Abstract
The type IV P-type ATPases (P4-ATPases) thus far characterized are lipid flippases that transport specific substrates, such as phosphatidylserine (PS) and phosphatidylethanolamine (PE), from the exofacial leaflet to the cytofacial leaflet of membranes. This transport activity generates compositional asymmetry between the two leaflets important for signal transduction, cytokinesis, vesicular transport, and host-pathogen interactions. Most P4-ATPases function as a heterodimer with a β-subunit from the Cdc50 protein family, but Neo1 from Saccharomyces cerevisiae and its metazoan orthologs lack a β-subunit requirement and it is unclear how these proteins transport substrate. Here we tested if residues linked to lipid substrate recognition in other P4-ATPases also contribute to Neo1 function in budding yeast. Point mutations altering entry gate residues in the first (Q209A) and fourth (S457Q) transmembrane segments of Neo1, where phospholipid substrate would initially be selected, disrupt PS and PE membrane asymmetry, but do not perturb growth of cells. Mutation of both entry gate residues inactivates Neo1, and cells expressing this variant are inviable. We also identified a gain-of-function mutation in the second transmembrane segment of Neo1 (Neo1[Y222S]), predicted to help form the entry gate, that substantially enhances Neo1’s ability to replace the function of a well characterized phospholipid flippase, Drs2, in establishing PS and PE asymmetry. These results suggest a common mechanism for substrate recognition in widely divergent P4-ATPases.
1. Introduction
The plasma membrane of eukaryotic cells can be characterized as a transversely asymmetric bilayer, whose properties arise from the differential composition of proteins, carbohydrates, and lipids on the extracellular and cytosolic leaflets. Asymmetry of lipid species in the membrane is established by the action of P4-ATPases, transporters that catalyze translocation of specific phospholipids, such as PS and PE, unidirectionally from the extracellular leaflet to the cytosolic leaflet [1]. These pumps are typically heterodimers formed from a catalytic α subunit (the P4-ATPase) and a noncatalytic β subunit from the Cdc50 protein family [2, 3]. P4-ATPases localize to the Golgi, plasma membrane, and endosomes to help control the composition and organization of this entire endomembrane system. The resulting lipid asymmetry is crucial in the maintenance of various cellular activities, such as signal transduction, cell division and vesicular transport [4-7]. Disruption of PS asymmetry has been implicated as a key regulatory step in blood coagulation [8] as well as the recognition and phagocytosis of apoptotic cells [9-11].
The Saccharomyces cerevisiae genome encodes five P4-ATPases, including Neo1, Drs2, Dnf1, Dnf2, and Dnf3 [12]. Of these, Neo1, a Golgi and endosome localized P4-ATPase, is normally essential for cell viability, with its activity required for protein trafficking in the early secretory pathway, endosome to Golgi protein transport, regulation of Golgi and vacuolar pH, and homotypic fusion of vacuolar membranes [12-15] [16]. Studies using temperature sensitive alleles of NEO1 (neo1-ts) have shown that Neo1 contributes to the establishment of PS and PE asymmetry of the plasma membrane but seems to play a more substantial role in preventing exposure of PE [17].
While Neo1 does not interact with a Cdc50-related β-subunit, it does form complexes with Dop1 and Mon2, and these functionally important interactions are well conserved [16,18-21]. The essential role of NEO1 can be bypassed by deletion of ANY1, encoding a PQ-loop protein that interacts with Neo1, and like Dop1 and Mon2, has no known function [22]. Viability of neo1Δ any1Δ cells requires Drs2, and in the any1Δ background, Neo1 and Drs2 become functionally redundant for supporting cell growth [23]. Drs2 and its mammalian orthologs (ATP8A1 and ATP8A2) primarily transport PS and to a lesser extent PE [24-27]. The influence of Neo1 on PS/PE membrane asymmetry and the partially redundant function with Drs2 suggest that Neo1 can directly catalyze translocation of these phospholipids [17, 23].
Insight into the mechanism for how P4-ATPases recognize and flip their phospholipid substrate initially emerged from mutagenic studies [27-33]. For P-type ATPases, recognition of transport substrate is a function of the transmembrane domain, which is most commonly composed of 10 transmembrane segments (M1-M10). Cation transporters (P2-ATPases), such as the Na+/K+-ATPase and Ca++-ATPase, use positionally conserved residues in M4, M5, M6, M8 and M9 membrane segments to form ion binding sites in the middle of the membrane domain [34, 35]. In contrast, residues important for phospholipid selection by P4-ATPases have been mapped to M1, M2, M3, M4 and M6, suggesting a different transport mechanism for these pumps [27-33]. Many of these residues cluster in two regions of the membrane domain in structural models threaded onto the Ca++-ATPase or Na+/K+-ATPase: an entry gate at the exofacial leaflet where substrate is initially selected and an exit gate at the cytosolic leaflet where transported lipid is deposited [29, 36].
For the entry gate, a strong determinant of substrate specificity was mapped to the luminal side of M1 and is a QQ in Drs2 or a GA in Dnf1, which normally flips lysophosphatidylcholine (lyso-PC), lyso-PE, and the sphingolipid glucosylceramide [27-30, 37, 38]. A Dnf1 variant carrying the QQ in place of the GA sequence acquires the ability to flip PS, loses the ability to flip GlcCer but is unaffected for PC or PE transport [29]. Reciprocally, a Drs2 variant carrying GA in M1 in place of QQ displays a loss of PS recognition while maintaining the ability to transport PE [28]. Another critical entry gate residue is in M4, four residues N-terminal to a highly conserved proline (P − 4 position] that breaks the M4 helix. A P − 4 Gln is required for glucosylceramide recognition by Dnf1, Dnf2, and the orthologous human ATP10A and ATP10D [30]. Similarly, an Asn in the P − 4 position in M4 of ATP8A1 is critical for PS recognition [33].
Recent structural studies support this noncanonical mode of substrate recognition by P4-ATPases. Cryo-EM structures of Drs2-Cdc50 reveal a hydrophilic cleft formed by M1, 2, 4 and 6, and bounded by entry gate residues at the base of M1 and the P − 4 position near the middle of the membrane domain [39]. Moreover, a cryoEM structure of Atp8A1-Cdc50a in a E2~P conformational state has a phosphatidylserine residue bound in this entry gate position, with the serine head group coordinated by the first M1 Gln (Q88), along with the M4 P − 4 (N352) and P − 3 (N353) residues [40]. Backbone amide groups from the P + 1 and P + 2 positions in the conserved PISL sequence help coordinate the phosphate group, and the Leu may form a hydrophobic gate separating the entry and exit sites [33, 40].
Thus, the mechanism of substrate recognition appears to be well-conserved between orthologous P4-ATPases, such as budding yeast Drs2 and human Atp8A1, and between divergent P4-ATPase paralogs like Dnf2 and Atp8A1. In this study, we sought to test whether residues in positions important for phospholipid substrate recognition in other P4-ATPases are important for the function of Neo1.
2. Materials and methods
2.1. Reagents
Papuamide A was purchased from the University of British Columbia Depository. Duramycin, monoclonal FLAG antibody, and EzView FLAG beads were purchased from Sigma Aldrich. 5-fluoroorotic acid (5-FOA) was purchased from US Biologicals. Monoclonal GFP antibody (clone 1C9A5) was purchased from Vanderbilt Antibody and Protein Resource core. Polyclonal GFP antibody was purchased from Torrey Pines Biolabs. The anti-Arf1 antibody has been previously described [41]. IRDye® 800CW goat anti-rabbit IgG (H+L) and 680RD goat anti-mouse IgG (H+L) secondary antibodies were purchased from LiCOR Biosciences.
2.2. Strains and plasmid construction
Strains and plasmids used were listed in the Supplementary Table 1. Yeast strains were grown and transformed with plasmids using standard media and transformation techniques [42-44]. For growth and plasmid shuffling assays, 0.1 OD600 of cells and 10-fold serial dilutions were spotted onto synthetic media or synthetic media containing 5-FOA and other toxic agents. All images of yeast colonies are representative of three biological replicates (three independently isolated strains of same genotype). Yeast knock-out and knock-in strains were generated as previously described [45]. For co-IP experiments, 9XGLY-5XFLAG::HPHMX6 cassette was amplified from pFA6a-G9-5FLAG-HPHMX6 and integrated in the carboxyl terminus of ANY1 gene in strain BY4742. pRS313-NEO1-9XGLY-5XFLAG was generated by Gibson assembly mediated insertion of 9XGLY-5XFLAG tag into pRS313-PNEO1-NEO1 [23]. pRS313-NEO1-9XGLY-5XFLAG was transformed into YWY10 and shuffled as a replacement of pRS416-PNEO1-NEO1. Mutant plasmids were generated using PCR-mediated mutagenesis followed by 3-fragment or 2-fragment Gibson assembly according to the manufacturer’s instructions (New England Biolabs) [46]. Mutagenic primers were between 40 and 60 pairs in length and centered on the mutation site.
2.3. Toxin sensitivity assays
For toxin sensitivity assays, mid-log cells were diluted to 0.1 OD600 in fresh YPD medium and 100μl of cells were distributed to each well of 96 well-plate with or without the toxin in 100μl of YPD. Toxin dilutions were calculated based on final concentrations in a total well volume of 200μl. Plates were incubated at 30°C for 20 hours. Concentrations of the cells were measured in OD600/ml with a Multimode Plate Reader Synergy HT (Biotek). Sigmoidal curve fitting modality from Graphpad Prism8 was used to fit the data point from all samples, when R2 values are equal or greater than 0.8 for all samples. All values are an average of at least three biological replicates +/− standard deviation.
