Abstract
Purinergic signaling plays important roles in bone. P2X5, a member of ligand-gated ion channel receptors, has been demonstrated to regulate osteoclast maturation. However, the molecular mechanism of P2X5-mediated osteoclast regulation remains unclear. Here, we identified methylosome protein 50 (MEP50), a critical cofactor of the protein arginine methyltransferase 5 (PRMT5), as a P2X5-associating molecule. RNAi-mediated knockdown of MEP50 results in decreased formation of mature osteoclasts. MEP50 associates with P2X5, and this association requires the C-terminal intracellular region of P2X5. Additionally, impaired maturation of P2X5-deficient osteoclasts could be restored by transduction of full-length P2X5, but not a C-terminal deletion mutant of P2X5. These results indicate that P2X5 associates with MEP50 and suggest a link between the PRMT5 complex and P2X5 signaling in osteoclast maturation.
Keywords: Osteoclast, maturation, P2X5, RNAi-mediated knockdown, MEP50, PRMT5, Methylosome
Introduction
Bone homeostasis is maintained by the balanced functions of bone-forming osteoblasts and bone-resorbing osteoclasts [1]. Osteoclasts are vital for maintaining a healthy skeleton, but excessive osteoclast activity is associated with numerous pathophysiological processes in bone [2, 3]. Understanding the processes that control osteoclast biology is necessary to address these disease conditions. Significant progress has been made characterizing key factors, such as the receptor activator of NF-κB ligand (RANKL) - receptor activator of NF-κB (RANK) pathway, that act as major regulators of osteoclast differentiation [4, 5]. It is now important to identify and characterize additional molecular mechanisms employed by osteoclasts to fine-tune processes of differentiation and maturation.
The P2X family of purinergic receptors form ligand-gated ion channels [6]. Purinergic signaling plays a key role not only in the physiologic regulation of bone homeostasis but also in the pathophysiological processes leading to bone loss and bone fragility [7]. Seven P2X receptors (P2X1-P2X7) have been identified, and multiple P2X purinergic receptors have been detected in the major bone cell types (i.e., osteoclasts, osteoblasts, and osteocytes). In osteoclasts, P2X7 has been the most closely studied [8–12]. However, P2X7-deficient mice exhibit relatively unremarkable bone-related phenotypes both in vitro and in vivo [13]. Recently, we have demonstrated a critical role for P2X5 in osteoclast maturation [14], with P2X5 deficiency resulting in impaired osteoclast maturation in vitro. Although P2X5-deficient mice exhibit normal bone phenotypes under homeostatic conditions in vivo, P2X5 deficiency was found to provide protection against LPS-induced inflammatory bone loss [14]. Additionally, using an experimental periodontitis model, we found that P2X5 deficiency prevented inflammation-induced alveolar bone loss [15]. We further demonstrated that P2X5-mediated inflammasome activation leads to mature IL-1β production [14], but it remains unclear whether this represents the only, or even primary mechanism by which P2X5 controls osteoclast maturation. Therefore, in this study, we sought to identify novel molecules that associate with P2X5, and identified methylosome protein 50 (MEP50, also known as p44 and WDR77), a critical cofactor in the protein arginine methyltransferase 5 (PRMT5) methylosome complex [16, 17]. Recently, it has been shown by both chemical inhibition and siRNA knockdown that PRMT5 activity is required for optimal RANKL-induced osteoclast differentiation [18]. We show here that knockdown of the PRMT5 cofactor MEP50 results in defective osteoclast differentiation, and we demonstrate that MEP50 associates with P2X5 in a manner that requires the P2X5 C-terminal intracellular region. Impaired osteoclast maturation in P2X5-deficient cells was restored by retroviral transduction of full-length P2X5, but not by transduction of a C-terminal deletion mutant of P2X5 (P2X5Δ268). Taken together, our results suggest possible interaction between the methylosome signaling and purinergic receptor signaling during osteoclast maturation.
Materials and methods
Cell culture and mice.
