Abstract
Researchers have discovered associations between elements of the intestinal microbiome (including specific microbes, signaling pathways, and microbiota-related metabolites) and risk of colorectal cancer (CRC). However, it is unclear whether changes in the intestinal microbiome contribute to development of sporadic CRC or result from it. Changes in the intestinal microbiome can mediate or modify the effects of environmental factors on risk of CRC. Factors that affect risk of CRC also affect the intestinal microbiome, including overweight and obesity; physical activity; and dietary intake of fiber, whole grains, and red and processed meat. These factors alter microbiome structure and function, along with the metabolic and immune pathways that mediate CRC development. We review epidemiologic and laboratory evidence for the influence of the microbiome, diet, and environmental factors on CRC incidence and outcomes. Based on these data, features of the intestinal microbiome might be used for CRC screening and modified for chemoprevention and treatment. Integrated prospective studies are urgently needed to investigate these strategies.
Keywords: Dysbiosis, colitis, fecal microbiota transplant, infection
Colorectal cancer (CRC) is the third most common cancer and leading cause of cancer death in men and women in the United States (US), despite the increasing uptake of colonoscopy screening.1 In 2019, approximately 145,600 new cases of CRC and 51,020 deaths were estimated to occur. Moreover, although CRC incidence and mortality have decreased steadily in the past few decades among adults older than 65 years, an opposing trend has occurred in adults younger than 50 years,2 for whom routine screening has not been recommended. The incidence of colon cancer has increased by 2.4% per year in adults 20–29 years old and by 1.0% per year in adults 30–39 years old from the mid-1980s through 2013, and began increasing in adults 40–49 years old (1.3% per year) and 50–54 years old (0.5% per year) since mid-1990s.2 A prolonged and steeper increase has been observed for rectal cancer cases. This alarming trend in young adults coupled with the continued burden of CRC in the overall population indicates a need to develop new prevention strategies to complement screening.
Over the past few decades, migration studies and prospective cohort studies have established the important effects of diet and lifestyle in the development of CRC.3 Approximately 50%–60% of incident cases of CRC in the US are estimated to be attributable to modifiable risk factors4, 5 such as smoking; heavy consumption of alcohol; overweight and obesity; physical inactivity; high consumption of red and processed meat; and low consumption of dietary fiber, whole grains, and other healthful nutrients.
The microbiome (including bacteria, virus, fungi et al) regulates health and alterations can contribute to disease. Increasing data indicate that changes in the intestinal microbiome allow environmental risk factors to initiate and promote CRC.6, 7 This could be because changes of the microbiome affect metabolism and immune function. The intestinal microbiome might therefore be modified as part of CRC preventative strategies. Studies have identified differences in compositions of intestinal microbiomes between CRC cases and healthy individuals (controls) as well as individual microbes that are enriched or depleted in microbiomes of patients with CRC. Moreover, there is evidence that changes in the gut microbiome occur during early stages of colorectal carcinogenesis and can be used to identify individuals at risk for colorectal adenoma, the precursor lesion to CRC. Changes in the microbiome might therefore be used as biomarkers for early detection of CRC, to improve screening strategies. The intestinal microbiome can also influence the efficacy or toxicity of therapeutic agents, including immunotherapies.8
Although there have been numerous reviews of association between the gut microbiome and CRC, most have focused on specific microbes,9 mechanistic pathways,10–12 or individual risk factors.6 We review the interactions among the gut microbiome, environmental risk factors, and CRC based on findings from epidemiologic and laboratory research. We also discuss the potential of integrating analyses of the gut microbiome into CRC screening, chemoprevention, and treatment.
Intestinal Dysbiosis in Patients With CRC
The number of microbial species in human intestine is estimated to exceed 2000.13 The human intestinal microbiome primarily comprises Firmicutes, Bacteroidetes, Actinobacteria, and Proteobacteria. Intestinal microbes metabolize indigestible ingredients from food, synthesize nutrients such as vitamins, detoxify metabotypes, modulate the immune response, provide signals for epithelial cell renewal and maintenance of mucosal integrity, and secrete antimicrobial products.14 Dysbiosis is defined as pathogenic changes in microbiome profile and functions. Alterations in abundance of healthy intestinal microbes can promote chronic inflammatory conditions and production of carcinogenic metabolites, leading to neoplasia.
Patients with CRC have a less diverse microbiome than healthy individuals.15 However, a meta-analysis of metagenomic data from different cohorts and populations found higher richness in microbiomes of CRC than controls, partly due to expansions of species typically derived from the oral cavity.16, 17 Differences in abundance of individual microbes have been observed in comparisons of tumor and adjacent non-tumor mucosa, and between stool specimens collected from patients with CRC vs controls. Specific changes in the microbiome and metabolome occur during different stages of colorectal neoplasia, from adenomatous polyps to early-stage cancer to metastatic disease, supporting an etiological and diagnostic role for the microbiome.18–20 We summarize results from epidemiologic studies of microbes that have been associated with CRC (Tables 1 and 2).
Table 1.
Association of the Intestinal Microbiome with CRC
| Author, Year | Country | Study design | Sample Size | Biospecimen type | Sequencing method | Main findings comparing cases to controls |
|---|---|---|---|---|---|---|
| Scanlan, 2008211 | Belgium | case–control | 20 cancers / 20 polyps / 20 controls | Stool | 16s rRNA sequencing | ↑ diversity of the Clostridium leptum and C. coccoides subgroups |
| Sobhani, 2011212 | France | case–control | 60 cancers / 119 controls | Stool | 16s rRNA sequencing | ↑ Bacteroides/Prevotella |
| Castellarin, 201221 | Canada | case–control | 11 cancers with adjacent normal tissue | Tissue | RNA sequencing | ↑ Fusobacterium |
| Kostic, 201222 | USA | case– control | 95 cancers with adjacent normal tissue | Tissue | 16s rRNA sequencing | ↑ Fusobacterium |
| Sanapareddy, 2012213 | USA | case–control | 33 adenomas / 38 controls | Tissue | 16s rRNA sequencing | ↑ bacteria from 87 taxa, including potential pathogens such as Pseudomonas,Helicobacter and, Acinetobacter; ↓ bacteria from 5 taxa. |
| Chen, 2012214 Wang, 201245 |
China | case–control | 46 cancers / 56 controls | Tissue, stool, and swab | 16s rRNA sequencing | ↓ diversity; ↑ Bacteroides fragilis, Lactobacillales, Fusobacterium, Porphyromonas, Peptostreptococcus, and Mogibacterium; ↓ Bifidobacterium, Faecalibacterium, Blautia butyrate-producing bacteria |
| Ahn, 201315 | USA | case–control | 47 cancers / 94 controls | Stool | 16s rRNA sequencing | ↓ diversity; ↓ Clostridia; ↑ Fusobacterium, Porphyromonas; |
| Dejea, 2014215 | USA | case–control | 23 cancers and 2 adenomas with paired normal tissues, and 22 controls | Tissue | 16s rRNA sequencing | ↑ Fusobacterium; difference by biofilm status |
| Zackular, 201435 | USA | case–control | 30 cancers / 30 adenomas / 30 controls | Stool | 16s rRNA sequencing | ↑ Bacteroides fragilis, Fusobacterium, Porphyromonas; ↓ butyrate-producing bacteria |
| Zeller, 201436 | France | case–control | 91 cancers / 42 adenomas / 358 controls | Stool | Metagenomic sequencing | ↑ Bacteroidetes, Fusobacteria and Proteobacteria; ↓ Actinobacteria and Firmicutes |
| Burns, 2015216 | USA | case–control | 44 cancers with adjacent normal tissue | Tissue | 16s rRNA sequencing | ↑ diversity, Fusobacterium and Providencia; ↑ virulence-related genes |
| Feng, 201518 | Austria | case– control | 41 cancers / 42 adenomas / 55 controls | Stool | Metagenomic sequencing | ↑ B dorei, B vulgatus, E coli, Fusobacterium; ↓ Lactobacillus and Bifidobacterium |
| Lu, 2016217 | China | case– control | 31 adenomas / 20 controls | Tissue | 16s rRNA sequencing | ↑ diversity; ↑Lactococcus and Pseudomonas; ↓ Enterococcus, Bacillus, and Solibacillus. Similar composition between adenomatous and adjacent nonadenoma tissues |
| Vogtmann, 201637 | USA, France | case–control | 52 cancers / 52 controls | Stool | Metagenomic sequencing | ↑ Fusobacterium, Porphyromonas, Clostridia |
| Baxter, 2016187 | USA, Canada | case–control | 120 cancers / 198 adenomas / 172 no colonic lesions | Stool | 16s rRNA sequencing | ↑ Porphyromonas assaccharolytica, Peptostreptococcus stomatis, Parvimonas micra, and F nucleatum; ↓Lachnospiraceae |
| Hale, 2017218 | USA | case–control | 233 adenomas / 547 controls | Stool | 16s rRNA sequencing | ↑ Bilophila, Desulfovibrio, inflammatory bacteria in the genus Mogibacterium; ↓ Veillonella, Clostridia order, and Bifidobacteriales family |
| Yu, 201738 | Denmark, France, Austria | case–control | 74 cancers / 54 controls | Stool | Metagenomic sequencing | ↑ Peptostreptococcus stomatis, F nucleatum, Parvimonas micra, Solobacterium moorei |
| Liang, 201739 | China | case– control | 203 cancers / 236 controls | Stool | 16s rRNA sequencing | ↑ F nucleatum, Clostridium hathewayi; ↓ B clarus |
| Flemer, 201855 | Ireland | case–control | 43 cancers / 37 controls | Stool and tissue | 16s rRNA sequencing | ↓ Lachnospiraceae incertae sedis and Coprococcus |
Note: only studies with at least 1o cases (either CRC or precursors) are included. Pooled analyses of published individual studies are not included.
Table 2.
Microbes Associated With Increased or Reduced Risk of CRC
| Microbe | Epidemiologic Evidence | Potential mechanisms | References |
|---|---|---|---|
| Associated with higher risk of CRC | |||
| Fusobacterium nucleatum | Enriched tumor tissue; higher fecal abundance in patients with colorectal neoplasia than controls; associated with advanced cancer stage, lower infiltration by T cells, higher risk of recurrence, and poorer patient survival; correlated with the molecular characteristics of the serrated pathway. | Promotion of a tumor-permissive microenvironment through recruitment of myeloidderived suppressor cells and inhibition of antitumor defense by NK or T cells; modulation of Ecadherin/β-catenin. | 21, 22, 140, 219 |
| Enterotoxigenic Bacteroides fragilis (ETBF) | Enriched in tumor tissue and fecal samples of CRC patients; associated with advanced cancer stage and proximal colon tumor. | DNA damage | 40, 220, 221 |
| pks+ Escherichia coli | More frequently detected in individuals with than without CRC, more frequently in tumors than in normal flanking tissue, and more frequently in late-stage tumors than in early-stage tumors. | Promotion of intestinal inflammation | 49, 222 |
| Associated with lower risk of CRC | |||
| SCFA-producing bacteria | Lower abundance in CRC patients than bacteria controls; higher abundance in Native Americans with a low CRC incidence; associated with improved immune response and better metabolic parameters. | Metabolic and immune modulation of SCFAs that protects against CRC. | 124, 128, 223 |
Fusobacterium nucleatum
Two independent studies reported increased levels of Fusobacterium DNA and RNA sequences in tumor compared with non-tumor specimens.21, 22 Numerous studies, of multiple cohorts, of patients with CRC worldwide have found similar associations.23–27 In support of the colorectal carcinogenic effect of Fusobacterium nucleatum, a higher abundance of F nucleatum (present in approximately 10%–15% of tumors) has been associated with advanced disease stage, higher risk of recurrence, and shorter patient survival times.23, 26, 28 Moreover, levels of F nucleatum in tumor tissue have been associated with lower infiltration by T cells,29, 30 supporting studies reporting that F nucleatum reduces the anti-tumor immune response. Epidemiologic studies of patients with CRC or premalignant lesions have associated F nucleatum with specific clinical and molecular features, such as right-sided anatomic location, mutations in BRAF, and hypermutation with microsatellite instability.24–27 Given that these features characterize serrated neoplasia,31 F nucleatum might contribute to the serrated pathway of CRC development.
