Abstract
Role of GADD34, a protein that is induced following cellular stress, in HIV-1 replication was investigated. GADD34 was induced during the late phase of HIV-1 infection. siRNA-knockdown of GADD34 stimulated whereas overexpression of GADD34 inhibited HIV-1 replication. GADD34 N-terminal ER-binding-helix amino acid region 1-192 alone was found to be sufficient for the inhibition of HIV-1 replication whereas protein-phosphatase -1-binding domain and eIF-2α-phosphatase activity of GADD34 were not crucial for anti-HIV-1 activity. GADD34 did not alter the HIV-1 RNA levels but reduced the viral protein expression suggesting that GADD34 interferes in HIV protein synthesis. Studies on the effect of HIV-1-5'-UTR and its mutants on a human promoter-driven luciferase expression indicated that GADD34-inhibition was mediated by 5’-UTR/TAR RNA, probably by modulating TAR RNA structure. In summary, our data support a novel function of GADD34 as a putative anti-HIV-1 restriction factor.
Keywords: HIV-1, GADD34, HIV-1 TAR RNA, HIV-1 5’-UTR, Translation
Introduction
Host cellular proteins play critical roles in modulating nearly all the steps of HIV-1 biology. While disrupting cellular protein functions critical for virus replication may provide novel antiviral mechanisms, similar goals may be achieved by identifying new host anti-viral restriction factors whose expression levels can be manipulated to obtain desired anti-viral effects. Although some of the restriction factors like APOBEC3G, BST-2/Tetherin, SAMHD, TRIM5α, MX2, and SLFN11 (Ballana and Este, 2015; Fletcher and Towers, 2013; Haller et al., 2015; Harris and Liddament, 2004; Li et al., 2012; Neil et al., 2008) have been described, it is likely that more are still unknown.
HIV-1 5'-UTR RNA represents a highly structured motif that contains a number of important functional elements including trans-activation responsive (TAR) RNA region. Apart from Tat, the TAR region also binds a number of cellular proteins that are crucial for efficient HIV-1 mRNA translation (Charnay et al., 2009; Guerrero et al., 2015; Lai et al., 2013). In addition to the primary mechanism of HIV-1 translation that involves the recognition of the 5′-cap structure of mRNAs by host translation initiation machinery, HIV-1 also utilizes a cap-independent mechanism that bypasses the conventional ribosomal scanning pathway and uses an RNA element called the internal ribosome entry site (IRES) located in the 5'-UTR RNA (Berkhout et al., 2011; Monette et al., 2013; Ricci et al., 2008; Vallejos et al., 2012). IRES-dependent pathways have also been known to be operative in a number of other viruses (Balvay et al., 2009; Lee et al., 2017; Yamamoto et al., 2017) and in the synthesis of cellular proteins especially during extreme physiological conditions such as endoplasmic reticulum (ER) stress (Spriggs et al., 2010).
The ER is essential for the folding and trafficking of proteins. Biological ER stressors like amino acid starvation, disturbed calcium homeostasis, hypoxia, and viral infections are known to induce protein synthesis to the levels that burden the ER capacity. This leads to the misfolding of proteins and the accumulation of unfolded or malfolded proteins in the ER and the activation of the unfolded protein response (UPR) pathway (Fullwood et al., 2012; Ron and Walter, 2007; Walter and Ron, 2011). One of the highlights of the UPR pathway is the activation of PERK, a kinase that phosphorylates the translation initiation factor eIF-2α, the accumulation of which inhibits global protein synthesis (Proud, 2005). Viruses also induce global protein shutdown through the integrated stress response (ISR) pathway, which is modulated by the activation of PKR or GCN2 kinases that phosphorylate eIF-2α (He, 2006; Tsalikis et al., 2013). The accumulation of phosphorylated-eIF-2α (p-eIF-2α) triggers the expression of GADD34 (PPP1R15A) (growth arrested DNA damage- induced protein), a member of a group of genes that are induced following stressful growth arrest conditions and treatment with DNA-damaging agents (Dalton et al., 2012). GADD34 is the regulatory subunit of protein phosphatase 1 (PP1) that forms a highly specific phosphatase holoenzyme in association with the catalytic subunit of PP1 and dephosphorylates the p-eIF-2α leading to the restoration of protein homeostasis (Chen et al., 2015; Choy et al., 2015; Connor et al., 2001; Novoa et al., 2001; Novoa et al., 2003). Recent studies have shown that GADD34 belongs to the family of intrinsically disordered proteins that lack ordered 3D structure (Choy et al., 2015). GADD34 is expressed at low levels in unstressed cells (Novoa et al., 2003), and its half-life is ≤1 h (Choy et al., 2015). We and others have shown that GADD34 is induced during innate immune response to viral pathogens or to double stranded RNA (dsRNA) (Ishaq and Natarajan, 2016; Minami et al., 2007). The role of GADD34 in regulating HIV-1 replication is unknown. In this report, we explored the role of GADD34 in modulating HIV-1 replication and found that overexpression of the GADD34 inhibits viral replication by transcriptional-independent mechanisms but is dependent on inhibition of HIV-1 5'-UTR TAR RNA mediated translation. The antiviral activity of GADD34 was located in the N-terminal terminal region of the protein and was independent of the p-eIF-2α phosphatase activity of the protein.
Results
HIV-1 infection induces GADD34 expression in the late phase of infection and overexpression of GADD34 inhibits HIV-1 replication.
During the physiological conditions GADD34 protein is expressed at low levels, and only during ER stress its expression is induced to detectable levels (Novoa et al., 2003). GADD34 is also an interferon-stimulated gene (ISG) hard-wired to the innate immune response and induced during immune response to viral pathogens or dsRNA (Ishaq and Natarajan, 2016; Minami et al., 2007). The role of GADD34 in regulating HIV-1 biology is unknown. To study the effect of HIV-1 infection on the cellular GADD34 expression, we monitored the effect of HIV-1 infection on the GADD34 protein and mRNA levels in MT-2 cells. These cells express low levels of endogenous GADD34 and can be readily infected with pNL4-3 derived HIV- 1 virus. Western blot analysis (Fig. 1A) of HIV-1 infected MT-2 cell lysates showed the induction of GADD34 when the HIV- 1 protein expression reached the peak levels. Similar results were obtained with the GADD34 mRNA levels (Fig. 1B). These data suggest that host response to HIV-1 infection to induce GADD34 is lacking during early infection. These results are not surprising because HIV-1 is known to evade innate immune recognition and elicits a poor host interferon response and ISG induction (Rasaiyaah et al., 2013).
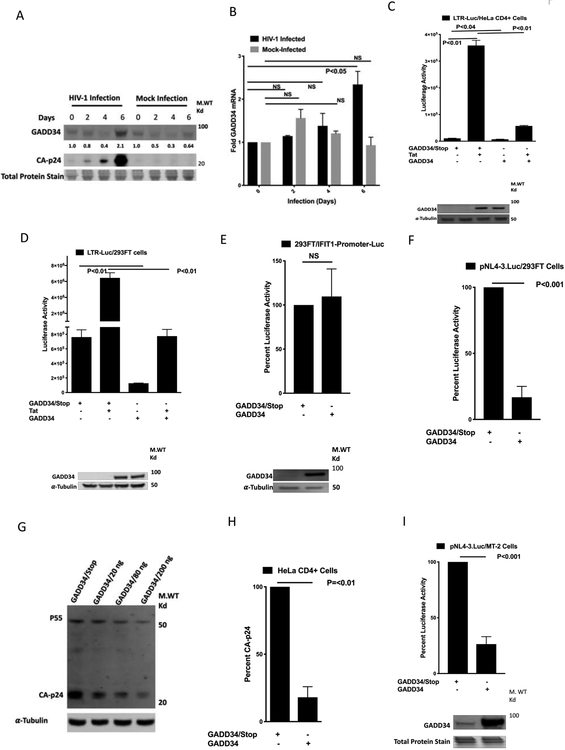
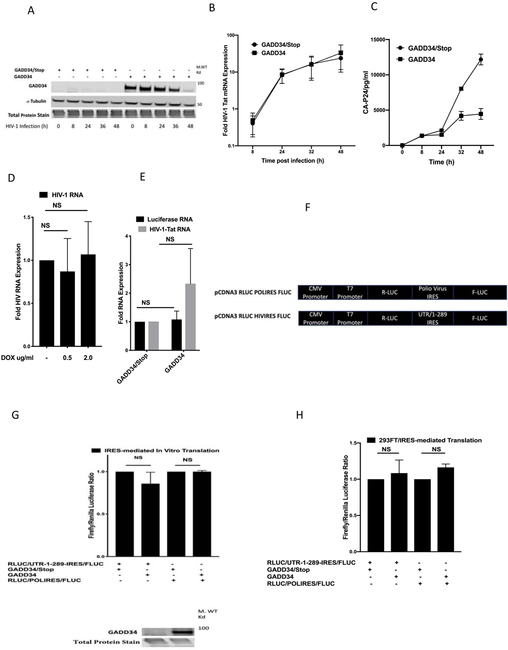
Figure 1. Overexpression of Human GADD34 inhibits Tat-mediated HIV1-LTR activity and HIV-1 replication.