HEK293T cells were maintained in DMEM containing 10% fetal bovine serum. P2rx5−/− mice were generated as previously described [14]. IL-1R−/− and Myd88−/− mice were purchased from the Jackson Laboratory. Wild-type, P2rx5−/−, IL-1R−/−, and Myd88−/− mouse bone marrow-derived monocytes (BMMs) and osteoclasts were prepared as described previously [14]. In brief, whole bone marrow cells were extracted from the femurs and tibias of mice and incubated in 100 mm petri dishes in α-MEM medium containing 10% fetal bovine serum and Macrophage Colony-Stimulating Factor 1 (M-CSF) (5 ng/ml) overnight. Non-adherent cells were collected and cultured for 3 days in 100 mm petri dishes in α-MEM medium containing 10% fetal bovine serum with M-CSF (60 ng/ml) to generate BMMs. For osteoclast differentiation, BMMs were washed with PBS and detached using enzyme-free cell dissociation buffer (Millipore) at 37 °C for 5 min. After generation of single-cell suspensions, cells were counted and suspended in α-MEM medium containing 10% fetal bovine serum with M-CSF (60 ng/ml) and RANKL (150 ng/ml), and cultured at 2 × 104/well in 96-well cell culture plates for 3 days. After culture for 3 days, the cells were fixed with 3.7% formaldehyde in PBS for tartrate-resistant acidic phosphatase (TRAP) activity and stained for TRAP using the Acid, Phosphatase, Leukocyte (tartrate-resistant acid phosphatase Kit (387A-1KT, Sigma)) following the manufacturer’s instructions. All mice were maintained and used in accordance with guidelines approved by the Institutional Animal Care and Use Committee (IACUC) at the University of Pennsylvania.
Mass spectrometry
HEK293T cells transiently transfected with a pCDNA vector encoding C-terminally Flag-tagged mouse P2X5 were lysed with ice cold lysis buffer (25 mM Tris-HCl pH7.4, 150 mM NaCl, 1% NP-40, 1 mM EDTA, 5% glycerol, and protease and phosphatase inhibitor cocktail (Roche)), and total cell lysates were pull-downed with anti-Flag antibody-conjugated beads followed by elution with 3x Flag peptide. Eluates were fractionated by SDS-PAGE on 4–12% gradient gels, stained with Coomassie blue R-250, and gel section ranges in size from 30 to 160 kDa were excised. The excised gel sections were subjected to MALDI-MS/MS analysis by Applied Biomics.
Real-time PCR (QPCR) analysis
Total RNA was extracted from cells with Trizol reagent (Invitrogen), and 1–5 μg of total RNA was reverse transcribed by using random hexamer primers and SuperScript III reverse transcriptase (Invitrogen). cDNA was analyzed by q-PCR using Quant Studio3 (Applied Biosystems) with the following TaqMan probes: MEP50 (Mm01296590_g1), Nuclear Factor of Activated T Cells 1 (Nfatc1) (Mm00479445_m1), Acid Phosphatase 5 Tartrate Resistant (Acp5, also known as TRAP5a) (Mm00475698_m1), and Cathepsin K (Ctsk) (Mm00484036_m1). 18s (Hs99999901_s1) was used as a reference gene for data normalization.
Transfection and retrovirus infection
HEK293T cells were cultured at 7 × 105/well in 6 well plates in DMEM containing 10% fetal bovine serum on the day before transfection. On the day of transfection, HEK293T cells were transfected with 1.5 μg each of the following pCDNA expression vectors using PEImax (Polysciences) at a DNA/PEImax ratio of 1:4; N-terminally HA-tagged mouse P2X5, N-terminally HA-tagged human P2X5, C-terminally Flag-tagged mouse P2X5, C-terminally Flag-tagged mouse P2X5Δ268, C-terminally Flag-tagged C-terminal intracellular region of P2×5 fused with human CD3 extracellular-transmembrane domain (mP2×5C-intra), C-terminally Flag-tagged mouse MEP50, and C-terminally Myc-tagged mouse MEP50. After 2 days, cells were used for Co-IP experiments. To prepare retroviral particles, Plat-E packaging cells were cultured at 7 × 106 in 100 mm dish in α-MEM medium containing 10% fetal bovine serum on the day before transfection. On the day of transfection, Plat-E cells were transfected with 10 μg each of the pSuper vector encoding siRNA targets for mouse MEP50 (5’-GGTGTCACTAGACTGGTAT-3’), pMX vectors encoding C-terminally Flag-tagged mouse P2X5, and mouse P2X5Δ268 using PEImax at a DNA/PEImax ratio of 1:4. Empty pSuper and pMX vectors were used as a negative control. After 3 days, medium containing retrovirus was harvested and passed through a syringe filter (0.45 μm pore diameter). P2rx5−/− BMMs were cultured at 1 × 107 in 100 mm dish in α-MEM medium containing 10% fetal bovine serum with M-CSF (60 ng/ml) on the day before retrovirus transduction. On the day of transduction, medium containing retrovirus was mixed with hexadimethrine bromide (8 μg/ml) and M-CSF (60 ng/ml), added in the P2rx5−/− BMMs, and incubated for 16 h. After washing with fresh medium, infected BMMs were selected by culturing for 2 days in the presence of puromycin (2 μg/ml) with M-CSF (60 ng/ml). Puromycin-resistant BMMs were used for induction of osteoclast.