2.4. Lipid uptake assays
Lipid uptake assays were performed with approximately one OD600 of mid-log phase dnf1,2Δ (PFY3275F) cells expressing wild-type Dnf1, empty vector and Dnf1 mutants and using 1-palmitoyl-2,6-[(7-nitro-2-1,3-benzoxadiazol4yl)amino]hexanoyl)-sn-glycero-3-phospholipids (NBD-PLs) for 30 min at 4°C as described previously [28, 29]. At least, three independent transformants were assayed for each experiment. Lipid uptake activities were plotted as the percentage of NBD-PC lipid activity of wild-type Dnf1.
2.5. Fluorescence microscopy
Strains expressing fluorescent fusion proteins were grown to mid-log in synthetic media, harvested, and resuspended in 1X PBS+2% glucose. Cells were mounted on glass slides and observed immediately at room temperature. Colocalization images were acquired using a DeltaVision Elite workstation (Applied Precision) based on an inverted microscope (IX-70; Olympus) equipped with a 100×, 1.4 NA oil immersion lens and deconvolved using an iterative-constrained algorithm and the measured point spread function (softWoRx 5.5 software; Applied Precision). Images were analyzed in FIJI (FIJI Is Just ImageJ) using the JaCOP (Just another Colocalization Plugin) add-on. Amount of colocalization was quantified through the computation of the Manders’ coefficient in JaCOP as the fraction of overlap between Neo1 protein and the various markers. Multiple images (at least 100 cells) were analyzed for calculating the Manders’ correlation coefficient.
2.6. Immunoprecipitation and western blot analysis
Mid-log phase cells were lysed and subjected to Western blot analysis as described previously [47]. For expression analysis, polyclonal anti-GFP (1:2000) and monoclonal FLAG (1:5000) antibodies were used. Total protein concentrations were assessed by Bio-RAD stain-free gel system. The preparation of crude and Golgi-enriched membranes was performed as described previously [48]. Golgi-enriched membrane fractions were solubilized with lysis buffer (10mM Tris-HCl pH7.5, 150mM NaCl, 2mM EDTA) supplemented with 1% CHAPS. Protein concentrations of Golgi-enriched membranes were quantified using a using a bicinchoninic acid assay (BCA). Immunoprecipitations of FLAG-tagged bait were performed from equal amount of total input using 50 μl of EzView FLAG beads for 2 hours at 4°C. To assess the immunoprecipitations (IP), 1% of total input and 10% total elute were loaded onto SDS-PAGE gel and immunoblotted for GFP-tagged and FLAG-tagged bait protein levels. IP(s) were quantified as a percentage of input.
3. Results
3.1. Q209 is required for Neo1 function in controlling membrane asymmetry
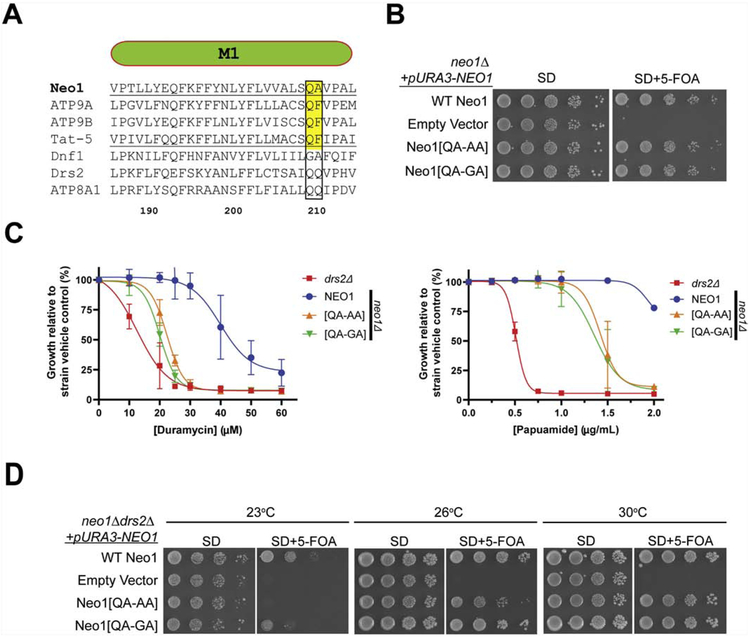
Neo1 has a QA motif in the M1 entry gate position with Q209 conserved in its metazoan orthologs (Tat-5, Atp9A, Atp9B) and in the PS flippases Drs2, ATP8A1 and ATP8A2 (Fig. 1A, Supplemental Fig. 1A). This Gln coordinates the serine headgroup of phosphatidylserine in ATP8A1 [40] and is required for PS recognition by Drs2 [29]; therefore, Q209 was predicted to facilitate PS transport by Neo1. We mutated Q209 to Ala or Gly and tested whether these Neo1 variants could support viability of a neo1Δ strain using a plasmid shuffle assay (these variants are described as Neo1[QA-AA] and Neo1[QA-GA] for consistency with prior studies comparing QQ and GA motifs in M1 of Drs2 and Dnf1, respectively [29, 30]). The parental neo1Δ strain harboring wild-type (WT) NEO1 on a URA3-marked plasmid was transformed with LEU2-marked plasmids expressing either WT Neo1, no insert (empty vector), Neo1[QA-AA], or Neo1[QA-GA]. The strains were replica plated on synthetic defined (SD) media to select for both plasmids, or SD + 5-fluoro-orotic acid (5-FOA) to counterselect against the URA3 plasmid (Fig. 1B). Because neo1Δ cells are inviable, the empty vector control strain could not lose the URA3-NEO1 plasmid and failed to grow on 5-FOA as expected. In contrast, the Neo1[QA-AA] and Neo1[QA-GA] mutants supported viability as well as the WT Neo1 control. Thus, the highly conserved Q209 can be altered without disrupting Neo1’s essential function.
Figure 1. Neo1 Q209 is critical for control of membrane asymmetry.
(A) Sequence alignment of transmembrane segment 1 (M1) from Neo1, Atp9a, Atp9b, Tat-5, Dnf1 and Drs2. Numbers are Neo1 residues and the QQ/QA motif is boxed. (B) Neo1[QA-AA] and Neo1[QA-GA] support growth of neo1Δ cells as well as WT Neo1 (image representative of 3 biological replicates). 5-FOA kills cells that are unable to lose the pURA3-NEO1 plasmid. (C) Cells expressing Neo1[QA-AA] and Neo1[QA-GA] are hypersensitive to duramycin and papuamide relative to NEO1 (WT) cells but are not as sensitive as drs2Δ (n = 3, +/− SD). (D) Expression of Neo1 variants in neo1Δ drs2Δ cells. Partial loss of function for Neo1[QA-AA] is revealed in this background by poor growth at 23°C and 26°C relative to Neo1.
We then tested whether strains carrying the Q209A/G variants displayed a loss of plasma membrane asymmetry using cytotoxic peptides that target cells exposing PS (papuamide) or PE (duramycin) in the outer leaflet (Fig. 1C). The neo1Δ strain complemented with a WT copy of NEO1 was resistant to duramycin and papuamide relative to a drs2Δ strain known to expose both lipids, and cells expressing Neo1[QA-GA] or Neo1-[QA-AA] displayed an intermediate sensitivity to both toxins. These results indicate Neo1-Q209 mutants display a partial loss of function in maintaining PS/PE asymmetry.
We also tested the Neo1-Q209 mutants for their ability to support growth of a drs2Δ neo1Δ double mutant across a range of temperatures using the plasmid shuffle assay (Fig. 1D). drs2Δ single mutants grow well at 30°C, but are cold-sensitive for growth and grow slowly at 26°C or 23°C. WT Neo1 and both Neo1-Q209 variants supported growth of drs2α neo1Δ equivalently at 30°C, and the cold-sensitive growth defects caused by drs2Δ was apparent in the cells expressing WT Neo1 (these cells are genotypically drs2Δ NEO1). However, loss of function phenotypes for the Neo1-Q209 variants were apparent at the lower temperatures. The double mutant expressing Neo1[QA-AA] failed to grow at 23°C, comparable to the empty vector control, while Neo1[QA-GA] cells grew more slowly than the WT Neo1 control. Neo1[QA-AA] cells also displayed a growth defect at 26°C, but Neo1[QA-GA] supported WT growth at this temperature. Thus, growth defects caused by mutation of Q209 were revealed in the absence of Drs2, the primary PS flippase in the cell, and Q209A perturbed Neo1 function more substantially than Q209G.
