Abstract
PilB is the assembly ATPase for the bacterial type IV pilus (T4P), and as a consequence, it is essential for T4P-mediated bacterial motility. In some cases, PilB has been demonstrated to regulate the production of exopolysaccharide (EPS) during bacterial biofilm development independently of or in addition to its function in pilus assembly. While the ATPase activity of PilB resides at its C-terminal region, the N terminus of a subset of PilBs forms a novel cyclic-di-GMP (cdG)-binding domain. This multi-domain structure suggests that PilB binds cdG and adenine nucleotides through separate domains which may influence the functionality of PilB in both motility and biofilm development. Here, Chloracidobacterium thermophilum PilB is used to investigate ligand binding by its separate domains and by the full-length protein. Our results confirm the specificity of these individual domains for their respective ligands and demonstrate communications between these domains in the full-length protein. It is clear that when the N- and the C-terminal domains of PilB bind to cdG and ADP, respectively, they mutually influence each other in conformation and in their binding to ligands. We propose that the interactions between these domains in response to their ligands play critical roles in modulating or controlling the functions of PilB as a regulator of EPS production and as the T4P assembly ATPase.
Keywords: ATPase, cyclic-di-GMP, motility, biofilm, PilB, type IV pili
Introduction
The bacterial type IV pilus (T4P) is a cell surface appendage with versatile biological functions [1]. A single pilus, a filamentous polymer of thousands of pilins, extends several microns from the cell with a diameter of 6–9 nm [2]. It is known to function in many bacterial processes including biofilm formation and surface motility [3–7]. During biofilm formation, T4P has been demonstrated to facilitate bacterial attachment to biotic or abiotic surfaces during the early stages of biofilm development. Its retraction is known to power surface motility in a variety of Gram-negative bacteria including Myxococcus xanthus and Thermus thermophilus as well as Pseudomonas aeruginosa and Neisseria gonorrhea [5,8–12]. The current model is that the distal end of an extended pilus can attach or tether to the surface of a substratum [5,12,13]. The retraction or disassembly of the pilus at the proximal end of the cell body leads to the physical displacement of the bacterium [12,13]. The recurrent cycles of extension and retraction or assembly and disassembly result in sustained bacterial motility on solid surfaces [1,3,13]. It is known that the retraction of a single T4P can pull a force up to 150 pN, making T4P the strongest biological motor currently known [12,14]. The roles of T4P in both biofilm development and motility in a same bacterium are somewhat paradoxical, because motility is associated with the motile lifestyle, while biofilm represents sessility. These two lifestyles are hence considered to be mutually exclusive as alternate states of existence for bacteria. How the functions of T4P in these seemingly non-compatible processes are regulated in the same organism remains largely tenuous and enigmatic [15,16].
The type IV pilus machinery (T4PM) in Gram-negative bacteria is made of a dozen T4P or Pil proteins including the T4P assembly ATPase PilB (or PilF in T. thermophilus and a few others) [10,17–19]. Most of these proteins form a stable supramolecular complex that spans the bacterial envelope from the cytoplasm to the outer membrane [17]. PilB is proposed to dock with this stable T4PM complex on the cytoplasmic side and hydrolyze ATP to power the assembly of pilin monomers into the base of a growing T4P filament [17,20,21]. PilB and all proteins in this stable complex are required for T4P assembly and deletions of their genes result in a non-piliated and non-motile phenotype [10,18]. The C-terminus of PilB contains the structure of an AAA+ ATPase which crystallizes as a hexamer [22–25]. Based on the hexameric structures of these ATPase domains, it is proposed that PilB utilizes a symmetric rotary mechanism to hydrolyze ATP [23–26]. That is, two ATP molecules are hydrolyzed to ADP simultaneously by two protomers at the opposite corners of the hexameric ring. Another pair adjacent to these two would hydrolyze ATP next. It is proposed that the hydrolysis of ATP by the C-terminal ATPase domain of PilB results in the movement of the N-terminal domain which is mechanically coupled to the insertion of pilins into the pilus through other components in the T4PM [22]. More recently, the full-length PilB of Chloracidobacterium thermophilum (CtPilB) was shown to be a hexamer in solution with robust ATPase activity in vitro [27]. All available evidence supports the model that the ATPase activity of PilB endowed by its C terminus energizes T4PM for T4P assembly as a prerequisite for T4P-mediated bacterial surface motility.
There is evidence from T. thermophilus and M. xanthus that PilB functions as a signaling protein to regulate biofilm development in addition to or independently of its role as the T4P assembly ATPase [28,29]. In both cases, PilB was shown to regulate the production of exopolysaccharides (EPS) which is the major constituent of bacterial biofilm matrix [30]. A phosphoproteomic study discovered T. thermophilus PilF (TtPilF) to be phosphorylated on a Thr and a Ser residue [29]. Non-phosphorylatable mutations of these two residues were found to have no effect on T4P assembly or T4P-dependent motility. Instead, they were found to result in defects in biofilm formation and reduced EPS production [29]. In M. xanthus, a genetic screen led to the revelation that a PilB mutant variant, while lacking ATPase activity to support T4P assembly and motility, constitutively signals EPS production [28]. These observations support a direct role of PilB in biofilm and EPS regulation that is distinct from its function as the T4P assembly ATPase. It is unclear how the paradoxical functions of PilB in T4P-dependent motility for active bacterial translocation and EPS signaling for biofilm development are managed at the molecular level in these organisms.