Co-Immunoprecipitation (Co-IP) and Western blot
Transfected HEK293T cells in 6 well plates were washed with ice-cold phosphate-buffered saline (PBS) and lysed with 200 μl of ice-cold lysis buffer (25 mM Tris-HCl pH7.4, 150 mM NaCl, 1% NP-40, 1 mM EDTA, 5% glycerol, and protease and phosphatase inhibitor cocktail (Roche)) and incubated for 5 min with periodic mixing. The lysates were transferred to a microcentrifuge tube and centrifuge at 13000 x g for 10 min at 4°C to remove debris, the supernatants were collected, and protein concentrations were determined using the Bradford assay. Equal amounts of lysates (2 mg of protein) were pull-downed using protein G beads conjugated with anti-epitope antibodies or control IgG for overnight. The beads were washed and fractionated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) on 4–12% gradient gels and transferred onto polyvinyl difluoride (PVDF) membranes. Western blotting was performed with the following antibodies: anti- Flag: M2 (Sigma-Aldrich), anti-HA: sc805 (Santacruz), anti-Myc: sc-40 (Santacruz), and anti-Actin: sc-47778 (Santacruz).
Statistical Analysis
Experiments were analyzed using one-way ANOVA by Prism 8.1 (GraphPad). P < 0.05 was considered statistically significant.
Results
Inflammasome-induced cytokine is dispensable for P2X5-mediated osteoclast maturation
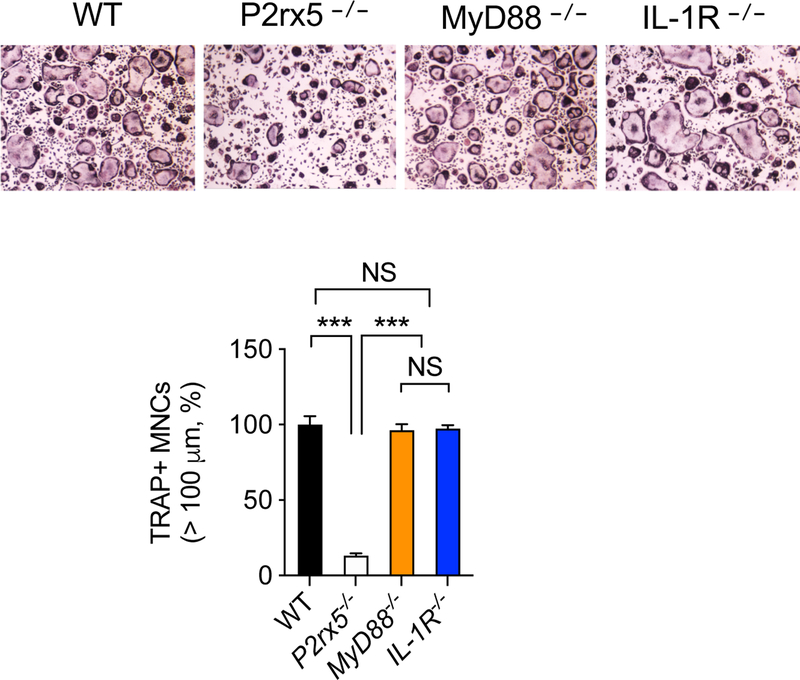
We have previously reported that P2X5 deficiency results in impaired osteoclast maturation that is associated with impaired inflammasome-induced IL-1β production. The impaired multinucleation could be rescued by adding exogenous IL-1β, suggesting a critical role of IL-1R signaling in P2X5-mediated osteoclast maturation [14]. To examine whether IL-1R signaling is required for osteoclast maturation, we prepared BMMs from IL-1R−/− mice and cultured with M-CSF + RANKL to generate osteoclasts in vitro, and found normal TRAP+ hyper-multinucleated (>100 μm) cells (i.e., mature osteoclasts) (Figure 1). In addition to IL-1β, inflammasome activation results in IL-18 production. Using BMMs lacking MyD88, the critical signaling mediator for both IL-1R and IL-18R (Figure 1), we found similar results as with IL-1R−/− osteoclasts (Figure 1), suggesting that although P2X5-dependent inflammasome-induced cytokine production may enhance osteoclast maturation in the absence of P2X5, it is not per se required for P2X5-regulated osteoclast maturation. Together, these results suggest the involvement of an additional signaling mechanism(s) in P2X5-mediated osteoclast maturation.
Figure 1.
