher, catalog no. 24600) or “colloidal blue” staining (ThermoFisher, catalog no. LC6025).
2.6. LC-MS/MS analyses and data processing
Liquid chromatography tandem mass spectrometry (LC-MS/MS) analysis was performed by the Proteomics and Metabolomics Facility at the Wistar Institute, Philadelphia, PA, using a Q Executive HF mass spectrometer (ThermoFisher Scientific) coupled with a Nano-ACQUITY UPLC system (Waters). Samples were digested in-gel with trypsin and injected onto a UPLC Symmetry trap column (180 μm i.d. × 2 cm packed with 5 μm C18 resin; Waters). Tryptic peptides were separated by reversed phase HPLC on a BEH C18 Nanocapillary analytical column (75 μm i.d. × 25 cm, 1.7 μm particle size; Waters) using a 95 min gradient formed by solvent A (0.1% formic acid in water) and solvent B (0.1% formic acid in acetonitrile). A 30-min blank gradient was run between sample injections to minimize carryover. Eluted peptides were analyzed by the mass spectrometer set to repetitively scan m/z from 400 to 2000 in positive ion mode. The full MS scan was collected at 60,000 resolution followed by data-dependent MS/MS scans at 15,000 resolution on the 20 most abundant ions exceeding a minimum threshold of 10,000. Peptide match was set as preferred; exclude isotopes option and charge-state screening was enabled to reject unassigned charged ions.
2.7. Data processing for Agno-interacting proteins using “FunRich” software and “STRING” database
AP/MS data was generated as a result of two independent affinity purification of the Agno-interacting proteins from HEK293T cells, which were then analyzed using “FunRich software program” (Pathan et al., 2015, 2017) and “STRING database” (https://string-db.org). We first generated a list of Agno-interacting proteins with minimum of 2 significant peptides with no background (a total of 124 out of 501 of these proteins show some background in the first run, but none in the second run) based on the combination of two AP/MS runs. A list of 501 Agno-binding proteins were used as an input into FunRich program to analyze our data. We primarily focused on the Agno targets that contain interesting domain types, the type of protein functions and the localization of the Agno-interacting proteins. Second, we turned our attention to the selected protein networks, organelles and clinical phenotypes in our analysis. “FunRich” is a standalone software to enrich the AP/MS data and to make a network analysis (Pathan et al., 2015, 2017), which uses the input data to build bar graphs based on a number of criteria and classifies the data into several categories including cellular components, molecular functions, biological processes, biological pathways, protein domains, site of expressions, transcription factors and clinical phenotypes. “STRING database” is a comprehensive protein-protein interaction data base, based on the experimentally proven and predicted interactions and was used to build number of interaction networks in which Agno is involved.
Using our AP/MS data, we first compiled the components or subunits of selected protein complexes showing interactions with Agno without background. This list was then used as an input into the “STRING program” to generate a number of protein networks that Agno targets.
2.8. GST-pull down assays
GST-Agno L33D + E34L mutant was generated with Quik Change™ mutagenesis kit using pGEX-1λt-JCVAgno as a template and specific mutagenic oligos. The mutations were confirmed by DNA sequencing (Genewiz, www.genewiz.com). GST-tagged proteins were expressed in E.coli and affinity purified as described previously (Saribas et al., 2011). Two micrograms of either GST alone or GST-Agno or GST-Agno mutant (L33D + E34L immobilized on glutathione-4B Sepharose beads (Glutathione Sepharose ™ 4B, GE healthcare, catalog no. 17-0756-01) were incubated with 0.5 mg of whole-cell extract prepared from SVG-A cells transfected with pcDNA3.1-Importin-flag (GenScript, catalog number: OHu09638D) or with pcDNA3.1-Rab11B-flag (GenScript, catalog number: OHu24337D) plasmids, at 4 °C overnight in lysis buffer containing 50 mM Tris-HCl (pH 7.4), 150 mM NaCl, and 0.5% NP-40 and supplemented with a cocktail of proteinase inhibitors (Sigma, catalog no. P 8340–5 ml). Formed protein complexes bound to the affinity beads were washed extensively with lysis buffer, resolved on a SDS-10% PAGE, and analyzed by Western blotting using an anti-flag antibody (Invitrogen, catalog no. MA1–91878) directed against the flag-tag of either Importin or Rab11B. In addition, 2 μg of either GST alone or GST-Exportin/Crm-1 (pGEX-TEV-hCRM-1, a kind gift from Dr. Yuh Min Chook, University of Texas Southwestern) immobilized on glutathione-4B Sepharose beads were incubated with 0.5 mg of WCE prepared from SVG-A cells infected with JCV Mad-1 strain (14 day post-infection) at 4 °C, overnight, in lysis buffer as described above. Then the formed protein complexes bound to the affinity beads were washed with lysis buffer, resolved on a SDS-10% PAGE and analyzed by Western blotting using an anti-Agno antibody.
2.9. Replication assays
Replication assays were carried out similar to the method previously described (Sariyer et al., 2011) with minor modifications. Bluescript KS-JCV Mad-1 Agno L33D + E34L mutant was generated with Ouik Change™ mutagenesis kit using a Bluescript KS-JCV Mad-1 WT plasmid as template and specific oligonucleotides. The mutations were confirmed by DNA sequencing (Genewiz). Briefly, the plasmid constructs (Bluescript KS-JCV Mad-1 WT, Bluescript KS-JCV Mad-1 Agno mutant (L33D + E34L) were digested with BamHI to liberate the inserts from the vector (Bluescript KS+). SVG-A cells (2 × 106 cells/75 cm2 flask) were then separately transfected/infected with each of the digested DNA (10 μg each) using lipofectamine® 3000 according to manufacturer’s recommendations. After 18 hrs, the cells were trypsinized, transferred into 167 cm2 flasks and fed with DMEM supplemented with 10% heat inactivated fetal bovine serum (FBS, Gemini, catalog no 100–106) and antibiotics [penicillin-streptomycin (100 μg/ml) (Gemini, catalog no 400–1001), ciprofloxacin (MP biochemical, catalog no. 199020, 10 μg/ml)]. Two thirds of the media were replenished every three days posttransfection. At the indicated time points, low-molecular-weight DNA containing both input and replicated viral DNA was isolated as follows. Cells were harvested by trypsinization, washed with PBS by centrifugation at 1500 RPM (Eppendorf Centrifuge, 5810R) for 3 min. Cells were then resuspended in P1 buffer (250 μl, 50 mM Tris-HCl pH 8.0, 10 mM EDTA, 100 μg/ml RNaseA), lysed with P2 buffer (250 μl, 200 mM NaOH, 1% SDS) and neutralized with P3 buffer (250 μl, 3.0 M potassium acetate pH 5.5). The cell lysates were spun for 4 min at 15000 RPM (Eppendorf Centrifuge 5424) and the supernatant (650 μl) was transferred into new Eppendorf tube without taking any cell debris. DNA was precipitated by isopropanol (650 μl) by centrifugation of the samples at 15000 RPM for 5 min at room temperature. DNA pellet was resuspended in 100 μl dH2O containing RNase A (1 μg total/sample). Ten microliters of each sample were digested with BamHI and DpnI enzymes, resolved on 1% agarose gel, and transferred onto nitrocellulose membrane (Amersham hybond-N, catalog no RPN 203N). Southern blots were analyzed using JCV-specific probes as describe previously (Sariyer et al., 2011).
2.10. Co-immunoprecipitation (Co-IP) and immunocytochemistry (ICC)
Transfection/infection of SVG-A cells were carried out in SVG-A cells by using Bluescript KS-JCV Mad-1 WT and Bluescript KS-JCV Mad-1 Agno mutant (L33D + E34L) as described under replication assays above. At 14 d posttransfection/infection, whole-cell extracts were prepared in TNN lysis buffer [50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1 mM EDTA and 1.0% NP-40] in the presence of protease inhibitors (Sigma, catalog no. P8340)] on a rocking platform at 4 °C for 30 min (2 × 106 cells/1000 μl lysis buffer). The cell lysates were then cleared by centrifugation at 17,000×g for 10 min at 4 °C and stored at - 80 °C until use. For co-immunoprecipitation assays, 300 μg of protein extract (NP-40 concentration was adjusted to 0.3% in along with Protein G Sepharose Fast Flow beads (20 μl) (GE, catalog no 17061801) for 16 h in a rotating platform at 4 °C. Beads were then washed 3 times with lysis incubation buffer and protein complexes were resolved on a SDS-8% PAGE, transferred onto a nitrocellulose membrane with 0.2 μm pore size (LI-COR Odyssey, catalog no. P/N 926–31092) for 3 h at 250 mA. Membranes were first blocked with 5% bovine serum albumin prepared in TBST (50 mM Tris-HCL, pH 7.4, 150 mM NaCl, 0.1% Tween 20) buffer for 30 min at room temperature. Membranes were then probed with the primary mouse monoclonal α-Crm-1 (Santa Cruz, catalog no. sc-74457) and secondary goat anti-mouse IRDye 680LT (LI-COR, catalog no. 926–68070) antibodies. Finally, membranes were washed twice with 1 × PBS and scanned using Odyssey® CLx Infrared Imaging System (LI-COR) to detect the protein of interest.