MT-2 cells were mock infected or infected by spinoculation with 20 ng of p24-CA- antigen equivalent HIV-1-supernatant for indicated days. Cells were harvested for Western blot analysis using antibodies to GADD34, and CA-p24 (A) or RNA isolation and RT-PCR with GADD34 and GAPDH specific primers (B). C) HeLa CD4+ cells were transfected with 100 ng of indicated plasmid along with 100 ng of pBlue3'LTR-luc-A plasmid and 38 h later were harvested for luciferase measurement. D and E) 293FT cells were transfected with 100 ng of pBlue3'LTR-luc-A plasmid in the presence or absence of 50 ng of pcDNA3.1-TAT-1-101-FLAG (D) or 200 ng of IFIT1-Promoter-Luc plasmid (E) in the presence of 0.5 μg of indicated plasmids. Cells were harvested 38 h later for luciferase measurement. F) 293FT cells were transfected with 100 ng of pNL4-3.Luc.R-E- plasmid in the presence of 200 ng of indicated plasmids.. Cells were harvested 38 h later for luciferase measurement. G) 293FT cells were transfected with 0.25 μg of pNL4-3 plasmid in the presence of 200 ng of GADD34/Stop plasmid or indicated concentrations of GADD34 plasmid. Cells were harvested 38 h later for Western blot analysis using antibodies to HIV-1 CA-p24. H) HeLa CD4+ cells were transfected with 0.5 μg of indicated plasmids. Cells were infected 24 h later with 20 ng of CA-p24 antigen equivalent pNL.4-3/HIV-1-supernatant for 2h. The virus was removed, cells were washed with medium and infection continued for an additional 40 h before CA-p24 quantitation. I) MT-2 cells were electroporated with 2.5 μg of pNL4-3.Luc.R-E- plasmid in the presence of 5.0 μg indicated plasmids and harvested 36 h later for luciferase measurement and Western blot analysis with GADD34 specific antibodies.
Next, we explored the question whether cells overexpressing GADD34 early-on could interfere with HIV-1 replication. The effect of GADD34 overexpression on HIV-1 replication was studied by monitoring the effect of expression on 1) HIV-1-LTR-reporter (a luciferase reporter plasmid containing HIV-1 promoter and 5’-UTR/TAR region) activity or pNL4-3.Luc.R-E-reporter (an HIV-1 NL4-3 ΔEnv Vpr luciferase reporter plasmid) activity, 2) viral production in transfected cells using pNL4-3 (an HIV-1 NL4-3 infectious molecular clone expressing plasmid), and 3) viral replication in HIV-1-infected HeLa CD4+ cells. Initial studies on the effect of GADD34 on HIV-1 gene expression were carried out in HeLa and HEK-293 cells because these cells express low levels of GADD34 (Brush and Shenolikar, 2008) and are easily transfectable. Whereas in HeLa CD4+ cells, basal HIV-LTR activity was inhibited by GADD34 by nearly 30% (Fig. 1C), an 80% inhibition of basal HIV-LTR activity was observed in HEK-293FT cells expressing GADD34 (Fig. 1D). However, the Tat-mediated HIV-1-LTR luciferase activity was strongly inhibited in both cell types in the presence of GADD34 (Fig. 1C and 1D). The effect of GADD34 overexpression on a eukaryotic promoter reporter plasmid IFIT1-Promoter-Luc was studied as a control. No significant effect of GADD34 expression on luciferase activity was observed in cells transfected with IFIT1-Promoter-Luc (Fig. 1E). These data clearly showed that overexpression of GADD34 strongly interfered in Tat-mediated HIV-1-LTR activity while moderately affecting basal HIV-1-LTR activity, depending on the cell type. Studies with TZM-bl cells (HeLa CD4+ cells that contain integrated copies of the luciferase reporter under the control of the HIV-1 promoter) yielded similar results as with HeLa CD4+ cells (Fig. S1). Together, these data demonstrate that GADD34 overexpression interferes in HIV-1 LTR activity from unintegrated as well as integrated forms of HIV-1 LTR promoter. Next, we tested the effect of GADD34 on pNL4-3.Luc.R-E-plasmid, a full-length HIV proviral reporter in which firefly luciferase replaces the Nef gene and two frameshift mutations render it Env− and Vpr− (He et al., 1995). As shown in the Fig.1F, GADD34 strongly inhibited the pNL4-3.Luc-driven luciferase activity. To test the effect of GADD34 on virus production, 293FT cells were transfected with pNL4-3 plasmid, an infectious clone of HIV-1, in the presence of GADD34 expressing plasmid. A dose-dependent inhibition of cellular CA-p24 expression was observed in the presence of GADD34 (Fig. 1G). The quantitation of HIV-1 CA-p24 protein in cell supernatants indicated a strong dose-dependent inhibition of viral production by GADD34 (Fig. S2). To study the effect of GADD34 overexpression on HIV replication, a widely used HeLa CD4+ cell model (Mondor et al., 1998) was utilized. GADD34 transfected HeLa CD4+ cells were in infected with HIV-1. GADD34 strongly inhibited the virus replication in infected HeLa CD4+ cells as measured by CA-p24 assay in the cell supernatants (Fig. 1H).
Effect of GADD34 on HIV-1 gene expression in CD4+ T cells, MT-2 and Jurkat cells was also analyzed. As seen in Fig. 1I (MT-2 cells) and Fig. S3 (Jurkat cells), transfection in the presence of GADD34 strongly inhibited luciferase activity from pNL4-3.Luc as compared to transfection in the presence of a control GADD34 non-expressing plasmid. Together, these data indicate that GADD34 also inhibits HIV-1 gene expression in CD4+ T cell lines.
Tetracycline (doxycycline)-mediated induction of cloned GADD34 in HeLa cells inhibits HIV-1 LTR activity and viral gene expression.
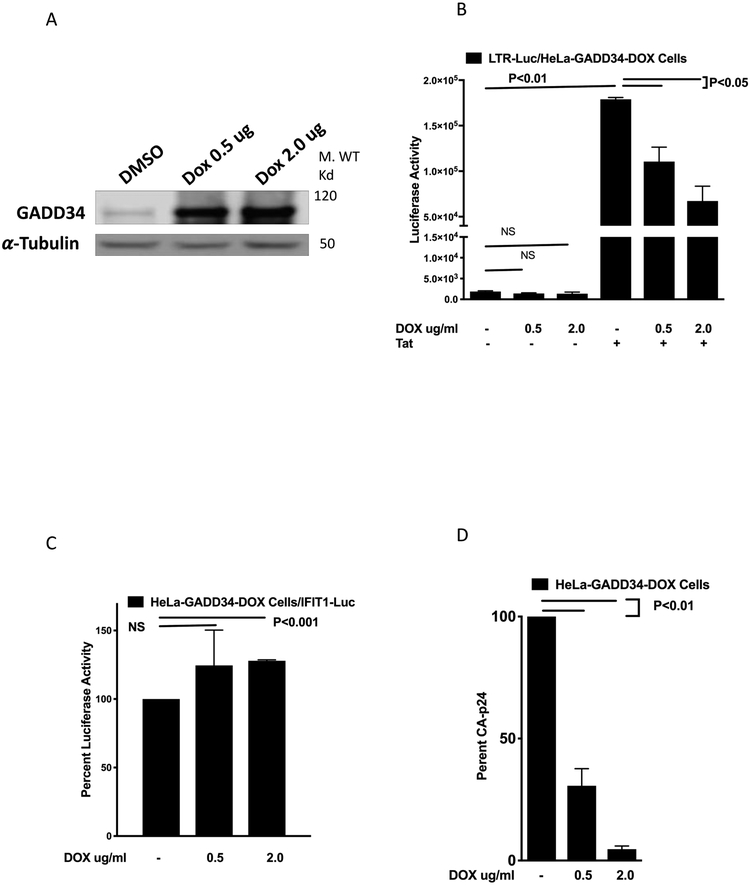
To explore the use of an inducible expression of GADD34, a tetracycline dependent transcriptional activation system in which the presence of tetracycline (doxycycline (DOX)) induces the expression of cloned GADD34 was employed to express GADD34 in HeLa cells and study its effect on HIV1-LTR-Luc activity and HIV-1 production. The treatment of HeLa-GADD34/DOX cells with DOX induced a dose dependent increase in GADD34 expression (Fig. 2A). HeLa-GADD34/DOX cells were transfected with HIV-1-LTR-reporter plasmid in the presence or absence of HIV-1 Tat plasmid and treated with DOX to induce GADD34 expression. Data (Fig. 2B) show that while there was no significant effect on Tat-independent luciferase activity, DOX treatment resulted in a dose-dependent reduction in Tat-mediated HIV-LTR promoter activity. The effect of DOX treatment on the activity of a cellular gene promoter (IFIT-Promoter-Luc), was studied as a control. DOX- induced GAAD34 did not inhibit the luciferase activity of this promoter but instead showed about 25% increase in the activity at 2.0 μg/ml dose (Fig. 2C). Next, the effect of DOX treatment on the HIV-1 production was studied by transfecting cells with pNL4-3 plasmid followed by treatment with DOX. A dose dependent decrease in CA-p24 levels in the cell supernatants was seen in cells following treatment with DOX (Fig. 2D). These results show that overexpression of GADD34 in an induced system also interferes in HIV-1-LTR-reporter activity and viral production.