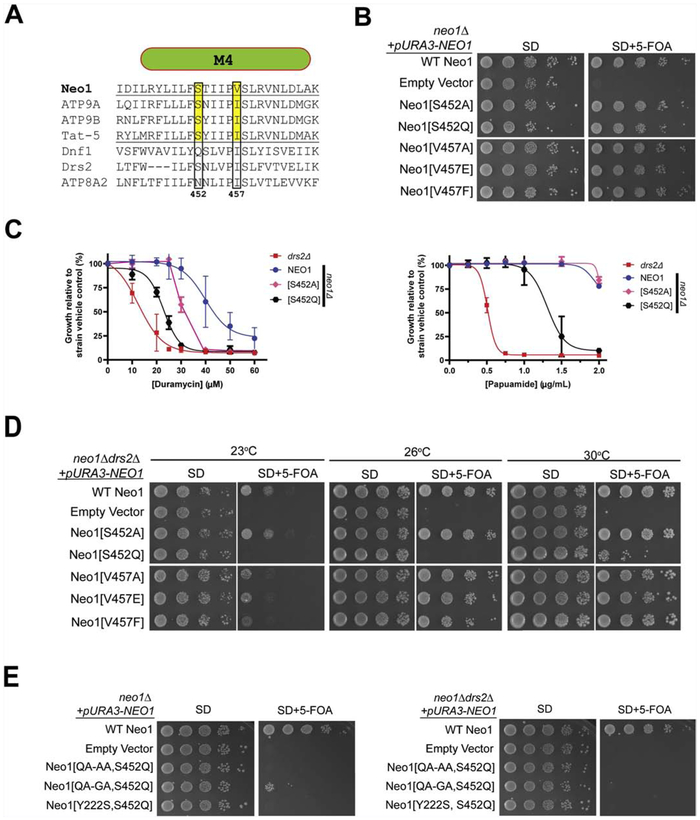
3.2. P − 4 Ser in M4 is required for Neo1 function in controlling membrane asymmetry
The P − 4 residue in TM4 of Dnf1, Dnf2 and ATP8A1 is also important for substrate selection, helping to coordinate the serine headgroup along with the M1 Gln residue in the case of ATP8A1 [30, 33, 40]. While the P − 4 residue is an Asn in ATP8A1, it is a Ser in Neo1 (S452), ATP9A, ATP9B, Tat-5 and Drs2 (Fig. 2A). To test the influence of this residue on Neo1 function, S452 was mutated to an Ala or Gln (the P − 4 residue in Dnf1 and Dnf2) and assayed for the ability to support viability in a neo1Δ strain. We also mutated the P + 1 Val to Ala, Glu or Phe to test the requirement of this potential hydrophobic gate residue on Neo1 function. Mutations at this position partially disrupt the function of ATP8A2, and a P + 1 Ile to Met mutation in ATP8A2 causes cerebellar atrophy, mental retardation, disequilibrium (CAMRQ) syndrome in humans [33, 49].
Figure 2. M1 and M4 entry gate residues are essential for Neo1 function.
(A) Sequence alignment of transmembrane segment 4 (M4) from divergent P4-ATPases. Numbers are Neo1 residues and the P − 4 and P + 1 residues are boxed. (B) Neo1[S452] and Neo1[V457] variants support growth of neo1Δ cells as well as WT Neo1 (image representative of 3 biological replicates). (C) Cells expressing Neo1[S452Q] and Neo1[S452A] are hypersensitive to duramycin relative to NEO1 (WT) cells, but only Neo1[S452Q] is sensitive to papuamide. (D) Expression of Neo1 variants in neo1Δ drs2Δ cells. Partial loss of function for Neo1[S452Q] is revealed in this background by poor growth at all temperatures relative to cells expressing WT Neo1. In contrast, Neo1[S452A] and the Neo1[V457X] variants all supported growth equivalently to WT Neo1. (E) Combining entry gate mutations inactivates Neo1. Neo1 variants carrying mutation in both M1 (QA-AA) and M4 (S452Q) fail to support growth of either neo1Δ (left) or neo1Δ drs2Δ (right) strains. Neo1[QQ-GQ, S452Q] very weakly sustains viability of neo1Δ, but not neo1Δ drs2Δ. The M2 entry gate mutation Y222S is also lethal when combined with S452Q. All images are representative of three biological replicates.
Surprisingly, all of these Neo1 M4 variants complemented neo1Δ and supported growth as well as WT Neo1 (Fig. 2B). However, we found that cells expressing Neo1-S452A displayed a partial loss of PE asymmetry as measured by duramycin sensitivity, but did not expose PS any more than cells expressing WT Neo1 (Fig. 2C). In contrast, Neo1-S452Q caused a partial loss of PE and PS asymmetry. Moreover, Neo1-S452Q weakly supported viability of neo1Δ drs2Δ cells at 30°C, and failed to support growth at 23°C or 26°C; but Neo1-S452A supported growth of the double mutant as well as WT Neo1 (Fig. 2D). We conclude that the P − 4 Ser in Neo1 is important for function, and the loss of Neo1’s ability to support PS asymmetry correlates with growth defects revealed by the absence of Drs2. This conclusion is consistent with prior studies suggesting a PS flippase activity in Golgi membranes is required for budding yeast viability [23].
Neo1 bearing mutations in the P + 1 position of M4, (V457A, V457E, V457F) supported growth of neo1Δ and neo1Δ drs2Δ cells nearly as well as WT Neo1, with a very modest growth delay observed at 23°C in the double mutant (Fig. 2B and 2D). These mutants all displayed an equivalent degree of duramycin hypersensitivity with an IC50 of 27 – 30 μM (Supplemental Fig. 1B). We did not measure the papuamide sensitivity of these mutants because of the limited supply of this reagent. However, the near WT ability to support growth of the neo1Δ drs2Δ double mutant (Fig. 2D) suggests that these P + 1 variants can prevent PS exposure.
Coordination of PS in the entry gate of ATP8A1 is achieved by multiple interactions and it is possible that no single side-chain interaction with substrate would be essential for transport [40]. Therefore, we tested if mutating both the P − 4 Ser in M4 and Q207 in M1 of Neo would give a more complete loss of function phenotype. Consistent with this idea, the Neo1[QA-AA, S452Q] mutant failed to support growth of either neo1Δ or neo1Δ drs2Δ strains (Fig. 2E). The Neo1[QA-GA, S452Q] double mutant weakly supported growth of neo1Δ, but could not support viability of neo1Δ drs2Δ cells. We conclude that the M1 and M4 entry gate residues in Neo1 are essential for the function of this P4-ATPase, supporting the role of Neo1 in flipping PE and PS across Golgi membranes.
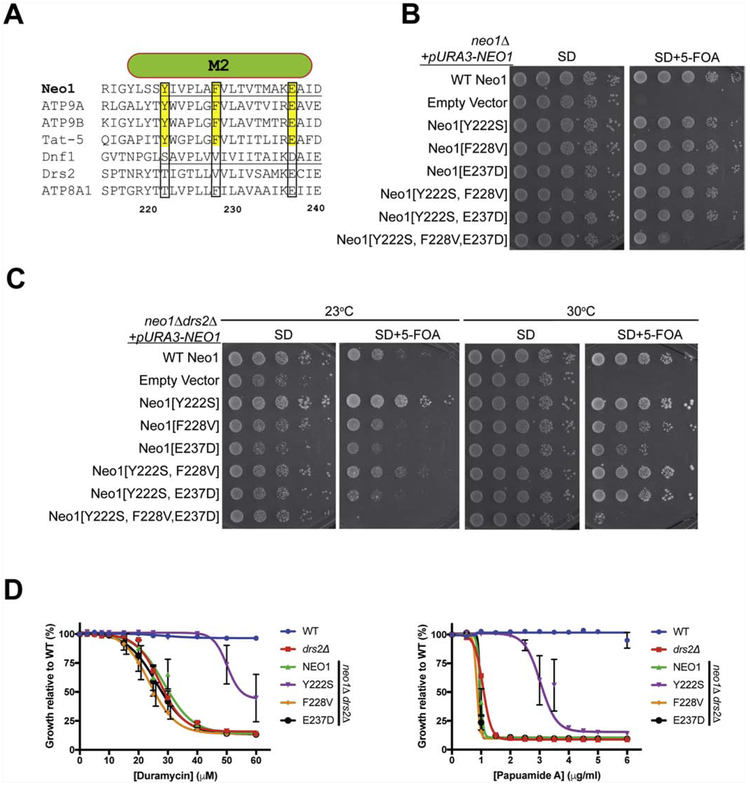
3.3. Conserved M2 residues important for Neo1 function
We previously found that D258 in M2 of Dnf1 played a role in substrate specificity by suppressing PS recognition [28]. This M2 residue is part of the exit gate and is a conserved Glu in Neo1 (E237) and its metazoan orthologs, as well as in ATP8A1 and Drs2 (Fig. 3A), which suggests that a Glu in this position is important for PS recognition. We also noted that Y222 and F228 in M2 are well conserved in the Neo1/ATP9/Tat-5 group, but are Ser/Thr and Val, respectively in Dnf1 and Drs2 (Fig. 3A). To explore the functional significance of these conserved M2 residues in Neo1, we mutated them to their corresponding Dnf1 residues (Y222S, F228V, and E237D). The single mutants and two double mutant combinations all supported growth of neo1Δ cells comparably to WT Neo1. However, the Neo1[Y222S, F228V E237D] triple mutant displayed a significant growth defect (Fig. 3B). The F228V single mutant supported growth of neo1Δ drs2Δ cells as well as WT Neo1, but a partial growth defect was observed for the E237D mutant in this background. Thus, these conserved Neo1 residues in M2 can individually be mutated to Dnf1 residues without substantially perturbing Neo1’s essential function, but collectively these Neo1 residues are critical.
Figure 3. Gain of function and loss of function mutations in M2 of Neo1.