Immunocytochemistry assays were performed as previously described (Coric et al., 2014). Briefly, in parallel to the Co-IP assays described above, transfected/infected cells were transferred to glass slide chambers (Nunc, catalog no. 154461) at six day posttransfection/infection and incubated for an additional 24 h. The next day, the cells were washed twice with 1 × PBS, fixed in 4% formaldehyde + 0.05% Triton X-100 prepared in 1xPBS and incubated with 5% bovine serum albumin in PBS for 2 h. Chamber slides were then incubated with a combination of α-Crm-1 monoclonal antibody (1:200 dilution) (Santa Cruz, catalog no. sc-74457) and α-Agno polyclonal primary antibodies overnight. Cells were washed three times with TBST buffer for 10 min intervals and subsequently incubated either with a fluorescein isothiocyanate (FITC)-conjugated goat anti-rabbit (Abcam, catalog no. Ab6717) and Rhodamine-conjugated goat anti-mouse (Millipore, catalog no. AP124R) secondary antibodies for 45 min. Cells were then washed with TBST buffer three times for 3 min each, incubated with DAPI (ThermoFisher, 4′,6-Diamidino-2-Phenylindole, Dihydrochloride, catalog no. D1306) (300 ng/ml prepared in 1xPBS) to stain the nucleus, mounted using “ProLong® Gold Antifade” mounting medium (Life Technologies, catalog no. P36934) and dried overnight. Slides were then examined under a fluorescence microscope (Leica, DMI-6000B, objective: HCX PL APD 60×/1.25 oil, employing LAS AF operating software) for visualization of the protein of interests.
3. Results and discussion
3.1. Construction of an efficient affinity purification system to pull down Agno-binding cellular proteins
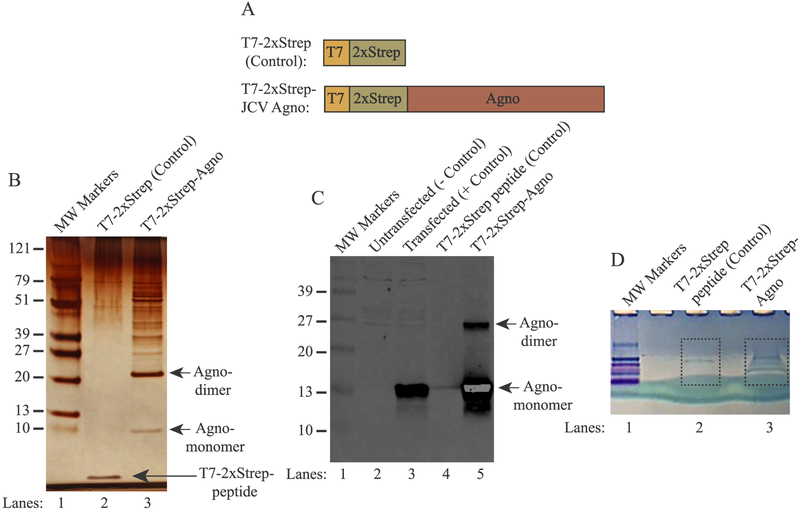
Short peptide affinity tags are widely used for affinity purification of proteins due to their highly desirable features such as providing highly pure and functional proteins at physiological conditions after a rapid and one step elution. The “Strep tag II” is one of those and selectively binds to its commercially available target, “Strep Tactin®“, an engineered streptavidin protein. Recently a third generation of Strep Tactin system was developed designated as “Twin Strep Tag”, through which its binding affinity significantly increased (~100-fold) to its target (Strep-Tactin XT), which is linked to magnetic beads, creating an even more efficient purification system. The “Strep-Tag II” has several advantages over several other short peptide affinity tags, such as Flag and 6xHis, as follows: It is biologically inert, proteolytically stable, does not interfere with membrane translocation or protein folding. Therefore, we believe that this tag does not interfere with Agno function in cells. More importantly, Strep-tagged proteins can be eluted under physiological and mild buffer conditions using biotin, which even make it a more attractive purification system to be utilized in many low background-requiring applications, such as the construction of an interactomic map of a protein with its cellular targets. We took advantage of all these attractive features of the “Strep-tag” and Strep-tactin affinity purification system to map the cellular targets of JCV Agno. Note that Agno is an unstable protein when expressed alone in the absence of the viral infection cycle and its expression can be stabilized when fused to a T7 tag its N-terminus. Due to this special requirement by Agno, we have modified the “Twin Strep Tag” by fusing the T7 tag to the N-terminus of the tag system (Fig. 1A) as described in materials and methods.
Fig. 1. Analysis of the Agno-associated host proteins by silver and colloidal blue staining.
(A) Graphical representation of the T7–2xStrep-Agno affinity purification system. (Upper panel) the T7–2xStrep sequences were cloned into the HindIII/BamHI restriction sites of pcDNA3.1(+) vector. (Lower panel) JCV Agno coding sequences were then cloned into the BamHI/XhoI restriction sites in-frame with the T7–2xStrep tag as described in materials and methods. (B) Analysis of the Agno-associated proteins by silver staining. HEK293T cells were transfected with either pcDNA3.1(+)-T7–2xStrep-Stop-Agno (control, expresses only the tag) (lane 2) or pcDNA3.1(+)-T7–2xStrep-Agno plasmids (lane 3). Then the whole-cell extracts (10 mg) prepared from these transfectants were subjected to affinity purification using MagStrep “Type 3” XT magnetic beads. After elution with biotin, protein samples were resolved on a NUPAGE 4–12% Bis-Tris protein gel and stained with silver staining reagents as described in materials and methods. Note that 1 μg of a synthetic T7–2xStrep peptide was also incubated along with the extracts prepared from the control cells as a control to demonstrate the non-specific binding to the tag alone (lane 2). The migration pattern of the Agno monomers and homodimers were indicated by the arrows. (C) Western blot analysis of the affinity purified Agno and Agno-interacting proteins. In parallel to the experiments described in panel B, the affinity purified protein samples were analyzed by Western blotting using anti-Agno antibody. In lanes 2 and 3, whole cell extracts (20 μg/lane) prepared from the HEK293T cells transfected with control and experimental plasmids were loaded on a SDS-15% PAGE gel as a negative and positive control respectively. MW: Molecular weight. (D) In parallel to the affinity purification protocols described for panel B, 10 mg of whole cell extracts prepared from the HEK293T cells transfected with either control plasmid plus incubated with T7–2xStrep peptide (lane 2) or transfected with the experimental plasmid (lane 3) were resolved shortly on a NUPAGE 4–12% gradient gel and stained with colloidal blue. The corresponding bands encased by the dash-lined rectangles were excised from the gel, in-gel digested with trypsin and analyzed by LC-MS/MS.
3.2. Analysis of Agno-binding proteins by silver staining prior to mass spectrometry analysis
Two expression plasmids [pcDNA3.1(+)-T7–2xStrep-JCVAgno (experimental) and pcDNA3.1(+)-T7–2xStrep-Stop-JCVAgno (control)] were separately transfected into the human embryonic kidney epithelial cells (HEK293T) due to their high transfection efficiency by calcium-phosphate transfection method and the ability to express various glial specific genes (Shaw et al., 2002). Whole-cell extracts prepared from both control and experimental transfectants (10 mg each) were incubated with “MagStrep Type 3 XT” magnetic beads to capture Agno and thereby Agno-associated proteins as described in materials and methods. Next day, protein samples were washed, eluted and resolved on a NUPAGE 4–12% Bis-Tris gradient gel and visualized by silver staining (Fig. 1B). Note that, control extracts were also concomitantly incubated with a highly pure and commercially synthesized T7–2xStrep peptide in order to subtract out any non-specific binding to the tag portion of Agno, which allowed us to have a reliable and highly low background system for our proteomic studies. As demonstrated on Fig. 1B, the T7–2xStrep peptide is readily detectable on the silver staining gel system (lane 2). In addition, the comparison of the control and experimental sample lanes shows a clear difference among them showing that Agno binds to a considerably high number of the host proteins (compare lane 2 and 3). Moreover, two distinct Agno-containing bands were also visibly detectable on the gel, which represent Agno monomers and homodimers as indicated. As we previously reported, Agno forms homodimers/oligomers mediated through its major α-helix domain (Fig. 2) (Saribas et al., 2011). In parallel, affinity purified Agno-associated proteins were also analyzed by Western blotting (2 mg extract) using anti-Agno antibody (Fig. 1C, lane 5). As observed in a silver staining gel, as expected, the monomeric and dimeric forms of Agno were also detected on the Western blot (Fig. 1C, lane 5). Moreover, in parallel to the silver staining analysis, the same set of samples were also partially resolved on a NUPAGE 4–12% Bis-Tris gradient gel to remove small contaminants such as “avidin” from the eluted samples and prepare them for trypsin digestion followed by mass spectroscopy analysis. As shown in Fig. 1D, the gel areas encased by the dash-lined rectangles were excised from the gel for trypsin digestion followed by LC-MS/MS analysis. Of note, a flow chart provided in Fig. 2 summarizes each step in the experimental design during the affinity purification of Agno-associated proteins followed by AP/MS analysis.
3.3. Data enrichment and analysis
Whole-cell extracts prepared from these transfected cells were subjected to the affinity purification of the Agno-associated proteins under two different buffer conditions. In the first round of affinity purification cycle, the incubation buffer, TNN, contained 150 mM NaCl, but the salt concentration was increased to 250 mM in the second round of purification steps to create a more stringent binding condition for Agno-associated proteins. However, the final NP40 detergent concentration in the incubation buffer in both cases remained constant at 0.3%. In addition, the AP/MS data obtained from both purification cases were subjected to a further enrichment process based on several data exclusion criteria as follows: a) Interaction between Agno and its targets has to be present in both AP/MS data runs with more than 2 significant peptides. b) No background is allowed in data enrichment. c) The second AP/MS data was chosen as a base line in enrichment process in order to select any data with 1 significant peptide from the second AP/MS run “if and only if” the data from the first AP/MS run contains at least 2 significant interaction peptides.
Two independent proteomics data sets were obtained from two independent affinity purification processes followed by AP/MS runs. The first proteomics data run provided with a total of 1629 interactions with Agno with at least with 2 significant peptides, 1186 of which had no background (Supplement 1). The second data set provided with 687 interactions with Agno with 2 significant peptides, 560 of which had no background (Supplement 2). The reason why a relatively less number of interactions observed for the second data run is that we set a more stringent condition for the second affinity purification by increasing NaCl concentration from 150 mM to 250 mM in the incubation buffer. In addition, interaction with NUP88, SCFD1, and SEC13 proteins provided only 1 significant peptide in the second AP/MS data, but more than 2 in the first. Similarly, interaction of Agno with GCN1L1 provided 1 peptide in first AP/MS but 2 in the second. These protein interactions were also included in the enrichment process (Supplement 3). We then compared both data sets based on the enrichment criteria established above, which resulted in 377 common interaction between two data sets with no background (Supplement 3). An additional 124 host proteins also showed a common interaction with Agno between two data sets, which had some degree of low background in the first AP/MS but none in the second, which were also included in the enrichment process (Supplements 4 and 5).