Figure 2. Tetracycline (doxycycline)-mediated induction of cloned GADD34 in HeLa cells inhibits HIV-LTR activity and HIV replication.
A) Tet-On HeLa-GADD34/DOX were treated with DMSO or 0.5 μg /ml and 2 μg /ml of DOX for 18 h and cells were harvested for Western blot analysis using antibodies against GADD34 and α-Tubulin. B) Tet-On HeLa-GADD34/DOX cells were transfected with 100 ng pBlue3'LTR-luc-A plasmid in the presence or absence of 100 ng pcDNA3.1-TAT-1-101-FLAG plasmid. After 18 h DOX was added as indicated and cells were incubated for another 18 h before harvest and luciferase measurement. C) Tet-On HeLa-GADD34/DOX cells were transfected with 200 ng of IFIT1-Promoter-Luc plasmid. After 24 h cells were treated with DOX as indicated for 18 h before harvest and luciferase measurement. D) Tet-On HeLa-GADD34/DOX cells were transfected with 1.0 μg of pNL4-3 plasmid and treated with DOX as indicated. Cells were harvested 38 h later for CA-p24 quantitation.
siRNA-mediated knockdown of GADD34 expression induces HIV-1 replication.
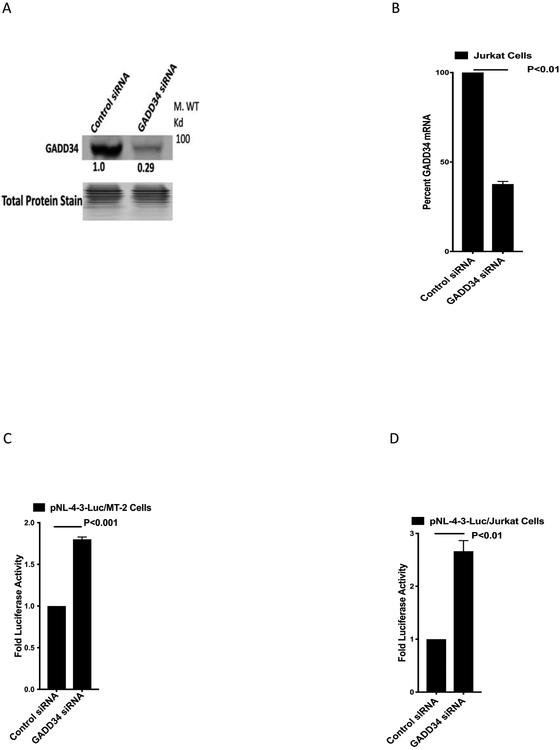
Because GADD34 overexpression inhibits HIV-1 replication and HIV-1 infection does not induce GADD34 expression till late in the infection, the data suggests that HIV-1 replication could be induced in cells deprived of GADD34. To test this hypothesis, we knocked down GADD34 expression in MT-2 and Jurkat cells by GADD34-specific siRNA. Knockdown of GADD34 in MT-2 cells by GADD34 specific siRNA resulted in about 70% reduction in the protein levels (Fig 3A). Jurkat cells did not express detectable levels of GADD34 (data not shown). However, RT-PCR analysis showed the presence of GADD34 mRNA in Jurkat cells and it was reduced by 65% following the knockdown with GADD34-specific siRNA (Fig 3B). A significant increase in the pNL4-3.Luc.R-E-reporter vector mediated luciferase activity was seen GADD34 knocked down MT-2 and Jurkat cells as compared to the control siRNA transfected cells (Fig. 3C,D). These data clearly indicate that the loss of endogenous levels of GADD34 enhances the levels of HIV-1 gene expression confirming the inhibitory role of GADD34 in HIV-1 replication.
Figure 3. siRNA-mediated GADD34 knockdown induces HIV-1 gene expression.
A-B) MT-2 and Jurkat cells were transfected by electroporation with 1.0 uM siRNA as indicated in the presence of 5.0 μg of pNL4-3.Luc.R-E- plasmid for 36 h. Cells were harvested for Western blot analysis (A) with GADD34 specific antibodies (MT2 cells) or RNA isolation for RT-PCR (B) with GADD34 and GAPDH specific primers (Jurkat cells). C-D) Cell lysates from transfected MT-2 (C) and Jurkat (D) were processed for luciferase measurement.
Anti-HIV activity resides in the N-terminal region of GADD34 and the PP1-binding phosphatase domain is not essential for the inhibition of HIV-1 replication.
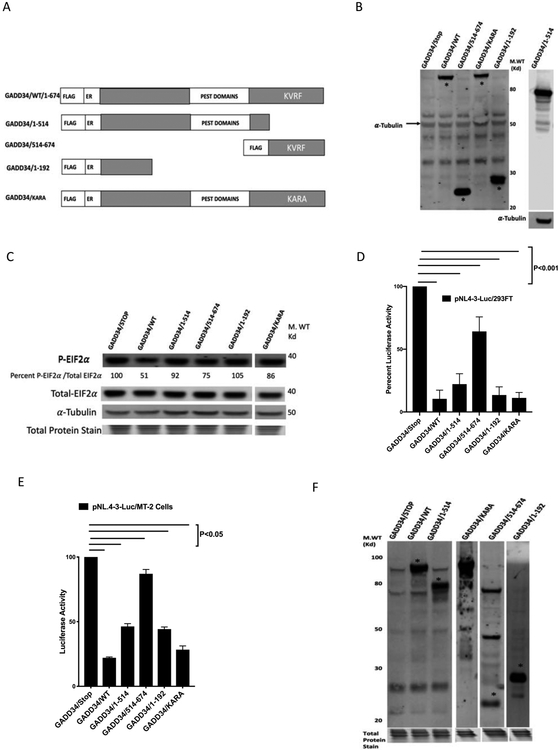
GADD34 is a 73.5 Kd protein with an N-terminal ER binding domain, four central PEST repeats, and a C-terminal PP1-binding domain that contains a PP1 binding KVRF motif (Choy et al., 2015; Connor et al., 2001; Novoa et al., 2001). To identify the GADD34 domains involved in HIV-1 inhibition, a number of GADD34 mutants (Fig. 4A, 4B) were tested for their ability to inhibit viral production. The relative p-eIF-2α-phosphatase activities of various GADD34 constructs is shown in Fig. 4C and is represented as the percent p-eIF-2α levels in the cell lysates. As expected, mutations in PP1 binding (KVRF) domain inhibited phosphatase activity, while as N-terminal deletion (514-674) and some C-terminal deletions also compromised the phosphatase activity of GADD34. Luciferase activity was monitored following the transfection of 293FT cells with pNL4-3.Luc.R-E-plasmid in the presence of various GADD34 plasmid constructs. The data (Fig. 4D) showed that GADD34/KARA retained full anti-HIV activity (compared to GADD34/WT), whereas GADD34/514-674 mutant was found to have considerably less anti-HIV activity as compared to GADD34/WT. These results reveal that PP1-binding activity is not crucial for the anti-HIV activity of GADD34 and the activity is confined mostly to the N-terminal (1-514) region of the protein. Because PP1-binding is critical for the GADD34-mediated dephosphorylation of p-eIF-2α, our data suggests that the p-eIF-2α-phosphatase activity of GADD34 is not needed for anti-HIV-1 activity. C-terminal deletion mutants 1-514 (containing N-terminal ER-binding-helix and a central region with four PEST repeats) and 1-192 (containing N-terminal ER-binding-helix) (Fig. 4D) retained nearly full anti-HIV activity comparable to GADD34/WT. These data also reveal that the 1-192 amino acid region of GADD34 that contains N-terminal ER-binding-helix domain is sufficient for the anti-HIV-1 activity of GADD34. Similar results were obtained with transfection of T cell lines MT-2 (Fig. 4E, 4F) and Jurkat (data not shown). Together, the results presented show that N-terminal ER-binding-helix region of GADD34 inhibits HIV-1 replication whereas protein-phosphatase −1-binding domain and eIF-2α-phosphatase activity of GADD34 are not crucial for the anti-HIV-1 activity.