Recent findings suggested that the functionality of MxPilB and TtPilF, as well as their close orthologues, network directly with cyclic-di-GMP (c-di-GMP or cdG), a ubiquitous bacterial second messenger that regulates the transition between sessile and motile states of bacteria [31,32]. Vibrio cholerae MshE (VcMshE) is the PilB equivalent in the T4P system known as the mannose-sensitive hemagglutinin (MSHA) pilus [33,34]. A systematic study to identify cdG receptors in V. cholerae discovered the N terminus of MshE (MshEN) to constitute a novel cdG binding domain [33]. The crystal structure of an MshEN-cdG binary complex delineated MshEN to contain a tandem repeat of two conserved cdG binding motifs (labeled Motif I and Motif II in Supplementary Figure S1). A single motif is contained within a sequence of 24 residues long. Its consensus is RLGx2(L/V/I)2x2G(L/V/I)2x4Lx3Lx2Q, where x could be any residue while L/V/I could be any of these three specified amino acids (Supplementary Figure S1) [34]. The MshEN domain has been demonstrated to bind cdG with high affinity and mutations within these motifs severely impacted cdG binding and the production of MSHA pilus in V. cholera [29,33]. The conserved features of this domain are found at the N-termini of a subset of PilB including MxPilB, TtPilF, and CtPilB, which may be referred to as the MshEN+-type of T4P assembly ATPases. Although crystal structures of both the MshEN and the ATPase domains of PilB are available, there is yet no structure of the full-length protein. At present, it is unclear how the MshEN domain in PilB proteins interacts with their C-terminal ATPase domain and how ligand binding may affect such interactions and PilB functions.
In this report, we used CtPilB, the first and only known member of PilB ATPase family amicable to biochemical characterization in vitro, as the model to study MshEN+-type PilB ATPases in vitro. We demonstrate that, while the N- and the C-terminal domains of CtPilB are able to specifically bind cdG and adenine nucleotides, the full-length protein binds both types of ligands with higher affinity. Our results further demonstrate that ligand binding by one domain can significantly influence the mechanisms of ligand binding by the other. These results indicate effective communications and mutual allosteric effects between the N- and the C-terminal domains of PilB. We suggest that ligand binding by both the N- and C-terminal domains of PilB may impact its function in EPS signaling as well as T4P assembly.
Experimental procedures
Expression and purification protein
Three CtPilB variants were expressed in Escherichia coli and purified in this study. The E. coli strains XL1-Blue (Stratagene) and BL21 (DE3) (Novagen) were used for the construction of expression plasmids and/or protein expression. All E. coli strains were grown in LB media at 37 °C. When appropriate, 100 μg/ml of ampicillin was supplemented for the selection and maintenance of expression plasmids. Plasmid pKD129 was used for the expression of PilBN. For its construction, a PCR fragment was first obtained using primers PilBN_F (GCCATATGTCAGCAAAACTTGGTGA) and PilBN_R (GCCTCGAGTTCCGGGGCGTAGTAG). This fragment was digested with NdeI and XhoI and cloned into the same sites in pET-22b (Novagen) to construct pKD129. BL21 (DE3) was used as the host for pKD129 for the expression of PilBN. Plasmid pAS104 was constructed for the expression of PilBΔN. A PCR was performed using primers CctPilB_trunc#1_F (GCGGATCCGCCCCGGAAAAGGGACTTGTT) and C_R-HindIII (TGACAAGCTTAGTGATGGTGATGGTGATG) with pAS101 [24] as the template. This fragment was digested with BamHI and HindIII and cloned into the same sites of pQE16 (Qiagen) to produce pAS104. Full-length PilB and PilBΔN were expressed in XL1-Blue, All three proteins were purified by FPLC using Ni-NTA and SEC columns as was described [27], except that the activity buffer with 10% glycerol and no Zn2+ was used as the gel filtration buffer. The apparent molecular weights (MWs) of proteins were determined using analytical size exclusion chromatography [27].
ITC experiments
All isothermal titration calorimetry (ITC) experiments were performed using a MicroCal PEAQ-ITC calorimeter (Malvern) at 25 °C. All titrations were carried with a reference power of 10 μCal·s−1 and a 300 rpm stirring speed. All protein samples were dialyzed into activity buffer [27] using dialysis tubing with an 8 kDa molecular weight cut-off (MWCO) before their use in ITC experiment. 300 μl of a protein sample with or without a specified effector was loaded into the sample cell and a ligand was titrated into the sample cell from the syringe. Whenever an effector was included in the sample cell with the protein, it was incubated with the protein on ice for 15 min prior to being placed into the sample cell. The concentrations of the effectors used were 100 μM for cdG and 50 μM for both ATPγS and ADP. Three or more independent experiments were performed for each titration and the data from one representative experiment are presented in this paper. All samples and titrants were degassed at 4 °C prior to their use in ITC experiments. cdG, ATPγS, and ADP were purchased from InvivoGen, Cayman Chemicals, and Research Products International, respectively.
When cdG was the ligand or titrant in the syringe, the following were the concentrations used for the ITC experiments. PilBΔN and full-length PilB which are hexameric were diluted to a final concentration of 5.3 μM with activity buffer after dialysis and before their use in the sample cell. PilBN, which is monomeric, was diluted to a final concentration of 30 μM. The syringe contained cdG at 0.5 mM in activity buffer. Titrations were performed with a single 0.4 μl injection followed by 29 injections of 1 μl. The time interval between injections was 150 s.
When adenine nucleotides were used as titrants, the following were the concentrations used in the ITC experiments. After dialysis and before their use in the sample cell, PilBΔN and full-length CtPilB were diluted to a final concentration of 15 μM. The syringe was loaded with either ATPγS or ADP at 1 mM concentration. Titrations were carried out with a single 0.4 μl injection followed by 39 injections of 1 μl of the ligand with time intervals of 150 and 120 s for PilBΔN and full-length CtPilB, respectively.