We are aware of the fact that since Agno is overexpressed in HEK293T cells, some of these interactions may result from the protein overexpression or totally from non-specific interactions of host protein with Agno due to their sticky nature and/or higher quantities present in cells (Engeland et al., 2014). Therefore, we analyzed the AP/MS data with greater scrutiny. The non-specific-interacting partners of Agno or contaminants may include ribosomal proteins, translation initiation and elongation factors, heat shock proteins, ribonucleoproteins, desmin, peripherin, vimentin, keratin, myosin, tubulin, cofilin, actin and others as described by Engeland et al. (2014). We discarded the majority of these assumed artifacts and contaminating proteins which showed interaction with Agno but exhibited high background. However, it should be taken in consideration that some of these proteins may still be genuine binding partners to Agno, such as those including translation initiation factors, exportins, and ubiquitin enzymes, some of which exhibited strong interaction with Agno with zero background. Of note, several host proteins appeared in our AP/MS data were previously reported to interact with Agno by means of various protein-protein interaction assays (Table 1).
Table 1.
Current and previously reported host-cell proteins that interact with agnoprotein.
| Gene | Function | Reference |
|---|---|---|
| YB-1 | Transcriptional activation | Safak et al. (2002) |
| p53 | Cell cycle regulation | Darbinyan et al. (2002) |
| Ku70 | DNA repair | Darbinyan et al. (2004) |
| FEZ | Axonal growth | Suzuki et al. (2005) |
| PP2A | Dephosphorylation | Sariyer et al. (2008) |
| α-SNAP | Vesicular trafficking | Johannessen et al. (2011) |
| AP3 | Vesicular trafficking | Suzuki et al. (2013) |
| Importin | Transport of the proteins containing NLS into the nucleus | Current study |
| Rab11 | Regulation of the different intracellular trafficking pathways | Current Study |
| Exportin | Nuclear export of cellular | Current study |
| (Crm-1) | proteins and RNA |
3.4. The use of FunRich web tool (http://www.funrich.org) in analysis of Agno-associated proteins revealed that Agno primarily targets the host proteins with distinct domains including those with coiled-coil, ARM, WD40, TPR, AAA, PINT, WHEP-TRS, MCM, eIF7C, IBN_N, HELCc and DEXDc motifs
The analysis AP/MS data by FunRich with respect to “Protein Domain” category revealed that the overwhelming majority of the host proteins targeted by Agno contain “coiled-coil” domains among the first 11 protein groups with statistically significant p-values (Fig. 4A). However, such an analysis also revealed that some other host proteins with various motifs, including armadillo repeat motif (ARM), W and D dipeptide repeating units (WD40), Tetratricopeptide (TPR) repeat unit, AAA (ATPases Associated with diverse cellular Activities), PINT (motif in proteosome subunits, Int-6, Nip-1 and TRIP-15), WHEP-TRS (a conserved domain of amino acids, called WHEP-TRS present in a number of higher eukaryote aminoacyl-transfer RNA synthetases), MCM (mini-chromosome maintenance proteins with DNA-dependent ATPases required for the initiation of eukaryotic DNA replication), eIF5C (a domain at the C-termini of the GCD6, eIF-2B epsilon, eIF-4 gamma and eIF-5), HELICc (a domain in DNA and RNA helicase superfamily c-terminal) and DEXDc (DEAD-(asp-glu-ala-asp)-like helicases) motifs also showed binding affinity to Agno (Fig. 4A) (Supplements 3 and 4). Rather than explaining the biological and functional characteristics of every motif containing protein provided above, we focused our attention to only selected ones as detailed below. “coiled-coil” domain containing proteins contain two to five α-helices wrapped around each other to form a supercoil structure. They are primarily found in proteins that are responsible for “maintenance of the organelle architecture” (Mason and Arndt, 2004). The coiled-coil domains play critical roles in assembly and disassembly of the protein complexes and are involved in “vesicle transport” in Golgi as well. They are also functional in motor proteins to regulate the cargo binding and processivity. Moreover, the protein-protein interactions mediated by the “coiled-coil” domains are highly critical for the biological activity of the transcription factors and tRNA synthetases (Truebestein and Leonard, 2016). The coiled-coil domain interactions occur through the heptad repeats designated as (a b c d e f g)n where “a and d” are hydrophobic amino acids such as Leu and Ile, and “e and g” are charged residues such as Glu, Asp, Arg and Lys. Former residues (a and d) can generate “hydrophobic interactions”, while latter ones (e and g) can build “salt bridges” ultimately forming a highly stable dimeric and oligomeric protein structure (Apostolovic et al., 2010). Since the major α-helix domain of Agno contains not only Leu/Ile/Phe rich hydrophobic residues but also harbors a number of negatively and positively charged amino acids (Glu, Asp, Lys and Arg) at the opposite site of the hydrophobic surfaces of the α-helix surface (Coric et al., 2014) (Figs. 2A and 7B), it can potentially establish hydrophobic and ionic interactions with host coiled-coil domain-containing proteins. Thus, we believe that the major α-helix domain of Agno facilitates not only homo-dimer/oligomer formation within itself but also plays a role in establishing heterodimers/oligomers with host proteins that contain the coiled-coil domains.
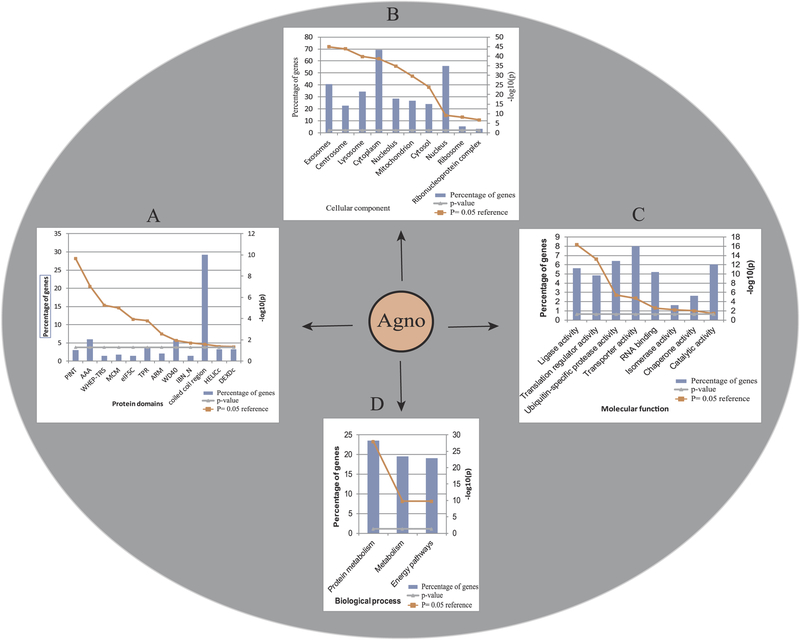
Fig. 4. Enrichment and classification of Agno-targeted host proteins using FunRich software.
Agno-targeted host proteins were further analyzed by using “FunRich”, based on the p = 0.05 reference value. Agno-interacting proteins were grouped together on the bases of having specific (A) “protein domains”, (B) as those integrated into various “cellular components” (exosomes, centrosomes, lysosomes, cytoplasm, nucleolus, mitochondrion, cytosol, nucleus, ribosome and nucleoprotein complexes), (C) those associated with various “molecular functions” (ligase activity, translation regulator activity, ubiquitin specific protease activity, transporter activity, RNA binding, isomerase activity, chaperone activity and catalytic activity) and (D) those involved in various “biological processes” (protein metabolism, metabolism and energy pathways). Percentage of genes is defined as the ratio of the host proteins that interact with Agno to the available number of proteins present in the FunRich data in that specific category.
The interaction profile of Agno with various arrays of protein motifs is highly consistent with its distribution pattern in cells. It has been consistently observed that Agno is primarily localized to the cytoplasmic compartment of the infected cells with high concentrations accumulating at the perinuclear region (Fig. 8) (Saribas et al., 2012). However, a small amount of the protein has also been consistently detected in the nucleus (15–20%) (Saribas et al., 2012). Agno has a weak bipartite nuclear localization signal located towards the N-terminal region of the protein which appears to be the main contributor to its nucleus localization (Dingwall and Laskey, 1986, 1991; Saribas et al., 2012, 2019). Consistent with these findings, it was also reported to co-localize with endoplasmic reticulum (ER) markers (Suzuki et al., 2010) and lipid droplets (LD)” in infected cells (Unterstab et al., 2010). Such a broad distribution profile of JCV Agno in different regions of the cell suggests diverse roles for this protein in all those distributed regions (Fig. 8).