Figure 4. Anti-HIV activity of GADD34 is independent of PP1-binding phosphatase domain.
A) A schematic representation of GADD34 mutants analyzed in this study, with blocks highlighting the N-terminal tag, ER box, the central PEST repeats, and the consensus KVRF PP1-binding motif. B) 293FT cells were transfected with indicated GADD34 plasmids and subject to Western blot analysis with antibodies to FLAG-tag and α-Tubulin (left panel) or antibodies to GADD34 (Proteintech) (specific for GADD34 amino acids 307-572) and α-Tubulin (right panel). The * indicates the position of GADD34 protein. C) 293FT cells cell lysates from B were analyzed by Western blot using antibodies to total eIF-2α, p-eIF-2α, and α-tubulin antibodies. The numbers indicate ratio of the normalized intensities of p-eIF-2α and total eIF-2α bands. D) 293FT cells were transfected with 100 ng of pNL4-3.Luc.R-E- plasmid in the presence of 200 ng of indicated plasmids. Cells were harvested 38 h later for luciferase measurement. E and F) MT-2 were electroporated with 2.5 μg of pNL4-3.Luc.R-E- plasmid in the presence of 5.0 μg of indicated plasmids. The cells were harvested for luciferase measurement (E) and Western blot analysis (F) with GADD34 specific antibody from Proteintech (specific for amino acids 307-572) (first four lanes starting from left) or Thermofisher (specific for full length protein) (lanes five and six starting from left). The * indicates the position of GADD34 protein.
GADD34 does not reduce HIV-1 RNA synthesis but inhibits 5’-UTR RNA-mediated translation.
The effect of GADD34 on the time course of HIV-1 RNA expression in HeLa-CD4+ cells infected with pNL4-3 virus was studied by quantitating HIV-1 transcripts by RT-PCR. RNAs isolated from the HeLa CD4+ cells transfected with GADD34/Stop or GADD34 plasmids (Fig. 5A) followed by infection with HIV-1 for diffferent time periods (8-48 h) were subject to RT-PCR using primers specific for spliced HIV-1 Tat RNA (Table 1). As seen in Fig 5B, there was a robust time dependent increase in the expression of HIV-1 Tat mRNA following infection with virus, the levels of which did not decrease over time in the presence of GADD34. There was instead a minor but not significant increase in the levels of RNA observed at 48 h of infection in GADD34 overexpressing cells. Interestingly, even though the levels of GADD34 protein (peak expression at 24-48 h after transfection) had reduced considerably at 48 h post-infection (most likely due to the instability and short half life-of the GADD34 protein), HIV RNA levels were not affected. In contrast, a marked decrease in the CA-p24 protein levels was observed 24- 48h after infection in cells overexpressing GADD34 (Fig. 5C).
Figure 5. GADD34 overexpression does not inhibit HIV-1 RNA transcription.
A-C) HeLa CD4+ cells were transfected with 2.0 μg of indicated plasmids. Cells were infected 24 h later with 20 ng of CA-p24 antigen equivalent HIV-1-supernatant for 2 h. The virus was removed, cells were washed with medium and infection continued for the time indicated before harvest for Western analysis using anti-GADD34 (Proteintech, specific for amino acids 307-572) and anti-α-Tubulin antibodies (A), RT-PCR using HIV-1 Tat specific primers and normalized to cellular RNaseP mRNA levels (B), and quantitation of the CA-p24 in the medium using ELISA kit (C). D) Tet-On HeLa-GADD34/DOX cells were transfected with 0.4 μg of pNL4-3 plasmid. After 2 h DMSO or DOX was added as indicated and cells were harvested 38 h later for RNA isolation. RT- PCR was performed using HIV-1 RNA specific primers and the levels were normalized to cellular RNaseP mRNA levels. E) 293FT cells were transfected with 500 ng of pNL4-3.Luc.R-E- plasmid in the presence of 2.5 μg of indicated plasmids and RNA isolated after 36h for RT-PCR with HIV-1 Tat and luciferase specific primers. The data were normalized to cellular RNaseP mRNA levels. F) Schematic representation of pCDNA3 RLUC POLIRES FLUC plasmid in which polio IRES sequences were replaced with HIV-1 5’-UTR (1-289 nt). G) 1.0 μg pCDNA3 RLUC POLIRES FLUC or pCDNA3 RLUC HIVIRES FLUC plasmids were mixed with Quick TNT lysate in the presence of 1.0 μg of GADD34/Stop or GADD34 plasmids and incubated for 1.5 h at 30°C. Firefly and renilla luciferase activities were quantitated using the Dual-Luciferase Reporter Assay System and plotted as ratio of Firefly and Renilla luciferase activities. The expression of GADD34 protein (lower panel) was monitored by Western blot analysis using antibody to GADD34 (Proteintech, specific for amino acids 307-572). H) 293FT cells were transfected with 0.5 ug of pCDNA3 RLUC POLIRES FLUC or pCDNA3 RLUC HIVIRES FLUC plasmids in the presence of 0.5 μg of GADD34/Stop or GADD34 plasmids for 36 h. Firefly and renilla luciferase activities were quantitated using the Dual-Luciferase Reporter Assay System and plotted as ratio of Firefly and Renilla luciferase activities.
TABLE 1.
List of Primers.
| Primer Name | Sequence 5’-3’ |
|---|---|
| TAT-F | AGACAGCGACGAAGAGCTCATCAG |
| TAT-R | CCACCTTCTTCTTCTATTCCTTCGG |
| HIVRNA-F | GTGCCCGTCTGTTGTGTGAC |
| HIVRNA-R | GGCGCCACTGCTAGAGATTT |
| LUCIFERASE-F | GTTTCCAAAAAGGGGTTGC |
| LUCIFERASE-R | GGGAGGTAGATGAGATGTGACGAAC |
| RNASEP-F | AGATTTGGACCTGCGAGCG |
| RNASEP-R | GAGCGGCTGTCTCCACAAGT |
| GAPDH-F | TGACAACTTTGGTATCGTGG |
| GAPDH-R | ATGATGTTCTGGAGAGCCC |
Similar results were obtained with RT-PCR using HIV-1 RNA primers (Table 1) from RNAs isolated from HeLa-GADD34/DOX cells that were transfected with pNL4-3.Luc plasmid followed by treatment with DOX to induce GADD34 expression (Fig. D). There was no significant effect of DOX-mediated GADD34 expression on HIV-1 RNA expression. Similarly, no significant effect of GADD34 expression was observed on HIV-1 Tat and luciferase mRNAs in 293FT cells transfected with pNL4-3.Luc and GADD34 plasmids (Fig. 5E). These results reveal that GADD34 overexpression does not inhibit HIV-1 RNA transcription and suggest that post-transcriptional mechanisms are involved in the ability of GADD34 to inhibit HIV-1. Based on the above data that the overexpression of GADD34 does not inhibit HIV-1 RNA synthesis whereas HIV-1 protein level is significantly attenuated, we next explored the whether GADD34 has an effect on the translation of HIV-1 mRNAs. All of the HIV-1 viral mRNAs contain a common non-coding exon of 289 nucleotide (nt) length at the 5’ end (5’-UTR). HIV-1 5’-UTR represents a highly structured motif that contains a number of important functional elements including the TAR RNA region that binds Tat and a number of cellular proteins that are crucial for efficient HIV-1 mRNA translation. HIV-1 5’-UTR also contains IRES elements for the cap-independent translation that HIV-1 utilizes in addition to cap-dependent translation (Berkhout et al., 2011; Monette et al., 2013; Ricci et al., 2008; Vallejos et al., 2012). To quantitate cap-independent IRES mediated translation and study the effect of GADD34 on such translation, we used pCDNA3 RLuc POLIRES FLuc plasmid (Poulin et al., 1998) that expresses renilla luciferase by cap-dependent translation and firefly luciferase by polio IRES-mediated translation. We replaced polio virus IRES sequences in this plasmid with HIV-1 UTR 1- 289 nt region to create pCDNA3 RLUC HIVIRES FLUC plasmid (Fig 5F) and tested the effect of GADD34 expression on pCDNA3 RLUC POLIRES FLUC and pCDNA3 RLUC HIVIRES FLUC plasmids in a T7-driven rabbit reticulocyte-based in vitro translation assay. As can be seen from the Fig. 5G, HIV-1 UTR 1- 289 exhibited significant IRES-dependent activity as has been reported earlier (Berkhout et al., 2011; Monette et al., 2013; Ricci et al., 2008; Vallejos et al., 2012). GADD34 had no effect on polio-driven or HIV-1 UTR 1- 289-driven IREs mediated- translation indicating that GADD34 does not inhibit HIV-1 UTR-mediated translation by IRES-dependent mechanisms. To exclude the possibility that the experiments with T7-driven rabbit reticulocyte-based in vitro translation assay may not account for the post-translational modifications or the cellular localization requirements for GADD34 that may not occur in these lysates, we studied the effect of GADD34 overexpression on IRES-mediated translation in vivo in HEK293FT cells transfected with pCDNA3 RLUC POLIRES FLUC and pCDNA3 RLUC HIVIRES FLUC constructs. The data (Fig. 5H) showed that GADD34 did not alter the IRES-mediated translation driven by either of these constructs confirming the in vitro data obtained with TNT lysates.