Control titrations were performed for each ligand into activity buffer containing no protein. The heat from these control injections were subtracted from the data before the final analysis and curve fitting. All ITC data analyses were performed using the MicroCal PEAQ-ITC Analysis Software (Malvern) with a model of single set of identical ligand binding sites.
ATPase assays
The ATPase activities of PilBΔN and PilB were analyzed by a malachite green (MLG)-based endpoint assay as previously described [27]. Assays were carried out in triplicates in activity buffer for 15 min at 54 °C with 75 nM of PilBΔN or PilB and ATP at specified concentrations. Controls had ATP at various concentrations but with no protein. The results shown were the averages and standard deviations from three independent experiments. If two samples had a P-value of <0.05 from the student t-test, they were considered to be significantly different statistically.
Results
N- and C-terminal domains of CtPilB bind cdG and adenine nucleotides individually
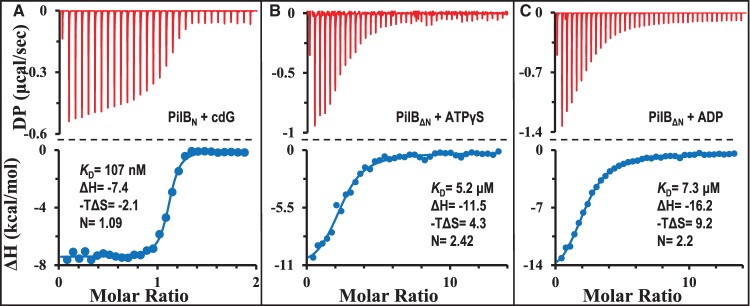
Based on sequence alignment [34] (Supplementary Figure S1), many PilB proteins contain the MshEN-type cdG binding domain at their N termini besides the ATPase domain at their C termini [22–26,35]. Here, the N- and C-terminal domains of CtPilB were examined for their ability to individually bind their respective ligands. First, the MshEN equivalent of CtPilB was analyzed for its binding of cdG. Residues 1–146 of CtPilB (PilBN), corresponding to the cdG binding domain of MshE (Supplementary Figure S1), was expressed in E. coli with a 6×His tag at its C-terminus and purified by FPLC using Ni-NTA and gel filtration columns (Supplementary Figure S2). Analytical size exclusion chromatography (SEC) estimated that the purified PilBN has an apparent MW of 21 kDa (Supplementary Figure S2). This is in comparison with 19.5 kDa, the theoretical MW of the PilBN with the 6×His affinity tag, indicating that PilBN is monomeric in solution. As shown in Figure 1A, the binding of PilBN with cdG, which was analyzed by ITC [36], was exothermic as observed for that of MshEN [34]. Analysis of the ITC results indicated that PilBN binds cdG with an apparent KD of 107 nM and a 1 : 1 stoichiometry. This association has a ΔH of −7.4 kcal/mol and a calculated −TΔS value of −2.1 kcal/mol (Table 1). These negative values indicate that the binding interactions between PilBN and cdG are driven by favorable changes in both enthalpic (ΔH) and entropic (ΔS) forces [37–39]. This is consistent with both the hydrophilic and hydrophobic interactions of MshEN with cdG observed in their co-crystal structure [18]. The change in free energy (ΔG) for PilBN-cdG binding is −9.5 kcal/mol (Table 1), indicating a fairly favorable interaction thermodynamically. For the purpose of comparison in scale, the free energy (ΔG) released from the hydrolysis of ATP is −8.48 kcal/mol or −35 kJ/mol [40].
Figure 1. N- and C-terminal domains of CtPilB bind to their respective ligands specifically.
ITC results and their analysis are shown for the binding of (A) PilBN with cdG, (B) PilBΔN with ATPγS, and (C) PilBΔN with ADP. In each panel, the ITC thermogram is shown on the top and the isotherm from curve fitting at the bottom. DP stands for power differential and the time interval between injections was 150 s. Shown with the isotherm is the thermodynamic parameter calculated from curve fitting. These include ΔH and −TΔS in kilocalorie per mole as well as the dissociation constant (KD) and the number of ligand binding sites (N) per protein. Standard deviations for the relevant parameters from curve fitting are listed in Table 1.
Table 1. Summary of parameters for PilB and ligand binding with or without effectors.
| PilBN | PilBΔN | Full-length PilB | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Ligand1 | cdG | ATPγS | ADP | cdG | ATPγS | ADP | cdG | cdG | ATPγS | ADP |
| Effector2 | None | None | None | None | None | None | ATPγS | ADP | cdG | cdG |
| KD3 | 107±14* | 5.2±0.7 | 7.3±0.5 | 53±10* | 3.8±0.5 | 4.3±1.0 | 58±15* | 18±6* | 4.0±0.5 | 9.0±0.7 |
| ΔG | −9.5 | −7.2 | −7.0 | −9.9 | −7.4 | −7.3 | −9.9 | −10.6 | −7.4 | −6.9 |
| ΔH | −7.4±0.5 | −11.5±0.6 | −16.2±0.4 | −14.5±0.13 | −10.3±0.3 | −15.4±1.0 | −14.0±0.3 | −12.0±0.09 | −10.3±0.3 | −46.2±1.6 |
| −TΔS | −2.1 | 4.3 | 9.2 | 4.6 | 2.9 | 8.1 | 4.1 | 1.5 | 2.9 | 39.4 |
| N | 1.09±0.03 | 2.42±0.07 | 2.20±0.03 | 5.83±0.01 | 2.8±0.06 | 2.20±0.02 | 6.33±0.08 | 5.94±0.09 | 2.91±0.04 | 1.79±0.06 |
Ligand represents the compound that was titrated into the sample cell with the specified protein;
Effector refers the chemical that was pre-incubated with the protein;
The apparent dissociation constant (KD) is in micromolar (μm) except those for cdG (*) which is in nanomolar (nM). ΔG, ΔH, and TΔS are in kilocalorie per mole (kcal/mol). N is the number of ligand binding site per protein.