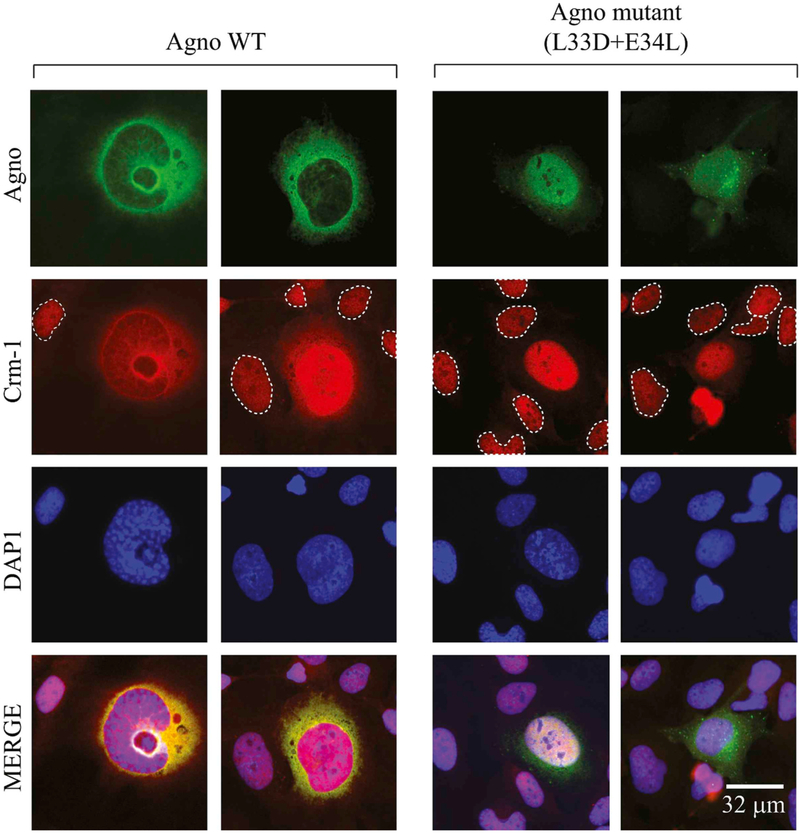
Fig. 8. Analysis of Agno - Crm-1 interactions by immunocytochemistry.
SVG-A cells (Major et al., 1985) were transfected/infected with JCV Mad-1 strain. At 6th day post-infection, the infected cells were fixed with 4% formaldehyde containing 0.05% Triton X-100 and processed for immunocytochemistry in glass slide chambers using primary polyclonal α-Agno and monoclonal α-Crm-1 antibodies and with appropriate secondary antibodies as described in material and methods. The uninfected cells were demarcated by the dashed circles. Cells were finally mounted using mounting medium and examined under a fluorescence microscope (Leica, DMI-6000B, objective: HCX PL APD 60×/1.25 oil, LAS AF operating software) for visualization of the cellular distribution of Agno and Crm-1. Scale bar: 32 μm.
Additionally, we also attempted to classify the Agno-associated proteins into various categories using FunRich web tool. Beside the “Protein Domains” (Fig. 4A) category, FunRich currently supports categorizing the input data into 7 more different categories. We only utilized several of those in FunRich analysis including “Cellular Component” (4B) and “Molecular Function” (Fig. 4C) and “Biological Process” (4D) to highlight the significance of Agno interaction with its host targets. We will further discuss some of these interesting Agno-associated proteins with aforementioned domains in details with respect to their role in different protein networks and organelles in upcoming sections based on the “STRING data base” analysis of the AP/MS data.
3.5. Building a map of Agno interactome using STRING web tool
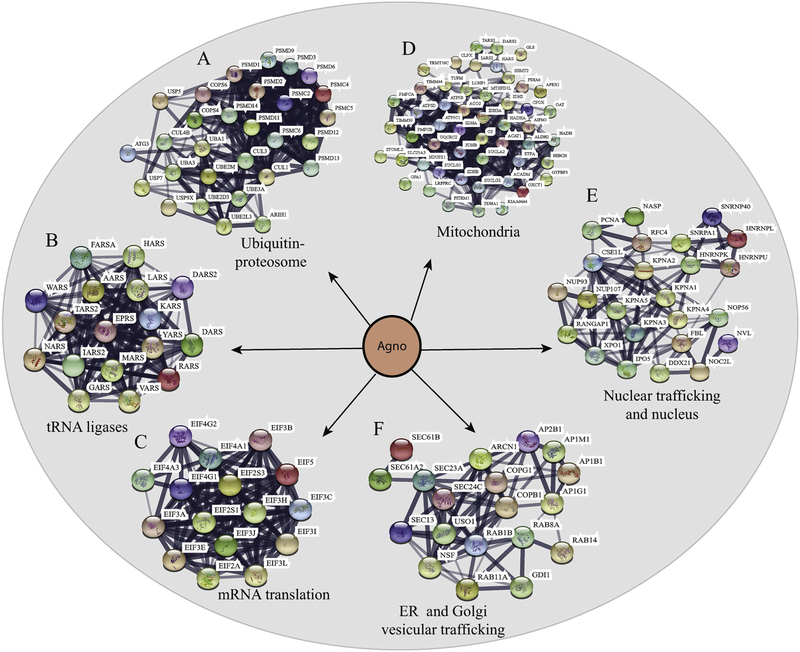
We next analyzed Agno proteomics data to build a map of Agno interactome using STRING web tool, which revealed that Agno primarily targets a number of protein networks involved in various cellular functions, including protein degradation (Fig. 5A), tRNA ligases involved in protein synthesis (Fig. 5B) RNA degradation (supplement 3 and 4), and mRNA translation complexes (Fig. 5C) (supplements 1, 2, 3,4 and 5). Moreover, the mapping analysis also showed that Agno interacts with various organelles, including mitochondria (Fig. 5D), nuclear and organelle trafficking (Fig. 5E), mitochondria associated endoplasmic reticulum membranes (MAMs) (supplement 3 and 4), ER and Golgi vesicular trafficking (Fig. 5F) (Supplement 1, 2, 3 and 4). Rather than providing a survey on those interaction pathways, we focused our attention to only two Agno-targeted systems, (mitochondria; and nuclear and cellular membrane trafficking) for which we have supporting and validation data that correlate with the Agno proteomics.
Fig. 5. Interactome maps for the selected host-cell protein networks targeted by Agno using “STRING database”.
The host-cell protein networks involved in (A) protein degradation pathways (ubiquitinproteasome complexes), (B) synthesis of tRNA (tRNA synthases) and (C) mRNA translation pathways. (DEF) Establishment of the interactome maps for the organelle-integrated proteins targeted by Agno using lism, m databases), (B)mitochondria-integrated proteins, (E) nuclear proteins and the proteins involved in nuclear trafficking networks, (F) endoplasmic reticulum (ER)- and Golgi network vesicular trafficking.
3.5.1. Agno targets “the mitochondrial protein import pathways” and several other mitochondria-integrated protein complexes involved in “electron transport”
Mitochondrion is the powerhouse of each cell and contains its own genome and translational machinery, although the majority of its proteins is encoded by the nuclear genome and imported into this organelle. The protein import is mediated by their own 10–70 amino acid long mitochondrial targeting sequences (MTS) which are generally located at their N-terminal regions. Many viral proteins target and alter the function of mitochondria. Most of those viral proteins do not possess the MTS sequence in their N-terminus, instead, some utilize their amphipathic helices located in the other regions of the protein with proapoptotic features. The others use carboxy terminal MTS which is classified as anti-apoptotic (Boya et al., 2004; Castanier and Arnoult, 2011). Interestingly, 3D NMR structure and computer prediction studies revealed that the N-terminus (aa 1–14) of Agno contains a MTS with a small positively charged α-helix region (Fig. 3). In addition, the current AP/MS data showed that Agno exhibits interactions with nearly 50 mitochondrial proteins including mitochondrial import machinery, electron transport chain (ETC) complexes and metabolic enzymes, which corroborates with our predictions (Fig. 5D) and NMR structure data (Fig. 3). Classical mitochondrial protein imports are carried out by the outer (TOM) and inner (TIM) membrane translocase complexes, which are essential for the mitochondrial maturation and survival. Majority of the mitochondrial proteins were synthesized along with MTS in the cytosol and imported into mitochondria via TOM/TIM complexes (Becker et al., 2012; Harbauer et al., 2014; Rehling et al., 2004). Our data further revealed that Agno particularly targets the TOM70 and TOM22 in the TOM complex and several components of TIM23 complex (Fig. 5D, Supplement 3 and 4), suggesting that Agno may dysregulate the mitochondrial functions by targeting its protein import pathways.
Fig. 3. Presentation of 3D NMR of JCV Agno (Coric et al., 2014, 2017).
(A) The major (Arg24 through Phe39) and minor (Leu6 through Lys13) α-helical domains are indicated. The rest of the protein forms an intrinsically disordered conformation. In other words, amino acids 1–5, 14–23 and 40–71 do not display a defined tertiary structure.
Furthermore, our AP/MS data also showed the interaction of Agno with the mitochondrial electron transport chain (ETC) complexes including subunits of complex I, II, III and ATPase (Fig. 5D, Supplements 3 and 4). To support these findings, we examined the effect of Agno on the mitochondrial energy production by the transient Agno expression in cells, which resulted in a significant reduction in ATP production by these cells (unpublished data). In addition to the energy production, Agno was also found to significantly increase the Ca2+ influx into the mitochondria, the mechanism of which is currently unknown (unpublished data), which also corroborates with its interaction with cal-reticuline, calnerexin and Ca-ATPase in ER.
3.5.2. Agno interacts with the host proteins involved in nuclear and membrane trafficking pathways
Agno contains a weak bipartite nuclear localization signal (NLS) (Saribas et al., 2019) as well as a nuclear export signal (NES) (Fig. 7A) within its structure indicating that it can shuttle between cytoplasm and nucleus. Previously, we reported that a small fraction of Agno can be found in nucleus (Saribas et al., 2012). In principle, the nuclear trafficking is mediated by Importin and Exportin/Crm-1(Xpo1) complexes in a GTP dependent manner in host cells (Cautain et al., 2015) and many viruses target these complexes in order to succeed in their life cycle. One of the prime examples of a viral protein that targets the host exporting complexes is the HIV-1 Rev protein, which targets the host Crm-1 protein. Rev is known to mediate the transport of the HIV transcripts from nucleus to cytoplasm in a Crm-1-dependent manner (Rausch and Le Grice, 2015). Our proteomics (5E) and validation (Fig. 6) data showed that Agno interacts with both Importin (Fig. 6C) and Crm-1 (Fig. 6E, Supplement 3 and 4). Since these nuclear trafficking complexes use nuclear pores to complete their task, Agno was also found to interaction with several subunits of nuclear pore complexes (Fig. 5E, Supplement 3 and 4), further supporting a possible role of Agno in modulation of the nuclear import/export system.