A study associated F nucleatum with the consensus molecular subtype 1 of CRC,32 which is characterized by microsatellite instability and upregulation of immune pathways.33 More recently, among CRC patients with distant metastases, nearly identical, viable strains of Fusobacterium were found at similar relative abundances in paired primary tumors and metastases. So, Fusobacterium appears to be an important component of the tumor microenvironment.34 In addition to studies of tumor tissues, studies of fecal microbiomes that used either 16s rRNA or shotgun metagenomic sequencing found that F nucleatum to be increased in fecal samples from patients with CRC or adenoma compared with controls (Table 1).15, 18, 35–39
Bacteroides fragilis
Enterotoxigenic Bacteroides fragilis (ETBF), which produces the Bacteroides fragilis toxin, has been associated with CRC. Although there is strong preclinical evidence for the association between ETBF and CRC, there is little evidence from epidemiologic studies. Only a few studies have examined ETBF in human tumor tissues.40–43 Among them, 2 found significant enrichment of ETBF in tumor tissues compared to tissues from controls or adjacent normal tissues40, 43. However, the proportions of ETBF-positive colorectal tumors differed between these studies (26% vs 89%), possibly due to the differences in assays or sample processing methods.40 Significantly higher proportions of late-stage vs early stage, and of right-side vs left-side, tumors were ETBF positive.40, 43 Higher proportions of patients with familial adenomatous polyposis42 or sporadic premalignant lesions had intestinal mucosa that tested positive for ETBF than of controls.41
Compared to mucosa, the abundance of ETBF in fecal samples is lower, because ETBF colonizes colon epithelial crypts.40, 44 Moreover, different isotypes of ETBF colonize stool (bft-1) vs mucosa (bft-2). However, ETBF was reported to be enriched in fecal samples of patients with CRC compared with controls.35, 44, 45 A meta-analysis of 4 case–control studies of metagenomes of patients with CRC found that Bacteroides fragilis was the only species that was consistently enriched in intestinal microbiomes of patients with CRC worldwide.46
Escherichia coli
Increases in mucosa-associated E coli have been observed in patients with inflammatory bowel diseases (IBD) and in patients with CRC, compared with healthy individuals. In patients with CRC, E coli invade the colonic mucosa and become intracellular.47, 48 E coli strains with the polyketide synthase gene complex (pks) gene, which mediates production of genotoxin colibactin (called pks+ E coli), are found at higher frequency in individuals with than without CRC,49–52 in tumors than in adjacent non-tumor tissue,51 and in late-stage tumors compared with early-stage tumors.51 Levels of mucosa-associated and internalized E coli associate with the cell’s proliferation index, assessed by Ki-67 expression.51 However, only 1 fecal microbiome study found E coli to be enriched in samples of CRC patients.18 This might be because E coli colonize the mucosa and reside within intestinal cells, rather than the lumen, so they are not shed into feces.
Oral microbiome
Besides Fusobacterium, other oral bacteria and markers of periodontal disease are enriched in colorectal tumors and feces of patients, including the genera of Porphyromonas and Peptostreptococcus, and Parvimonas micra.15, 35, 37, 38, 53 Moreover, oral microbiome profiling studies identified several members of the oral biofilms with different abundances in patients with CRC compared with controls, including Haemophilus, Parvimonas, Prevotella, Alloprevotella, Lachnoanaerobaculum, Neisseria, and Streptococcus.54, 55 These bacteria have been associated with distinct mucosal gene expression profiles, which might contribute to development of CRC. Interestingly, similar networks of oral bacteria were found samples from oral and colonic mucosal surfaces of individuals with colonic neoplasia and controls.55
These findings support the reported association of periodontal disease with CRC risk.38, 56, 57 In addition, as a major risk factor for CRC, smoking might change the oral microbiome composition. Smokers have a decreased relative abundance of Neisseria and increased relative abundance of Veillonellaceae families.58 Also, bacterial metabolic activities have pathophysiological consequences on oral and systemic health.58 So, alterations to the oral and intestinal microbiomes might each contribute to colorectal carcinogenesis, via microbe dissemination and induction of inflammation.54, 55, 59,60.
Bacteria
Streptococus bovis or gallolyticus has been associated with CRC (see ref 61). Serologic epidemiology studies (based on immunoassays) found the incidence of non-typhoid Salmonella infection to be much higher (about 600-fold ) than reported,62 ranging from 56 per 1000 person-years in Finland to 547 per 1000 person-years in Poland, from of 2003 through 2008 (ref 63), and increasing from 13 per 1000 person-years to 217 per 1000 person-years in Denmark, from 1983 through 1999.64 Furthermore, Salmonellosis, primarily caused by its major serotypes, S ser. Typhimurium and S ser. Enteritidis, has also been associated with development of disorders such irritable bowel syndrome65 and IBD.66 Studies from Scandinavian countries found that the probability of a new diagnosis of IBD following an episode of non-typhoid Salmonella infection (all subspecies combined), particularly within the first 10 years, increased significantly (2–3 fold) compared with general population.67, 68 Antibody titers to Salmonella ser. Typhimurium were higher in CRC cases than controls in the US and the Netherlands. Smoking and dietary iron were identified as potential risk factors, indicating a link between non-typhoid Salmonella infection and intestinal tumoigenesis69. This observation was confirmed in an independent population-based linkage study in the Netherlands.70 The investigators analyzed cancer registry and public health surveillance data and found an increased incidence of CRC, compared with the general population, among residents who with reported enteric (not systemic) salmonellosis. Moreover, epidemiology studies associated a serologic response to Helicobacter pylori with increased risk of CRC among African Americans.71
Infection-associated CRC might develop via transformation of cells to premalignant lesions, adenomas, to malignancies. Strategies to detect specific bacteria or bacterial DNA sequences might be used in screening for early-stage colorectal adenomas or carcinomas.60, 61
Virome and mycobiome
The enteric virome and mycobiome (fungal microbiome) have also been linked with CRC.72–74 Compared to controls, CRC cases had increased diversity in bacteriophage viromes, associated with reduced bacterial diversity, and modest enrichment of specific viral taxa, such as members of Inovirus and Tunalikevirus, whose bacterial hosts have been implicated in CRC (such as ETBF, F nucleatum, and pks+ E coli). These findings indicate an interaction between the virome and bacteriome in risk of CRC.72, 74 Distinct mycobiome profiles were identified in patients with CRC vs controls and in patients with early- vs late-stage CRC. Ecological analysis revealed more co-exclusive correlations between fungi and bacteria in fecal samples from patients with CRC than controls,73 supporting the preclinical findings for an antagonistic relationship between bacteria and fungi during CRC development.75 However, given the limited data, further studies are needed to investigate how interactions among the enteric virome, mycobiome, and bacteriome might contribute to development of CRC.
Environmental Factors
The composition of microbiota is determined by genetic, environmental, and dietary factors. For example, variants in the gene encoding the vitamin D receptor affect the composition of the microbiome,76 but it is still not clear whether genetic or environmental factors have a bigger effect on the gut microbiome.77 Although there is no direct evidence, CRC-related risk factors have been associated with specific changes in the intestinal microbiota.
Overweight and obesity
A meta-analysis found that each 5-kg/m2 increase in body mass index (BMI, calculated as body weight in kilograms divided by square of height in meters) is associated with 5% increase in risk of CRC.78 Obesity might also be contributing to the increasing incidence of young-onset CRC.79 Epidemiology studies have provided evidence for an association between obesity-related metabolic and inflammatory factors, including changes in insulin-like growth factor 1 signaling, adipokines, sex hormones, and systemic inflammation, and CRC risk.80 Obesity-associated changes in the intestinal microbes and their metabolites may also contribute to carcinogenesis.
Obesity has been associated with a significant decrease in the diversity of the gut microbiota.81, 82 Some83 but not all81, 84 cross-sectional studies found an enrichment of the phylum Firmicutes and depletion of the phylum Bacteroidetes in obese individuals compared with lean individuals. Although it is not clear whether alterations to the intestinal microbiome are a cause or consequence of obesity, dietary intervention studies have shown that changes in body weight affect the gut microbiota.85 Obese individuals who lost weight on a fat-restricted or carbohydrate-restricted low-calorie diet had increased abundance of Bacteroidetes and decreased Firmicutes, irrespective of diet type.83 A decrease in the ratio of Firmicutes:Bacteroidetes was also observed in individuals who lost weight in other trials.86
Bacteria that produce short-chain fatty acids (SCFAs) are important regulators of metabolic homeostasis.87, 88 A lower abundance of SCFA-producing bacteria has been associated with higher risk of type 2 diabetes.89 Some dietary intervention studies, paradoxically, associated weight loss with reduced abundance of SCFA-producing bacteria.84, 90–93 However, most of the studied diets were low in total calorie and carbohydrate, an important substrate for SCFA synthesis, so the observed reduction in SCFA-producing bacteria might have resulted from carbohydrate restriction in the diet, rather than the weight loss itself. In patients on a non-hypocaloric low-fat intervention, weight loss was associated with an increase in SCFA-producing bacteria including Clostridium Cluster IV, Bifidobacterium spp., and Faecalibacterium prausnitzii.86 A bidirectional Mendelian randomization analysis provided genetic evidence that increased intestinal production of SCFAs improves patient responses to insulin and reduces abnormalities in the production or absorption of SCFAs and risk of type 2 diabetes.87 Obesity might therefore increase risk of CRC by reducing the abundance of SCFA-producing bacteria and SCFA production in the intestine 94. Amino acids such as glutamate and deoxycholate have also been associated with obesity, intestinal dysbiosis, and metabolic disorders.20, 95
Akkermansia muciniphila might also provide a link between obesity and CRC,96 although there is controversy over the role of A muciniphila in colorectal carcinogensis.53, 97 Several studies of different diets associated weight loss with enrichment of A muciniphila.86, 93, 98, 99 The bacteria correlated with better metabolic parameters, including lower fasting level of plasma glucose, lower level of plasma triglycerides, and improved insulin response.100
Obesity might contribute to systemic inflammation by altering intestinal barrier function. Leakage of microbial products, such as the endotoxin lipopolysaccharide (LPS), causes metabolic endotoxemia.101 Higher BMIs have been associated with increased blood levels of LPS and LPS-binding protein (LBP), whereas weight loss reduces circulating LPS and LBP levels.102–104 A cross-sectional study found that patients with adenomas had higher blood levels of LPS than controls, and that patients whose adenomas had villous features had higher levels of LPS than patients with only tubular adenomas.105 A polymorphism in the LBP gene (rs2232596) was associated with higher risk of CRC.106
A large prospective study of mostly Caucasian persons associated higher levels of LPS with increased risk of CRC in men,107 whereas a prospective study in a racially diverse cohort observed that, compared to individuals in the first quartile of plasma LBP, those in the third, but not fourth, quartile had an increased risk of CRC.108 Given these limited data, further studies, preferably pooled analyses of multiple cohorts, are needed to examine the relationship of LPS with CRC risk, according to sex and adiposity.
A recent meta-analysis assessed the effect of the gut microbiome on the relationship between obesity and increased CRC risk.109 The study reported that the association between BMI and CRC risk was only slightly attenuated when several CRC-associated taxa were added to the analytic model, indicating a weak effect of these taxa. Although this meta-analysis was the first effort to analytically assess the mediating effect of the gut microbiome on the relationship between obesity and CRC, it could not establish that these taxa contributed to development of CRC, due to the cross-sectional design of the included studies—particularly the contemporary assessment of BMI and gut microbiome in patients with CRC vs controls. Prospective studies that assess BMI and the gut microbiome and then follow patients to see which ones develop CRC are needed to better understand the role of the gut microbiome in obesity-associated increase in CRC risk.