Inflammasome-induced cytokine production is dispensable for P2X5-mediated osteoclast maturation. BMMs derived from wild-type, P2rx5−/−, Myd88−/−, and IL-1R−/− were cultured with M-CSF + RANKL for three days to induce osteoclasts. Frequency of TRAP+ hyper-multinucleated cells (>100 μm) are shown. Data are shown as mean ± S.D. ***; p<0.001, NS; not significant.
Identification of MEP50 as a P2X5-associating molecule
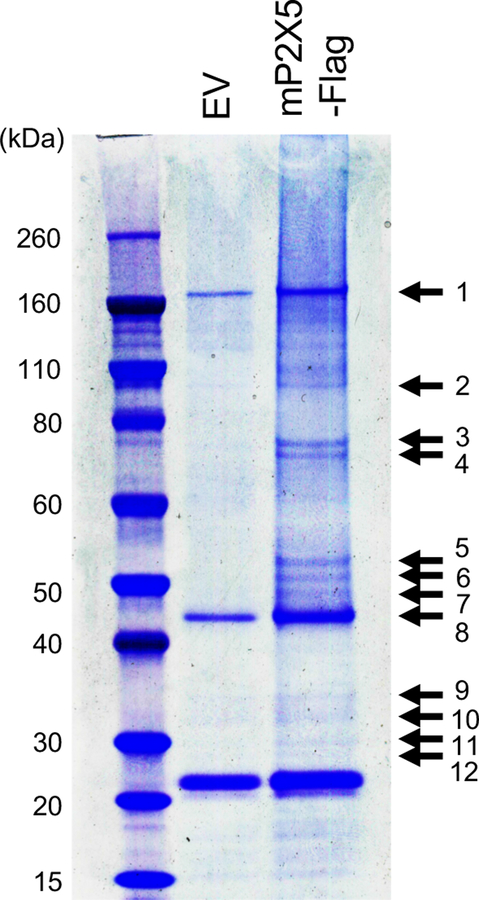
In order to identify novel P2X5-associated signaling mechanisms, we sought to first identify molecules that associate with P2X5, and therefore employed a Co-IP approach. We prepared lysates from HEK293T cells transfected with C-terminally Flag-tagged mouse full-length P2X5 (mP2X5), and pulled down mP2X5 and associating molecules with anti-Flag antibody. Flag peptide-eluates were separated by SDS-PAGE and stained with Coomassie blue R-250. We detected multiple mP2X5-associating proteins ranging in size from 30 to 160 kDa (Figure 2). To identify these molecules, we excised stained bands from the gel and subjected them to MALDI (matrix-assisted desorption/ionization) MS/MS mass spectrometry. Mascot analysis revealed two proteins, identified by multiple peptides with both high-confidence protein and ion scores (~100%), as human P2X5 in excised band 5, and human MEP50 in multiple excised bands (4, 5, 9 and 6) (Table 1). Identification of human P2X5 as an association molecule with mP2X5 suggested that mouse and human P2X5 could form oligomers.
Figure 2.
Identification of MEP50 as a P2X5-associating molecule. HEK293T cells transiently expressed C-terminal Flag-tagged mP2X5 (mP2×5-Flag) were lysed and subjected to immunoprecipitation with anti-Flag beads. HEK293T cells transfected with empty vector (EV) were used as a control. Elutes were separated by SDS-PAGE and visualized with Coomassie blue R-250. Arrows indicate bands subjected to MALDI-MS/MS mass spectrometry.
Table 1.
Identification of P2X5-associated proteins by MALDI-MS/MS mass spectrometry
| Band No. | Protein Name | Accession No. | Gene name | MW (kDa) | Matched Peptides | Protein Score C. I. % |
|---|---|---|---|---|---|---|
| 5 | P2Xpurinoceptor5 | Q93086 | P2RX5 | 47 | 14 | 100 |
| 4 | Methylosome protein 50 | Q9BQA1 | MEP50 | 37 | 10 | 100 |
| 9 | Methylosome protein 50 | Q9BQA1 | MEP50 | 37 | 9 | 100 |
| 5 | Methylosome protein50 | Q9BQA1 | MEP50 | 37 | 10 | 100 |
| 6 | Methylosome protein 50 | Q9BQA1 | MEP50 | 37 | 7 | 94.925 |
MEP50 expression is required for optimal osteoclast differentiation
To investigate whether MEP50 has biological relevance to osteoclast maturation, we first examined gene expression dynamics of MEP50 along with osteoclast differentiation marker genes, Nfatc1, Acp5, and Ctsk, by temporal qPCR analysis using in vitro-induced osteoclasts treated with M-CSF + RANKL. We found that expression of MEP50 was induced by RANKL and gradually increased during differentiation as with Nfatc1, Acp5 and Ctsk (Figure 3A). Next, we performed RNAi-mediated knockdown of mouse MEP50 in wild-type BMMs. BMMs retrovirally transduced with shRNA encoding mouse MEP50 showed significant reduction of RANKL-induced formation of TRAP+ hyper-multinucleated cells (Figure 3B, 3C). The expression of Nfatc1, Acp5 and Ctsk were also decreased in MEP50 knockdown cultures (Figure 3D). These results suggest involvement of MEP50 in osteoclast maturation, and therefore support further characterization of the relationship between MEP50 and P2X5 as a potential foundation for a novel mechanism of controlling P2X5-mediated osteoclast maturation.