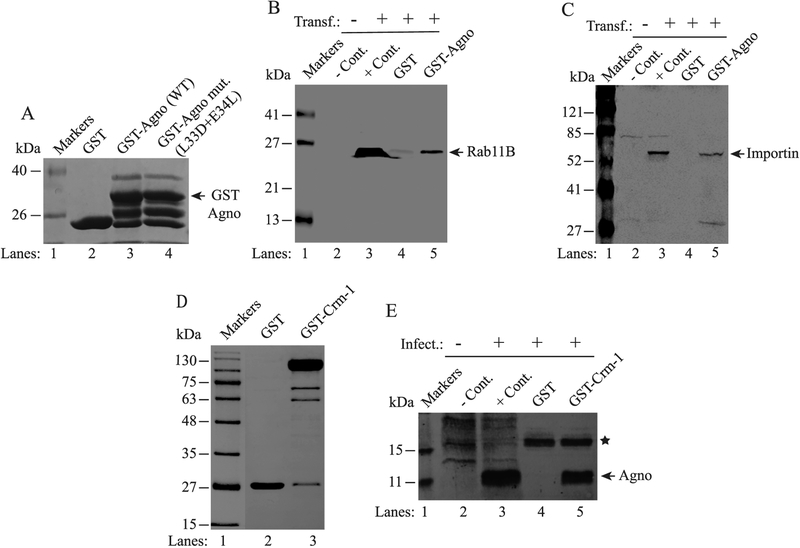
Fig. 6. Validation of Rab11B, Importin and Crm-1 interactions with Agno.
(A) Analysis of GST, GST-Agno and GST-Agno mutant (L33D + E34L) fusion proteins by SDS-10% PAGE. GST and GST-Agno were produced in bacteria and affinity purified as previously described (Saribas et al., 2011). Five micrograms of each protein was loaded on the gel, resolved and stained by Coomassie blue. (B and C) Analysis of the interaction of Agno with Rab11B and Crm-1 by GST pull-down assays. Whole-cell extracts (0.5 mg) prepared from SVG-A cells transfected with either flag-tagged-Rab11B or flag-tagged Crm-1 expression plasmids (lanes 4, and 5 on each panel) were incubated with either GST alone (lane 4) or GST-Agno (lane 5). After washing, proteins interacting with GST or GST-Agno were analyzed by Western blotting using an anti-flag antibody for detection of flag-tagged Rab11B and flag-tagged Crm-1. (D) Analysis of bacterially-produced GST and GST-Crm-1 fusion protein by SDS-10% PAGE. GST-Crm-1 was produced as described previously (Dong et al., 2009). Two micrograms of GST and GST-Crm-1 were loaded on the gel, resolved and stained by Coomassie blue. (E) Interaction of Agno with Crm-1. Whole-cell extracts prepared from SVG-A cells infected with JCV Mad-1 strain (14th day post-infection) were incubated with either GST alone (lane 4) or GST-Crm-1 (lane 5). After washing, proteins interacting with GST or GST-Crm-1 were analyzed by Western blotting using an anti-Agno antibody. On panels B, C and E, whole-cell extracts prepared from untransfected or uninfected or transfected or infected cells were loaded as negative (− Cont.) and positive (+Cont.) controls as indicated. A star on panel E points to a nonspecific interaction of a host protein with GST. Transf.: Transfection. Infect.: Infection.
As mentioned above, Agno also interacts with the membrane trafficking pathways. A typical eukaryotic cell contains a large number of membranous vesicles, which are destined to deliver a broad range of factors between the organelles in order to maintain cellular homeostasis. One group of proteins that are closely involved in the regulation of such transport system is the family of Ras like (Rab) GTPases. They regulate endocytosis, secretion, and Golgi trafficking. Their deficiency can result in a range of disease states including Parkinson’s, Huntington’s diseases, and Charcot-Marie Tooth disease [see review (Hutagalung and Novick, 2011),]. Two independent runs of our proteomic data consistently showed that Agno targets the Rab GTPase family members, including GDI alpha, RAB8A, RAB11A, RAB11B and RAB14 (Fig. 5F, Supplement 3, 4 and 5). Among those, Rab11B was shown to play a critical role in distribution of several plasma membrane proteins including receptors such as transferrin to their specialized final destinations (Guichard et al., 2014). Many neurological diseases show defects in recycling of the endosomes (Li and DiFiglia, 2012; Mitra et al., 2011). Particularly, deregulation of Rab11 activity is found to be associated with a number of neurodegenerative diseases (Schlierf et al., 2000; Zerial and McBride, 2001), including Huntington (Giorgini and Steinert, 2013), Alzheimer’s (Dumanchin et al., 1999; Udayar et al., 2013), Prion diseases (Massignan et al., 2010) and intellectual disability (Lamers et al., 2017). Taken together, such a targeting by Agno deserves a further investigation to determine whether there is effect of Agno on “vesicular trafficking” in cells.
3.6. Further validation of the Agno proteomics for several host proteins
The current proteomics data validates several previously identified and functionally characterized host-cell proteins that interact with Agno. That is, JCV and BK virus (BKV) Agnoproteins were previously reported to interact with several host and viral proteins [see for review, (Saribas et al., 2019)] and the current AP/MS data validated some of these interactions. For example, a stress inducible transcription factor, Y-box binding protein, Yb-1 (Safak et al., 2002), an adaptor protein complex 3 delta subunit (AP3-delta) (Suzuki et al., 2013), a soluble N-ethylmaleimide sensitive fusion attachment protein (α-SNAP) (Johannessen et al., 2011), protein phosphatase 2A (Sariyer et al., 2008), were all validated to be interacting with Agno (Supplement 3 and 4, Table 1). However, the interaction of a tumor suppressor and a stress inducible protein, p53 (Darbinyan et al., 2002), fasciculation and elongation protein zeta 1 (FEZ1) (Suzuki et al., 2005), and a DNA repair protein, Ku70 that binds to the DNA double-strand break ends (Darbinyan et al., 2004) were not detected by our AP/MS data, suggesting that each may require a special interaction condition with Agno (Table 1). All these interactions were functionally characterized with respect to the viral life cycle or that the impact of Agno on the host-cell biology.
3.6.1. Interaction of Agno with Rab11B, Importin and Crm-1 was validated by protein-protein interaction assays
Interaction of Agno with Rab11B, Importin and Crm-1 was validated by GST pull down assays (Figs. 6 and 7). For this purpose, whole-cell extracts prepared from SVG-A cells (Major et al., 1985) expressing a flag-tagged Rab11B (Fig. 6B) or flag-tagged-Importin (Fig. 6C) were incubated with GST or GST–Agno fusion (Fig. 6A) proteins immobilized on glutathione-sepharose 4B beads. After washing, the host proteins that interact with Agno were analyzed by SDS-PAGE/Western using an anti-flag antibody. As shown in Fig. 6B and C, GST-pull down experiments resulted in a strong binding of Rab11B (Fig. 6B) and Importin (Fig. 6C) with Agno in separate experiments but not with GST, indicating the specificity of the interaction of both proteins with Agno. In parallel, we also performed a similar GST pull down assays to validate the interaction of Agno with Crm-1 as shown in Fig. 6E, using the whole-cell extracts prepared from the JCV-infected SVG-A cells. Again, Agno showed a strong interaction with Crm-1 (lane 5) but not with GST alone, suggesting a specific interaction of Agno with Crm-1.
3.6.2. Importance of Agno L33 and E34 residues in interaction with Crm-1 revealed by GST-pulldown and co-immunoprecipitation assays
In order to provide further evidence that this interaction is specific, we utilized a specific mutant of Agno (L33D + E34L) in JCV infection assays. As shown in Fig. 7A, Agno contains a nuclear export signal (NES) within its major α-helix domain (Saribas et al., 2019) with high homology to HIV-1 Rev NES (Fig. 7A). HIV Rev protein plays an essential role in transport of viral transcripts from nucleus to cytoplasm by interacting with Crm-1 through its NES. A trans-dominant NES mutant of Rev termed M10 (L78D-E79L) cannot bind to Crm-1 and therefore makes HIV-1 Rev nonfunctional (Behrens et al., 2017; Malim et al., 1989a, 1989b). Due to high homology between Agno and REV NES sequences, we also created an M10-like mutant of the Agno within its NES in the viral background to provide evidence as to whether Agno interacts with Crm-1 through its NES. Note that, the major α-helix domain of Agno has four different surfaces (two hydrophobic surfaces, one aromatic surface and one hydrophilic surface) as shown in Fig. 7C. In Agno M10-like mutant, two amino acid substitutions were made in two different surfaces, one in the hydrophilic surface and the other in the hydrophobic surface I as indicated (Fig. 7B and C). Of note, Fig. 6A illustrates the coomassie blue staining of the SDS-PAGE gel analysis of the full-length GST-Agno and GST-Agno M10-like mutant. Fig. 6D illustrates the coomassie blue-staining of the SDS-PAGE gel analysis of the full-length GST-Crm-1.
First, we analyzed the efficiency of the interaction of Agno M10-like mutant employing GST-pull down assays using extracts prepared from the transfected cells. As shown in Fig. 7D, the interaction efficiency of Agno M10-like mutant with Crm-1 is significantly attenuated (reduced) compared to that of WT, suggesting that a specific interaction between Agno and Crm-1 occurs through its NES region similar to that observed for HIV-1 Rev (Behrens et al., 2017; Malim et al., 1989a, 1989b). Note that, the quality and amount of GST-Agno WT and GST-Agno mutant (L33D + E34L) proteins used in the experiments were highly similar (Fig. 6A). In addition, the expression levels of Agno WT and Agno M10-like mutant in transfected cells were also similar (Fig. 7E). Therefore, such a reduction observed in the binding efficiency of Agno M10-like mutant to Crm-1 cannot be attributed to the low-level of proteins used in GST-pull down assays.