Physical activity
Individuals with the highest level of physical activity have a 19% lower risk of colon cancer than individuals with the lowest level, but physical activity has not been associated with rectal cancer.78 Several cross-sectional studies have reported the effects of exercise on the composition of the intestinal microbiota and its functions.
Two studies compared the fecal microbiome and metabolomes of 40 elite professional rugby players with those of 46 male controls.96, 110 Because the athletes tended to have a higher BMI, to minimize the influence of BMI, the study included 2 control groups: 1 with BMIs ≤25 and the other with BMIs >28. The athletes were found to have a more diverse gut microbiome than the controls. Among the individual species that differed between athletes and controls, A muciniphila was found to be enriched by 16s rRNA and metagenomic sequencing analyses, as well as in the metabolic pathway analysis. These findings support the role of A muciniphila in metabolic regulation. Although dietary factors are often correlated with the metagenomic pathways, exercise and high protein intake also correlate with the metagenomic profiles of athletes. For example, levels of SCFAs were significantly higher in athletes compared with controls, although it is not clear whether this is due to higher fiber intake or the more intensive exercise by athletes. Cardiorespiratory fitness has also been associated with higher fecal levels of butyrate and increased abundances of butyrate-producing taxa, including Clostridiales, Roseburia, Lachnospiraceae, and Erysipelotrichaceae.111 Similar findings were observed in a study that compared active vs sedentary women.112
Two intervention studies reported effects of exercise on intestinal microbiomes.113, 114 In the first study, 32 sedentary adults underwent 2 weeks of baseline analysis, 6 weeks of endurance-based exercise intervention, and 6 weeks of washout, during which participants were instructed to refrain from exercising.113 Exercise training increased fecal concentrations of SFCAs in lean but not obese participants, and increased butyrate-producing bacterial taxa that associated with parallel shifts in body composition in lean individuals. Although these findings support the role of exercise in regulation of SCFA production, it is unclear why the benefit was restricted to lean individuals. It could be that lean participants are more likely to comply with the intervention and achieve a higher intensity of exercise than obese individuals. Notably, the study also found that a return to sedentary lifestyle for 6 weeks reversed changes in the gut microbiota observed after exercise training, again indicating the sensitivity of the gut microbiome to physical activity.
In the second study, 8 weeks of mixed aerobic and resistance exercise training improved cardiorespiratory fitness and body composition, but did not produce significant changes in the fecal microbiome114 or metabolites. Interestingly, exercise was associated with reduced levels of phenylacetylglycine and trimethylamine N-oxide (TMAO) in urine. TMAO is a gut microbiota-derived metabolite of dietary choline and L- carnitine obtained from consumption of red and other types of meat. Give the finding of metagenomic enrichment of choline-metabolizing pathways in patients with CRC, 16 these findings support the benefits of physical activity in reducing production of TMAO and risk of CRC.
Dietary fiber and whole grains
The hypothesis that higher intake of dietary fiber protects against CRC originated from the observation of the substantially low rates of CRC in Africans, who consume a high-fiber diet.115 Although numerous epidemiologic studies have tested this hypothesis, none have produced conclusive results. According to the recent meta-analysis of 21 prospective studies, no linear association was found between fiber intake and CRC risk.78 However, there was substantial heterogeneity among studies. In contrast to most US studies, which reported no association,116–119 the European Prospective Investigation into Cancer and Nutrition cohort consistently found an association between fiber intake and reduced risk CRC.120–122 This might be due to differences in the food sources of fiber between European and American diets (mostly cereals vs fruits and vegetables) and the relatively low fiber intake in the US cohorts, which might not have achieved an effective threshold.3 In contrast, several studies have reported associations between whole grains and reduced risk of CRC. A meta-analysis found a 17% reduction in CRC risk per 90 g/day increase in consumption of whole grains.78
The anti-CRC effects of fiber could be related to its effects on the gut microbiota.6 Fiber is fermented by bacteria to produce SCFAs, which regulate the immune system and metabolism and reduce risk of CRC.6, 88, 94, 123–125 Some but not all cross-sectional studies associated higher fiber intake with increased fecal levels of SCFAs and enrichment of SCFA-producing bacteria, such as Eubacterium rectale, Roseburia spp., and Faecalibacterium prausnitzii.126, 127 Randomized controlled trials have examined the effect of supplementation with fiber or related prebiotics on the gut microbiota and metabolome in healthy adults. These findings were summarized in a meta-analysis128 of 64 trials of various types of supplements (such as resistant starch, inulin, arabinoxylan-oligosaccharide, and short-chain fructo-oligosaccharides) with sample sizes ranging from 8 to 84 participants and durations of 1 to 6 weeks. Fiber supplementation was found to enrich Bifidobacterium and Lactobacillus spp., whereas no effect was found on the other common SCFA producers except for increases in Roseburia spp. and F prausnitzii in parallel (rather than crossover) trials.
In support of the relevance of these findings to CRC, several studies have shown that compared with controls, patients with CRC had lower abundances of Bifidobacterium and Lactobacillus spp,16, 18 and other SCFA-producing bacteria, such as the genera Clostridium and Roseburia, the family Lachnospiraceae, and Faecalibacterium prausnitzii.15, 35, 37, 45, 53, 129 Bifidobacterium and Lactobacillus spp. produce lactate and acetate and can also increase production of butyrate through cross-feeding interactions with the butyrate-producing species, such as Eubacterium rectale.130–132 Fiber supplementation increased the concentration of butyrate in fecal samples.128 Furthermore, in support of the benefit of butyrate for CRC, a few cross-sectional studies reported that patients with CRC had a lower abundance of butyrate-producing species and lower fecal levels of butyrate than controls (Table 1).97, 133–135
In some136, 137 but not all138, 139 randomized controlled trials, increased consumption of whole grains was associated with higher abundance of SCFA-producing bacteria, such as Roseburia and Lachnospira, lower abundance of proinflammatory Enterobacteriaceae, and higher levels of fecal SCFAs through anti-inflammaion.136 Whole grains contain other beneficial nutrients, including polyphenols and flavonoids, which are also important modulators of the gut microbiome. Consumption of whole-grain wheat increases the fecal concentration of ferulic acid (the most abundant phenolic compound in whole grains) and serum concentration of dihydroferulic acid (a metabolite derived from ferulic acid by Lactobacillus and Bifidobacterium).136 Given the shared microbes in the synthesis of SCFA and dihydroferulic acid, synergistic mechanisms might augment the production of beneficial metabolites. These might account for ability of whole grains to reduce CRC risk compared with other food sources of dietary fiber. A large prospective study found that the association of dietary fiber with reduced risk of CRC had a larger effect on tumors with detectable F nucleatum than those without F nucleatum.140
Red and processed meat
Higher intake of red and processed meat has been associated with increased incidence of CRC. A meta-analysis associated each 100 g/day intake of red and processed meat with a 12% higher risk of CRC.78 The carcinogenic effect might be mediated by preservatives in red and processed (such as nitrates and nitrites), other additives (such as emulsifiers), chemicals produced during meat processing and cooking (such as heterocyclic amines and polycyclic aromatic hydrocarbons), or nutrients enriched in meats (such as heme iron, sulfur, and choline).3 Some of these elements can be metabolized by the gut bacteria to produce metabolites that have been implicated in CRC.
Meat, particularly red meat, has high content of choline and carnitine, which are precursors to gut microbiota-mediated formation of TMA and TMAO.141 A randomized controlled trial found that chronic ingestion of red meat, but not white meat or non-meat, increased plasma and urine levels of TMAO.142 High levels of TMAO have been associated with increased risk of cardiovascular disease and mortality.143–145 Several lines of evidence indicate the potential role of choline–TMAO pathway in development of CRC. A meta-analysis of metagenomes of fecal samples146 found patients with CRC to have higher levels of 2 bacterial genes that regulate TMA synthesis: choline TMA-lyase (cutC) and choline TMA-lyase-activating enzyme (cutD). Several taxonomic features associated with sequence variants of cutC were also found to be enriched in patients with CRC, including Hungatella hathewayi, Clostridium asparagiforme, Klebsiella oxytoca, and E coli.
Some,147, 148 but not all,149–151 prospective studies have associated higher dietary and plasma levels of choline with increased risk of colorectal neoplasia. A higher plasma level of TMAO was associated with an increased risk of CRC,151 but this finding was not replicated.152 Choline is required for DNA methylation and synthesis, along with other nutrients (such as folate and vitamin B12),3 so the ultimate effect of choline might depend on its distribution in the circulation for 1-carbon metabolism vs in the gut for microbial production of TMAO. In support of this hypothesis, the association between plasma level of TMAO and risk of CRC was restricted to patients with low plasma levels of vitamin B12.151 Higher levels of choline and TMAO have been linked to a spectrum of metabolic disturbances that might increase CRC risk, including higher BMI, greater visceral adiposity, insulin resistance, diabetes, and fatty liver.153, 154 Changes in TMAO and choline associated with increased insulin sensitivity in a weight-loss intervention study of obese individuals.155
Studies in the 1990s reported higher circulating levels of deoxycholic acid in patients with colorectal adenomas than controls.156, 157 A large prospective study of serum metabolomes associated concentration of glycochenodeoxycholic acid with CRC risk in women but not in men.158 Moreover, the bai operon, encoding 7α-dehydroxylation in Clostridium spp. required for synthesis of secondary bile acids, was found to be enriched in metagenomes of fecal samples from patients with CRC.146 Patients with gallstone disease and cholecystectomy, who are believed to have higher concentrations of secondary bile acids due to continuous flow of bile acids to the bowel, have an increased risk of CRC.159 Secondary bile acids might contribute to development of CRC because they generate reactive oxygen and nitrogen species that cause DNA damage and promote resistance to apoptosis.160 Changes in bile acid composition and concentrations have been associated with metabolic disorders and IBD, which increases risk for CRC.161, 162 A recent analysis of 8 geographically and technically diverse fecal shotgun metagenomic studies found evidence for increased production of secondary bile acids in patients with CRC, indicating a metabolic link between cancer-associated microbes and a fat- and meat-rich diet. 146
Hydrogen sulfide (H2S) is generated in the gut either by sulfur-reducing bacteria from inorganic sulfur (sulfate and sulfite) that is routinely used as a preservative in processed meat or by fermentative bacteria that metabolize organic sulfur compounds that are enriched in animal products such as red meat.3 Higher intakes of sulfur and sulfate were associated with increased risk of IBD163, and fecal samples from patients with colon cancer have higher concentrations of H2S than controls.164 Several sulfidogenic bacteria were found to be enriched in tissue samples from patients with CRC, including Fusobacterium, Bilophila wadsworthia, and the genera Lactococcus, Porphyromonas, Odoribacter, Bilophila, and Pyramidobacter.165, 166 H2S-producing pathways are increased in fecal samples from patients with CRC.20 Interestingly, African Americans have substantially higher abundances of sulfur-reducing bacteria and B wadsworthia than Caucasians, even after adjusting for dietary variables. Sulfidogenic bacteria might therefore contribute to the higher incidence of CRC in African Americans than Caucasians.166
In a cohort of elderly men (mean age of 71 years), higher dietary intake of organic sulfur was associated with increased fecal abundance of H2S-producing Clostridium clostridioforme.167 Consistent with the finding that processed red meat increases risk for distal colon cancer, in particular,168, 169 we associated dietary patterns with higher fecal abundances of sulfur-producing and increased risk of distal CRC, but not proximal colon cancer.167
Mechanisms
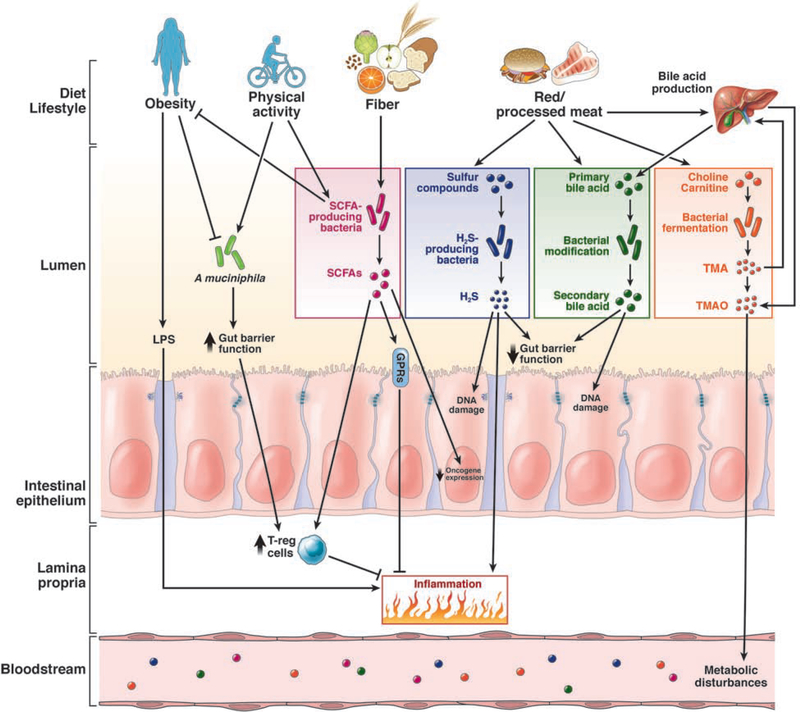
The intestine must maintain commensal microbes and a high load of bacterial products, but swiftly respond to pathogens that threaten its integrity. Analyses of gut microbiota of mice raised in germ-free or gnotobiotic conditions and transgenic mice, using techniques such as next-generation sequencing, microbial gene mutations, and microbial RNA-sequencing, have identified mechanisms by which the intestinal microbiota might contribute to or prevent development of CRC (Figure 1). For example, obesity might promote colorectal carcinogenesis via LPS-mediated systemic inflammation and depletion of SCFA-producing bacteria. Red and processed meat might increase CRC risk by increasing bacterial production of secondary bile acids, H2S, and TMAO. Physical activity and dietary fiber might reduce risk of CRC by increasing the abundance of A muciniphila and SCFA-producing bacteria. For reviews on the mechanisms by which the gut microbiome contributes to colorectal carcinogenesis, see refs 9, 11, 12.