(A) Sequence alignment of transmembrane segment 2 (M2) from divergent P4-ATPases. Numbers are Neo1 residues and targeted residues are boxed. (B) Neo1 mutants Y222S, F228V, E237D and double mutant combinations tested support growth of neo1Δ cells as well as WT Neo1. In contrast, the triple mutant displays a substantial growth defect. (C) Growth of neo1Δ drs2Δ cells expressing Neo1 M2 variants at 23°C and 30°C). Neo1-Y222S improves growth of this strain relative to WT Neo1 while Neo1[E237D] causes a partial growth defect. Y222S suppression of low temperature growth defects is attenuated by F228V and E237D in the double mutants. The Neo1 triple mutant is unable to support growth of this strain. (D) Neo1-Y222S suppresses loss of membrane asymmetry as indicated by increased resistance to duramycin and papuamide relative to WT Neo1. F228V and E237D had no measurable influence on Neo1 activity by these assays.
3.4. Neo1[Y222S] is a dominant gain of function suppressor of drs2Δ
Surprisingly, neo1Δ drs2Δ cells expressing Neo1[Y222S] grew significantly better than neo1Δ drs2Δ cells expressing WT Neo1 at 23°C and even 30°C (Fig. 3C). This phenotype indicates Neo1[Y222S] not only complemented neo1Δ, but it also suppressed the drs2Δ cold-sensitive growth defect. The ability of the Y222S to confer suppression of drs2Δ was attenuated by the F228V mutation and eliminated by the E237D mutation in the double mutants (compare double mutants to WT Neo1 and the corresponding single mutants). Over the course of these studies, a number of mutations were introduced into the membrane domain of Neo1 (Fig. 1-3, supplemental Fig. 2), and only the Q209A (QA-AA), S452Q and E237D single mutants consistently caused a loss of function growth phenotype in the neo1Δ drs2Δ background, and Y222S was the only gain-of-function mutation detected. By contrast, an ATPase-dead mutant (Neo[D503N]) failed to support growth of neo1Δ or neo1Δ drs2Δ, indicating Neo1 catalytic activity is essential for its function in vivo (Supplemental Fig. 2).
The growth phenotypes shown in Figure 3C suggested that E237D perturbed PS recognition by Neo1 and Y222S enhanced PS recognition. To test this, we probed the neo1Δ drs2Δ strains harboring Neo1 with M2 single mutations (Fig. 3D). However, no significant difference was observed for the papuamide or duramycin sensitivity of the F228V and E237D strains relative to neo1Δ drs2Δ expressing WT Neo1. It is possible that the influence of E237D on substrate selection was too subtle to detect by this toxin sensitivity assay. In contrast, Neo1[Y222S] conferred greater resistance to papuamide and duramycin than WT Neo1 in this background. Thus, Neo1[Y222S] fully complemented neo1Δ and partially suppressed the loss of PS and PE asymmetry caused by drs2Δ. The corresponding Thr at this position in ATP8A1 (T101) is within the entry gate and is proximal to Q88 in M1, and the P − 4 Asn in M4 that coordinates the serine headgroup ([40] and Supplemental Fig. 3). We conclude that Y222 is similarly part of the entry gate of Neo1 and Y222S likely enhances substrate loading.
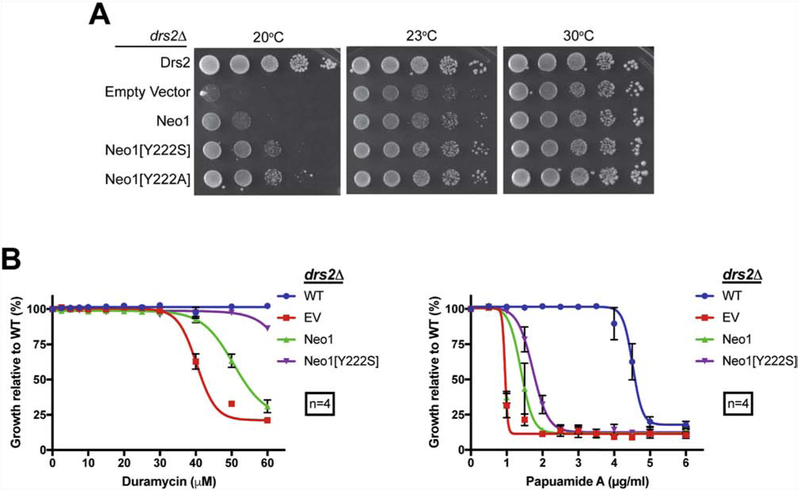
We used wild-type Neo1, Neo1[Y222S] and Neo1[Y222A] variants to transform drs2Δ NEO1 cells to test whether the mutation is dominant or recessive, and if the loss of tyrosine or presence of serine at Y222 was responsible for the gain-of-function phenotype. An extra copy of WT Neo1 weakly suppressed drs2Δ cold-sensitive growth as well as the papuamide and duramycin sensitivity (Fig. 4A-C). Neo1[Y222S] and Neo1[Y222A] suppressed drs2Δ cold-sensitivity at 20°C and 23°C to a greater extent than Neo1, but not as completely as restoring Drs2 (Fig. 4A). Thus, Y222S and Y222A mutations in Neo1 are dominant alleles and both allow suppression of drs2Δ. Neo1[Y222S] restored PE asymmetry in drs2Δ cells to near WT levels but weakly suppressed PS exposure in drs2Δ cells (Fig. 4B). Thus, Neo1[Y222S] seemed to have a greater influence on duramycin sensitivity than papuamide sensitivity in the drs2Δ background, where an additional copy of WT NEO1 is present.
Figure 4. Neo1[Y222S] and Neo1[Y222A] suppress drs2Δ growth and membrane asymmetry defects.
(A) Growth of drs2Δ cells expressing additional copies of Neo1, Neo1[Y222S] or Neo1[Y222A] at temperatures indicated. (B) Neo1[Y222S] suppresses duramycin sensitivity and weakly suppresses papuamide sensitivity of drs2Δ cells.
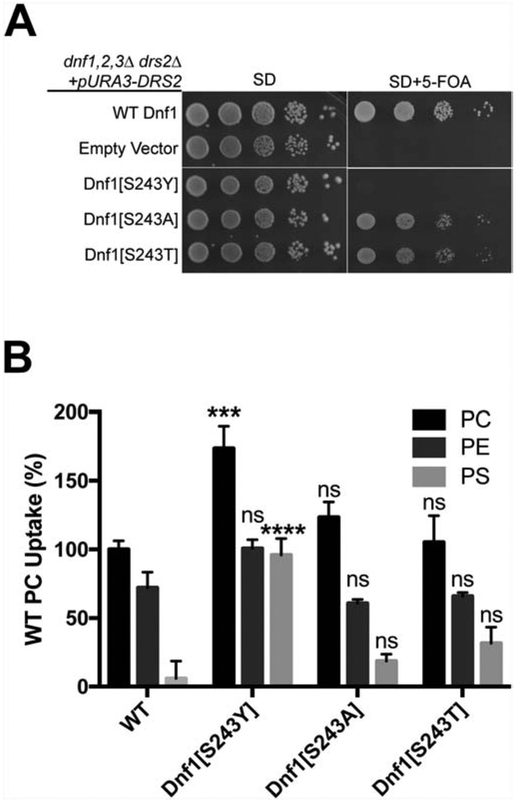
3.5. Influence of the S243Y reciprocal mutation in Dnf1
To test the influence of the reciprocal substitutions in M2 of Dnf1 we mutated S243 to tyrosine, threonine or alanine (Fig. 3A, Neo1-Y222 aligns with Dnf1-S243). Dnf1[S243A], Dnf1[S243Y] and Dnf1[S243T] were tested for their ability to complement dnf1,2,3Δdrs2Δ and support viability of this strain. WT Dnf1 supports growth of this strain at 30°C, but Dnf1[S243Y] failed to complement dnf1,2,3Δdrs2Δ (Fig. 5A). By contrast, Dnf1[S243A] and Dnf1[S243T] complemented dnf1,2,3Δdrs2Δ nearly as well as wild-type Dnf1 (Fig. 5A). We tested the NBD-PL uptake activities of Dnf1[S243] mutants in a dnf1Δ dnf2Δ background. Surprisingly, Dnf1[S243Y] was active on the plasma membrane and displayed a significant increase (p<0.01) in the NBD-PC and NBD-PS transport activities and no change in NBD-PE transport relative to WT Dnf1 (Fig. 5B). NBD-phospholipid transport activities of Dnf1[S243T] and Dnf1[S243A] were similar to that of wild-type Dnf1 (Fig. 5B). These unexpected results indicate that the S243Y substitution attenuates the ability of Dnf1 to provide the essential function of this group of P4-ATPases, while retaining activity at the plasma membrane and actually enhancing the ability of Dnf1 to transport NBD-PS. Similarly, the presence of tyrosine at position 222 in Neo1 (the WT residue) attenuates its ability to substitute for Drs2 in vivo, while Neo1[Y222S] can partially substitute for Drs2 (Fig. 4).
Figure 5. Influence of the reciprocal Dnf1[S243Y] mutation on complementation and transport activity.
(A) Dnf1[S243Y] failed to suppress dnf1,2,3Δdrs2Δ synthetic lethality, but Dnf1[S234A] and Dnf1[S243T] supported growth of this strain nearly as well as wild-type Dnf1. (B) Dnf1[S243Y] displays a significant increase in NBD-PC and NBD-PS transport relative to wild-type Dnf1, Dnf1[S243A] and Dnf1[S243T]. Lipid uptake assays were performed with dnf1,2Δ cells expressing wild-type Dnf1, Dnf1[S243Y], Dnf1[S243A] and Dnf1[S243T] on a single copy plasmid grown at 30°C using fluorescent-labeled (NBD) phospholipids. Lipid uptake activities were plotted as percentage of NBD-PC uptake for wild-type cells. (***p < 0.001. ****p < 0.0001; student T-test comparisons between WT and mutants, ns = not significant).