Figure 3.
MEP50 expression is required for optimal osteoclast differentiation. (A) Expression dynamics of MEP50, Acp5, Nfatc1, and Ctsk during osteoclast differentiation was quantified by Q-PCR. (B, C, D) Wild-type BMMs retrovirally transduced with the indicated shRNAs were cultured with M-CSF + RANKL for three days to induce osteoclasts. (B) Knockdown efficiency of MEP50 was determined by Q-PCR. (C) Cells were stained for TRAP. TRAP activity, frequency of TRAP+ multinucleated cells (3 nuclei or more per cell), and TRAP+ hyper-multinucleated cells (>100 μm) are shown. (D) Relative expression of Acp5, Nfatc1, and Ctsk was determined by Q-PCR. Data are shown as mean ± S.D. **; p<0.01, ***; p<0.001.
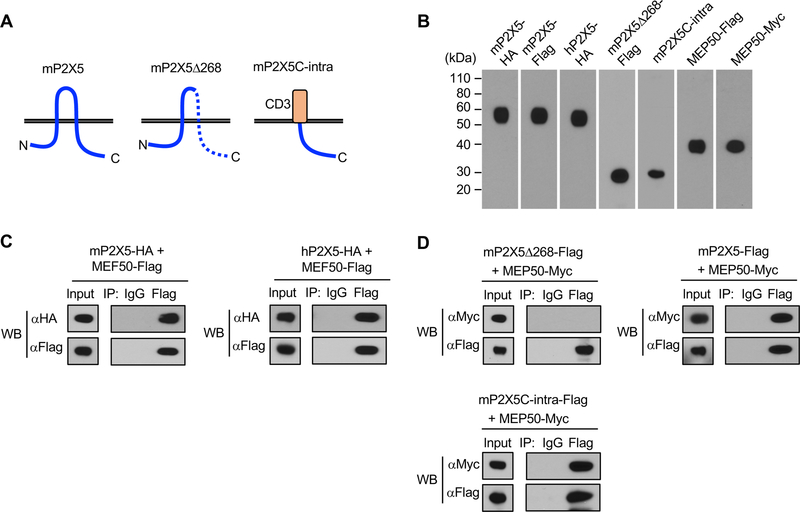
P2X5 associates with MEP50 through its C-terminal region
To confirm and further characterize the association of P2X5 with MEP50, we generated mouse MEP50 (mMEP50) expression vector, and transiently co-transfected into HEK293T cells with mouse mP2X5 expression vector (Figure 4A, 4B). By Co-IP and western blot analysis, we detected association of mP2X5 with mMEP50 (Figure 4C, 4D). We also detected association of human P2X5 (hP2X5) with mMEP50 (Figure 4C). To gain insight into the structural requirement for association with MEP50, we made expression vectors which encode a C-terminal deletion mutant of mouse P2X5 (mP2X5Δ268) or a chimeric protein in which the mouse P2X5 C-terminal intracellular region (mP2X5C-intra) is fused to the human CD3 extracellular-transmembrane region in order to stabilize expression (Figure 4A, 4B), and performed Co-IP experiments. As a result, we found association of mP2X5C-intra with mMEP50 (Figure 4D). By contrast, full human CD3 alone did not associate with MEP50 (supplemental figure 1), indicating the association of mP2X5C-intra with mMEP50 is mediated by P2X5 intracellular region. There was also no association observed between mP2X5Δ268 and mMEP50 (Figure 4D). Therefore, MEP50 interaction with P2X5 can be mapped to a specific C-terminal region of P2X5.
Figure 4.