Figure 1.
Pathways by which dietary and environmental factors affect the intestinal microbiome and their roles in colorectal carcinogenesis. Obesity may promote CRC through LPS-mediated systemic inflammation and depletion of A muciniphila and SCFA-producing bacteria, whereas physical activity might protect against CRC by increasing the abundance of A muciniphila and SCFA-producing bacteria. The benefit of dietary fiber might be mediated by enrichment of SCFA-producing bacteria and increased production of SCFAs that inhibit CRC development, modulation of the immune and metabolic response. Red and processed meat may increase CRC risk by increased bacterial production of secondary bile acids, H2S, and TMAO.
Abbreviations: H2S, hydrogen sulfide; T-reg cell, T-regulatory cells.
Metabolites
The pathogenic mechanisms of bacteria associated with CRC (such as F nucleatum. Bacteroides fragilis, E coli, and inflammatory Enterobacteriaceae) have been investigated in animal models. Chronic inflammation, dysfunction of immunity, increased serum levels of LPS, secondary bile acids, and leaky gut are representative pathways that link the gut microbiota with CRC (Figure 1).
Although many types of bacteria have been identified that might contribute to colorectal carcinogenesis, we have also identified bacterial metabolites that affect CRC risk. High levels of secondary bile acids and SCFAs have opposing effects on colon inflammation.170 High fat content and consumption of red and processed meats increase secretion of primary bile acids that can be metabolized by the gut bacteria to secondary bile acid, including deoxycholic acid, lithocholic acid, and glycochenodeoxycholic acid. Sulfidogenic bacteria are increased in tissue samples from patients with CRC.165, 166 Sulfur is another compound in red and processed meat that is closely linked to the gut microbiota.
SCFA-producing bacteria, such as Roseburia and Lachnospira, reduce risk of CRC. Higher levels of fecal SCFAs were associated with increased numbers of memory T cells in blood,171 reduced levels of inflammatory cytokines (such as tumor necrosis factor), and increased levels of anti-inflammatory cytokines (such as interleukin 10).136 Butyrate increases intestinal expression of vitamin D receptor mRNA and protein, reduces intestinal dysbiosis, and activates the autophagy response of Paneth cells to inhibit chronic inflammation.172
Contribution of diet to colitis and colon cancer
Fiber regulates the immune response and levels of F nucleatum. It is possible that higher intake of fiber could reduce the carcinogenic effects of F nucleatum by restoring effective immunosurveillance.173
In Il10−/− mice and mice without disruption of this gene, a diet high in saturated (milk-derived) fat (MF) promoted expansion of the immunogenic sulfite-reducing pathobiont Bilophila wadsworthia, a member of the Deltaproteobacteria.174 The Bilophila wadsworthia expansion resulted from a MF diet-induced shift in hepatic conjugation of bile acids, from glycocholic to taurocholic acid, which helps solubilize the more hydrophobic MF diet. H2S-producing bacteria might promote intestinal inflammation to increase the risk for colitis-associated cancer.
Infection
Although dysbiosis is associated with chronic inflammation and production of carcinogenic metabolites, 175, 176 there is limited evidence to support a direct link between specific intestinal bacteria, their virulence factors, and sporadic CRC. Increases in population-wide exposure to antibiotics 177 might contribute to high rates of bacterial infection.178 More than 1 million people in the US acquire Salmonella infection annually as a foodborne illness mainly from eggs, meats, dairy, and other contaminated non-animal foods.178 In C57BL/6J mice, recurrent infection with Salmonella enterica promoted intestinal inflammation and progressively disabled protective mechanisms, inducing endogenous neuraminidase activity to reduce the abundance and protective effects of intestinal alkaline phosphatase.179 The protein AvrA, produced by Salmonella, promotes inflammation and colon tumor development by activating beta-catenin signaling to STAT3.36, 180 Expression of AvrA protein, detected by immunohistochemistry, was significantly higher in pre-cancer colon tissues than in normal human colon or tumor tissues, based on pathology differences.181 Mice with Citrobacter rodentium-induced colonic crypt hyperplasia have alterations in beta-catenin and Notch signaling, intestinal barrier function, and fecal dysbiosis, resulting in development of colon tumors.182, 183 F nucleatum induce expression of microRNA 21 by CRC cells by activating TLR4 signaling via MYD88 and nuclear factor-kappa B. This increased proliferation of the CRC cells.184
Specific strains of bacteria therefore appear to disrupt the intestinal microbiome and promote inflammation to increase risk for IBD and colon carcinogenesis. The epithelial and metabolic changes that occur during development of CRC might provide a competitive advantage to a subset of intestinal bacteria.185 Genetic variants, along with environmental factors, also contribute to contribute to dysbiosis and development of CRC. Microbial pathogens and chronic inflammation can compromise barrier function and enhance permeability. Translocation of microbial products, metabolites, increased serum LPS, and immune activation promote dysbiosis, barrier failure, and inflammation.
Screening
Alterations in the fecal microbiomes of patients with CRC have also been observed in patients with colorectal adenoma—these might be used in screening for individuals at risk for CRC. Fecal microbiome analysis identifies patients with adenomas with reasonable levels of accuracy (area under receiver operating curves ranging from 0.55 to 0.67 in validation studies), although this is a lower value than for detection of CRC.16, 18, 186–188 Combining the fecal microbiome data with scores from risk factor-based models or results of screening tests (such as fecal occult blood test and fecal immunochemical test) increases the accuracy of detection for advanced adenomas.35, 186, 187 For example, addition of fecal F nucleatum quantitation to fecal immunochemical test, which has suboptimal sensitivity in detecting adenomas, doubled the sensitivity for detection of advanced adenomas. A similar improvement was observed in an independent validation cohort.186
Several questions must be answered before fecal microbiomes can be used in CRC screening. First, studies have identified and used different microbial features to construct their analysis models. It is unclear to what extent the heterogeneity among studies reflects the true differences in the ability to detect CRC based on different microbial patterns or variations in the technical aspects of studies (such as stool collection methods, timing of bowel preparation for colonoscopy, and sequencing and analysis methods). Therefore, it is not clear whether there is one specific microbial signature that can be used to identify individuals with CRC or its precursors in diverse populations. Second, despite the discriminatory accuracy, the reliability and predictivity of the gut microbiome-based classifiers must be established in prospective studies. Most screening methods have limited abilities to detect proximal lesions (such as the fecal immunohistochemical test, flexible sigmoidoscopy, and colonoscopy), so it is important to determine whether analyses of fecal microbiomes can improve the sensitivity of detection of proximal colon neoplasias. Other practical issues must be evaluated before fecal microbiome analysis can be used in CRC screening, such as determination of cost effectiveness, affordability, and acceptability by patients and physicians, compared with established screening strategies.
Therapy
In addition to affecting CRC development, the gut microbiota modulates the response to cancer therapy and susceptibility to toxic side effects, although there is only limited evidence from patients (for reviews, see 8, 189). In 2013, Iida et al190 reported that alterations in the intestinal microbiota can affect the efficacy of an immunotherapy (CpG-oligonucleotide) and oxaliplatin, a platinum compound used in chemotherapy for CRC and other cancers. Both therapies had reduced efficacy in mice given antibiotics or germ-free mice, which had lower production of cytokines and less tumor necrosis after CpG-oligonucleotide administration and reduced production of reactive oxygen species and cytotoxicity after chemotherapy compared. Bacterial metabolism was found to affect the efficacy of the anti-pyrimidine drugs 5-fluorouracil and 5-fluoro-20-deoxyuridine and the topoisomerase I inhibitor camptothecin in Caenorhabditis elegans 191, 192. Different bacterial species increased the response to 1 drug and decreased the effect of another. Bacterial ribonucleotide metabolism affected the cytotoxic effects 5-fluorouracil and 5-fluoro-20-deoxyuridine by altering production of regulatory metabolites had synergistic effects with drug-induced DNA damage. However, no patients or mouse models of CRC were used in these studies.
F nucleatum was promotes resistance of CRC cells to chemotherapy.28 Patients with post-chemotherapy recurrence had a higher abundance of F nucleatum in tumor tissues.28, 193 Bioinformatic and functional studies indicated that F nucleatum activates innate immune responses, via TLR4 and MYD88, resulting in loss of specific microRNAs. This resulted in activation of autophagy activation and promotion of chemoresistance in patients with CRC.28
Several studies have indicated that the gut microbiota can determine the efficacy of cancer immunotherapy, although there is no evidence for its effects on immunotherapy for CRC. For example, Bacteroides thetaiotaomicron and B fragilis have been associated with higher efficacy of agents that block cytotoxic T-lymphocyte associated protein 4 (CTLA4), possibly by affecting interleukin 12-dependent T-helper 1 cell-mediated immune responses. Bifidobacterium increased the response of tumors to antibodies against programmed cell death 1 (PDCD1), which increased dendritic cell function, priming of CD8+ T cells, and their accumulation in the tumor microenvironment.194 A muciniphila195 and Faecalibacterium196 have been associated with greater efficacy of PDCD1 blocking agents—possibly by increasing recruitment of T cells to tumors and their functions there. Moreover, specific bacteria have been associated with resistance to the development of immune checkpoint inhibitor-associated colitis,197 and fecal microbiota transplants might be used to treat this form of colitis.198
Dietary interventions can be used to modulate the gut microbiota in patients receiving cancer therapy. Higher intake of nutrients such as fiber,199 marine omega-3 fatty acid,200–202 vitamin D,203–205 or calcium,203 or coffee206, 207 or a plant-based low-carbohydrate diet207 has been associated with increased survival times of patients with CRC. Many of these factors have immunomodulatory and microbiota-modifying effects, so they might increase the efficacy and reduce the adverse effects of immunotherapies or other therapeutic agents.208
Future Directions
Epidemiologic evidence for the effects of the gut microbiome on CRC risk comes from retrospective case–control studies of biospecimens collected from patients with established CRC or precursor lesions. It is not clear whether the identified microbial alterations are a cause or consequence of colorectal carcinogenesis.209 Moreover, clinical studies have been limited by small sample sizes (mostly <100 cases), likely underpowered to identify predictive signals from hundreds to thousands of reads counts generated by high-throughput sequencing. Prospective studies, with large-scale microbial biospecimen collection and long-term follow up, are urgently needed to determine which microbes or collections of microbes contribute to CRC development discover and identify signatures that can be used in screening.210
Data on environmental factors that alter gut microbiome are mostly derived from cross-sectional or short-term interventional studies. Cross-section studies cannot determine whether changes in diet and lifestyle cause changes in the intestinal microbiota, its metabolism, or its effects on the immune response. Correlations among individual dietary and lifestyle factors and the gut microbiome are difficult to make due to confounding factors. The short duration of most intervention studies makes it impossible to examine the effects of alterations to the gut microbiome of long-term exposures that affect risk for CRC. Prospective studies with detailed diet and lifestyle data collected long before participants develop CRC are needed to better characterize the long-term influence of environmental exposures on the gut microbiome and their effects on CRC prevention. Furthermore, we need to uncover the specific mechanisms by which these diet and lifestyle factors influence the gut microbiome and risk of CRC.