Next, the ATPase domain of CtPilB at the C terminus was examined for its interaction with adenine nucleotides. A truncated CtPilB (PilBΔN) without the first 139 residues (Supplementary Figure S1) was expressed in E. coli with a 6×His affinity tag fused to its C-terminus. It was purified by FPLC and examination by analytical SEC estimated its apparent MW to be 305 kDa (Supplementary Figure S2). This is close to six times of the calculated MW of 53.8 kDa for the fusion protein, indicating that PilBΔN without the cdG binding domain still exists as a hexamer like the full-length CtPilB [27]. Initially, ATP was used as a ligand to analyze its binding to PilBΔN by ITC. The results from the titration (Supplementary Figure S3A), however, suggested that PilBΔN is likely active as an ATPase (also see later sections), and the heat released from ATP hydrolysis under our experimental conditions interfered with productive data analysis. To circumvent this issue, the ATP analog ATPγS was used as a ligand instead (Figure 1B). The thermogram showed no detectable hydrolysis of ATPγS and curve fitting indicated an apparent KD of 5.2 μM with ∼2 ATPγS binding sites per PilBΔN hexamer. The change in free energy is −7.2 kcal/mol, the sum of a ΔH of −11.5 kcal/mol and −TΔS of 4.3 kcal/mol (Table 1). Binding of ADP to PilBΔN was similarly analyzed (Figure 1C). Curve fitting resulted in an apparent KD of 7.3 μM and ∼2 ADP binding sites per PilBΔN hexamer. The change in free energy (ΔG) is −7.0 kcal/mol, a value similar to that for the binding of ATPγS. These results indicate that the C-terminal ATPase domain of PilB, which is likely active as a hexameric ATPase (Supplementary Figure S2), can bind adenine nucleotides on its own without the cdG binding domain. It is noted that there are differences between the binding of ATPγS and ADP with PilB mechanistically. The respective ΔH values for these two nucleotides, −11.5 and −16.2 kcal/mol, are significantly different statistically (Figure 1B,C and Table 1), and so are the −TΔS values at 4.3 and 9.2 kcal/mol. These differences suggest that the binding of ADP is driven by favorable hydrophilic or enthalpic (H) interactions accompanied by more pronounced conformational or unfavorable entropic changes (ΔS) in comparison with ATPγS [37–39]. The binding of ATPγS and ADP to two of the six available binding sites in the hexamer is consistent with the symmetrical rotary model of catalysis proposed based on the crystal structures of the ATPase domains of PilB [23–25].
Lastly, both PilBN and PilBΔN were examined to determine whether they could associate with the ligand(s) of the other domain. The thermograms (Supplementary Figure S4) from ITC titrations and data analysis indicated no detectable binding between PilBN with either ADP (Supplementary Figure S4A) or ATPγS (Supplementary Figure S4B). Nor was there any detectable binding between PilBΔN with cdG (Supplementary Figure S4C). The collective results from ITC analysis with truncated CtPilB variants here (Figure 1 and Supplementary Figure S4) indicate that the N- and C-terminal domains of PilB are specific for their respective ligands. That is, the N terminus of PilB binds cdG while its C-terminal ATPase domain interacts with adenine nucleotides as expected for an ATPase. It is inferred from the ITC analysis that the monomeric PilBN binds cdG with a 1 : 1 stoichiometry while the hexameric PilBΔN has two binding sites for ATPγS and ADP under our experimental conditions. The thermodynamic signatures suggest that ATP and ADP bind to PilB through distinct binding processes or mechanisms with concurrent but unique changes in protein conformation [37–39].
Full-length PilB binds cdG and adenine nucleotides with higher affinity
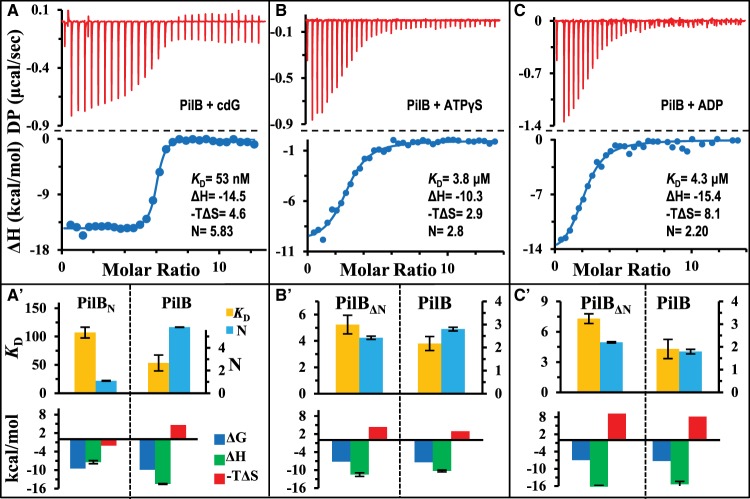
While the individual N- and C-terminal domains of PilB can bind to their respective ligands specifically, it is not known if the presence of one domain influences ligand binding by the other. Here, the full-length PilB was expressed and purified for the examination of its binding of cdG and adenine nucleotides by ITC. As shown in Figure 2A, the binding of cdG to the full-length PilB has a ΔG of −9.9 kcal/mol, which is as exergonic as PilBN–cdG binding with a ΔG of −9.5 kcal/mol (Table 1). Analysis and curve fitting of the data indicated a KD of 53 nM with a binding stoichiometry ∼6 cdG molecules per PilB hexamer (Table 1). This KD value is about half of that by PilBN (Table 1 and Figure 2A), indicating that the presence of the ATPase domain enhances the binding of cdG to the MshEN domain in the full-length PilB protein. The comparison of thermodynamic signatures (ΔG, ΔH, and −TΔS) of binding (Figure 2A′ and Table 1) also suggests differences between the binding process of PilBN and full-length PilB to cdG. While the ΔG remains largely unaffected, the ΔH here is −14.5 kcal/mol, which is significantly higher than −7.4 kcal/mol for PilBN. In even starker contrast, the values for −TΔS were negative for PilBN but positive for full-length CtPilB (Table 1). This suggests that the binding of cdG to full-length PilB involves or brings about additional and more favorable hydrophilic interactions than its binding to the N-terminal domain alone [37–39]. These favorable interactions help to overcome the unfavorable changes in entropy (ΔS) and likely explain the increase in affinity of the full-length PilB for cdG.