Next, we further examined the interaction of Agno with Crm-1 during the viral infection cycle by Co-IP assays. As shown in Fig. 7F, Crm-1 co-immunoprecipitates with Agno validating its interaction during the viral infection cycle (lane 5). Of note, although there is no significant difference between transiently expressed Agno WT and Agno M10-like mutant in transfected cells (Fig. 7E), the expression levels of Agno M10-like mutant in infected cells were detected to be significantly low. It is undetectable by Western analysis (Fig. 7G, lane 4) and barely detectable by immunoprecipitation (Fig. 7G, lane 7). Although we can detect low level expression of Agno M10-like mutant in infected cells by ICC (Fig. 8), it apparently becomes highly unstable when infected cells are processed for whole-cell extract preparation, even though a sufficient amount of the protease inhibitors are included in the lysis buffer. Therefore, we could not perform Co-IP for the extracts prepared from the Agno mutant-infected cells.
3.6.3. Agno lost its predominant cytoplasmic distribution pattern and became a more nuclear protein when mutations were made within its NES (L33D + E34L)
In parallel, we performed ICC assays to support our findings from the Co-IP experiments in infected cells. As shown in Fig. 8, Agno shows a typical cytoplasmic distribution pattern, mostly accumulating at the perinuclear areas of the infected cells. In contrast, Crm-1 exhibits a nuclear localization pattern as observed in the uninfected cells (demarcated by dashed circles). Surprisingly, the distribution pattern of Crm-1 altered in infected cells and became slightly cytoplasmic in addition to its main nuclear localization pattern (73% of the cells shows this type of phenotype, n = 32). ICC studies also showed that Agno colocalizes with Crm-1 mostly at the perinuclear area and in the nucleus of the infected cells, suggesting interaction between these two proteins (Fig. 8). It was also surprising that the distribution pattern of Agno M10-like mutant dramatically altered compared to that of Agno WT in infected cells. It became more nuclear than cytoplasmic. This suggests that when the Agno mutant protein translocates into nucleus, which is mediated by its weak nuclear localization signal (NLS), (Saribas et al., 2019), it cannot relocate back to cytoplasm most likely due to its attenuated interaction with Crm-1. All cells infected with Agno mutant showed a nuclear/cytoplasmic distribution pattern for Agno. However, the distribution pattern of Crm-1 in those cells remained similar to that observed for the uninfected cells. Functional relevance of Agno-Crm-1 interaction remains to be further investigated. However, similar to HIV-1 Rev protein, Agno contains a cluster of positively charged residues located towards its N-terminus. Whether these residues are involved transport of JCV transcripts from nucleus to cytoplasm in a Crm-1 dependent manner, as seen in HIV-1 Rev case, is yet to be determined.
3.6.4. Analysis of the effect of the Agno M10-like mutant on JCV replication
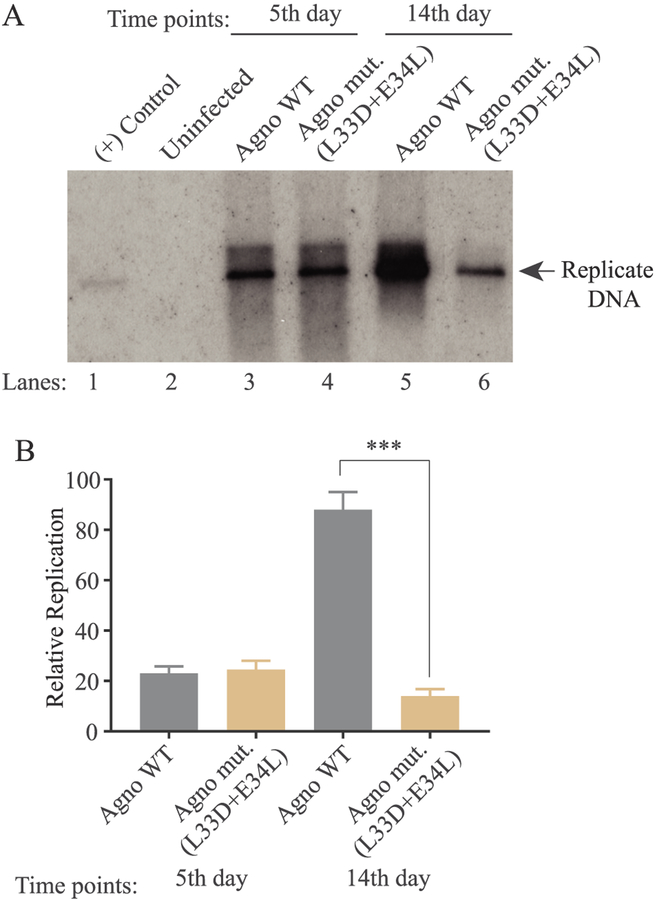
Next, we finally investigated the functional consequences of the impact of Agno M10-like mutant on the viral replication cycle by performing viral replication assays. Note that, JCV is a slow growing virus and completes its first round of replication cycle in 6–7 days and then the second round of replication cycle starts. As shown in Fig. 9A and B, there was no significant difference between the replication efficiency of JCV WT and that of Agno M10-like mutant during the first round of replication. However, this pattern dramatically changed by 14th day replication and the replication efficiency substantially reduced (~5-fold reduction), suggesting the importance of the interaction of Agno with Crm-1 during the JCV life cycle. We have previously reported the negative effect of both E34A and L33A single mutants on the viral replication cycle (Coric et al., 2014). We now observed a more dramatic effect of this double mutant on the viral replication which correlates with our previous findings and further supports importance of these residues in functional interaction of Agno with Crm-1.
Fig. 9. Analysis of the replication efficiency of JCV mutant virus.
(A) Southern blot analysis of replicated viral DNA. The plasmid constructs [Bluescript KS-JCV Mad-1 WT and Bluescript KS-JCV Mad-1 Agno (L33D + E34L)] were separately transfected/infected into SVG-A cells. At the indicated time points, the low-molecular-weight DNA containing both input and replicated viral DNA was isolated and analyzed by Southern blotting as described in materials and methods. In lane 1, 2 ng of JCV Mad-1 WT linearized by BamHI digestion was loaded as positive control. Replication assays were run in duplicates and a representative data is shown here. (B) Quantitation analysis of Southern blots by a semi-quantitative densitometry method (using NIH Image J program) and presentation of the results in relative units. Results were statistically analyzed by using the GraphPad program using One-way ANOVA and data columns were compared by Sidak’s multiple comparison test. *** indicates the significant differences at the 14d data point between WT and mutant Agno with respect to replication efficiency.
4. Conclusions
Agno is one of the important regulatory proteins of the human polyomavirus, JCV, and plays critical roles during the viral replication cycle. That is, in the absence of its expression, JCV is unable to sustain its efficient replication cycle (Sariyer et al., 2011). Although a considerable progress has been made with respect to unrevealing the structural and functional features of this protein, the identity of its host-cell targets remained elusive. In this study, we identified the molecular targets of Agno employing an elegant affinity purification system followed by mass spectroscopy. Through the enrichment of the two independent data runs, we have identified 377 host proteins common to both experimental runs with no background and an additional 124 host-cell proteins also showed a common interaction pattern with a low background.
Consistent with its distribution pattern in cells (Fig. 8), our AP/MS data uncover the fact that Agno simultaneously targets numerous protein networks and organelles, highlighting its multifunctional characteristics. In addition, this proteomics data presents itself as a comprehensive data resource to the scientific community and therefore opens up new avenues for further characterization of those interactions with respect to the viral replication cycle and paves the ways for identification of new anti-viral targets. Target wise, one thing is clear in that Agno overwhelmingly targets the host proteins with “coiled-coil” domains. It is also important to note that its simultaneous targeting of numerous protein networks and organelles provides an in-depth molecular insights into how Agno aid to JCV to manipulate the host machinery for the successful completion of the viral life cycle.
Supplementary Material
Acknowledgments
This work was supported by a NIH grant (R01NS090949.) awarded to MS. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication. The authors also express deep gratitude to Dr. Hsin-Yao Tang and Thomas Beer for their assistance and analysis of the protein samples by LC-MS/MS at the Wistar Institute Proteomics and Metabolomics Facility, Philadelphia, PA, USA.
Footnotes
Declaration of competing interest
Authors declare no conflict of interest.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.virol.2019.10.005.