There are few human data on the role of effects of alterations in the gut microbiota in CRC treatment. Including microbiota specimen collection into clinical trials, along with assessments of diet and other environmental factors, could provide information on how they affect treatment outcomes and survival times of patients with CRC. It is important to elucidate the immune and metabolic pathways that mediate the effects of the gut microbiota and dietary factors on treatment for CRC and survival times.
Researchers have generated exciting preliminary data for the role of the microbiota in CRC development, prevention, and treatment. Integrated prospective studies will open new avenues for studies of intestinal microbiota and its manipulation in CRC screening, prevention, and treatment.
Acknowledgments
Funding Support
This work was supported by the U.S. National Institutes of Health grant (NIH) NIDDK R01 DK105118, R01DK114126, DOD BC160450P1, and the UIC Cancer Center support (to J. S.); the American Cancer Society (Grant Number MRSG-17-220-01 – NEC to M.S.), the NIH grants (K99 CA215314 and R00 CA215314 to M.S.; K24 DK098311, R01 CA137178, R01 CA202704, R01 CA176726 to A.T.C.). Dr. Chan is a Stuart and Suzanne Steele MGH Research Scholar.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Fedewa SA, et al. Colorectal cancer statistics, 2017. CA Cancer J Clin 2017;67:177–193. [DOI] [PubMed] [Google Scholar]
- 3.Song M, Garrett WS, Chan AT. Nutrients, foods, and colorectal cancer prevention. Gastroenterology 2015;148:1244–60 e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Islami F, Goding Sauer A, Miller KD, et al. Proportion and number of cancer cases and deaths attributable to potentially modifiable risk factors in the United States. CA Cancer J Clin 2018;68:31–54. [DOI] [PubMed] [Google Scholar]
- 5.Mima K, Cao Y, Chan AT, et al. Fusobacterium nucleatum in Colorectal Carcinoma Tissue According to Tumor Location. Clin Transl Gastroenterol 2016;7:e200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Song M, Chan AT. Environmental Factors, Gut Microbiota, and Colorectal Cancer Prevention. Clin Gastroenterol Hepatol 2019;17:275–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scott AJ, Alexander JL, Merrifield CA, et al. International Cancer Microbiome Consortium consensus statement on the role of the human microbiome in carcinogenesis. Gut 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roy S, Trinchieri G. Microbiota: a key orchestrator of cancer therapy. Nat Rev Cancer 2017;17:271–285. [DOI] [PubMed] [Google Scholar]
- 9.Tilg H, Adolph TE, Gerner RR, et al. The Intestinal Microbiota in Colorectal Cancer. Cancer Cell 2018;33:954–964. [DOI] [PubMed] [Google Scholar]
- 10.Dai Z, Zhang J, Wu Q, et al. The role of microbiota in the development of colorectal cancer. Int J Cancer 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keku TO, Dulal S, Deveaux A, et al. The gastrointestinal microbiota and colorectal cancer. Am J Physiol Gastrointest Liver Physiol 2015;308:G351–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang Y, Jobin C. Novel insights into microbiome in colitis and colorectal cancer. Curr Opin Gastroenterol 2017;33:422–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Almeida A, Mitchell AL, Boland M, et al. A new genomic blueprint of the human gut microbiota. Nature 2019;568:499–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun J, Chang EB. Exploring gut microbes in human health and disease: Pushing the envelope. Genes Dis 2014;1:132–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ahn J, Sinha R, Pei Z, et al. Human gut microbiome and risk for colorectal cancer. Journal of the National Cancer Institute 2013;105:1907–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thomas AM, Manghi P, Asnicar F, et al. Metagenomic analysis of colorectal cancer datasets identifies cross-cohort microbial diagnostic signatures and a link with choline degradation. Nat Med 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sze MA, Schloss PD. Leveraging Existing 16S rRNA Gene Surveys To Identify Reproducible Biomarkers in Individuals with Colorectal Tumors. MBio 2018;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feng Q, Liang S, Jia H, et al. Gut microbiome development along the colorectal adenoma-carcinoma sequence. Nat Commun 2015;6:6528. [DOI] [PubMed] [Google Scholar]
- 19.Nakatsu G, Li X, Zhou H, et al. Gut mucosal microbiome across stages of colorectal carcinogenesis. Nat Commun 2015;6:8727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yachida S, Mizutani S, Shiroma H, et al. Metagenomic and metabolomic analyses reveal distinct stage-specific phenotypes of the gut microbiota in colorectal cancer. Nature Medicine 2019. [DOI] [PubMed] [Google Scholar]
- 21.Castellarin M, Warren RL, Freeman JD, et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res 2012;22:299–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kostic AD, Gevers D, Pedamallu CS, et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res 2012;22:292–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Flanagan L, Schmid J, Ebert M, et al. Fusobacterium nucleatum associates with stages of colorectal neoplasia development, colorectal cancer and disease outcome. Eur J Clin Microbiol Infect Dis 2014;33:1381–90. [DOI] [PubMed] [Google Scholar]
- 24.Ito M, Kanno S, Nosho K, et al. Association of Fusobacterium nucleatum with clinical and molecular features in colorectal serrated pathway. Int J Cancer 2015;137:1258–68. [DOI] [PubMed] [Google Scholar]
- 25.Li YY, Ge QX, Cao J, et al. Association of Fusobacterium nucleatum infection with colorectal cancer in Chinese patients. World J Gastroenterol 2016;22:3227–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mima K, Nishihara R, Qian ZR, et al. Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis. Gut 2016;65:1973–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tahara T, Yamamoto E, Suzuki H, et al. Fusobacterium in colonic flora and molecular features of colorectal carcinoma. Cancer Res 2014;74:1311–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu T, Guo F, Yu Y, et al. Fusobacterium nucleatum Promotes Chemoresistance to Colorectal Cancer by Modulating Autophagy. Cell 2017;170:548–563 e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nosho K, Sukawa Y, Adachi Y, et al. Association of Fusobacterium nucleatum with immunity and molecular alterations in colorectal cancer. World J Gastroenterol 2016;22:557–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mima K, Sukawa Y, Nishihara R, et al. Fusobacterium nucleatum and T Cells in Colorectal Carcinoma. JAMA Oncol 2015;1:653–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rashtak S, Rego R, Sweetser SR, et al. Sessile Serrated Polyps and Colon Cancer Prevention. Cancer Prev Res (Phila) 2017;10:270–278. [DOI] [PubMed] [Google Scholar]
- 32.Purcell RV, Visnovska M, Biggs PJ, et al. Distinct gut microbiome patterns associate with consensus molecular subtypes of colorectal cancer. Sci Rep 2017;7:11590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guinney J, Dienstmann R, Wang X, et al. The consensus molecular subtypes of colorectal cancer. Nat Med 2015;21:1350–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bullman S, Pedamallu CS, Sicinska E, et al. Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science 2017;358:1443–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zackular JP, Rogers MA, Ruffin MTt, et al. The human gut microbiome as a screening tool for colorectal cancer. Cancer Prev Res (Phila) 2014;7:1112–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zeller G, Tap J, Voigt AY, et al. Potential of fecal microbiota for early-stage detection of colorectal cancer. Mol Syst Biol 2014;10:766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vogtmann E, Hua X, Zeller G, et al. Colorectal Cancer and the Human Gut Microbiome: Reproducibility with Whole-Genome Shotgun Sequencing. PLoS One 2016;11:e0155362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Momen-Heravi F, Babic A, Tworoger SS, et al. Periodontal disease, tooth loss and colorectal cancer risk: Results from the Nurses’ Health Study. Int J Cancer 2017;140:646–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liang Q, Chiu J, Chen Y, et al. Fecal Bacteria Act as Novel Biomarkers for Noninvasive Diagnosis of Colorectal Cancer. Clin Cancer Res 2017;23:2061–2070. [DOI] [PubMed] [Google Scholar]
- 40.Boleij A, Hechenbleikner EM, Goodwin AC, et al. The Bacteroides fragilis toxin gene is prevalent in the colon mucosa of colorectal cancer patients. Clin Infect Dis 2015;60:208–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Purcell RV, Pearson J, Aitchison A, et al. Colonization with enterotoxigenic Bacteroides fragilis is associated with early-stage colorectal neoplasia. PLoS One 2017;12:e0171602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dejea CM, Fathi P, Craig JM, et al. Patients with familial adenomatous polyposis harbor colonic biofilms containing tumorigenic bacteria. Science 2018;359:592–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Viljoen KS, Dakshinamurthy A, Goldberg P, et al. Quantitative profiling of colorectal cancer-associated bacteria reveals associations between fusobacterium spp., enterotoxigenic Bacteroides fragilis (ETBF) and clinicopathological features of colorectal cancer. PLoS One 2015;10:e0119462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Toprak NU, Yagci A, Gulluoglu BM, et al. A possible role of Bacteroides fragilis enterotoxin in the aetiology of colorectal cancer. Clin Microbiol Infect 2006;12:782–6. [DOI] [PubMed] [Google Scholar]
- 45.Wang T, Cai G, Qiu Y, et al. Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. ISME J 2012;6:320–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dai Z, Coker OO, Nakatsu G, et al. Multi-cohort analysis of colorectal cancer metagenome identified altered bacteria across populations and universal bacterial markers. Microbiome 2018;6:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Swidsinski A, Khilkin M, Kerjaschki D, et al. Association between intraepithelial Escherichia coli and colorectal cancer. Gastroenterology 1998;115:281–6. [DOI] [PubMed] [Google Scholar]
- 48.Martin HM, Campbell BJ, Hart CA, et al. Enhanced Escherichia coli adherence and invasion in Crohn’s disease and colon cancer. Gastroenterology 2004;127:80–93. [DOI] [PubMed] [Google Scholar]
- 49.Arthur JC, Perez-Chanona E, Muhlbauer M, et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science 2012;338:120–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Buc E, Dubois D, Sauvanet P, et al. High prevalence of mucosa-associated E. coli producing cyclomodulin and genotoxin in colon cancer. PLoS One 2013;8:e56964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bonnet M, Buc E, Sauvanet P, et al. Colonization of the human gut by E. coli and colorectal cancer risk. Clin Cancer Res 2014;20:859–67. [DOI] [PubMed] [Google Scholar]
- 52.Prorok-Hamon M, Friswell MK, Alswied A, et al. Colonic mucosa-associated diffusely adherent afaC+ Escherichia coli expressing lpfA and pks are increased in inflammatory bowel disease and colon cancer. Gut 2014;63:761–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shah MS, DeSantis TZ, Weinmaier T, et al. Leveraging sequence-based faecal microbial community survey data to identify a composite biomarker for colorectal cancer. Gut 2017. [DOI] [PubMed] [Google Scholar]
- 54.Yang Y, Cai Q, Shu XO, et al. Prospective study of oral microbiome and colorectal cancer risk in low-income and African American populations. Int J Cancer 2019;144:2381–2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Flemer B, Warren RD, Barrett MP, et al. The oral microbiota in colorectal cancer is distinctive and predictive. Gut 2018;67:1454–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hu JM, Shen CJ, Chou YC, et al. Risk of colorectal cancer in patients with periodontal disease severity: a nationwide, population-based cohort study. Int J Colorectal Dis 2018;33:349–352. [DOI] [PubMed] [Google Scholar]