3.6. The gain of function phenotype of Neo1[Y222S] is dependent on Any1
Any1 is an integral membrane protein of the Golgi that binds to Neo1 and appears to antagonize Neo1’s activity [23]. We considered the possibility that Y222S disrupted the interaction between Any1 and Neo1, thus allowing hyperactivity of Neo1 and its ability to suppress drs2Δ. To test this possibility, we immunoprecipitated FLAG-tagged Neo1 and Neo1[Y222S] from strains expressing GFP-tagged Any1. However, GFP-Any1 co-immunoprecipitated equally well with Neo1 and Neo1[Y222S] (Supplemental Fig. 4). Therefore, Y222S does not appear to disrupt the Neo1 interaction with Any1. Changes in localization of Neo1[Y222S] relative to Neo1 might contribute to the gain-of-function phenotype. Neo1 localizes broadly throughout the Golgi and endosomes while Drs2 localizes primarily to the TGN. To test if the Y222S mutation shifts Neo1 localization to the TGN, we examined localization of GFP-tagged Neo1 and Neo1[Y222S] relative to mCherry-Tlg1 (TGN/early endosome), Sec7-mKate2 (TGN), Aur1-mKate2 (medial-Golgi), and COPI-mKate2 (cis-Golgi) (Supplemental Fig. 5). We quantified the extent of co-localization of GFP-tagged and RFP-tagged proteins using Manders’ correlation coefficient. While there was no substantial difference in the localization of GFP-Neo1[Y222S] relative to GFP-Neo1, the Manders’ coefficients showed a small but statistically significant change in the distribution of Neo1[Y222S] towards later compartments of the Golgi marked by Sec7 and Tlg1 (Supplemental Figure 5A-C). Altered localization of Neo1[Y222S] to late Golgi compartments where Drs2 normally resides might be a factor contributing to the gain-of-function phenotype, but whether this small difference is biologically significant seems unlikely.
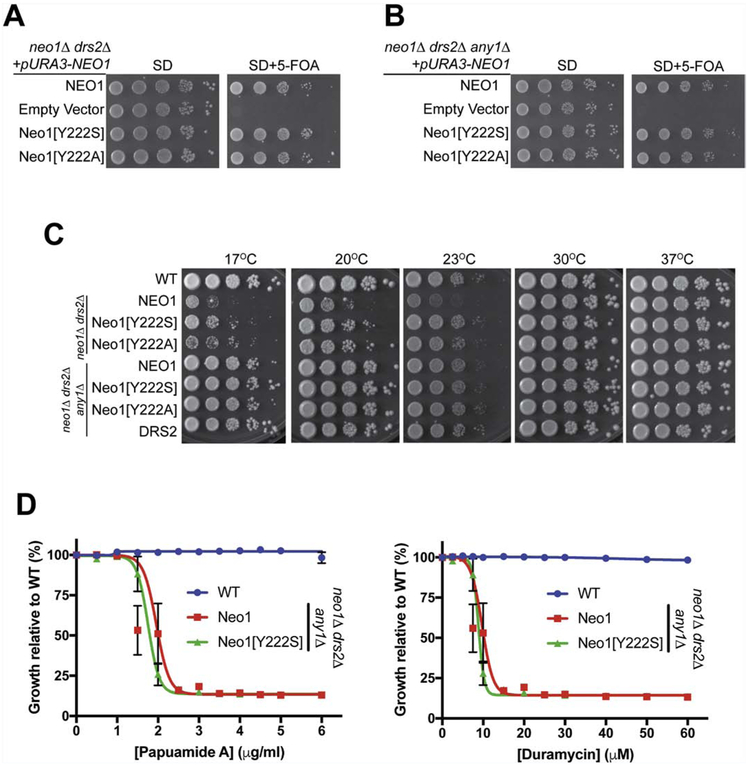
We next tested whether Neo1[Y222S] suppression of drs2Δ was influenced by the PQ-loop protein Any1. Neo1[Y222S] and Neo1[Y222A] fully supported growth of neo1Δdrs2Δ and neo1Δdrs2Δany1Δ equivalently to Neo1 (Fig. 6A,B). Growth of neo1ΔdrsΔ and neo1Δdrs2Δany1Δ strains expressing Neo1, Neo1[Y222S], Neo1[Y222A] or Drs2 were tested over a wide range of temperatures to examine suppression of drs2Δ cs growth. As shown before, Neo1[Y222S] and Neo1[Y222A] mutants partially suppressed the drs2Δ cold-sensitivity of neo1Δdrs2Δ at low temperatures (17°C, 20°C and 23°C) (Fig. 6C). For neo1Δany1Δdrs2Δ, WT Neo1 supported viability and any1Δ suppressed the cs growth defects caused by drs2Δ and so these cells grew well at all temperatures. No influence of Neo1[Y222S] or Neo1[Y222A] on growth relative to Neo1 could be observed in this background (Fig. 6C). To investigate the influence of Neo1[Y222S] and any1Δ on membrane asymmetry, we performed PapA and duramycin sensitivity assays in the neo1Δdrs2Δany1Δ background along with wild-type cells. Surprisingly, Neo1[Y222S] failed to suppress PS and PE exposure when Any1 was absent (Fig. 6D). There was no significant difference between cells expressing wild-type Neo1 or Neo1[Y222S] in the sensitivities to these toxins. Thus, Neo1[Y222S] requires the presence of Any1 to suppress PS/PE asymmetry defects caused by drs2Δ (compare Fig. 6D to Fig. 4D).
Figure 6. Loss of the PQ-loop protein Any1 abolishes the gain-of-function phenotype of Neo1[Y222S].
(A) Neo1[Y222S] and Neo1[Y222A] can complement neo1Δdrs2Δ and (B) neo1Δdrs2Δany1Δ lethality. (C) Suppression of drs2Δ cold-sensitive growth by Neo1[Y222S] is dependent on Any1. any1Δ effectively suppresses drs2Δ cold-sensitive growth masking any influence of Neo1[Y222S]. (D) Neo1[Y222S] cannot suppress hypersensitivity of neo1Δdrs2Δany1Δ cells to duramycin or papuamide relative to Neo1.
4. Discussion
Neo1 and its orthologs, for example human ATP9A/B and Tat-5 from C. elegans, form an evolutionarily distinct clade described as P4B-ATPases [50] and are the most enigmatic members of the P4-ATPase group. Only a few members of the P4A-ATPase family (e.g Drs2-Cdc50, ATP8A1-CDC50) have been biochemically reconstituted to definitively demonstrate their phospholipid flippase activity [25, 51, 52], and this has not yet been achieved with any P4B-ATPase. For all other P4-ATPases examined, a β-subunit from the Cdc50 protein family is essential for α-subunit function in vivo. In addition, the Drs2-Cdc50 structure shows multiple points of contact between cytosolic, membrane and luminal domains of each subunit, suggesting an intimate role for the β subunit in the transport mechanism [39]. The observation that P4B-ATPases do not require a β-subunit raised the question as to whether these enzymes could catalyze a phospholipid flippase activity. Here we present evidence supporting the phospholipid flippase activity of Neo1, and that the mechanism of phospholipid substrate recognition by Neo1 is conserved with well-established phospholipid flippases. We find that Neo1 mutations in residues predicted to form a phospholipid substrate-selecting entry gate in the membrane domain disrupt the function of Neo1 and cause a loss of membrane asymmetry. We also identify a gain-of-function mutation within the entry gate of Neo1 that substantially enhances its ability to replace the cellular function of the PS flippase Drs2.
Phospholipid flippases play an important role in establishing membrane asymmetry, characterized by the very low levels of PS and PE in the outer leaflet of the plasma membrane relative to the cytosolic leaflet. An expected phenotype for mutants deficient in flippase activity is the exposure of PS and PE in the outer leaflet of the plasma membrane, which can be detected by hypersensitivity to pore-forming toxins that specifically bind PS (papuamide) or PE (duramycin) [53, 54]. It is difficult to quantify the precise number of PS or PE molecules exposed in the outer leaflet in flippase mutants, and so we quantify hypersensitivity to duramycin and papuamide and benchmark these values relative to drs2Δ mutant cells. We previously found that inactivation of Neo1 activity (using a neo1-1 temperature-sensitive mutant) caused considerably greater sensitivity to duramycin than deleting Drs2 (duramycin IC50 of ~2μM for neo1-1 versus ~22μM for drs2Δ). In contrast, drs2Δ mutants displayed greater sensitivity to papuamide than neo1-1 cells (IC50 ~2 μg/ml for neo1-1 vs 0.5 μg/ml for drs2Δ) [17]. These results suggested that both P4-ATPases were PS/PE flippases, but Drs2 primarily flips PS while Neo1 primarily flips PE.