References
- Apostolovic B, Danial M, Klok HA, 2010. Coiled coils: attractive protein folding motifs for the fabrication of self-assembled, responsive and bioactive materials. Chem. Soc. Rev 39, 3541–3575. [DOI] [PubMed] [Google Scholar]
- Becker T, Bottinger L, Pfanner N, 2012. Mitochondrial protein import: from transport pathways to an integrated network. Trends Biochem. Sci 37, 85–91. [DOI] [PubMed] [Google Scholar]
- Behrens RT, Aligeti M, Pocock GM, Higgins CA, Sherer NM, 2017. Nuclear export signal masking regulates HIV-1 rev trafficking and viral RNA nuclear export. J. Virol 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger JR, 2000. Progressive multifocal leukoencephalopathy. Curr. Treat. Options Neurol 2, 361–368. [DOI] [PubMed] [Google Scholar]
- Berger JR, Kaszovitz B, Post MJ, Dickinson G, 1987. Progressive multifocal leukoencephalopathy associated with immunodeficiency virus infection. A review of the literature with a report of sixteen cases. Ann. Intern. Med 107, 78–87. [DOI] [PubMed] [Google Scholar]
- Boya P, Pauleau A-L, Poncet D, Gonzalez-Polo R-A, Zamzami N, Kroemer G, 2004. Viral proteins targeting mitochondria: controlling cell death. Biochim. Biophys. Acta Bioenerg 1659, 178–189. [DOI] [PubMed] [Google Scholar]
- Brito AF, Pinney JW, 2017. Protein–protein interactions in virus–host systems. Front. Microbiol 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castanier C, Arnoult D, 2011. Mitochondrial localization of viral proteins as a means to subvert host defense. Biochim. Biophys. Acta Mol. Cell Res 1813, 575–583. [DOI] [PubMed] [Google Scholar]
- Cautain B, Hill R, de Pedro N, Link W, 2015. Components and regulation of nuclear transport processes. FEBS J 282, 445–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coric P, Saribas AS, Abou-Gharbia M, Childers W, Condra JH, White MK, Safak M, Bouaziz S, 2017. Nuclear magnetic resonance structure of the human polyoma JCV agno. J. Cell. Biochem 118, 3268–3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coric P, Saribas AS, Abou-Gharbia M, Childers W, White MK, Bouaziz S, Safak M, 2014. Nuclear magnetic resonance structure revealed that the human polyomavirus JCV Agno contains an alpha-helix encompassing the Leu/Ile/Phe-rich domain. J. Virol 88, 6556–6575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craigie M, Cicalese S, Sariyer IK, 2018. Neuroimmune regulation of JCV by intracellular and extracellular agno. J. Neuroimmune Pharmacol 13, 126–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang X, Vidal JE, Oliveira A.C.P.d., Simpson DM, Morgello S, Hecht JH, Ngo LH, Koralnik IJ, 2012. JCV granule cell neuronopathy is associated with VP1 C terminus mutants. J. Gen. Virol 93, 175–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darbinyan A, Darbinian N, Safak M, Radhakrishnan S, Giordano A, Khalili K, 2002. Evidence for dysregulation of cell cycle by human polyomavirus, JCV, late auxiliary protein. Oncogene 21, 5574–5581. [DOI] [PubMed] [Google Scholar]
- Darbinyan A, Siddiqui KM, Slonina D, Darbinian N, Amini S, White MK, Khalili K, 2004. Role of JCV agno in DNA repair. J. Virol 78, 8593–8600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Valle L, Gordon J, Enam S, Delbue S, Croul S, Abraham S, Radhakrishnan S, Assimakopoulou M, Katsetos CD, Khalili K, 2002. Expression of human neurotropic polyomavirus JCV late gene product Agno in human medulloblastoma. J. Natl. Cancer Inst 94, 267–273. [DOI] [PubMed] [Google Scholar]
- Dingwall C, Laskey RA, 1986. Protein import into the cell nucleus. Annu. Rev. Cell Biol 2, 367–390. [DOI] [PubMed] [Google Scholar]
- Dingwall C, Laskey RA, 1991. Nuclear targeting sequences–a consensus? Trends Biochem. Sci 16, 478–481. [DOI] [PubMed] [Google Scholar]
- Dong X, Biswas A, Suel KE, Jackson LK, Martinez R, Gu H, Chook YM, 2009. Structural basis for leucine-rich nuclear export signal recognition by CRM1. Nature 458, 1136–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Pasquier RA, Corey S, Margolin DH, Williams K, Pfister LA, De Girolami U, Mac Key JJ, Wuthrich C, Joseph JT, Koralnik IJ, 2003. Productive infection of cerebellar granule cell neurons by JCV in an HIV+ individual. Neurology 61, 775–782. [DOI] [PubMed] [Google Scholar]
- Dumanchin C, Czech C, Campion D, Cuif MH, Poyot T, Martin C, Charbonnier F, Goud B, Pradier L, Frebourg T, 1999. Presenilins interact with Rab11, a small GTPase involved in the regulation of vesicular transport. Hum. Mol. Genet 8, 1263–1269. [DOI] [PubMed] [Google Scholar]
- Dyson HJ, Wright PE, 2005. Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol 6, 197–208. [DOI] [PubMed] [Google Scholar]
- Ellis LC, Koralnik IJ, 2015. JCV nucleotides 376–396 are critical for VP1 capsid protein expression. J. Neurovirol 21, 671–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo S, Okada Y, Orba Y, Nishihara H, Tanaka S, Nagashima K, Sawa H, 2003. JCV Agno colocalizes with tubulin. J. Neurovirol 9 (Suppl. 1), 10–14. [DOI] [PubMed] [Google Scholar]
- Engeland CE, Brown NP, Börner K, Schümann M, Krause E, Kaderali L, Müller GA, Kräusslich H-G, 2014. Proteome analysis of the HIV-1 Gag interactome. Virology 460–461, 194–206. [DOI] [PubMed] [Google Scholar]
- Ferenczy MW, Marshall LJ, Nelson CD, Atwood WJ, Nath A, Khalili K, Major EO, 2012. Molecular biology, epidemiology, and pathogenesis of progressive multifocal leukoencephalopathy, the JCV-induced demyelinating disease of the human brain. Clin. Microbiol. Rev 25, 471–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink AL, 2005. Natively unfolded proteins. Curr. Opin. Struct. Biol 15, 35–41. [DOI] [PubMed] [Google Scholar]
- Franzosa EA, Xia Y, 2011. Structural principles within the human-virus protein-protein interaction network. Proc. Natl. Acad. Sci 108, 10538–10543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisque RJ, Bream GL, Cannella MT, 1984. Human polyomavirus JCV genome. J. Virol 51, 458–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerits N, Moens U, 2012. Agno of mammalian polyomaviruses. Virology 432, 316–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorgini F, Steinert JR, 2013. Rab11 as a modulator of synaptic transmission. Commun. Integr. Biol 6, e26807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham FL, van der Eb AJ, 1973. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology 52, 456–467. [DOI] [PubMed] [Google Scholar]
- Guichard A, Nizet V, Bier E, 2014. RAB11-mediated trafficking in host-pathogen interactions. Nat. Rev. Microbiol 12, 624–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbauer AB, Zahedi RP, Sickmann A, Pfanner N, Meisinger C, 2014. The protein import machinery of mitochondria-a regulatory hub in metabolism, stress, and disease. Cell Metabol 19, 357–372. [DOI] [PubMed] [Google Scholar]
- Hutagalung AH, Novick PJ, 2011. Role of Rab GTPases in membrane traffic and cell physiology. Physiol. Rev 91, 119–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannessen M, Walquist M, Gerits N, Dragset M, Spang A, Moens U, 2011. BKV agno interacts with α-soluble N-Ethylmaleimide-Sensitive fusion attachment protein, and negatively influences transport of VSVG-EGFP. PLoS One 6, e24489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinschmidt-DeMasters BK, Tyler KL, 2005. Progressive multifocal leukoencephalopathy complicating treatment with natalizumab and interferon beta-1a for multiple sclerosis. N. Engl. J. Med 353, 369–374. [DOI] [PubMed] [Google Scholar]
- Lamers IJC, Reijnders MRF, Venselaar H, Kraus A, Study DDD, Jansen S, de Vries BBA, Houge G, Gradek GA, Seo J, Choi M, Chae JH, van der Burgt I, Pfundt R, Letteboer SJF, van Beersum SEC, Dusseljee S, Brunner HG, Doherty D, Kleefstra T, Roepman R, 2017. Recurrent De Novo mutations disturbing the GTP/GDP binding pocket of RAB11B cause intellectual disability and a distinctive brain phenotype. Am. J. Hum. Genet 101, 824–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langer-Gould A, Atlas SW, Green AJ, Bollen AW, Pelletier D, 2005. Progressive multifocal leukoencephalopathy in a patient treated with natalizumab. N. Engl. J. Med 353, 375–381. [DOI] [PubMed] [Google Scholar]
- Li X, DiFiglia M, 2012. The recycling endosome and its role in neurological disorders. Prog. Neurobiol 97, 127–141. [DOI] [PubMed] [Google Scholar]
- Major EO, Miller AE, Mourrain P, Traub RG, de Widt E, Sever J, 1985. Establishment of a line of human fetal glial cells that supports JCV multiplication. Proc. Natl. Acad. Sci. U. S. A 82, 1257–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malim MH, Bohnlein S, Hauber J, Cullen BR, 1989a. Functional dissection of the HIV-1 Rev trans-activator–derivation of a trans-dominant repressor of Rev function. Cell 58, 205–214. [DOI] [PubMed] [Google Scholar]
- Malim MH, Hauber J, Le SY, Maizel JV, Cullen BR, 1989b. The HIV-1 rev transactivator acts through a structured target sequence to activate nuclear export of unspliced viral mRNA. Nature 338, 254–257. [DOI] [PubMed] [Google Scholar]
- Mason JM, Arndt KM, 2004. Coiled coil domains: stability, specificity, and biological implications. Chembiochem 5, 170–176. [DOI] [PubMed] [Google Scholar]
- Massignan T, Biasini E, Lauranzano E, Veglianese P, Pignataro M, Fioriti L, Harris DA, Salmona M, Chiesa R, Bonetto V, 2010. Mutant prion protein expression is associated with an alteration of the Rab GDP dissociation inhibitor alpha (GDI)/Rab11 pathway. Mol. Cell. Proteom 9, 611–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra S, Cheng KW, Mills GB, 2011. Rab GTPases implicated in inherited and acquired disorders. Semin. Cell Dev. Biol 22, 57–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaco MC, Jensen PN, Hou J, Durham LC, Major EO, 1998. Detection of JCV DNA in human tonsil tissue: evidence for site of initial viral infection. J. Virol 72, 9918–9923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura S, Khoury G, Jay G, 1983. Subcellular localization of the simian virus 40 Agno. J. Virol 45, 428–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada Y, Endo S, Takahashi H, Sawa H, Umemura T, Nagashima K, 2001. Distribution and function of JCV agno. J. Neurovirol 7, 302–306. [DOI] [PubMed] [Google Scholar]