- 57.Michaud DS, Lu J, Peacock-Villada AY, et al. Periodontal Disease Assessed Using Clinical Dental Measurements and Cancer Risk in the ARIC Study. J Natl Cancer Inst 2018;110:843–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kato I, Vasquez AA, Moyerbrailean G, et al. Oral microbiome and history of smoking and colorectal cancer. J Epidemiol Res 2016;2:92–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lamont RJ, Koo H, Hajishengallis G. The oral microbiota: dynamic communities and host interactions. Nat Rev Microbiol 2018;16:745–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sun J, Kato I. Gut microbiota, inflammation and colorectal cancer. Genes Dis 2016;3:130–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Abdulamir AS, Hafidh RR, Abu Bakar F. The association of Streptococcus bovis/gallolyticus with colorectal tumors: the nature and the underlying mechanisms of its etiological role. J Exp Clin Cancer Res 2011;30:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kuhn KG, Falkenhorst G, Ceper TH, et al. Detecting non-typhoid Salmonella in humans by ELISAs: a literature review. J Med Microbiol 2012;61:1–7. [DOI] [PubMed] [Google Scholar]
- 63.Falkenhorst G, Simonsen J, Ceper TH, et al. Serological cross-sectional studies on salmonella incidence in eight European countries: no correlation with incidence of reported cases. BMC Public Health 2012;12:523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Simonsen J, Strid M A, MØLbak K, et al. Sero-epidemiology as a tool to study the incidence of Salmonella infections in humans. Epidemiology and Infection 2008;136:895–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Thabane M, Kottachchi DT, Marshall JK. Systematic review and meta-analysis: the incidence and prognosis of post-infectious irritable bowel syndrome. Alimentary Pharmacology & Therapeutics 2007;26:535–544. [DOI] [PubMed] [Google Scholar]
- 66.Keithlin J, Sargeant JM, Thomas MK, et al. Systematic review and meta-analysis of the proportion of non-typhoidal Salmonella cases that develop chronic sequelae. Epidemiology and Infection 2015;143:1333–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ternhag A, Törner A, Svensson Å, et al. Short- and Long-term Effects of Bacterial Gastrointestinal Infections. Emerging Infectious Diseases 2008;14:143–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jess T, Simonsen J, Nielsen NM, et al. Enteric Salmonella or Campylobacter infections and the risk of inflammatory bowel disease. Gut 2011;60:318–324. [DOI] [PubMed] [Google Scholar]
- 69.Kato I, Boleij A, Kortman GA, et al. Partial associations of dietary iron, smoking and intestinal bacteria with colorectal cancer risk. Nutr Cancer 2013;65:169–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mughini-Gras L, Schaapveld M, Kramers J, et al. Increased colon cancer risk after severe Salmonella infection. PLoS One 2018;13:e0189721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Butt J, Varga MG, Blot WJ, et al. Serologic Response to Helicobacter pylori Proteins Associated With Risk of Colorectal Cancer Among Diverse Populations in the United States. Gastroenterology 2019;156:175–186 e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nakatsu G, Zhou H, Wu WKK, et al. Alterations in Enteric Virome Are Associated With Colorectal Cancer and Survival Outcomes. Gastroenterology 2018;155:529–541 e5. [DOI] [PubMed] [Google Scholar]
- 73.Coker OO, Nakatsu G, Dai RZ, et al. Enteric fungal microbiota dysbiosis and ecological alterations in colorectal cancer. Gut 2019;68:654–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hannigan GD, Duhaime MB, Ruffin MTt, et al. Diagnostic Potential and Interactive Dynamics of the Colorectal Cancer Virome. MBio 2018;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Malik A, Sharma D, Malireddi RKS, et al. SYK-CARD9 Signaling Axis Promotes Gut Fungi-Mediated Inflammasome Activation to Restrict Colitis and Colon Cancer. Immunity 2018;49:515–530 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang J, Thingholm LB, Skieceviciene J, et al. Genome-wide association analysis identifies variation in vitamin D receptor and other host factors influencing the gut microbiota. Nat Genet 2016;48:1396–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rothschild D, Weissbrod O, Barkan E, et al. Environment dominates over host genetics in shaping human gut microbiota. Nature 2018;555:210–215. [DOI] [PubMed] [Google Scholar]
- 78.World Cancer Research Fund / American Institute for Cancer Research. Continuous Update Project Report: Diet, Nutrition, Physical Activity and Colorectal Cancer. Available at: wcrf.org/colorectal-cancer-2017 All CUP reports are available at wcrf.org/cupreports, 2017.
- 79.Liu PH, Wu K, Ng K, et al. Association of Obesity With Risk of Early-Onset Colorectal Cancer Among Women. JAMA Oncol 2019;5:37–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Renehan AG, Zwahlen M, Egger M. Adiposity and cancer risk: new mechanistic insights from epidemiology. Nat Rev Cancer 2015;15:484–98. [DOI] [PubMed] [Google Scholar]
- 81.Turnbaugh PJ, Hamady M, Yatsunenko T, et al. A core gut microbiome in obese and lean twins. Nature 2009;457:480–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Greenblum S, Turnbaugh PJ, Borenstein E. Metagenomic systems biology of the human gut microbiome reveals topological shifts associated with obesity and inflammatory bowel disease. Proc Natl Acad Sci U S A 2012;109:594–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ley RE, Turnbaugh PJ, Klein S, et al. Microbial ecology: human gut microbes associated with obesity. Nature 2006;444:1022–3. [DOI] [PubMed] [Google Scholar]
- 84.Duncan SH, Lobley GE, Holtrop G, et al. Human colonic microbiota associated with diet, obesity and weight loss. Int J Obes (Lond) 2008;32:1720–4. [DOI] [PubMed] [Google Scholar]
- 85.Seganfredo FB, Blume CA, Moehlecke M, et al. Weight-loss interventions and gut microbiota changes in overweight and obese patients: a systematic review. Obes Rev 2017;18:832–851. [DOI] [PubMed] [Google Scholar]
- 86.Remely M, Tesar I, Hippe B, et al. Gut microbiota composition correlates with changes in body fat content due to weight loss. Benef Microbes 2015;6:431–9. [DOI] [PubMed] [Google Scholar]
- 87.Sanna S, van Zuydam NR, Mahajan A, et al. Causal relationships among the gut microbiome, short-chain fatty acids and metabolic diseases. Nat Genet 2019;51:600–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Koh A, De Vadder F, Kovatcheva-Datchary P, et al. From Dietary Fiber to Host Physiology: Short-Chain Fatty Acids as Key Bacterial Metabolites. Cell 2016;165:1332–1345. [DOI] [PubMed] [Google Scholar]
- 89.Zhao L, Zhang F, Ding X, et al. Gut bacteria selectively promoted by dietary fibers alleviate type 2 diabetes. Science 2018;359:1151–1156. [DOI] [PubMed] [Google Scholar]
- 90.Pataky Z, Genton L, Spahr L, et al. Impact of Hypocaloric Hyperproteic Diet on Gut Microbiota in Overweight or Obese Patients with Nonalcoholic Fatty Liver Disease: A Pilot Study. Dig Dis Sci 2016;61:2721–31. [DOI] [PubMed] [Google Scholar]
- 91.Simoes CD, Maukonen J, Scott KP, et al. Impact of a very low-energy diet on the fecal microbiota of obese individuals. Eur J Nutr 2014;53:1421–9. [DOI] [PubMed] [Google Scholar]
- 92.Cotillard A, Kennedy SP, Kong LC, et al. Dietary intervention impact on gut microbial gene richness. Nature 2013;500:585–8. [DOI] [PubMed] [Google Scholar]
- 93.Walker AW, Ince J, Duncan SH, et al. Dominant and diet-responsive groups of bacteria within the human colonic microbiota. ISME J 2011;5:220–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bultman SJ. Molecular pathways: gene-environment interactions regulating dietary fiber induction of proliferation and apoptosis via butyrate for cancer prevention. Clin Cancer Res 2014;20:799–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Liu R, Hong J, Xu X, et al. Gut microbiome and serum metabolome alterations in obesity and after weight-loss intervention. Nat Med 2017;23:859–868. [DOI] [PubMed] [Google Scholar]
- 96.Clarke SF, Murphy EF, O’Sullivan O, et al. Exercise and associated dietary extremes impact on gut microbial diversity. Gut 2014;63:1913–20. [DOI] [PubMed] [Google Scholar]
- 97.Weir TL, Manter DK, Sheflin AM, et al. Stool microbiome and metabolome differences between colorectal cancer patients and healthy adults. PLoS One 2013;8:e70803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Candela M, Biagi E, Soverini M, et al. Modulation of gut microbiota dysbioses in type 2 diabetic patients by macrobiotic Ma-Pi 2 diet. Br J Nutr 2016;116:80–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Louis S, Tappu RM, Damms-Machado A, et al. Characterization of the Gut Microbial Community of Obese Patients Following a Weight-Loss Intervention Using Whole Metagenome Shotgun Sequencing. PLoS One 2016;11:e0149564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Shoaie S, Ghaffari P, Kovatcheva-Datchary P, et al. Quantifying Diet-Induced Metabolic Changes of the Human Gut Microbiome. Cell Metab 2015;22:320–31. [DOI] [PubMed] [Google Scholar]
- 101.Cani PD, Amar J, Iglesias MA, et al. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes 2007;56:1761–72. [DOI] [PubMed] [Google Scholar]
- 102.Yang B, Bostick RM, Tran HQ, et al. Circulating Biomarkers of Gut Barrier Function: Correlates and Nonresponse to Calcium Supplementation among Colon Adenoma Patients. Cancer Epidemiol Biomarkers Prev 2016;25:318–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Moreno-Navarrete JM, Ortega F, Serino M, et al. Circulating lipopolysaccharide-binding protein (LBP) as a marker of obesity-related insulin resistance. Int J Obes (Lond) 2012;36:1442–9. [DOI] [PubMed] [Google Scholar]
- 104.Lassenius MI, Pietilainen KH, Kaartinen K, et al. Bacterial endotoxin activity in human serum is associated with dyslipidemia, insulin resistance, obesity, and chronic inflammation. Diabetes Care 2011;34:1809–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kang M, Edmundson P, Araujo-Perez F, et al. Association of plasma endotoxin, inflammatory cytokines and risk of colorectal adenomas. BMC Cancer 2013;13:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chen R, Luo FK, Wang YL, et al. LBP and CD14 polymorphisms correlate with increased colorectal carcinoma risk in Han Chinese. World J Gastroenterol 2011;17:2326–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kong SY, Tran HQ, Gewirtz AT, et al. Serum Endotoxins and Flagellin and Risk of Colorectal Cancer in the European Prospective Investigation into Cancer and Nutrition (EPIC) Cohort. Cancer Epidemiol Biomarkers Prev 2016;25:291–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Citronberg JS, Wilkens LR, Le Marchand L, et al. Plasma lipopolysaccharide-binding protein and colorectal cancer risk: a nested case-control study in the Multiethnic Cohort. Cancer Causes Control 2018;29:115–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Greathouse KL, White JR, Padgett RN, et al. Gut microbiome meta-analysis reveals dysbiosis is independent of body mass index in predicting risk of obesity-associated CRC. BMJ Open Gastroenterol 2019;6:e000247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Barton W, Penney NC, Cronin O, et al. The microbiome of professional athletes differs from that of more sedentary subjects in composition and particularly at the functional metabolic level. Gut 2018;67:625–633. [DOI] [PubMed] [Google Scholar]