Figure 2. Full-length CtPilB binds cdG, ATPγS, and ADP with higher affinity.
Shown are ITC results and their analysis for the binding of CtPilB with (A) cdG, (B) ATPγS, and (C) ADP as presented in Figure 1. The time interval between injections was 150 s in (A) and 120 s in (B) and (C). In addition, panels (A′), (B′), and (C′) show graphical comparisons of the thermodynamic signatures for the binding of the same ligand to the full-length PilB and its relevant ligand-binding domains individually (either PilBN or PilBΔN). The comparisons for KD and N are on the top and those for ΔG, ΔH, and −TΔS at the bottom. The unit for KD is nanomolar in (A′) and micromolar in (B′) and (C′).
The full-length PilB was similarly examined for its binding with ATPγS and ADP by ITC (Figure 2B,C). The thermograms indicated clear exergonic reactions for the binding of both nucleotides to the full-length PilB with ΔG values similar as those for PilBΔN (Table 1). Curve fitting of the data resulted in KD values of 3.8 and 4.3 μM for the binding of ATPγS and ADP to PilB, respectively. In comparison, the respective KD's for the binding of these nucleotides by PilBΔN are ∼40% and 80% higher (Table 1), suggesting that the presence of the MshEN domain increases the binding affinity of these nucleotides to the ATPase domains. The binding signatures (Figure 2B,C and Table 1) suggest that the increase in the affinity of full-length PilB for these nucleotides may be attributable to less unfavorable entropic or hydrophobic forces. That is, the binding by the full-length protein tends to have slightly less change in −TΔS values as compared with PiBΔN. The 2 : 6 stoichiometry of ADP and PilB binding here (Table 1) differs from the 4 : 6 ratio reported for Geobacter sulfurreducens PilB (GsPilB) [25], perhaps because differences in the catalytic mechanism of these PilBs and/or the particular experimental conditions in these experiments. It is interesting to note that signs of ATP hydrolysis were again observed with the full-length CtPilB protein as indicated by the wider opening near or slower return of the power differential (DP) to the baseline in ITC experiment (Supplementary Figure S3B). However, the reduced interference by the heat released from ATP hydrolysis in comparison with PilBΔN (Supplementary Figure S3A) allowed an isotherm to be fitted to the data set with a KD of 8.5 μM and a stoichiometry of 2 ATP molecules per PilB hexamer (Supplementary Figure S3B). While this fitting should be taken with a grain of salt, these results provided indications that the full-length PilB may be less active than PilBΔN as an ATPase.
In summary, the results here indicated that the full-length PilB binds both cdG and adenine nucleotides with higher affinity than its separate domains in isolation. The most significant change in affinity occurs for cdG, followed by that for ADP. The binding of cdG results in more drastic change in the full-length protein than in PilBΔN with regard to both hydrophilic interactions (ΔH) and protein conformation (ΔS). In contrast, the binding of ATPγS and ADP may result in less conformational alterations in the full-length protein than in PilBΔN. All six cdG binding sites in the PilB hexamer are accessible for ligand binding while only two to three of the adenine nucleotide-binding sites are occupied under equilibrium conditions in our experiment.
Presence of ADP readies PilB for cdG binding
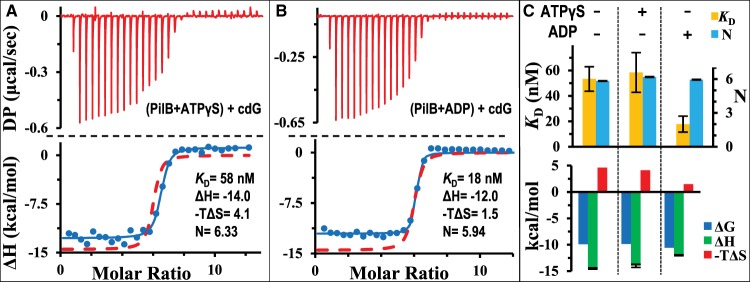
There is evidence that the binding of adenine nucleotides or lack thereof can influence the activity of MxPilB in signaling or promoting the production of EPS [28]. It is, therefore, possible that the presence of adenine nucleotides may impact the binding between PilB and cdG as the latter is the key signaling second messenger in biofilm regulation [31]. Here we examined the effect of ATPγS and ADP at saturating concentrations on the binding of cdG to PilB. Experimentally, PilB was pre-incubated with an effector, either ATPγS or ADP, and cdG was then titrated into the sample cell in ITC analysis. In these experiments, the effector was present at 50 μM, which is more than 10 times of their apparent KD value (Table 1 and Figure 2).