P2X5 associates with MEP50 through its C-terminal region. (A) Schema of mP2X5, mP2X5Δ268, and mP2×5C-intra used for Co-IP. (B) Expression of the indicated vectors in 293T cells was confirmed by western blot. (C, D) Cell lysates transiently transfected with the indicated vectors were used for Co-IP with the indicated antibodies (IP) and detected with the indicated antibodies (WB). Input shows expression in whole cell lysates. (C) Association of mP2X5 with MEP50 (left) and hP2X5 with MEP50 (right) was observed by western blot. (D) Association of mP2×5C-intra with MEP50 was observed, while association of mP2X5Δ268 with MEP50 was not observed by western blot.
The P2X5 C-terminal region is required for osteoclast maturation
In order to determine whether the region of P2X5 required for interaction with MEP50 correlates with P2X5-dependent osteoclast maturation, we prepared BMMs from P2X5−/− mice, and retrovirally transduced with mP2X5 or mP2X5Δ268, followed by M-CSF and RANKL treatment to induce osteoclasts. Exogenous gene expression was confirmed by western blot (Figure 5A). As previously reported [14], P2X5−/− cells transduced with control retrovirus showed impaired formation of TRAP+ hyper-multinucleated cells, and retroviral transduction of mP2X5 completely rescued impaired formation of hyper-multinucleated cells (Figure 5B). By contrast, retroviral transduction of mP2X5Δ268 failed to rescue impaired hyper-multinucleation (Figure 5B). Taken together, these results suggest that P2X5 association with MEP50 requires the C-terminal region of P2X5, and the C-terminal region of P2X5 is required for osteoclast hyper-multinucleation. Our results support MEP50 as a potential mediator of P2X5-dependent signaling required for osteoclast maturation.
Figure 5.
The P2X5 C-terminal region is required for osteoclast maturation. (A) WT and P2rx5−/− BMMs were retrovirally transduced with the indicated vectors. Overexpression of transduced genes (mP2×5 and mP2×5Δ268) was confirmed by western blot. (B) WT and P2X5−/− BMMs retrovirally transduced with the indicated vectors were cultured with M-CSF + RANKL for three days to induce osteoclasts. TRAP activity, frequency of TRAP+ multinucleated cells (3 nuclei or more per cell), and frequency TRAP+ hyper-multinucleated cells (>100 μm) are shown. Data are shown as mean ± S.D. **; p<0.01, ***; p<0.001, NS; not significant.
Discussion
The purinergic receptor P2X5 has been shown to be a critical actor in regulating osteoclast maturation and associated hyper-multinucleation [7, 19]. Although P2X5-mediated regulation of the inflammasome and IL-1β production is associated with osteoclast maturation, our findings suggest the presence of an additional signaling mechanism(s) utilized by P2X5 to regulate osteoclast maturation (Figure 1). In this study, we identified the PRMT5 methylosome protein MEP50 as a P2X5-associated molecule. Using RNAi-mediated gene knockdown, we showed a role for MEP50 in osteoclast maturation. We also showed that the P2X5 C-terminal region is required for association with MEP50. Furthermore, impaired multinucleation observed in P2X5 deficiency was restored by retroviral transduction with full-length P2X5 but not a C-terminal deletion mutant. Our findings are consistent with the involvement of MEP50 in P2X5-mediated signaling in osteoclast maturation. These results also potentially represent a novel interaction between the purinergic signaling pathway and PRMT5 methylosome activity.
MEP50 was initially identified as an essential component of the 20 S protein arginine methyltransferase complex, termed the methylosome [20, 21]. MEP50 forms a tight complex with PRMT5, a type II PRMT enzyme which catalyzes symmetric demethylation by transferring the methyl group from S-adenosyl-L-methionine (SAM) to two of the three guanidine nitrogen atoms of an arginine residue. MEP50 binds PRMT5 to stabilize and increase its activity and also functions to recruit substrate proteins for methylation [16, 22]. Given that PRMT5 functions in protein complexes that invariably contain the MEP50, it is plausible that the PRMT5 methylosome complex is involved in P2X5-mediated purinergic signaling during osteoclast maturation. The PRMT5 methylosome complex has a vital role in a wide spectrum of cellular processes by methylating substrate proteins [23, 24], including NF-κB p65, SREBP1a, E2F1, and EGFR, which are all known to have important functions in osteoclasts [20, 25–28]. Interestingly, PRMT5 has been reported to be induced by RANKL stimulation, and involved in osteoclastogenesis through regulation of expression of chemokine C-X-C motif ligand 10 (Cxcl10) and antiviral protein RSAD2 [18]. Further studies will be required to confirm involvement of the PRMT5 methylosome complex in P2X5-mediated signaling, and to characterize how P2X5 regulates activation of the methylosome during osteoclast maturation.