- 111.Estaki M, Pither J, Baumeister P, et al. Cardiorespiratory fitness as a predictor of intestinal microbial diversity and distinct metagenomic functions. Microbiome 2016;4:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bressa C, Bailen-Andrino M, Perez-Santiago J, et al. Differences in gut microbiota profile between women with active lifestyle and sedentary women. PLoS One 2017;12:e0171352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Allen JM, Mailing LJ, Niemiro GM, et al. Exercise Alters Gut Microbiota Composition and Function in Lean and Obese Humans. Med Sci Sports Exerc 2018;50:747–757. [DOI] [PubMed] [Google Scholar]
- 114.Cronin O, Barton W, Skuse P, et al. A Prospective Metagenomic and Metabolomic Analysis of the Impact of Exercise and/or Whey Protein Supplementation on the Gut Microbiome of Sedentary Adults. mSystems 2018;3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Burkitt DP. Epidemiology of cancer of the colon and rectum. Cancer 1971;28:3–13. [DOI] [PubMed] [Google Scholar]
- 116.Kunzmann AT, Coleman HG, Huang WY, et al. Dietary fiber intake and risk of colorectal cancer and incident and recurrent adenoma in the Prostate, Lung, Colorectal, and Ovarian Cancer Screening Trial. Am J Clin Nutr 2015;102:881–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Higginbotham S, Zhang ZF, Lee IM, et al. Dietary glycemic load and risk of colorectal cancer in the Women’s Health Study. J Natl Cancer Inst 2004;96:229–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Michels KB, Fuchs CS, Giovannucci E, et al. Fiber intake and incidence of colorectal cancer among 76,947 women and 47,279 men. Cancer Epidemiol Biomarkers Prev 2005;14:842–9. [DOI] [PubMed] [Google Scholar]
- 119.He X, Wu K, Zhang X, et al. Dietary intake of fiber, whole grains and risk of colorectal cancer: An updated analysis according to food sources, tumor location and molecular subtypes in two large US cohorts. Int J Cancer 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Bingham SA, Day NE, Luben R, et al. Dietary fibre in food and protection against colorectal cancer in the European Prospective Investigation into Cancer and Nutrition (EPIC): an observational study. Lancet 2003;361:1496–501. [DOI] [PubMed] [Google Scholar]
- 121.Bingham SA, Norat T, Moskal A, et al. Is the association with fiber from foods in colorectal cancer confounded by folate intake? Cancer Epidemiol Biomarkers Prev 2005;14:1552–6. [DOI] [PubMed] [Google Scholar]
- 122.Murphy N, Norat T, Ferrari P, et al. Dietary fibre intake and risks of cancers of the colon and rectum in the European prospective investigation into cancer and nutrition (EPIC). PloS one 2012;7:e39361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ploger S, Stumpff F, Penner GB, et al. Microbial butyrate and its role for barrier function in the gastrointestinal tract. Ann N Y Acad Sci 2012;1258:52–9. [DOI] [PubMed] [Google Scholar]
- 124.Makki K, Deehan EC, Walter J, et al. The Impact of Dietary Fiber on Gut Microbiota in Host Health and Disease. Cell Host Microbe 2018;23:705–715. [DOI] [PubMed] [Google Scholar]
- 125.Bhatt AP, Redinbo MR, Bultman SJ. The role of the microbiome in cancer development and therapy. CA Cancer J Clin 2017;67:326–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.O’Keefe SJ, Li JV, Lahti L, et al. Fat, fibre and cancer risk in African Americans and rural Africans. Nature communications 2015;6:6342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Chen HM, Yu YN, Wang JL, et al. Decreased dietary fiber intake and structural alteration of gut microbiota in patients with advanced colorectal adenoma. Am J Clin Nutr 2013;97:1044–52. [DOI] [PubMed] [Google Scholar]
- 128.So D, Whelan K, Rossi M, et al. Dietary fiber intervention on gut microbiota composition in healthy adults: a systematic review and meta-analysis. Am J Clin Nutr 2018;107:965–983. [DOI] [PubMed] [Google Scholar]
- 129.Flemer B, Lynch DB, Brown JM, et al. Tumour-associated and non-tumour-associated microbiota in colorectal cancer. Gut 2017;66:633–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Flint HJ, Duncan SH, Scott KP, et al. Links between diet, gut microbiota composition and gut metabolism. Proc Nutr Soc 2015;74:13–22. [DOI] [PubMed] [Google Scholar]
- 131.Belenguer A, Duncan SH, Calder AG, et al. Two routes of metabolic cross-feeding between Bifidobacterium adolescentis and butyrate-producing anaerobes from the human gut. Appl Environ Microbiol 2006;72:3593–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Duncan SH, Louis P, Flint HJ. Lactate-utilizing bacteria, isolated from human feces, that produce butyrate as a major fermentation product. Appl Environ Microbiol 2004;70:5810–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Lin Y, Ma C, Bezabeh T, et al. (1) H NMR-based metabolomics reveal overlapping discriminatory metabolites and metabolic pathway disturbances between colorectal tumor tissues and fecal samples. Int J Cancer 2019. [DOI] [PubMed] [Google Scholar]
- 134.Lin Y, Ma C, Liu C, et al. NMR-based fecal metabolomics fingerprinting as predictors of earlier diagnosis in patients with colorectal cancer. Oncotarget 2016;7:29454–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Monleon D, Morales JM, Barrasa A, et al. Metabolite profiling of fecal water extracts from human colorectal cancer. NMR Biomed 2009;22:342–8. [DOI] [PubMed] [Google Scholar]
- 136.Vitaglione P, Mennella I, Ferracane R, et al. Whole-grain wheat consumption reduces inflammation in a randomized controlled trial on overweight and obese subjects with unhealthy dietary and lifestyle behaviors: role of polyphenols bound to cereal dietary fiber. Am J Clin Nutr 2015;101:251–61. [DOI] [PubMed] [Google Scholar]
- 137.Martinez I, Lattimer JM, Hubach KL, et al. Gut microbiome composition is linked to whole grain-induced immunological improvements. ISME J 2013;7:269–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Vuholm S, Nielsen DS, Iversen KN, et al. Whole-Grain Rye and Wheat Affect Some Markers of Gut Health without Altering the Fecal Microbiota in Healthy Overweight Adults: A 6-Week Randomized Trial. J Nutr 2017;147:2067–2075. [DOI] [PubMed] [Google Scholar]
- 139.Roager HM, Vogt JK, Kristensen M, et al. Whole grain-rich diet reduces body weight and systemic low-grade inflammation without inducing major changes of the gut microbiome: a randomised cross-over trial. Gut 2019;68:83–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Mehta RS, Nishihara R, Cao Y, et al. Association of Dietary Patterns With Risk of Colorectal Cancer Subtypes Classified by Fusobacterium Nucleatum in Tumor Tissue. JAMA oncology 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Brown JM, Hazen SL. Microbial modulation of cardiovascular disease. Nat Rev Microbiol 2018;16:171–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Wang Z, Bergeron N, Levison BS, et al. Impact of chronic dietary red meat, white meat, or non-meat protein on trimethylamine N-oxide metabolism and renal excretion in healthy men and women. Eur Heart J 2019;40:583–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Shan Z, Sun T, Huang H, et al. Association between microbiota-dependent metabolite trimethylamine-N-oxide and type 2 diabetes. Am J Clin Nutr 2017;106:888–894. [DOI] [PubMed] [Google Scholar]
- 144.Heianza Y, Ma W, Manson JE, et al. Gut Microbiota Metabolites and Risk of Major Adverse Cardiovascular Disease Events and Death: A Systematic Review and Meta-Analysis of Prospective Studies. J Am Heart Assoc 2017;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Schiattarella GG, Sannino A, Toscano E, et al. Gut microbe-generated metabolite trimethylamine-N-oxide as cardiovascular risk biomarker: a systematic review and dose-response meta-analysis. Eur Heart J 2017;38:2948–2956. [DOI] [PubMed] [Google Scholar]
- 146.Wirbel J, Pyl PT, Kartal E, et al. Meta-analysis of fecal metagenomes reveals global microbial signatures that are specific for colorectal cancer. Nat Med 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Nitter M, Norgard B, de Vogel S, et al. Plasma methionine, choline, betaine, and dimethylglycine in relation to colorectal cancer risk in the European Prospective Investigation into Cancer and Nutrition (EPIC). Ann Oncol 2014;25:1609–15. [DOI] [PubMed] [Google Scholar]
- 148.Cho E, Willett WC, Colditz GA, et al. Dietary choline and betaine and the risk of distal colorectal adenoma in women. J Natl Cancer Inst 2007;99:1224–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Myte R, Gylling B, Schneede J, et al. Components of One-carbon Metabolism Other than Folate and Colorectal Cancer Risk. Epidemiology 2016;27:787–96. [DOI] [PubMed] [Google Scholar]
- 150.Lee JE, Giovannucci E, Fuchs CS, et al. Choline and betaine intake and the risk of colorectal cancer in men. Cancer Epidemiol Biomarkers Prev 2010;19:884–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Bae S, Ulrich CM, Neuhouser ML, et al. Plasma choline metabolites and colorectal cancer risk in the Women’s Health Initiative Observational Study. Cancer Res 2014;74:7442–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Guertin KA, Li XS, Graubard BI, et al. Serum Trimethylamine N-oxide, Carnitine, Choline, and Betaine in Relation to Colorectal Cancer Risk in the Alpha Tocopherol, Beta Carotene Cancer Prevention Study. Cancer Epidemiol Biomarkers Prev 2017;26:945–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Randrianarisoa E, Lehn-Stefan A, Wang X, et al. Relationship of Serum Trimethylamine N-Oxide (TMAO) Levels with early Atherosclerosis in Humans. Sci Rep 2016;6:26745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Barrea L, Annunziata G, Muscogiuri G, et al. Trimethylamine-N-oxide (TMAO) as Novel Potential Biomarker of Early Predictors of Metabolic Syndrome. Nutrients 2018;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Heianza Y, Sun D, Li X, et al. Gut microbiota metabolites, amino acid metabolites and improvements in insulin sensitivity and glucose metabolism: the POUNDS Lost trial. Gut 2019;68:263–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Bayerdorffer E, Mannes GA, Ochsenkuhn T, et al. Unconjugated secondary bile acids in the serum of patients with colorectal adenomas. Gut 1995;36:268–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Bayerdorffer E, Mannes GA, Richter WO, et al. Increased serum deoxycholic acid levels in men with colorectal adenomas. Gastroenterology 1993;104:145–51. [DOI] [PubMed] [Google Scholar]
- 158.Cross AJ, Moore SC, Boca S, et al. A prospective study of serum metabolites and colorectal cancer risk. Cancer 2014;120:3049–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Shabanzadeh DM, Sorensen LT, Jorgensen T. Association Between Screen-Detected Gallstone Disease and Cancer in a Cohort Study. Gastroenterology 2017;152:1965–1974 e1. [DOI] [PubMed] [Google Scholar]
- 160.Bernstein H, Bernstein C, Payne CM, et al. Bile acids as endogenous etiologic agents in gastrointestinal cancer. World J Gastroenterol 2009;15:3329–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Staels B, Fonseca VA. Bile acids and metabolic regulation: mechanisms and clinical responses to bile acid sequestration. Diabetes Care 2009;32 Suppl 2:S237–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Franzosa EA, Sirota-Madi A, Avila-Pacheco J, et al. Gut microbiome structure and metabolic activity in inflammatory bowel disease. Nat Microbiol 2019;4:293–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Owczarek D, Rodacki T, Domagala-Rodacka R, et al. Diet and nutritional factors in inflammatory bowel diseases. World J Gastroenterol 2016;22:895–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Kanazawa K, Konishi F, Mitsuoka T, et al. Factors influencing the development of sigmoid colon cancer. Bacteriologic and biochemical studies. Cancer 1996;77:1701–6. [DOI] [PubMed] [Google Scholar]
- 165.Yazici C, Wolf PG, Carroll TP, et al. 511 Bilophila wadsworthia Is More Abundant in the Colonic Microbiome of Colorectal Cancer Cases Compared to Healthy Controls. Gastroenterology 2015;148:S-100. [Google Scholar]