The thermogram for the experiment in the presence of ATPγS is shown in Figure 3A. Analysis indicated a good fit of an isotherm to the data with a KD of 58 nM for cdG. This is similar with the KD of 53 nM without ATPγS (Table 1), showing that ATPγS has no obvious effect on the affinity of PilB for cdG. Likewise, the binding stoichiometry and other thermodynamic properties of PilB–cdG interactions are not significantly affected by the presence of ATPγS. For example, the ΔG remains at 9.9 kcal/mol, regardless of the presence or absence of ATPγS. ΔH values were similar at −14.5 and −14.0 kcal/mol in the absence and presence of ATPγS, respectively (Table 1). These results indicate that the presence of ATPγS has negligible effect on the binding of PilB to cdG thermodynamically and stoichiometrically.
Figure 3. ADP, but not ATPγS, enhances the binding PilB with cdG.
Shown are the ITC results and their analysis for the titration of cdG into CtPilB pre-incubated with 50 μM of (A) ATPγS or (B) ADP presented in the same format as in Figure 1. The time interval between injections was 150 s. For the purpose of comparison, the isotherm for the binding of cdG and CtPilB without any adenine nucleotide from Figure 2 is included as the red dashed lines in both panels. Panel (C) shows the graphical comparison of thermodynamic signatures of cdG and CtPilB binding with (+) or without (−) pre-incubation with ADP and ATPγS as indicated at the top of the panel.
On the other hand, the results from ITC experiments in the presence of ADP indicate that ADP enhances the binding of PilB to cdG. As shown in Figure3B, an isotherm fitted to the data set resulted in a KD of 18 nM, one-third of the KD value of 53 nM in the absence of ADP (Table 1). The thermodynamic binding signature (Figure 3C) suggests that the increase in cdG affinity in the presence of ADP may be attributable to less unfavorable entropic or hydrophobic forces. While the values for both ΔH and ΔG shifted little (Table 1), there were significant differences in changes in entropy (ΔS) (Table 1). The −TΔS value is 1.5 and 4.6 kcal/mol with and without ADP, respectively. This three-fold change in ΔS correlates with the three-fold enhancement in the binding affinity of PilB for cdG in the presence of ADP. In the presence of both ADP and ATPγS, the binding parameters for cdG were essentially identical with those observed in the presence of ADP alone (data not shown). These results suggest that the presence of ADP enhances the binding of PilB to cdG, most likely by minimizing unfavorable changes in entropy correlated with protein conformation [37–39].
We, therefore, conclude here that it is the binding of ADP, but not ATP, that allosterically impacts or enhances the binding of PilB to the signaling molecule cdG. Thermodynamically, the binding of ADP to PilB seems to reduce the entropic barrier for the binding of cdG. Among the three PilB ligands examined, ADP results in the most drastic conformational changes in PilB when it binds. This is evident from the most negative TΔS value at −8.1 kcal/mol for the binding of ADP to PilB as compared with −2.9 and −4.6 kcal/mol for ATPγS and cdG. As has been mentioned, the −TΔS for the binding of cdG to PilB is reduced from 4.6 to 1.5 kcal/mol by the presence of ADP. In other words, it requires about two times less entropic energy for PilB to transition from a cdG-unbound state to a cdG-bound state in the presence of ADP. The binding of ADP, therefore, appears to pre-configure PilB to a conformational state more ready to accommodate cdG than a PilB in an apo state. This preparative or pre-configuration process of PilB for cdG binding appears only induced by the presence of ADP, but not ATPγS.
Presence of cdG decreases the affinity of PilB for ADP
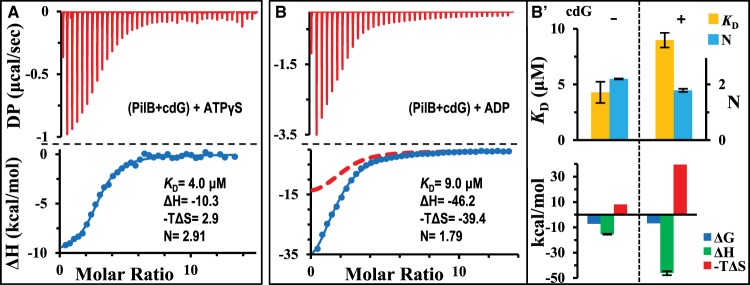
There is genetic evidence suggesting that the binding of cdG to PilB can modulate its function in T4P assembly [31,33,34] and possibly its interactions with adenine nucleotides. Here, we determined if cdG could influence the binding of PilB to adenine nucleotides by the PilB ATPase. These experiments were conducted by the pre-incubation of PilB with cdG at 100 μM, a concentration more than 40× of the KD for its binding to CtPilB (Table 1). The thermogram for the binding of ATPγS is shown in Figure 4A, which indicated that cdG did not affect the binding of PilB to ATPγS. Curve fitting produced a KD of 4.0 μM (Figure 4A), for example, which is indistinguishable from the KD of 3.8 μM without cdG (Table 1). In addition, the ΔG, ΔH, ΔS, and N values remain virtually unchanged regardless of the presence or absence of cdG (Table 1). These results indicate that cdG has no discernible thermodynamic effect on the equilibrium binding of ATPγS to PilB under our experimental conditions.
Figure 4. cdG inhibits the binding of PilB to ADP, but not ATPγS.
ITC results and their analysis for the titration of CtPilB pre-incubated with 100 μM cdG with (A) ATPγS and (B) ADP. The time interval between injections was 120 s. For comparison, the isotherm for the binding of ADP with CtPilB without cdG is included in (B) as a red dashed line. Panel (B′) shows the graphical comparison of thermodynamic signatures for the titration of ADP into PilB without (−) or with (+) pre-incubation with cdG as indicated at the top of the panel.