In this study, we showed that the P2X5 C-terminal region is important for osteoclast multinucleation, and that the C-terminal region is required for association with MEP50. However, we cannot conclude that the importance of the C-terminal region is simply due to association with MEP50, because the C-terminal region may also be required for activation of the inflammasome and IL-1β production. Future studies will be required to determine the structural requirements within P2X5 for methylosome association versus inflammasome activation.
Taken together, we provide evidence here of association of the purinergic receptor P2X5 with the PRMT5 methylosome-related factor MEP50, potentially identifying a novel mechanism of cross-talk between purinergic signaling and the methylosome. Although a full understanding of how MEP50 regulates P2×5-mediated signaling required for osteoclast maturation remains unclear, these findings represent a novel link between the PRMT and P2X signaling pathways, and further, should contribute to clarification of the mechanism(s) of P2X5-mediated regulation of osteoclast maturation.
Supplementary Material
Full human CD3 does not associate with MEP50. (A) Expression of full human CD3 expression vector in 293T cells was confirmed by western blot. (B) Cell lysates transiently transfected with the indicated vectors were used for Co-IP with the indicated antibodies (IP) and detected with the indicated antibodies (WB). Input shows expression in whole cell lysates.
Acknowledgements
This study was supported in part by NIH grants (AR067726 and AR069546 to YC).
Footnotes
Conflicts of interest
The authors have no conflicting interests.
References
- 1.Teitelbaum SL (2000) Bone resorption by osteoclasts, Science. 289, 1504–8. [DOI] [PubMed] [Google Scholar]
- 2.Moutsopoulos NM, Konkel J, Sarmadi M, Eskan MA, Wild T, Dutzan N, Abusleme L, Zenobia C, Hosur KB, Abe T, Uzel G, Chen W, Chavakis T, Holland SM & Hajishengallis G (2014) Defective neutrophil recruitment in leukocyte adhesion deficiency type I disease causes local IL-17-driven inflammatory bone loss, Sci Transl Med. 6, 229ra40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Souza PP & Lerner UH (2013) The role of cytokines in inflammatory bone loss, Immunol Invest. 42, 555–622. [DOI] [PubMed] [Google Scholar]
- 4.Boyle WJ, Simonet WS & Lacey DL (2003) Osteoclast differentiation and activation, Nature. 423, 337–42. [DOI] [PubMed] [Google Scholar]
- 5.Boyce BF & Xing L (2007) The RANKL/RANK/OPG Pathway, Curr Osteoporos Rep. 5, 98–104. [DOI] [PubMed] [Google Scholar]
- 6.Zimmermann H (2016) Extracellular ATP and other nucleotides-ubiquitous triggers of intercellular messenger release, Purinergic signalling. 12, 25–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jorgensen NR (2019) Role of the purinergic P2X receptors in osteoclast pathophysiology, Curr Opin Pharmacol. 47, 97–101. [DOI] [PubMed] [Google Scholar]
- 8.Rumney RM, Wang N, Agrawal A & Gartland A (2012) Purinergic signalling in bone, Front Endocrinol (Lausanne). 3, 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gombault A, Baron L & Couillin I (2012) ATP release and purinergic signaling in NLRP3 inflammasome activation, Front Immunol. 3, 414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mariathasan S, Weiss DS, Newton K, McBride J, O’Rourke K, Roose-Girma M, Lee WP, Weinrauch Y, Monack DM & Dixit VM (2006) Cryopyrin activates the inflammasome in response to toxins and ATP, Nature. 440, 228–32. [DOI] [PubMed] [Google Scholar]
- 11.Orriss IR, Burnstock G & Arnett TR (2010) Purinergic signalling and bone remodelling, Curr Opin Pharmacol. 10, 322–30. [DOI] [PubMed] [Google Scholar]
- 12.Korcok J, Raimundo LN, Ke HZ, Sims SM & Dixon SJ (2004) Extracellular nucleotides act through P2X7 receptors to activate NF-kappaB in osteoclasts, Journal of bone and mineral research : the official journal of the American Society for Bone and Mineral Research. 19, 642–51. [DOI] [PubMed] [Google Scholar]
- 13.Gartland A, Buckley KA, Hipskind RA, Perry MJ, Tobias JH, Buell G, Chessell I, Bowler WB & Gallagher JA (2003) Multinucleated osteoclast formation in vivo and in vitro by P2X7 receptor-deficient mice, Crit Rev Eukaryot Gene Expr. 13, 243–53. [DOI] [PubMed] [Google Scholar]