- 166.Yazici C, Wolf PG, Kim H, et al. Race-dependent association of sulfidogenic bacteria with colorectal cancer. Gut 2017;66:1983–1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.L.H. N, Abu-Ali G, Mehta RS, et al. Dietary patterns, sulfur intake, and the abundance of sulfate-reducing bacteria. Digestive Disease Week; San Diego, CA, 2019. [Google Scholar]
- 168.Etemadi A, Abnet CC, Graubard BI, et al. Anatomical subsite can modify the association between meat and meat compounds and risk of colorectal adenocarcinoma: Findings from three large US cohorts. Int J Cancer 2018;143:2261–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Bernstein AM, Song M, Zhang X, et al. Processed and Unprocessed Red Meat and Risk of Colorectal Cancer: Analysis by Tumor Location and Modification by Time. PLoS One 2015;10:e0135959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Zeng H, Umar S, Rust B, et al. Secondary Bile Acids and Short Chain Fatty Acids in the Colon: A Focus on Colonic Microbiome, Cell Proliferation, Inflammation, and Cancer. Int J Mol Sci 2019;20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Vanegas SM, Meydani M, Barnett JB, et al. Substituting whole grains for refined grains in a 6-wk randomized trial has a modest effect on gut microbiota and immune and inflammatory markers of healthy adults. Am J Clin Nutr 2017;105:635–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Wu S, Zhang YG, Lu R, et al. Intestinal epithelial vitamin D receptor deletion leads to defective autophagy in colitis. Gut 2015;64:1082–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Mehta RS, Abu-Ali GS, Drew DA, et al. Stability of the human faecal microbiome in a cohort of adult men. Nat Microbiol 2018;3:347–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Devkota S, Wang Y, Musch MW, et al. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10−/− mice. Nature 2012;487:104–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Joossens M, Huys G, Cnockaert M, et al. Dysbiosis of the faecal microbiota in patients with Crohn’s disease and their unaffected relatives. Gut 2011;60:631–637. [DOI] [PubMed] [Google Scholar]
- 176.Marchesi JR, Dutilh BE, Hall N, et al. Towards the Human Colorectal Cancer Microbiome. PLoS ONE 2011;6:e20447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Fleming-Dutra KE, Hersh AL, Shapiro DJ, et al. Prevalence of inappropriate antibiotic prescriptions among us ambulatory care visits, 2010–2011. JAMA 2016;315:1864–1873. [DOI] [PubMed] [Google Scholar]
- 178.Cianflone NFC. Salmonellosis and the GI Tract: More than Just Peanut Butter. Current gastroenterology reports 2008;10:424–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Yang WH, Heithoff DM, Aziz PV, et al. Recurrent infection progressively disables host protection against intestinal inflammation. Science 2017;358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Lu R, Wu S, Zhang YG, et al. Salmonella Protein AvrA Activates the STAT3 Signaling Pathway in Colon Cancer. Neoplasia 2016;18:307–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Lu R, Bosland M, Xia Y, et al. Presence of Salmonella AvrA in colorectal tumor and its precursor lesions in mouse intestine and human specimens. Oncotarget 2017;8:55104–55115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Ahmed I, Chandrakesan P, Tawfik O, et al. Critical roles of Notch and Wnt/beta-catenin pathways in the regulation of hyperplasia and/or colitis in response to bacterial infection. Infect Immun 2012;80:3107–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Ahmed I, Roy BC, Raach RT, et al. Enteric infection coupled with chronic Notch pathway inhibition alters colonic mucus composition leading to dysbiosis, barrier disruption and colitis. PLoS One 2018;13:e0206701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Yang Y, Weng W, Peng J, et al. Fusobacterium nucleatum Increases Proliferation of Colorectal Cancer Cells and Tumor Development in Mice by Activating Toll-Like Receptor 4 Signaling to Nuclear Factor-kappaB, and Up-regulating Expression of MicroRNA-21. Gastroenterology 2017;152:851–866 e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Boleij A, Tjalsma H. The itinerary of Streptococcus gallolyticus infection in patients with colonic malignant disease. Lancet Infect Dis 2013;13:719–24. [DOI] [PubMed] [Google Scholar]
- 186.Wong SH, Kwong TNY, Chow TC, et al. Quantitation of faecal Fusobacterium improves faecal immunochemical test in detecting advanced colorectal neoplasia. Gut 2017;66:1441–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Baxter NT, Ruffin MTt, Rogers MA, et al. Microbiota-based model improves the sensitivity of fecal immunochemical test for detecting colonic lesions. Genome Med 2016;8:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Dadkhah E, Sikaroodi M, Korman L, et al. Gut microbiome identifies risk for colorectal polyps. BMJ Open Gastroenterol 2019;6:e000297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Alexander JL, Wilson ID, Teare J, et al. Gut microbiota modulation of chemotherapy efficacy and toxicity. Nat Rev Gastroenterol Hepatol 2017;14:356–365. [DOI] [PubMed] [Google Scholar]
- 190.Iida N, Dzutsev A, Stewart CA, et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science 2013;342:967–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Scott TA, Quintaneiro LM, Norvaisas P, et al. Host-Microbe Co-metabolism Dictates Cancer Drug Efficacy in C. elegans. Cell 2017;169:442–456 e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Garcia-Gonzalez AP, Ritter AD, Shrestha S, et al. Bacterial Metabolism Affects the C. elegans Response to Cancer Chemotherapeutics. Cell 2017;169:431–441 e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Deng X, Li Z, Li G, et al. Comparison of Microbiota in Patients Treated by Surgery or Chemotherapy by 16S rRNA Sequencing Reveals Potential Biomarkers for Colorectal Cancer Therapy. Front Microbiol 2018;9:1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Sivan A, Corrales L, Hubert N, et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science 2015;350:1084–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Routy B, Le Chatelier E, Derosa L, et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 2018;359:91–97. [DOI] [PubMed] [Google Scholar]
- 196.Gopalakrishnan V, Spencer CN, Nezi L, et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 2018;359:97–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Dubin K, Callahan MK, Ren B, et al. Intestinal microbiome analyses identify melanoma patients at risk for checkpoint-blockade-induced colitis. Nat Commun 2016;7:10391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Wang Y, Wiesnoski DH, Helmink BA, et al. Fecal microbiota transplantation for refractory immune checkpoint inhibitor-associated colitis. Nat Med 2018;24:1804–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Song M, Wu K, Meyerhardt JA, et al. Fiber Intake and Survival After Colorectal Cancer Diagnosis. JAMA Oncol 2018;4:71–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Cockbain AJ, Toogood GJ, Hull MA. Omega-3 polyunsaturated fatty acids for the treatment and prevention of colorectal cancer. Gut 2012;61:135–49. [DOI] [PubMed] [Google Scholar]
- 201.Song M, Zhang X, Meyerhardt JA, et al. Marine omega-3 polyunsaturated fatty acid intake and survival after colorectal cancer diagnosis. Gut 2017;66:1790–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Van Blarigan EL, Ou FS, Niedzwiecki D, et al. Dietary Fat Intake after Colon Cancer Diagnosis in Relation to Cancer Recurrence and Survival: CALGB 89803 (Alliance). Cancer Epidemiol Biomarkers Prev 2018;27:1227–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Yang W, Ma Y, Smith-Warner S, et al. Calcium Intake and Survival after Colorectal Cancer Diagnosis. Clin Cancer Res 2019;25:1980–1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Zgaga L, Theodoratou E, Farrington SM, et al. Plasma vitamin D concentration influences survival outcome after a diagnosis of colorectal cancer. J Clin Oncol 2014;32:2430–9. [DOI] [PubMed] [Google Scholar]
- 205.Vaughan-Shaw PG, Zgaga L, Ooi LY, et al. Low plasma vitamin D is associated with adverse colorectal cancer survival after surgical resection, independent of systemic inflammatory response. Gut 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Guercio BJ, Sato K, Niedzwiecki D, et al. Coffee Intake, Recurrence, and Mortality in Stage III Colon Cancer: Results From CALGB 89803 (Alliance). J Clin Oncol 2015;33:3598–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Hu Y, Ding M, Yuan C, et al. Association Between Coffee Intake After Diagnosis of Colorectal Cancer and Reduced Mortality. Gastroenterology 2018;154:916–926 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Song M, Chan AT. The Potential Role of Exercise and Nutrition in Harnessing the Immune System to Improve Colorectal Cancer Survival. Gastroenterology 2018;155:596–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Tjalsma H, Boleij A, Marchesi JR, et al. A bacterial driver-passenger model for colorectal cancer: beyond the usual suspects. Nature reviews. Microbiology 2012;10:575–82. [DOI] [PubMed] [Google Scholar]
- 210.Sinha R, Ahsan H, Blaser M, et al. Next steps in studying the human microbiome and health in prospective studies, Bethesda, MD, May 16–17, 2017. Microbiome; 2018;6:210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Scanlan PD, Shanahan F, Clune Y, et al. Culture-independent analysis of the gut microbiota in colorectal cancer and polyposis. Environ Microbiol 2008;10:789–98. [DOI] [PubMed] [Google Scholar]
- 212.Sobhani I, Tap J, Roudot-Thoraval F, et al. Microbial dysbiosis in colorectal cancer (CRC) patients. PLoS One 2011;6:e16393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Sanapareddy N, Legge RM, Jovov B, et al. Increased rectal microbial richness is associated with the presence of colorectal adenomas in humans. ISME J 2012;6:1858–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Chen W, Liu F, Ling Z, et al. Human intestinal lumen and mucosa-associated microbiota in patients with colorectal cancer. PLoS One 2012;7:e39743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Dejea CM, Wick EC, Hechenbleikner EM, et al. Microbiota organization is a distinct feature of proximal colorectal cancers. Proc Natl Acad Sci U S A 2014;111:18321–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Burns MB, Lynch J, Starr TK, et al. Virulence genes are a signature of the microbiome in the colorectal tumor microenvironment. Genome Med 2015;7:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Lu Y, Chen J, Zheng J, et al. Mucosal adherent bacterial dysbiosis in patients with colorectal adenomas. Sci Rep 2016;6:26337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Hale VL, Chen J, Johnson S, et al. Shifts in the Fecal Microbiota Associated with Adenomatous Polyps. Cancer Epidemiol Biomarkers Prev 2017;26:85–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Brennan CA, Garrett WS. Fusobacterium nucleatum - symbiont, opportunist and oncobacterium. Nat Rev Microbiol 2019;17:156–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Sears CL, Pardoll DM. Perspective: alpha-bugs, their microbial partners, and the link to colon cancer. J Infect Dis 2011;203:306–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Chung L, Orberg ET, Geis AL, et al. Bacteroides fragilis Toxin Coordinates a Procarcinogenic Inflammatory Cascade via Targeting of Colonic Epithelial Cells. Cell Host Microbe 2018;23:421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Wilson MR, Jiang Y, Villalta PW, et al. The human gut bacterial genotoxin colibactin alkylates DNA. Science 2019;363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Donohoe DR, Holley D, Collins LB, et al. A gnotobiotic mouse model demonstrates that dietary fiber protects against colorectal tumorigenesis in a microbiota- and butyrate-dependent manner. Cancer Discov 2014;4:1387–97. [DOI] [PMC free article] [PubMed] [Google Scholar]