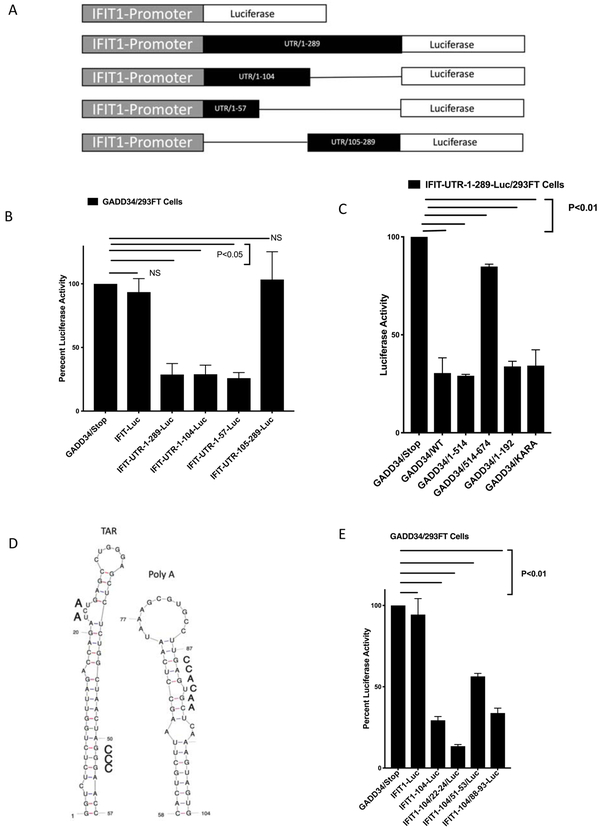
To study the role of cap-dependent translation and explore the role of 5’-UTR RNA sequences in GADD34-mediated inhibition of HIV-1 replication, the 289 nt 5’-UTR sequence and its truncated mutants were cloned between the transcription initiation site of IFIT1 promoter and luciferase gene of IFIT1-promoter-Luc plasmid. The resulting constructs represented a series of the IFIT1-promoter-UTR-Luc plasmids, IFIT1-promoter-UTR/1-289-Luc, IFIT1- promoter-UTR/1-104-Luc, IFIT1- promoter-UTR/1-57-Luc, and IFIT1- promoter-UTR/105-289-Luc (Fig. 6A). We tested the effect of GADD34 on these constructs by transfecting these plasmids in the presence of GADD34/Stop or GADD34 plasmids. The data show that while GADD34 did not have any significant effect on IFIT1-Promoter-Luc activity, it significantly inhibited the luciferase activity of IFIT1-promoter-UTR/1-289-Luc, IFIT1- promoter-UTR/1-104-Luc, and IFIT1- promoter-UTR/1-57-Luc plasmids (Fig. 6B). However, the luciferase activity of IFIT1-UTR/105-289-Luc was not significantly inhibited by GADD34. These results show that the introduction of HIV-1 5’-UTR elements make an otherwise GADD34-unresponsive IFIT1- promoter-Luc into a GADD34-sensitive reporter suggesting that GADD34 inhibits HIV-1 replication by 5’-UTR-mediated translational inhibition. These data also demonstrate that 5’-UTR nucleotides 1-104 are crucial for GADD34-mediated inhibition whereas nucleotides 105-289 are not important for inhibition. The data reveal that 5’-UTR nucleotides 1-57 that represent TAR region is sufficient to mediate GADD34 inhibition. To explore whether the effect of different GADD34 mutants on IFIT1-UTR-1-289-Luc activity was similar to that seen on pNL.4-3-Luc activity (Fig. 4D), cells were transfected with IFIT1-UTR-1-289-Luc plasmid in the presence of GADD34/Stop, GADD34/WT, and various mutants described in Fig. 4A. The data (Fig. 6C) reveal that the inhibition levels obtained with different GADD34 mutants using IFIT1-UTR-1-289-Luc reporter were similar to that seen with pNL.4-3-Luc suggesting that GADD34-mediated 5’-UTR-dependent translational inhibition is independent of PP1-binding and that the 1-192 amino acid region alone is sufficient for the inhibition.
Figure. 6. GADD34 inhibits HIV-1 protein expression by viral 5'-UTR TAR RNA-mediated translational inhibition.
A) Schematic representation of human IFIT1-promoter-Luc plasmid in which HIV-1 5’-UTR (1-289 nt) and its deletion mutants were cloned downstream of IFIT1 promoter RNA initiation site.
B) 293FT cells were transfected with 0.5 μg of IFIT1-promoter-Luc or indicated IFIT1-promoter-HIV-1- 5’-UTR-Luc chimeric plasmids in the presence or absence of 0.5 μg of GADD34/Stop or GADD34 plasmids for 36h. The effect of GADD34 transfection on the luciferase activity of IFIT1-promoter and the chimeric plasmids is plotted as a percentage of the effect of the control GADD34/Stop plasmid on the luciferase activity. The bar labeled GADD34/Stop represents luciferase activity of each IFIT1-Luc construct in the presence of GADD34/Stop set at 100 percent. C) 293FT cells were transfected with 0.5 μg of IFIT-UTR-1-289 plasmid in the presence of indicated plasmids. Cells were harvested 38 h later for luciferase measurement. The effect of transfection of indicated GADD34 plasmids on the luciferase activity of IFIT-UTR-1-289-Luc plasmid is plotted as a percentage of the effect of the control GADD34/Stop plasmid on the IFIT-UTR-1-289 plasmid- driven luciferase activity. The bar labeled GADD34/Stop represents luciferase activity of the IFIT-UTR-1-289 plasmid in the presence of GADD34/Stop set at 100 percent. D) HIV-1 TAR RNA and Poly A hairpin regions with mutations shown in bold letters. E) 293FT cells were transfected with 0.5 μg of the indicated plasmids in the presence or absence of 0.5 μg of GADD34/Stop or GADD34 plasmids. Cells were harvested 38 h later for luciferase measurement. The effect of GADD34 transfection on the luciferase activity of IFIT1-promoter and the chimeric plasmids is plotted as a percentage of the effect of the control GADD34/Stop plasmid on the luciferase activity. The bar labeled GADD34/Stop represents luciferase activity of each IFIT1-Luc construct in the presence of GADD34/Stop set at 100 percent.
GADD34 inhibits HIV-1 TAR RNA-mediated translation.