- Okada Y, Suzuki T, Sunden Y, Orba Y, Kose S, Imamoto N, Takahashi H, Tanaka S, Hall WW, Nagashima K, Sawa H, 2005. Dissociation of heterochromatin protein 1 from lamin B receptor induced by human polyomavirus Agno: role in nuclear egress of viral particles. EMBO Rep 6, 452–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otlu O, De Simone FI, Otalora YL, Khalili K, Sariyer IK, 2014. The Agno of polyomavirus JC is released by infected cells: evidence for its cellular uptake by uninfected neighboring cells. Virology 468–470, 88–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou HD, Kwiatkowski W, Deerinck TJ, Noske A, Blain KY, Land HS, Soria C, Powers CJ, May AP, Shu X, Tsien RY, Fitzpatrick JA, Long JA, Ellisman MH, Choe S, O’Shea CC, 2012. A structural basis for the assembly and functions of a viral polymer that inactivates multiple tumor suppressors. Cell 151, 304–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padgett BL, Zu Rhein GM, Walker DL, Echroade R, Dessel B, 1971. Cultivation of papova-like virus from human brain with progressive multifocal leukoencephalopathy. Lancet i, 1257–1260. [DOI] [PubMed] [Google Scholar]
- Pathan M, Keerthikumar S, Ang C-S, Gangoda L, Quek CYJ, Williamson NA, Mouradov D, Sieber OM, Simpson RJ, Salim A, Bacic A, Hill AF, Stroud DA, Ryan MT, Agbinya JI, Mariadason JM, Burgess AW, Mathivanan S, 2015. FunRich: an open access standalone functional enrichment and interaction network analysis tool. Proteomics 15, 2597–2601. [DOI] [PubMed] [Google Scholar]
- Pathan M, Keerthikumar S, Chisanga D, Alessandro R, Ang C-S, Askenase P, Batagov AO, Benito-Martin A, Camussi G, Clayton A, Collino F, Di Vizio D, Falcon-Perez JM, Fonseca P, Fonseka P, Fontana S, Gho YS, Hendrix A, Hoen E.N. t., Iraci N, Kastaniegaard K, Kislinger T, Kowal J, Kurochkin IV, Leonardi T, Liang Y, Llorente A, Lunavat TR, Maji S, Monteleone F, Øverbye A, Panaretakis T, Patel T, Peinado H, Pluchino S, Principe S, Ronquist G, Royo F, Sahoo S, Spinelli C, Stensballe A, Théry C, van Herwijnen MJC, Wauben M, Welton JL, Zhao K, Mathivanan S, 2017. A novel community driven software for functional enrichment analysis of extracellular vesicles data. J. Extracell. Vesicles 6, 1321455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausch JW, Le Grice SF, 2015. HIV rev assembly on the rev response element (RRE): a structural perspective. Viruses 7, 3053–3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehling P, Brandner K, Pfanner N, 2004. Mitochondrial import and the twin-pore translocase. Nat. Rev. Mol. Cell Biol 5, 519–530. [DOI] [PubMed] [Google Scholar]
- Rinaldo CH, Hirsch HH, 2007. Antivirals for the treatment of polyomavirus BK replication. Expert Rev. Anti Infect. Ther 5, 105–115. [DOI] [PubMed] [Google Scholar]
- Safak M, Barrucco R, Darbinyan A, Okada Y, Nagashima K, Khalili K, 2001. Interaction of JCV agno protein with T antigen modulates transcription and replication of the viral genome in glial cells. J. Virol 75, 1476–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safak M, Sadowska B, Barrucco R, Khalili K, 2002. Functional interaction between JCV late regulatory Agno and cellular Y-box binding transcription factor, YB-1. J. Virol 76, 3828–3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saribas AS, Abou-Gharbia M, Childers W, Sariyer IK, White MK, Safak M, 2013. Essential roles of Leu/Ile/Phe-rich domain of JCV Agno in dimer/oligomer formation, protein stability and splicing of viral transcripts. Virology 443, 161–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saribas AS, Arachea BT, White MK, Viola RE, Safak M, 2011. Human polyomavirus JC small regulatory Agno forms highly stable dimers and oligomers: implications for their roles in Agno function. Virology 420, 51–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saribas AS, Coric P, Bouaziz S, Safak M, 2019. Expression of novel proteins by polyomaviruses and recent advances in the structural and functional features of Agno of JCV, BK virus, and simian virus 40. J. Cell. Physiol 234, 8295–8315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saribas AS, Coric P, Hamazaspyan A, Davis W, Axman R, White MK, Abou-Gharbia M, Childers W, Condra JH, Bouaziz S, Safak M, 2016. Emerging from the unknown: structural and functional features of agno of polyomaviruses. J. Cell. Physiol 231, 2115–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saribas AS, White MK, Safak M, 2012. JCV Agno enhances large T antigen binding to the origin of viral DNA replication: evidence for its involvement in viral DNA replication. Virology 433, 12–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saribas AS, White MK, Safak M, 2018. Structure-based release analysis of the JCV Agno regions: a role for the hydrophilic surface of the major alpha helix domain in release. J. Cell. Physiol 233, 2343–2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sariyer IK, Akan I, Palermo V, Gordon J, Khalili K, Safak M, 2006. Phosphorylation mutants of JCV Agno are unable to sustain the viral infection cycle. J. Virol 80, 3893–3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sariyer IK, Khalili K, Safak M, 2008. Dephosphorylation of JCV Agno by protein phosphatase 2A: inhibition by small t antigen. Virology 375, 464–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sariyer IK, Saribas AS, White MK, Safak M, 2011. Infection by Agno-negative mutants of polyomavirus JC and SV40 results in the release of virions that are mostly deficient in DNA content. Virol. J 8, 255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlierf B, Fey GH, Hauber J, Hocke GM, Rosorius O, 2000. Rab11b is essential for recycling of transferrin to the plasma membrane. Exp. Cell Res 259, 257–265. [DOI] [PubMed] [Google Scholar]
- Schmidt TG, Skerra A, 2007. The Strep-tag system for one-step purification and highaffinity detection or capturing of proteins. Nat. Protoc 2, 1528–1535. [DOI] [PubMed] [Google Scholar]
- Schmidt TGM, Batz L, Bonet L, Carl U, Holzapfel G, Kiem K, Matulewicz K, Niermeier D, Schuchardt I, Stanar K, 2013. Development of the Twin-Strep-tag® and its application for purification of recombinant proteins from cell culture supernatants. Protein Expr. Purif 92, 54–61. [DOI] [PubMed] [Google Scholar]
- Shaw G, Morse S, Ararat M, Graham FL, 2002. Preferential transformation of human neuronal cells by human adenoviruses and the origin of HEK 293 cells. FASEB J 16, 869–871. [DOI] [PubMed] [Google Scholar]
- Soleimani-Meigooni DN, Schwetye KE, Angeles MR, Ryschkewitsch CF, Major EO, Dang X, Koralnik IJ, Schmidt RE, Clifford DB, Kuhlmann FM, Bucelli RC, 2017. JCV granule cell neuronopathy in the setting of chronic lymphopenia treated with recombinant interleukin-7. J. Neurovirol 23, 141–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Okada Y, Semba S, Orba Y, Yamanouchi S, Endo S, Tanaka S, Fujita T, Kuroda S, Nagashima K, Sawa H, 2005. Identification of FEZ1 as a protein that interacts with JCV Agno and microtubules: role of Agno-induced dissociation of FEZ1 from microtubules in viral propagation. J. Biol. Chem 280, 24948–24956. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Orba Y, Makino Y, Okada Y, Sunden Y, Hasegawa H, Hall WW, Sawa H, 2013. Viroporin activity of the JC polyomavirus is regulated by interactions with the adaptor protein complex 3. Proc. Natl. Acad. Sci. U. S. A 110, 18668–18673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Orba Y, Okada Y, Sunden Y, Kimura T, Tanaka S, Nagashima K, Hall WW, Sawa H, 2010. The human polyoma JCV Agno acts as a viroporin. PLoS Pathog 6, e1000801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Semba S, Sunden Y, Orba Y, Kobayashi S, Nagashima K, Kimura T, Hasegawa H, Sawa H, 2012. Role of JCV Agno in virion formation. Microbiol. Immunol 56, 639–646. [DOI] [PubMed] [Google Scholar]
- Truebestein L, Leonard TA, 2016. Coiled-coils: the long and short of it. Bioessays 38, 903–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udayar V, Buggia-Prevot V, Guerreiro RL, Siegel G, Rambabu N, Soohoo AL, Ponnusamy M, Siegenthaler B, Bali J, Aesg, Simons M, Ries J, Puthenveedu MA, Hardy J, Thinakaran G, Rajendran L, 2013. A paired RNAi and RabGAP overexpression screen identifies Rab11 as a regulator of beta-amyloid production. Cell Rep 5, 1536–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unterstab G, Gosert R, Leuenberger D, Lorentz P, Rinaldo CH, Hirsch HH, 2010. The polyomavirus BK Agno co-localizes with lipid droplets. Virology 399, 322–331. [DOI] [PubMed] [Google Scholar]
- Van Assche G, Van Ranst M, Sciot R, Dubois B, Vermeire S, Noman M, Verbeeck J, Geboes K, Robberecht W, Rutgeerts P, 2005. Progressive multifocal leukoencephalopathy after natalizumab therapy for Crohn’s disease. N. Engl. J. Med 353, 362–368. [DOI] [PubMed] [Google Scholar]
- White FA 3rd, Ishaq M, Stoner GL, Frisque RJ, 1992. JCV DNA is present in many human brain samples from patients without progressive multifocal leukoencephalopathy. J. Virol 66, 5726–5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuthrich C, Batson S, Anderson MP, White LR, Koralnik IJ, 2016. JCV infects neurons and glial cells in the Hippocampus. J. Neuropathol. Exp. Neurol 75 (8), 712–717. 10.1093/jnen/nlw050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuthrich C, Cheng YM, Joseph JT, Kesari S, Beckwith C, Stopa E, Bell JE, Koralnik IJ, 2009. Frequent infection of cerebellar granule cell neurons by polyomavirus JC in progressive multifocal leukoencephalopathy. J. Neuropathol. Exp. Neurol 68, 15–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerial M, McBride H, 2001. Rab proteins as membrane organizers. Nat. Rev. Mol. Cell Biol 2, 107–117. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.