In this work, we further explored the potential Neo1 PS flippase activity by targeting residues predicted to be important for PS recognition. A critical determinant of PS recognition by Drs2 and ATP8A1 is a QQ motif in the base of M1, and the first Gln is conserved in P4B-ATPases (Q209 of a QA motif in Neo1) [29]. Neo1[QA-AA] and Neo1[QA-GA] variants support growth of a neo1Δ strain as well as WT Neo1, but these mutations cause a loss of PE and PS asymmetry (Fig 1C). The papuamide sensitivity of Neo1[QA-AA] and Neo1[QA-GA] cells (~1.4 ug/ml), which reports PS exposure, is similar to what is observed in neo1-1 and neo1Δ any1Δ cells [17, 23], and presumably represents a complete loss of PS recognition by Neo1. In contrast, the duramycin sensitivity of these Q209 mutants (IC50 ~ 22 μM), indicating exposure of PE, was intermediate between WT (IC50 ~ 41 μM) and neo1-1 (IC50 ~ 2 μM) cells [17]. Thus, Neo1 Q209 mutations appear to preferentially disrupt PS recognition by Neo1 and more modestly perturb PE recognition. This is consistent with the influence of these M1 mutations on Drs2 (loss of PS recognition and no apparent disruption of PE recognition), and the reciprocal mutations in Ml of Dnf1 (GA-→QQ) which enhance PS transport without significantly altering PE transport. These results are also consistent with the structure of ATP8A1 bound to PS, in which Q88 in M1 hydrogen bonds to the PS carboxyl group, which is not present in PE [40] (see supplemental Fig. 3). While it remains speculative as to whether Q209 in Neo1 directly coordinates PS, our data clearly indicate this residue is important for Neo1 function.
We also find that the proline – 4 (P − 4) position in M4 of Neo1 is important for the function of this P4-ATPase in establishing membrane asymmetry, and to support growth in the absence of Drs2. For Dnf1 and Dnf2, a Gln in the P − 4 position and the M1 GA motif are critical for recognition and transport of glucosylceramide [30]. Mutations in the P − 4 Asn in ATP8A2 substantially disrupt the apparent affinity for phospholipid substrate (both PS and PE) [33], and this residue in ATP8A1, along with Q88 in M1, helps coordinate the PS headgroup within the entry gate [40]. Neo1, its metazoan orthologs, and Drs2 have a Ser in the P − 4 position, suggesting that this conserved residue may be important for substrate recognition. Indeed, we find Neo1[S452Q] causes a loss of PS and PE asymmetry equivalent to that caused by the Q209 mutations. Interestingly, Neo1[S452A] modestly perturbed PE asymmetry, but had no measurable influence on PS asymmetry. Both P − 4 mutations supported growth of a neo1Δ strain, but importantly, Neo1[S452Q] failed to support growth of neo1Δ drs2Δ cells at low temperatures, and these cells displayed a strong growth defect at 30°C as well. Growth defects caused by Neo1 Q209 mutations were also revealed when Drs2, the major PS flippase, was removed. In contrast, Neo1[S452A] maintained PS asymmetry and supported growth of neo1Δ drs2Δ cells at low temperature as well as WT Neo1. This correlation between Neo1 mutants deficient in supporting PS asymmetry and growth in the absence of Drs2, support the idea that PS transport is the critical overlapping function between Drs2 and Neo1 [23]. Further support for the importance of the P − 4 Ser in M4 and M1 Gln is the strong synergistic effect of mutating both residues. Neo1[QA-AA, S452Q] cannot support viability of neo1Δ cells, while each single mutant supports growth rates comparable to WT cells. We predict that Q209 and S452 coordinate phospholipid substrate in the entry gate of Neo1 and this is a highly conserved feature of P4-ATPases.
Additional support for the importance of Neo1 entry gate residues came from the serendipitous identification of a gain-of-function mutation in M2 (Y222S) that allows Neo1 to suppress drs2Δ phenotypes. We targeted three residues in M2 that are highly conserved between Neo1 orthologs (P4B-ATPases) but differ between Neo1 and Drs2/ATP8A1 or Dnf1 (P4A-ATPases). The Y222 position had not been previously implicated in substrate recognition in any P4-ATPase. However, we found that Neo1-Y222S could suppress drs2Δ cold-sensitive growth defects and strongly, but not completely, restored PS and PE asymmetry in the neo1Δ drs2Δ background. Drs2 and ATP8A1 have a Thr in this M2 position, and this sidechain projects into the entry gate near the P − 4 Ser/Asn and M1 Gln proximal to the PS headgroup (Supplemental figure 3)[39, 40]. We propose that Y222 is part of the entry gate that selects phospholipid substrate and Y222S in Neo1 enhances its transport activity. The bulky Tyr in this M2 position may partially occlude PS binding in the Neo1 entry gate while still allowing entry of the small PE headroup. The Neo1-Y222S and Y222A substitutions both allow suppression of drs2Δ, implying that the reduction in sidechain size at position 222, to open up the entry gate binding pocket, is the critical feature of these mutations that enhance PS transport. Another possibility is that Neo1-Y222S gains the ability to interact with Cdc50, the β-subunit for Drs2. However, the β subunit contacts are with the luminal loops, M10 and cytoplasmic domains of the alpha subunit and the M2 residue in question is far removed from these interfaces (see supplemental figure 3). Thus, it seems unlikely that the Y222S substitution would create the binding surfaces needed for interaction de novo.
Consistent with a role for Y222 in substrate selection, we find the reciprocal mutation in M2 of Dnf1 (S243Y) strongly enhances transport of NBD-PS (~8-fold increase) and NBD-PC (1.75-fold increase), with no significant influence on PE transport. No change of substrate preference is observed with S243A and S243T mutations in Dnf1, so the Tyr is needed at position 243 to exert this gain in PS transport. In this case, the requirement for the bulkier sidechain in Dnf1 M2 to allow PS recognition may be caused by the smaller entry gate residues in M1 (GA in Dnf1 as opposed to QQ in Drs2 and QA in Neo1). Surprisingly though, Dnf1[S243Y] failed to support growth of a dnf1,2,3Δ drs2Δ strain for reasons that are not understood. It is possible that S243Y perturbs recognition of an unknown substrate, or that this position influences the ability of Dnf1 to distinguish lyso-phospholipid relative to diacyl-phospholipid.
Further characterization of the Neo1-Y222S also indicate complexities in this gain-of-function phenotype that are not easily explained. Neo1-Y222S displays the strongest suppression of drs2Δ when it is expressed in neo1Δ drs2Δ as the sole copy of Neo1. Suppression of drs2Δ by Neo1-Y222S is apparent in a drs2Δ strain that carries a WT copy of NEO1 at its endogenous locus, but the influence of Y222S on PS asymmetry is attenuated. Neo1-Y222S suppresses exposure of PS and PE on the cell surface much better than WT Neo1 when expressed in neo1Δ drs2Δ cells, but fails to do so when expressed in neo1Δ any1Δ drs2Δ cells. This implies that the PQ-loop protein Any1 is somehow required for the Y222S gain-of-function phenotype. Because Any1 interacts with Neo1 and appears to attenuate Neo1 activity, we tested if Y222S disrupted this interaction. However, we found no difference in the ability of Neo1 or Neo1-Y222S to co-immunoprecipitate with Any1. The Y222S mutation does not appear to disrupt the interaction between Neo1 and Any1; however, it may alter the effect of this interaction on either Neo1 or Any1 activity. Neo1 engages in a complex network of interactions, which includes Any1, Mon2 and Dop1. A more comprehensive examination of all Neo1 interactions and a better understanding of interactor (Any1, Dop1 and Mon2) biochemical activity is needed to fully assess the impact of the Y222S gain-of-function mutation.
In summary, we find that residues critical for phospholipid substrate recognition in P4A-ATPases from yeast to man are essential for function of an evolutionarily distant P4B-ATPase (Neo1) from budding yeast. The P4B-ATPases lack a β-subunit from the Cdc50 family and it was not known if these proteins would transport substrate by the same mechanism as that employed by αβ heterodimers from the P4A-ATPase clade. This study provides strong evidence supporting a common mechanism of phospholipid substrate recognition by P4-ATPases and imply phospholipid flippase activity can be catalyzed by P4B-ATPases without assistance from a β subunit.
Supplementary Material
Supplemental Figure 1A. Snake plot of transmembrane segments 1, 2 and 4 of Neo1 with residues targeted for mutagenesis highlighted yellow.
Supplemental Figure 1B. Probing the influence of the P + 1 position in M4 of Neo1 on PE asymmetry. Mutations in the P + 1 position cause a partial loss of PE asymmetry when assayed in the neo1Δ background but are not substantially different from Neo1 when assayed in the neo1Δ drs2Δ background.
Supplemental figure 2. Growth phenotypes of neo1Δ (A) and neo1Δ drs2Δ (B and C) strains harboring Neo1 variants. Neo1[D503N] carries a mutation in the Asp that in the P domain that is phosphorylated during the catalytic cycle rendering this mutant catalytically dead. The remainder of the mutations are in membrane segments potentially involved in substrate recognition. Some of the Neo1 mutants appeared to alter growth phenotypes compared to WT Neo1 when assayed on SD+5-FOA (B); however, no differences were observed upon retesting growth in the neo1Δ drs2Δ background across a range of temperatures on rich media (YPD) (C). Most of these Neo1 variants were not further characterized in this study.
Supplemental figure 3. ATP8A1-CDC50 structure (PDB 6K7M) with bound phospholipid highlighting entry gate residues in Ml, M2 and M4. ATP8A1-T101 in M2 aligns with Neo1-Y222 and is positioned to coordinate substrate along with Q88 (M1) and N352 (M4). The phosphatidylserine (PS) carboxyl group is within 3 Å of the T101 hydroxyl group.
Supplemental figure 4. Any1 interaction with Neo1 is not disrupted by Y222S. (A) Neo1-5XFLAG and Neo1[Y222S]-5XFLAG were co-expressed with Any1-GFP and were immunoprecipitated with anti-FLAG antibodies. Cell lysates and immunoprecipitated (IP) samples were then blotted with anti-GFP, anti-FLAG and anti-Arf1 as previously described [23]. (B) Quantitation of the Any1-GFP co-immunoprecipitating with Neo1 and Neo1[Y222S].