Because the transfection studies revealed that both TAR RNA region (nt 1-57) and TAR RNA+ Poly A hairpin containing region (nt 1-104) mediated translational inhibition by GADD34, the role of various regions from nt 1-104 in GADD34-mediated inhibition was investigated. TAR RNA region is composed of a stem, a Tat binding bulge, and a loop. In addition to Tat, these sequences bind a number of cellular proteins that are crucial for efficient HIV-1 mRNA translation (Charnay et al., 2009; Guerrero et al., 2015; Lai et al., 2013). We constructed TAR stem, Tat binding-region, and Poly A hairpin stem mutants (Fig. 6D) of nt 1-104 region and tested in the transfection in the presence of GADD34/Stop or GADD34/expressing plasmids (Fig. 6E). The data show that the mutations in the TAR stem (nt 51-53) that lead to the disruption of the stem structure limited the ability of UTR/1-104 to mediate translational inhibition by GADD34. Mutations in the TAT binding bulge (nt 22-24), however, enhanced GADD34-mediated translational inhibition. These data indicate that GADD34 modulates translational inhibition via HIV-1 TAR RNA and may function by modulating the folding and remodeling of the TAR RNA structure. Mutations in the stem region of the Poly A hairpin (nt 88-93) did not affect the GADD34-mediated inhibition suggesting that the stem structure folding in this region is not crucial for GADD34 inhibition. However, this study does not rule out the importance of other structural elements in the Poly A hairpin region in mediating GADD34 inhibition.
Discussion
During the physiological conditions GADD34 protein is expressed at low levels, and only during ER stress its expression is induced to detectable levels (Novoa et al., 2003). GADD34 is also an ISG and hard-wired to the innate immune response and induced during immune response to viral pathogens or the transfection of cells with dsRNA (Ishaq and Natarajan, 2016; Minami et al., 2007). One of the major known roles attributed to GADD34 is to function as a regulatory subunit of PP1 and form a highly specific phosphatase holoenzyme capable of dephosphorylating p-eIF-2A and to restore protein homeostasis following ER stress (Chen et al., 2015; Choy et al., 2015; Connor et al., 2001; Novoa et al., 2001; Novoa et al., 2003). Here we have provided evidence that HIV-1 infection induces moderate levels of GADD34 expression late in the infection, indicating the lack of host response to induce to GADD34 during the early phase of infection.
We found that the overexpression of GADD34 inhibits whereas siRNA-mediated knockdown of GADD34 expression induces HIV-1 replication. The inhibition of HIV-1 replication was observed with both ectopic expression (transfection) with a GADD34 expressing plasmid and by endogenously mediated induction (Tet-On-induction system). Taken together, these observations point out to an inhibitory role of GADD34 in HIV-1 replication and also lack of adequate host response to express inhibitory levels of GADD34 during the HIV-1 infection especially in the early stages of infection. This is however, not surprising knowing that HIV-1 is known to evade innate immune recognition and elicits a poor host interferon response and ISG induction (Rasaiyaah et al., 2013). The inability to express GADD34 early in infection also points out to failure of HIV-1 to evoke adequate ISR or ER stress pathways during early stages of infection.
Earlier studies have shown that GADD34 restricts vesicular stomatitis virus (VSV) replication mediated by its phosphatase activity (Minami et al., 2007), and GADD34-deficient fibroblasts and neonate mice are susceptible to Chikungunya virus infection due to a lack of type I IFN and IL-6 production (Clavarino et al., 2012). Our data extends the anti-viral property of GADD34 to include HIV-1 and also reveals a role of GADD34 that is independent of its well-known p-eIF-2α-phosphatase activity. Surprisingly, this novel anti-HIV-1 function of GADD34 was confined to amino acid region 1-192 which contains the ER localization domain (Choy et al., 2015; Zhou et al., 2011). The C-terminal amino acid region 515-674, which contains the PP1 binding site, was not fully active, and a KARA mutant of GADD34 that is known to lack PP1 binding activity (Brush et al., 2003) was fully functional. The mechanism by which 1-192 amino acid region exhibits the anti-HIV-1 activity independent of other regions remains to be studied.
GADD34 failed to inhibit the HIV-1 RNA expression indicating that posttranscriptional mechanisms are responsible for the inhibition of HIV-1 protein expression. Using various IFIT1-promoter-HIV-1-UTR constructs we were able to find that 5’-UTR nucleotides 1-104 are crucial for GADD34-mediated inhibition whereas nucleotides 105-289 are not important for inhibition. Two possibilities could be predicted based on the ability of 5’-UTR/ nt-1-104 to mediate GADD34 inhibition. First, because this sequence encompasses the TAR region that is known to bind HIV Tat and a number of cellular proteins that are crucial for efficient HIV-1 mRNA translation (Charnay et al., 2009; Guerrero et al., 2015; Lai et al., 2013), GADD34 overexpression could disrupt the assembly of TAR-binding cellular activating factors in a way that inhibits translation without having any effect on transcription. Second, GADD34 inhibits translation driven by cap-independent mechanisms involving IRES elements located in HIV-1 UTR. Results from in vitro translation system and in vivo transfection studies ruled out a role for HIV-1/UTR IRES in mediating GADD34-mediated inhibition. The data, however, indicate that the TAR stem structure played a major role in TAR-mediated GADD34 inhibition. Mutations in the TAR region nt 51-53, that lead to the disruption of the stem structure, limited the ability of TAR to mediate GADD34 inhibition whereas mutations in the Poly (A) hairpin stem region (nt 88-93) did not have any effect on the ability of TAR to mediate GADD34 inhibition. Mutations in the TAT- binding bulge region, that perhaps enhanced the stem structure of the TAR and/or prevented the binding of cellular proteins, however, accentuated GADD34-mediated translational inhibition. Based on these findings, it is clear that GADD34 modulates translational inhibition primarily via HIV-1 TAR RNA perhaps by modulating the TAR RNA folding and structure. Does GADD34 physically interact with TAR RNA sequence? Electrophoretic mobility shift assays did not reveal any direct interaction between purified GADD34 protein and TAR RNA sequence (data not shown). Because GADD34 inhibited both Tat-dependent and Tat-independent HIV-1 gene expression, indirect interactions via HIV-1 Tat and/or other TAR RNA-binding proteins cannot be excluded and remains to be elucidated.
Conclusions
To summarize, we describe a novel function of GADD34 as a putative anti-HIV-1 restriction factor that inhibits HIV-1 translation independent of its PP1 binding and p-eIF-2α-phosphatase activity. We demonstrate that the GADD34 N-terminal region 1-192 that contains the ER-binding helix is sufficient for the inhibition. The inhibition is mediated primarily by TAR RNA and may involve GADD34 modulating the TAR RNA remodeling perhaps by targeting RNA helicases. GADD34 is expressed only in the conditions of stress and as an innate immune response to viruses or transfection with dsRNA (Ishaq and Natarajan, 2016; Minami et al., 2007), thus linking the innate immune response and ISR pathways. HIV-1 is known to evade innate immune recognition and elicits a poor interferon response (Rasaiyaah et al., 2013). We have shown that HIV-1 infection does not induce GADD34 expression during active phase of infection. This may perhaps be due to lack of an active innate immune or stress response against the virus, and/or for survival HIV-1 might possess a counteraction mechanism that prevents the accumulation of GADD34 in the cells. Future studies should reveal the identity of such a counteracting factor and the mechanism of GADD34 modulation of the TAR RNA structure.
Material and Methods
Cells and reagents.
HEK-293FT (Thermo Fisher Scientific, Waltham, MA), HeLa CD4+ (Maddon et al., 1986) and TZM-bl (Platt et al., 2009) (NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH) cells were maintained in Dulbecco's Modified Eagle's medium (DMEM) medium containing 10% fetal bovine serum and 500 μg/ml G418 (for HeLa CD4+ cells). Tet-On HeLa-GADD34/DOX (Dr. Stefan Marciniak, University of Cambridge, Cambridge, UK) cells were maintained in DMEM medium containing 10% tetracycline-free fetal bovine serum (Takara Bio, Mountain View, CA), 200 μg /ml of G418, and 200 uM of hygromycin. MT2 cells (Harada et al., 1985) (NIH AIDS Reagent Program) and Jurkat (ATCC, Manassas, VA) were maintained in RPMI 1640 medium containing 10% fetal bovine serum. Antibodies to α-tubulin, GADD34 specific for amino acids 307-572, GADD34 specific for full length protein, FLAG-tag, and HIV-1 CA-p24 were from Santa Cruz Biotechnology (Dallas, Texas), Proteintech (Rosemont, IL), ThermoFisher ( Waltham, MA), OriGene (Rockville, MD), and the NIH AIDS Reagent Program, respectively. Doxycycline hydrochloride (DOX), hygromycin, and G418 were from Millipore Sigma (St. Louis, MO).