In contrast, ITC results with ADP indicate that its binding to PilB is significantly impacted by the presence of cdG. As shown in Figure 4B, the fitting of an isotherm to the ITC data resulted in a KD of 9.0 μM with cdG as an effector. This is significantly different from the KD of 4.3 μM in the absence of cdG (Table 1). The thermodynamic signatures clearly indicate that the presence of cdG drastically influenced the binding process or mechanisms between PilB and ADP [37–39]. The change in enthalpy (ΔH) for ADP binding stands at −46.2 kcal/mol in the presence of cdG and −15.4 kcal/mol in its absence. These differences suggest that cdG binding significantly enhanced the hydrophilic and/or ionic interactions between PilB and ADP [37–39]. On the other hand, the changes in entropy (ΔS) in the presence of cdG made the binding of ADP much less favorable due to unfavorable entropic interactions (Table 1). The −TΔS is calculated to be 39.4 kcal/mol in the presence of cdG; this is in comparison with a −TΔS value of only 8.1 kcal/mol without cdG. In other words, the presence of cdG enhanced favorable hydrophilic interactions between PilB and ADP by two-fold as indicated by the more negative value of ΔH. This enhancement, however, is offset by a four-fold less favorable change in entropy which in essence correlates with hydrophobic and conformational changes in protein [37–39]. We may infer from these results that when PilB is bound to cdG, it assumes a conformation that requires more drastic changes to accommodate ADP. In other words, the binding of cdG may configure PilB to a state with a lower affinity for the binding of ADP because of a higher conformational barrier. This is in contrast with the earlier conclusion that the binding of ADP as an effector lowers the conformational barrier and raises the affinity for PilB to bind cdG.
Removal of the N-terminus increases ATPase activity of PilB
The early results from the ITC experiments with ATP suggested that the N-terminal truncated PilBΔN is likely an active enzyme, perhaps with higher ATPase activity than its full-length counterpart. We first analyzed the ATPase activity of PilBΔN in comparison with the full-length protein by an endpoint assay at ATP concentrations from 0.1 to 3.0 mM (Figure 5). The ATPase activity of full-length PilB increased with increasing ATP concentrations up to 1.0 mM and it declined at concentrations beyond 2.0 mM ATP as was previously observed [27]. The ATPase activity of the N-terminal truncated PilB showed a similar concentration dependency as the full-length protein. That is, the overall ATPase profile of PilBΔN has an incline, a plateau and a decline over the tested ATP concentration range. On the other hand, PilBΔN displayed consistently and statistically higher activities between 33% and 50% than the full-length protein in both the incline and the plateau phases at ≤2.0 mM ATP. In the declining phase at 2.5 and 3.0 mM ATP, the differences are no longer statistically significant although PilBΔN still appeared slightly more active under our experimental conditions. These results lead to the conclusion that the N-terminus of CtPilB has an inhibitory effect on the ATPase activity of the full-length protein. This conclusion, which is supported by the observations in the ITC experiments (Supplementary Figure S3) is consistent with the notion that the binding of cdG to the N-terminus of PilB can regulate the ATPase activity endowed by the C-terminus.
Figure 5. Removal of MshEN domain increases ATPase activity of CtPilB.
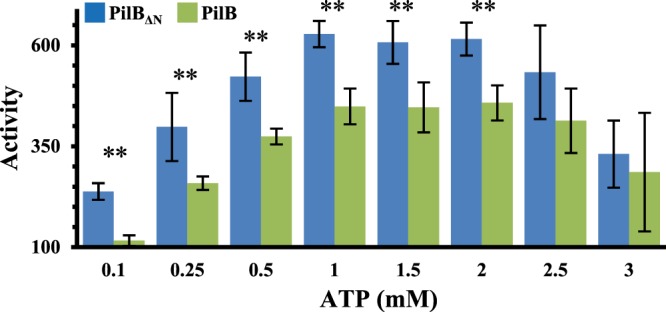
ATPase activity (nmol P/mg protein/min) of PilBΔN and full-length CtPilB was determined by MLG-based endpoint assays using 75 nM protein and ATP at specified concentrations. Shown are the averages and the standard deviations (error bar) from three independent experiments, each of which was conducted in triplicate samples. Pairs of stars (**) indicate the activities of PilBΔN and full-length CtPilB at the indicated ATP concentration are significantly different with a P-value <0.05 by unpaired student t-tests.
Since cdG binds to the N-terminus of CtPilB and influenced the binding of ADP, we next examined if this signaling molecule could regulate the ATPase activity of CtPilB. For this experiment, we used cdG concentrations at 0, 2, and 200 μM and observed no difference in ATPase activity by an endpoint assay (data not shown). We further analyzed the ATPase activity of PilB by kinetic experiments using ATP at concentrations of 0.5 and 1.0 mM (Supplementary Figure S5). Although it appears that the ATPase activity of PilB at both 0.5 and 1.0 mM ATP was slightly higher in the presence of cdG than in its absence, these small differences are not statistically significant. These results are consistent with published observations where only slight differences were observed in the ATPase activity of MshE in vitro in the absence and presence of cdG [33]. These results here (Figure 5 and Supplementary Figure S5) indicate that the N-terminal MshEN domain of CtPilB exerts an inhibitory effect on the ATPase activity of PilB, although cdG binding has negligible effect on the hydrolysis of ATP in vitro under our experimental conditions.