Supplemental figure 5. Neo1[Y222S] mutant is slightly more enriched in Drs2-positive compartments than wild-type Neo1. (A) Colocalization of wild-type GFP-tagged Neo1 with Cop1-mKate (cis-Golgi), Aur1-mKate (medial Golgi), Sec7-mKate (TGN) and mCherry-Tlg1 (endosome/TGN). (B) Colocalization of GFP-tagged Neo1[Y222S] with Cop1-mKate (cis-Golgi), Aur1-mKate (med-Golgi), Sec7-mKate (TGN) and mCherry-Tlg1 (endosome). (C) Manders’ overlap coefficient analyses were done for GFP-Neo1/Neo1[Y222S] with Golgi/endosomal markers. At least 100 cells (imaged on different days) were used to calculate the Manders’ overlap coefficient. (* p<0.05 , ** p<0.01, *** p<0.001, error bars ±SEM)
Highlights.
The P4-ATPase Neo1 transports phospholipid to control membrane asymmetry.
Positionally conserved residues in Neo1 are critical for substrate recognition.
A new entry gate residue (Y222) identified in Neo1 second membrane segment.
Neo1 mutations enhancing phosphatidylserine recognition suppress drs2Δ.
Acknowledgments
We would like to thank Davia Watkins for assistance with these studies and members of the Graham lab for helpful discussion during the course of this work. We also thank Chris Burd for strains expressing mKate-tagged Golgi proteins. This work was supported by NIH grant GM107978 to T.R.G..
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- [1].Hankins HM, Baldridge RD, Xu P, Graham TR, Role of flippases, scramblases and transfer proteins in phosphatidylserine subcellular distribution, Traffic, 16 (2015) 35–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Saito K, Fujimura-Kamada K, Furuta N, Kato U, Umeda M, Tanaka K, Cdc50p, a protein required for polarized growth, associates with the Drs2p P-type ATPase implicated in phospholipid translocation in Saccharomyces cerevisiae, Mol Biol Cell, 15 (2004) 3418–3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Lopez-Marques RL, Theorin L, Palmgren MG, Pomorski TG, P4-ATPases: lipid flippases in cell membranes, Pflugers Archiv : European journal of physiology, 466 (2014) 1227–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Best JT, Xu P, Graham TR, Phospholipid flippases in membrane remodeling and transport carrier biogenesis, Curr Opin Cell Biol, 59 (2019) 8–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Chen CY, Ingram MF, Rosal PH, Graham TR, Role for Drs2p, a P-type ATPase and potential aminophospholipid translocase, in yeast late Golgi function, J Cell Biol, 147 (1999) 1223–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Emoto K, Umeda M, An essential role for a membrane lipid in cytokinesis. Regulation of contractile ring disassembly by redistribution of phosphatidylethanolamine, J Cell Biol, 149 (2000) 1215–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].McEvoy L, Williamson P, Schlegel RA, Membrane phospholipid asymmetry as a determinant of erythrocyte recognition by macrophages, Proc Natl Acad Sci U S A, 83 (1986) 3311–3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Zwaal RF, Comfurius P, van Deenen LL, Membrane asymmetry and blood coagulation, Nature, 268 (1977) 358–360. [DOI] [PubMed] [Google Scholar]
- [9].Fadok VA, Voelker DR, Campbell PA, Cohen JJ, Bratton DL, Henson PM, Exposure of phosphatidylserine on the surface of apoptotic lymphocytes triggers specific recognition and removal by macrophages, J Immunol, 148 (1992) 2207–2216. [PubMed] [Google Scholar]
- [10].Nagata S, Suzuki J, Segawa K, Fujii T, Exposure of phosphatidylserine on the cell surface, Cell death and differentiation, 23 (2016) 952–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Bevers EM, Williamson PL, Getting to the Outer Leaflet: Physiology of Phosphatidylserine Exposure at the Plasma Membrane, Physiol Rev, 96 (2016) 605–645. [DOI] [PubMed] [Google Scholar]
- [12].Hua Z, Fatheddin P, Graham TR, An essential subfamily of Drs2p-related P-type ATPases is required for protein trafficking between Golgi complex and endosomal/vacuolar system, Mol Biol Cell, 13 (2002) 3162–3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Brett CL, Kallay L, Hua Z, Green R, Chyou A, Zhang Y, Graham TR, Donowitz M, Rao R, Genome-wide analysis reveals the vacuolar pH-stat of Saccharomyces cerevisiae, PloS one, 6 (2011) e17619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Dalton LE, Bean BDM, Davey M, Conibear E, Quantitative high-content imaging identifies novel regulators of Neo1 trafficking at endosomes, Mol Biol Cell, 28 (2017) 1539–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Wu Y, Takar M, Cuentas-Condori AA, Graham TR, Neo1 and phosphatidylethanolamine contribute to vacuole membrane fusion in Saccharomyces cerevisiae, Cellular logistics, 6 (2016) e1228791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Wicky S, Schwarz H, Singer-Kruger B, Molecular interactions of yeast Neo1p, an essential member of the Drs2 family of aminophospholipid translocases, and its role in membrane trafficking within the endomembrane system, Mol Cell Biol, 24 (2004) 7402–7418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Takar M, Wu Y, Graham TR, The Essential Neo1 Protein from Budding Yeast Plays a Role in Establishing Aminophospholipid Asymmetry of the Plasma Membrane, J Biol Chem, 291 (2016) 15727–15739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Barbosa S, Pratte D, Schwarz H, Pipkorn R, Singer-Kruger B, Oligomeric Dop1p is part of the endosomal Neo1p-Ys12p-Arl1p membrane remodeling complex, Traffic, 11 (2010) 1092–1106. [DOI] [PubMed] [Google Scholar]
- [19].Gillingham AK, Whyte JR, Panic B, Munro S, Mon2, a relative of large Arf exchange factors, recruits Dop1 to the Golgi apparatus, J Biol Chem, 281 (2006) 2273–2280. [DOI] [PubMed] [Google Scholar]
- [20].Beer KB, Rivas-Castillo J, Kuhn K, Fazeli G, Karmann B, Nance JF, Stigloher C, Wehman AM, Extracellular vesicle budding is inhibited by redundant regulators of TAT-5 flippase localization and phospholipid asymmetry, Proc Natl Acad Sci U S A, 115 (2018) E1127–E1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].McGough IJ, de Groot REA, Jellett AP, Betist MC, Varandas KC, Danson CM, Heesom KJ, Korswagen HC, Cullen PJ, SNX3-retromer requires an evolutionary conserved MON2:DOPEY2:ATP9A complex to mediate Wntless sorting and Wnt secretion, Nat Commun, 9 (2018) 3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].van Leeuwen J, Pons C, Mellor JC, Yamaguchi TN, Friesen H, Koschwanez J, Usaj MM, Pechlaner M, Takar M, Usaj M, VanderSluis B, Andrusiak K, Bansal P, Baryshnikova A, Boone CE, Cao J, Cote A, Gebbia M, Horecka G, Horecka I, Kuzmin E, Legro N, Liang W, van Lieshout N, McNee M, San Luis BJ, Shaeri F, Shuteriqi E, Sun S, Yang L, Youn JY, Yuen M, Costanzo M, Gingras AC, Aloy P, Oostenbrink C, Murray A, Graham TR, Myers CL, Andrews BJ, Roth FP, Boone C, Exploring genetic suppression interactions on a global scale, Science, 354 (2016) aag0838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Takar M, Huang Y, Graham TR, The PQ-loop protein Any1 segregates Drs2 and Neo1 functions required for viability and plasma membrane phospholipid asymmetry, J Lipid Res, 60 (2019) 1032–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Natarajan P, Wang J, Hua Z, Graham TR, Drs2p-coupled aminophospholipid translocase activity in yeast Golgi membranes and relationship to in vivo function, Proc Natl Acad Sci U S A, 101 (2004) 10614–10619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Coleman JA, Kwok MC, Molday RS, Localization, purification, and functional reconstitution of the P4-ATPase Atp8a2, a phosphatidylserine flippase in photoreceptor disc membranes, J Biol Chem, 284 (2009) 32670–32679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Paterson JK, Renkema K, Burden L, Halleck MS, Schlegel RA, Williamson P, Daleke DL, Lipid specific activation of the murine P4-ATPase Atp8a1 (ATPase II), Biochemistry, 45 (2006) 5367–5376. [DOI] [PubMed] [Google Scholar]