Plasmids and Transfections.
pBlue3'LTR-luc-A (Jeeninga et al., 2000), pNL4-3 (Adachi et al., 1986), and pNL4-3.Luc.R-E- (He et al., 1995) plasmids were from the NIH AIDS Reagent Program. pcDNA3.1-TAT-1-101-FLAG was from Addgene (Cambridge, MA). Human GADD34 expressing pSG5-Flag-GADD34 and pSG5-Flag-GADD34/KARA mutant plasmids were a gift from Dr. Shirish Shenolikar (Duke-NUS Medical School, Singapore). pSG5-Flag-GADD34/Stop non-expressing control plasmid was generated by introducing stop codon at amino acids 5 and 9 using the TagMaster® Site-Directed Mutagenesis Kit (GM Biosciences, Frederick, MD). Codon-optimized human GADD34 expressing pcDNA3.1-Flag-GADD34 and mutant plasmids pcDNA3.1-Flag-GADD34/514-674, pcDNA3.1-Flag-GADD34/1-514, and pcDNA3.1-Flag-GADD34/1-192 were custom constructed by Thermo Fisher Scientific. Dr. John Hiscott (Vaccine & Gene Therapy Institute of Florida, St. Lucie, FL) provided IFIT1 promoter-Luc plasmid. IFIT1-promoter-HIV-1- 5’-UTR-Luc chimeric plasmids containing HIV-1 5’-UTR (1-289) and its deletion mutants were constructed by cloning custom synthesized (Thermo Fisher) HIV-1 5’-UTR (1-289) and its deletion mutant DNA strings into the HindIII site of the 3’end of the IFIT1-promoter. For constructing pcDNA3 RLUC HIVIRES FLUC plasmid, poliovirus IRES sequences were deleted from the pcDNA3 RLUC POLIRES FLUC plasmid (Addgene, Watertown, MA) (Poulin et al., 1998) by mutagenesis and replaced with HIV-1 5’-UTR (1-289) sequence. All the plasmids were sequence verified. Control siRNA and GADD34-specific siRNA were purchased from Dharmacon (Lafayette, CO). Adherent cells were transfected in six or 12 well plates using FuGENE-HD (Promega, Maddison, WI). MT-2 and Jurkat cells were transfected by electroporation using a Gene Pulser II (Bio-Rad, Hercules, CA) at 0.250 kV and 975 μF. Luciferase activity was quantitated using the Luciferase Assay System (Promega) and normalized to protein concentrations in the extracts.
Virus and infections.
pNL4-3 plasmid was used for growing HIV-1 virus by transfecting 293FT cells in six well plates with 6 μg DNA/well using ProFection® Mammalian Transfection System (Promega). After 48 h, virus in supernatants was quantitated using the Alliance HIV-1 CA-p24 Antigen ELISA kit (PerkinElmer, New York, NY). The virus was amplified by infecting MT2 cells with 25ng of CA-p24 antigen equivalent HIV-1-supernatant by spinoculation. Cells were passaged four times every 3-4 days to minimize the concentration of input genomic DNA in the viral supernatants. Supernatants were titrated for CA-p24 antigen and stored in 1000 ng/ml aliquots at −80°C. To study the effect of HIV-1 infection on GADD34 expression, MT-2 cells were mock infected or infected by spinoculation with 20 ng of CA-p24 antigen equivalent HIV-1-supernatant for indicated days. Cells were harvested for GADD34 immunoblot. HeLa CD4+ cells in 12 well plates were transfected with 500 ng of GADD34 plasmids per well using FuGENE-HD and after 24 h infected with 20 ng of CA-p24 antigen equivalent HIV-1-supernatant for 2 h. The virus was removed, the cells were washed with medium, and infection continued for an additional 40 h before CA-p24 quantitation.
Real-time RT-PCR.
Total RNA was isolated using the Qiagen (Germantown, MD) RNeasy kit by following the manufacture's protocol. The RNA samples were treated with DNAse to ensure that there was no DNA contamination in the samples. For Real-Time RT-PCR total RNA was reverse transcribed with Superscript II reverse transcriptase (Thermo Fisher Scientific, Waltham, MA) in the presence of 2.5 μM random hexamers and cDNAs were subject to Real-Time PCR with gene-specific primers (Table 1) using a 7500 Fast Real-Time PCR System and Fast SYBR Green Master Mix from Applied Biosystems. Each experiment included reverse transcriptasenegative controls to ensure that there was no DNA contamination in the RNA samlples. The comparative threshold cycle method was used to calculate the relative gene expression.
Western blot.
Protein extracts were electrophoresed in a 4-12% NuPAGE Bis Tris Gel using NuPAGE MES SDS running buffer (Thermo Fisher Scientific, Waltham, MA), and transferred to a polyvinylidene difluoride (PVDF) membrane using XCell Blot Module (Thermo Fisher Scientific, Waltham, MA). After treatment with primary antibodies, protein was detected using fluorophore labeled secondary antibodies and the Odyssey Infrared Imaging System (Li-cor Biotechnology, Lincoln, Nebraska). Protein specific bands were normalized to either α-tubulin or total proteins. Total proteins were stained using SimplyBlue Safestain (ThermoFisher) using manufacturer’s intructions and the stained gels were scanned using Odyssey Infrared Imaging System (Li-cor Biotechnology).
In vitro translation.
T7 promoter-based TnT Quick Coupled Transcription/Translation System (Promega) kit was used for in vitro translation studies. Coupled Transcription/Translation was carried out with 1.0 μg of various luciferase plasmid constructs in the presence 1.0 μg of GADD34/Stop or GADD34 plasmids as per the manufacturer’s instructions. Luciferase activity was quantitated using the Dual-Luciferase Reporter Assay System or Luciferase Assay System (Promega).
Statistical analysis.
The results presented are from a representative experiment performed in 2-3 replicates and each experiment repeated independently three times and expressed as mean ± SD. Statistical analysis was performed using Student’s t-test. A value of p<0.05 was considered significant.
Supplementary Material
Acknowledgements
This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. This work was supported by the National Institute of Allergy and Infectious Diseases.
We thank Dr. Stefan Marciniak (University of Cambridge, Cambridge, UK) for providing Tet-On HeLa-GADD34/DOX cells, Dr. Shirish Shenolikar (Duke-NUS Medical School, Singapore) for providing pSG5-Flag-GADD34 and pSG5-Flag-GADD34/KARA mutant plasmids, and Dr. John Hiscott (Vaccine & Gene Therapy Institute of Florida, St. Lucie, FL) for a gift of IFIT1 promoter-Luc plasmid. We also thank NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH for providing pBlue3'LTR-luc-A (Dr. Reink Jeeninga and Dr. Ben Berkhou), pNL4-3 (Dr. Malcolm Martin), and pNL4-3.Luc.R-E- (Dr. Nathaniel Landau) plasmids; HeLa CD4+ (Dr. Richard Axel), TZM-bl (Dr. John C. Kappes, Dr. Xiaoyun Wu and Tranzyme Inc), and MT2 (Dr. Douglas Richman) cell lines; and anti-HIV-1 CA-p24 antibody.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adachi A, Gendelman HE, Koenig S, Folks T, Willey R, Rabson A, Martin MA, 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol 59, 284–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballana E, Este JA, 2015. SAMHD1: at the crossroads of cell proliferation, immune responses, and virus restriction. Trends in microbiology 23, 680–692. [DOI] [PubMed] [Google Scholar]
- Balvay L, Soto Rifo R, Ricci EP, Decimo D, Ohlmann T, 2009. Structural and functional diversity of viral IRESes. Biochimica et biophysica acta 1789, 542–557. [DOI] [PubMed] [Google Scholar]