- 14.Kim H, Walsh MC, Takegahara N, Middleton SA, Shin HI, Kim J & Choi Y (2017) The purinergic receptor P2X5 regulates inflammasome activity and hyper-multinucleation of murine osteoclasts, Scientific reports. 7, 196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim H, Kajikawa T, Walsh MC, Takegahara N, Jeong YH, Hajishengallis G & Choi Y (2018) The purinergic receptor P2X5 contributes to bone loss in experimental periodontitis, BMB Rep. 51, 468–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Antonysamy S, Bonday Z, Campbell RM, Doyle B, Druzina Z, Gheyi T, Han B, Jungheim LN, Qian Y, Rauch C, Russell M, Sauder JM, Wasserman SR, Weichert K, Willard FS, Zhang A & Emtage S (2012) Crystal structure of the human PRMT5:MEP50 complex, Proceedings of the National Academy of Sciences of the United States of America. 109, 17960–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ho MC, Wilczek C, Bonanno JB, Xing L, Seznec J, Matsui T, Carter LG, Onikubo T, Kumar PR, Chan MK, Brenowitz M, Cheng RH, Reimer U, Almo SC & Shechter D (2013) Structure of the arginine methyltransferase PRMT5-MEP50 reveals a mechanism for substrate specificity, PLoS One. 8, e57008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dong Y, Song C, Wang Y, Lei Z, Xu F, Guan H, Chen A & Li F (2017) Inhibition of PRMT5 suppresses osteoclast differentiation and partially protects against ovariectomy-induced bone loss through downregulation of CXCL10 and RSAD2, Cell Signal. 34, 55–65. [DOI] [PubMed] [Google Scholar]
- 19.Walsh MC, Takegahara N, Kim H & Choi Y (2018) Updating osteoimmunology: regulation of bone cells by innate and adaptive immunity, Nat Rev Rheumatol. 14, 146–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Antonysamy S (2017) The Structure and Function of the PRMT5:MEP50 Complex, Subcell Biochem. 83, 185–194. [DOI] [PubMed] [Google Scholar]
- 21.Hosohata K, Li P, Hosohata Y, Qin J, Roeder RG & Wang Z (2003) Purification and identification of a novel complex which is involved in androgen receptor-dependent transcription, Molecular and cellular biology. 23, 7019–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burgos ES, Wilczek C, Onikubo T, Bonanno JB, Jansong J, Reimer U & Shechter D (2015) Histone H2A and H4 N-terminal tails are positioned by the MEP50 WD repeat protein for efficient methylation by the PRMT5 arginine methyltransferase, The Journal of biological chemistry. 290, 9674–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bedford MT & Richard S (2005) Arginine methylation an emerging regulator of protein function, Mol Cell. 18, 263–72. [DOI] [PubMed] [Google Scholar]
- 24.Karkhanis V, Hu YJ, Baiocchi RA, Imbalzano AN & Sif S (2011) Versatility of PRMT5-induced methylation in growth control and development, Trends Biochem Sci. 36, 633–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Inoue K & Imai Y (2015) Fatostatin, an SREBP inhibitor, prevented RANKL-induced bone loss by suppression of osteoclast differentiation, Biochim Biophys Acta. 1852, 2432–41. [DOI] [PubMed] [Google Scholar]
- 26.Taverna S, Pucci M, Giallombardo M, Di Bella MA, Santarpia M, Reclusa P, Gil-Bazo I, Rolfo C & Alessandro R (2017) Amphiregulin contained in NSCLC-exosomes induces osteoclast differentiation through the activation of EGFR pathway, Scientific reports. 7, 3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murata K, Fang C, Terao C, Giannopoulou EG, Lee YJ, Lee MJ, Mun SH, Bae S, Qiao Y, Yuan R, Furu M, Ito H, Ohmura K, Matsuda S, Mimori T, Matsuda F, Park-Min KH & Ivashkiv LB (2017) Hypoxia-Sensitive COMMD1 Integrates Signaling and Cellular Metabolism in Human Macrophages and Suppresses Osteoclastogenesis, Immunity. 47, 66–79 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vaira S, Alhawagri M, Anwisye I, Kitaura H, Faccio R & Novack DV (2008) RelA/p65 promotes osteoclast differentiation by blocking a RANKL-induced apoptotic JNK pathway in mice, The Journal of clinical investigation. 118, 2088–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Full human CD3 does not associate with MEP50. (A) Expression of full human CD3 expression vector in 293T cells was confirmed by western blot. (B) Cell lysates transiently transfected with the indicated vectors were used for Co-IP with the indicated antibodies (IP) and detected with the indicated antibodies (WB). Input shows expression in whole cell lysates.