- [27].Baldridge RD, Graham TR, Identification of residues defining phospholipid flippase substrate specificity of type IV P-type ATPases, Proc Natl Acad Sci U S A, 109 (2012) E290–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Baldridge RD, Xu P, Graham TR, Type IV P-type ATPases distinguish mono- versus diacyl phosphatidylserine using a cytofacial exit gate in the membrane domain, J Biol Chem, 288 (2013) 19516–19527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Baldridge RD, Graham TR, Two-gate mechanism for phospholipid selection and transport by type IV P-type ATPases, Proc Natl Acad Sci U S A, 110 (2013) E358–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Roland BP, Naito T, Best JT, Arnaiz-Yepez C, Takatsu H, Yu RJ, Shin HW, Graham TR, Yeast and human P4-ATPases transport glycosphingolipids using conserved structural motifs, J Biol Chem, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Roland BP, Graham TR, Directed evolution of a sphingomyelin flippase reveals mechanism of substrate backbone discrimination by a P4-ATPase, Proc Natl Acad Sci U S A, 113 (2016) E4460–4466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Mikkelsen SA, Mogensen LS, Vilsen B, Molday RS, Vestergaard AL, Andersen JP, Asparagine 905 of the mammalian phospholipid flippase ATP8A2 is essential for lipid substrate-induced activation of ATP8A2 dephosphorylation, J Biol Chem, 294 (2019) 5970–5979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Vestergaard AL, Coleman JA, Lemmin T, Mikkelsen SA, Molday LL, Vilsen B, Molday RS, Dal Peraro M, Andersen JP, Critical roles of isoleucine-364 and adjacent residues in a hydrophobic gate control of phospholipid transport by the mammalian P4-ATPase ATP8A2, Proc Natl Acad Sci U S A, 111 (2014) E1334–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Toyoshima C, Inesi G, Structural basis of ion pumping by Ca2+-ATPase of the sarcoplasmic reticulum, Annu Rev Biochem, 73 (2004) 269–292. [DOI] [PubMed] [Google Scholar]
- [35].Palmgren MG, Nissen P, P-type ATPases, Annual review of biophysics, 40 (2011) 243–266. [DOI] [PubMed] [Google Scholar]
- [36].Roland BP, Graham TR, Decoding P4-ATPase substrate interactions, Critical reviews in biochemistry and molecular biology, 51 (2016) 513–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Riekhof WR, Wu J, Gijon MA, Zarini S, Murphy RC, Voelker DR, Lysophosphatidylcholine metabolism in Saccharomyces cerevisiae: the role of P-type ATPases in transport and a broad specificity acyltransferase in acylation, J Biol Chem, 282 (2007) 36853–36861. [DOI] [PubMed] [Google Scholar]
- [38].Riekhof WR, Voelker DR, Uptake and utilization of lyso-phosphatidylethanolamine by Saccharomyces cerevisiae, J Biol Chem, 281 (2006) 36588–36596. [DOI] [PubMed] [Google Scholar]
- [39].Timcenko M, Lyons JA, Januliene D, Ulstrup JJ, Dieudonne T, Montigny C, Ash MR, Karlsen JL, Boesen T, Kuhlbrandt W, Lenoir G, Moeller A, Nissen P, Structure and autoregulation of a P4-ATPase lipid flippase, Nature, 571 (2019) 366–370. [DOI] [PubMed] [Google Scholar]
- [40].Hiraizumi M, Yamashita K, Nishizawa T, Nureki O, Cryo-EM structures capture the transport cycle of the P4-ATPase flippase, Science, (2019). [DOI] [PubMed] [Google Scholar]
- [41].Liu K, Surendhran K, Nothwehr SF, Graham TR, P4-ATPase requirement for AP-1/clathrin function in protein transport from the trans-Golgi network and early endosomes, Mol Biol Cell, 19 (2008) 3526–3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Gietz RD, Schiestl RH, Quick and easy yeast transformation using the LiAc/SS carrier DNA/PEG method, Nat Protoc, 2 (2007) 35–37. [DOI] [PubMed] [Google Scholar]
- [43].Sikorski RS, Hieter P, A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae, Genetics, 122 (1989) 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Sherman F, Getting started with yeast, Methods in enzymology, 350 (2002) 3–41. [DOI] [PubMed] [Google Scholar]
- [45].Van Driessche B, Tafforeau L, Hentges P, Carr AM, Vandenhaute J, Additional vectors for PCR-based gene tagging in Saccharomyces cerevisiae and Schizosaccharomyces pombe using nourseothricin resistance, Yeast, 22 (2005) 1061–1068. [DOI] [PubMed] [Google Scholar]
- [46].Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA 3rd, Smith HO, Enzymatic assembly of DNA molecules up to several hundred kilobases, Nature methods, 6 (2009) 343–345. [DOI] [PubMed] [Google Scholar]
- [47].Takar M, Wu Y, Graham TR, The Essential Neo1 Protein from Budding Yeast Plays a Role in Establishing Aminophospholipid Asymmetry of the Plasma Membrane*, in: J Biol Chem, vol. 291, 2016, pp. 15727–15739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Chen S, Wang J, Muthusamy BP, Liu K, Zare S, Andersen RJ, Graham TR, Roles for the Drs2p-Cdc50p complex in protein transport and phosphatidylserine asymmetry of the yeast plasma membrane, Traffic, 7 (2006) 1503–1517. [DOI] [PubMed] [Google Scholar]
- [49].Emre Onat O, Gulsuner S, Bilguvar K, Nazli Basak A, Topaloglu H, Tan M, Tan U, Gunel M, Ozcelik T, Missense mutation in the ATPase, aminophospholipid transporter protein ATP8A2 is associated with cerebellar atrophy and quadrupedal locomotion, European journal of human genetics : EJHG, 21 (2012) 281–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Palmgren M, Osterberg JT, Nintemann SJ, Poulsen LR, Lopez-Marques RL, Evolution and a revised nomenclature of P4 ATPases, a eukaryotic family of lipid flippases, Biochim Biophys Acta Biomembr, 1861 (2019) 1135–1151. [DOI] [PubMed] [Google Scholar]
- [51].Lee S, Uchida Y, Wang J, Matsudaira T, Nakagawa T, Kishimoto T, Mukai K, Inaba T, Kobayashi T, Molday RS, Taguchi T, Arai H, Transport through recycling endosomes requires EHD1 recruitment by a phosphatidylserine translocase, EMBO J, 34 (2015) 669–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Zhou X, Graham TR, Reconstitution of phospholipid translocase activity with purified Drs2p, a type-IV P-type ATPase from budding yeast, Proc Natl Acad Sci U S A, 106 (2009) 16586–16591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Parsons AB, Lopez A, Givoni IE, Williams DE, Gray CA, Porter J, Chua G, Sopko R, Brost RL, Ho CH, Wang J, Ketela T, Brenner C, Brill JA, Fernandez GE, Lorenz TC, Payne GS, Ishihara S, Ohya Y, Andrews B, Hughes TR, Frey BJ, Graham TR, Andersen RJ, Boone C, Exploring the mode-of-action of bioactive compounds by chemical-genetic profiling in yeast, Cell, 126 (2006) 611–625. [DOI] [PubMed] [Google Scholar]
- [54].Aoki Y, Uenaka T, Aoki J, Umeda M, Inoue K, A novel peptide probe for studying the transbilayer movement of phosphatidylethanolamine, J Biochem, 116 (1994) 291–297. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1A. Snake plot of transmembrane segments 1, 2 and 4 of Neo1 with residues targeted for mutagenesis highlighted yellow.
Supplemental Figure 1B. Probing the influence of the P + 1 position in M4 of Neo1 on PE asymmetry. Mutations in the P + 1 position cause a partial loss of PE asymmetry when assayed in the neo1Δ background but are not substantially different from Neo1 when assayed in the neo1Δ drs2Δ background.
Supplemental figure 2. Growth phenotypes of neo1Δ (A) and neo1Δ drs2Δ (B and C) strains harboring Neo1 variants. Neo1[D503N] carries a mutation in the Asp that in the P domain that is phosphorylated during the catalytic cycle rendering this mutant catalytically dead. The remainder of the mutations are in membrane segments potentially involved in substrate recognition. Some of the Neo1 mutants appeared to alter growth phenotypes compared to WT Neo1 when assayed on SD+5-FOA (B); however, no differences were observed upon retesting growth in the neo1Δ drs2Δ background across a range of temperatures on rich media (YPD) (C). Most of these Neo1 variants were not further characterized in this study.
Supplemental figure 3. ATP8A1-CDC50 structure (PDB 6K7M) with bound phospholipid highlighting entry gate residues in Ml, M2 and M4. ATP8A1-T101 in M2 aligns with Neo1-Y222 and is positioned to coordinate substrate along with Q88 (M1) and N352 (M4). The phosphatidylserine (PS) carboxyl group is within 3 Å of the T101 hydroxyl group.
Supplemental figure 4. Any1 interaction with Neo1 is not disrupted by Y222S. (A) Neo1-5XFLAG and Neo1[Y222S]-5XFLAG were co-expressed with Any1-GFP and were immunoprecipitated with anti-FLAG antibodies. Cell lysates and immunoprecipitated (IP) samples were then blotted with anti-GFP, anti-FLAG and anti-Arf1 as previously described [23]. (B) Quantitation of the Any1-GFP co-immunoprecipitating with Neo1 and Neo1[Y222S].
Supplemental figure 5. Neo1[Y222S] mutant is slightly more enriched in Drs2-positive compartments than wild-type Neo1. (A) Colocalization of wild-type GFP-tagged Neo1 with Cop1-mKate (cis-Golgi), Aur1-mKate (medial Golgi), Sec7-mKate (TGN) and mCherry-Tlg1 (endosome/TGN). (B) Colocalization of GFP-tagged Neo1[Y222S] with Cop1-mKate (cis-Golgi), Aur1-mKate (med-Golgi), Sec7-mKate (TGN) and mCherry-Tlg1 (endosome). (C) Manders’ overlap coefficient analyses were done for GFP-Neo1/Neo1[Y222S] with Golgi/endosomal markers. At least 100 cells (imaged on different days) were used to calculate the Manders’ overlap coefficient. (* p<0.05 , ** p<0.01, *** p<0.001, error bars ±SEM)