- Berkhout B, Arts K, Abbink TE, 2011. Ribosomal scanning on the 5'-untranslated region of the human immunodeficiency virus RNA genome. Nucleic acids research 39, 5232–5244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brush MH, Shenolikar S, 2008. Control of cellular GADD34 levels by the 26S proteasome. Molecular and cellular biology 28, 6989–7000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brush MH, Weiser DC, Shenolikar S, 2003. Growth arrest and DNA damage-inducible protein GADD34 targets protein phosphatase 1 alpha to the endoplasmic reticulum and promotes dephosphorylation of the alpha subunit of eukaryotic translation initiation factor 2. Molecular and cellular biology 23, 1292–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charnay N, Ivanyi-Nagy R, Soto-Rifo R, Ohlmann T, Lopez-Lastra M, Darlix JL, 2009. Mechanism of HIV-1 Tat RNA translation and its activation by the Tat protein. Retrovirology 6, 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Rato C, Yan Y, Crespillo-Casado A, Clarke HJ, Harding HP, Marciniak SJ, Read RJ, Ron D, 2015. G-actin provides substrate-specificity to eukaryotic initiation factor 2alpha holophosphatases. eLife 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy MS, Yusoff P, Lee IC, Newton JC, Goh CW, Page R, Shenolikar S, Peti W, 2015. Structural and Functional Analysis of the GADD34:PP1 eIF2alpha Phosphatase. Cell reports 11, 1885–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clavarino G, Claudio N, Couderc T, Dalet A, Judith D, Camosseto V, Schmidt EK, Wenger T, Lecuit M, Gatti E, Pierre P, 2012. Induction of GADD34 is necessary for dsRNA-dependent interferon-beta production and participates in the control of Chikungunya virus infection. PLoS pathogens 8, e1002708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connor JH, Weiser DC, Li S, Hallenbeck JM, Shenolikar S, 2001. Growth arrest and DNA damage-inducible protein GADD34 assembles a novel signaling complex containing protein phosphatase 1 and inhibitor 1. Molecular and cellular biology 21, 6841–6850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton LE, Healey E, Irving J, Marciniak SJ, 2012. Phosphoproteins in stress-induced disease. Progress in molecular biology and translational science 106, 189–221. [DOI] [PubMed] [Google Scholar]
- Fletcher AJ, Towers GJ, 2013. Inhibition of retroviral replication by members of the TRIM protein family. Current topics in microbiology and immunology 371, 29–66. [DOI] [PubMed] [Google Scholar]
- Fullwood MJ, Zhou W, Shenolikar S, 2012. Targeting phosphorylation of eukaryotic initiation factor-2alpha to treat human disease. Progress in molecular biology and translational science 106, 75–106. [DOI] [PubMed] [Google Scholar]
- Guerrero S, Batisse J, Libre C, Bernacchi S, Marquet R, Paillart JC, 2015. HIV-1 replication and the cellular eukaryotic translation apparatus. Viruses 7, 199–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haller O, Staeheli P, Schwemmle M, Kochs G, 2015. Mx GTPases: dynamin-like antiviral machines of innate immunity. Trends in microbiology 23, 154–163. [DOI] [PubMed] [Google Scholar]
- Harada S, Koyanagi Y, Yamamoto N, 1985. Infection of HTLV-III/LAV in HTLV-I-carrying cells MT-2 and MT-4 and application in a plaque assay. Science (New York, N.Y.) 229, 563–566. [DOI] [PubMed] [Google Scholar]
- Harris RS, Liddament MT, 2004. Retroviral restriction by APOBEC proteins. Nature reviews. Immunology 4, 868–877. [DOI] [PubMed] [Google Scholar]
- He B, 2006. Viruses, endoplasmic reticulum stress, and interferon responses. Cell death and differentiation 13, 393–403. [DOI] [PubMed] [Google Scholar]
- He J, Choe S, Walker R, Di Marzio P, Morgan DO, Landau NR, 1995. Human immunodeficiency virus type 1 viral protein R (Vpr) arrests cells in the G2 phase of the cell cycle by inhibiting p34cdc2 activity. J Virol 69, 6705–6711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishaq M, Natarajan V, 2016. Integrated Stress Response Signaling Pathways Induced by Supraphysiological Concentrations of Thyroid Hormone Inhibit Viral Replication. Signal Transduction Insights 5, 25–37. [Google Scholar]
- Jeeninga RE, Hoogenkamp M, Armand-Ugon M, de Baar M, Verhoef K, Berkhout B, 2000. Functional differences between the long terminal repeat transcriptional promoters of human immunodeficiency virus type 1 subtypes A through G. J Virol 74, 3740–3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai MC, Wang SW, Cheng L, Tarn WY, Tsai SJ, Sun HS, 2013. Human DDX3 interacts with the HIV-1 Tat protein to facilitate viral mRNA translation. PloS one 8, e68665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KM, Chen CJ, Shih SR, 2017. Regulation Mechanisms of Viral IRES-Driven Translation. Trends in microbiology 25, 546–561. [DOI] [PubMed] [Google Scholar]
- Li M, Kao E, Gao X, Sandig H, Limmer K, Pavon-Eternod M, Jones TE, Landry S, Pan T, Weitzman MD, David M, 2012. Codon-usage-based inhibition of HIV protein synthesis by human schlafen 11. Nature 491, 125–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddon PJ, Dalgleish AG, McDougal JS, Clapham PR, Weiss RA, Axel R, 1986. The T4 gene encodes the AIDS virus receptor and is expressed in the immune system and the brain. Cell 47, 333–348. [DOI] [PubMed] [Google Scholar]
- Minami K, Tambe Y, Watanabe R, Isono T, Haneda M, Isobe K, Kobayashi T, Hino O, Okabe H, Chano T, Inoue H, 2007. Suppression of viral replication by stress-inducible GADD34 protein via the mammalian serine/threonine protein kinase mTOR pathway. J Virol 81, 11106–11115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondor I, Ugolini S, Sattentau QJ, 1998. Human immunodeficiency virus type 1 attachment to HeLa CD4 cells is CD4 independent and gp120 dependent and requires cell surface heparans. J Virol 72, 3623–3634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monette A, Valiente-Echeverria F, Rivero M, Cohen EA, Lopez-Lastra M, Mouland AJ, 2013. Dual mechanisms of translation initiation of the full-length HIV-1 mRNA contribute to gag synthesis. PloS one 8, e68108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neil SJ, Zang T, Bieniasz PD, 2008. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature 451, 425–430. [DOI] [PubMed] [Google Scholar]
- Novoa I, Zeng H, Harding HP, Ron D, 2001. Feedback inhibition of the unfolded protein response by GADD34-mediated dephosphorylation of eIF2alpha. The Journal of cell biology 153, 1011–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novoa I, Zhang Y, Zeng H, Jungreis R, Harding HP, Ron D, 2003. Stress-induced gene expression requires programmed recovery from translational repression. The EMBO journal 22, 1180–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt EJ, Bilska M, Kozak SL, Kabat D, Montefiori DC, 2009. Evidence that ecotropic murine leukemia virus contamination in TZM-bl cells does not affect the outcome of neutralizing antibody assays with human immunodeficiency virus type 1. J Virol 83, 8289–8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulin F, Gingras AC, Olsen H, Chevalier S, Sonenberg N, 1998. 4E-BP3, a new member of the eukaryotic initiation factor 4E-binding protein family. J Biol Chem 273, 14002–14007. [DOI] [PubMed] [Google Scholar]
- Proud CG, 2005. eIF2 and the control of cell physiology. Seminars in cell & developmental biology 16, 3–12. [DOI] [PubMed] [Google Scholar]
- Rasaiyaah J, Tan CP, Fletcher AJ, Price AJ, Blondeau C, Hilditch L, Jacques DA, Selwood DL, James LC, Noursadeghi M, Towers GJ, 2013. HIV-1 evades innate immune recognition through specific cofactor recruitment. Nature 503, 402–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricci EP, Soto Rifo R, Herbreteau CH, Decimo D, Ohlmann T, 2008. Lentiviral RNAs can use different mechanisms for translation initiation. Biochemical Society transactions 36, 690–693. [DOI] [PubMed] [Google Scholar]
- Ron D, Walter P, 2007. Signal integration in the endoplasmic reticulum unfolded protein response. Nature reviews. Molecular cell biology 8, 519–529. [DOI] [PubMed] [Google Scholar]
- Spriggs KA, Bushell M, Willis AE, 2010. Translational regulation of gene expression during conditions of cell stress. Molecular cell 40, 228–237. [DOI] [PubMed] [Google Scholar]
- Tsalikis J, Croitoru DO, Philpott DJ, Girardin SE, 2013. Nutrient sensing and metabolic stress pathways in innate immunity. Cellular microbiology 15, 1632–1641. [DOI] [PubMed] [Google Scholar]
- Vallejos M, Carvajal F, Pino K, Navarrete C, Ferres M, Huidobro-Toro JP, Sargueil B, Lopez-Lastra M, 2012. Functional and structural analysis of the internal ribosome entry site present in the mRNA of natural variants of the HIV-1. PloS one 7, e35031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P, Ron D, 2011. The unfolded protein response: from stress pathway to homeostatic regulation. Science (New York, N.Y.) 334, 1081–1086. [DOI] [PubMed] [Google Scholar]
- Yamamoto H, Unbehaun A, Spahn CMT, 2017. Ribosomal Chamber Music: Toward an Understanding of IRES Mechanisms. Trends in biochemical sciences 42, 655–668. [DOI] [PubMed] [Google Scholar]
- Zhou W, Brush MH, Choy MS, Shenolikar S, 2011. Association with endoplasmic reticulum promotes proteasomal degradation of GADD34 protein. J Biol Chem 286, 21687–21696. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.