Discussion
Here, we used C. thermophilum PilB to study its interactions with two types of ligands. The first is cdG, a critical bacterial second messenger that regulates biofilm formation. The second are adenine nucleotides, which are the substrate and product of the PilB ATPase. We demonstrate that the N- and C-terminal domains of CtPilB on their own are sufficient for the binding of their respective ligands. These bindings are specific such that the C terminus binds only adenine nucleotides but not cdG while the N terminus has a strong affinity for cdG without detectable binding to ADP or ATPγS. The full-length PilB binds all ligands with higher affinity than its individual domains in isolation, indicating communications between these domains and potential allosteric effects of the different ligands on the functionality of the protein. Results indicate that there is indeed a mutual or reciprocal effect between ADP and cdG binding, although no effect was detected between ATPγS and cdG. More specifically, CtPilB displayed an increased affinity for cdG in the presence of ADP but showed decreased affinity for ADP in the presence of cdG. Enzymatically, an N-terminus truncated variant of CtPilB is more active as an ATPase than its full-length counterpart, yet the presence of cdG had negligible effect on ATP hydrolysis by the wild-type CtPilB in vitro.
As has been discussed earlier, MxPilB and TtPilF possess the MshEN domain at their N-terminus and belong to the MshEN+-type category of T4P assembly ATPases (Supplementary Figure S1) [34]. In both cases, they are known to regulate the production of EPS, in addition to or independently of their function in T4P biogenesis and motility [28,29]. TtPilF was found to be phosphorylated on two residues, T368 and S372, which are within a conserved MshEN domain (Supplementary Figure S1) [29]. Non-phosphorylatable mutations of these two residues were found to result in changes in biofilm and EPS phenotypes without any effect on piliation or twitching motility [29]. MxPilB was discovered to be a signaling protein in the regulation of EPS production upstream of a chemotaxis-like signal transduction pathway [28,41]. More specifically, a MxPilB mutant variant with a substitution in the Walker A box (K327A) is constitutively active as a regulator of EPS production despite its lack of ATPase activity and inability to support pilus assembly or T4P-dependent motility [28]. In vitro studies suggested that it is the adenine nucleotide-free or the apo form of PilB that actively signals EPS production [28]. These results indicate that MshEN+-type PilB ATPases, besides their role in T4P assembly, can function as regulators of EPS production for the development of bacterial biofilm.
The locations of PilB mutations in T. thermophilus and M. xanthus with effects on EPS production suggest communications between the N- and the C-terminal domains of PilB. The two TtPilB residues implicated in EPS regulation reside within a conserved MshEN domain immediately N terminal to its ATPase domain (Supplementary Figure S1) [29,35]. In contrast, the two mutations in MxPilB that led to constitutive EPS signaling are located in the Walker boxes within the ATPase domain [28]. While the mechanism of PilB signaling remains to be elucidated, these observations suggest that alteration in either the N-terminal MshEN domain or the C-terminal ATPase domain can drastically change the signaling properties of PilB. In other words, whatever the signaling mechanism may be, there is close communication between the N and the C termini of the protein because changes in either can significantly alter the regulatory outcome of PilB in signaling EPS production.
Because the N- and the C-terminal domains of the MshEN+-type PilB can bind to their distinct ligands individually, it is reasonable to assume the binding of one ligand to one domain can allosterically affect the conformation of and the ligand binding by the other domain. This is indeed manifested by the mutual allosteric effects of cdG and ADP as determined by their binding to CtPilB. There are significant changes in the mechanisms of binding of one in the presence of the other as indicated by their respective thermodynamic binding signatures (Table 1). In the presence of ADP, the binding cdG to PilB displayed a three-fold decrease in −TΔS (1.5 vs 4.6 kcal/mol), indicating that the binding of ADP lowered the entropic barrier for the binding of cdG. On the other hand, when PilB was pre-incubated with cdG, the binding of ADP required about a five-fold increase in −TΔS (39.4 vs 8.1 kcal/mol). This indicates that the binding of cdG significantly increased the entropic barrier for ADP binding. These changes in ΔS are reflected in the binding affinity because ADP decreased the KD of PilB for cdG from 53 to 18 nM while cdG increased the KD of PilB for ADP from 4.3 to 9.0 μM. While cdG and ADP have the opposite effect on the binding of the other, these observations established a mutual allosteric relationship between the binding of these two ligands.
The dichotomy of the opposite effect by ADP and cdG on the binding of the other might appear surprising but it does not defy logic that governs biological processes. In the two-state model for the activity of a signaling GTPase, a G-protein exists in either a GTP-bound and actively signaling state or a GDP-bound and inactive conformation [42]. In the GDP-bound state, the protein tends to have a higher affinity for GDP/GTP exchange factors (GEFs) [42,43]. That is, the association with GDP increases the affinity of a G-protein for GEFs. On the other hand, once a GEF binds, it catalyzes the release of GDP from the G-protein to facilitate the binding of the same pocket by GTP. In other words, the binding of GDP to a G-protein favors the binding of a GEF, but the association of a GEF reduces the affinity of the G-protein for GDP [42,43]. The ultimate outcome from these interactions is the conversion of the GDP-bound inactive protein to the GTP-bound actively signaling state. This scenario is analogous to the opposite effect of cdG and ADP on the binding of each other we observed here. That is, the presence of ADP enhances the binding of cdG while the binding of cdG decreases the affinity of PilB for ADP (Figure 6) [44]. While the biological relevance of this dichotomy in the case of G-proteins is clear, the significance or relevance of the allosteric interactions between cdG and ADP in our case remains to be fully elucidated. It is pertinent to note here that the signaling conformation of MxPilB was proposed to correspond with the adenine nucleotide-free state of PilB [28]. One possible explanation for the dichotomous effect between cdG and ADP could be that the association of ADP primes PilB for cdG binding as GEF does a G-protein for GTP (Figure 6). The full occupancy of the cdG binding sites on PilB, in turn, leads to the release of ADP to configure PilB to its EPS signaling conformation. That is, for the regulation of EPS production, PilB may use its cdG-binding domain for signaling output while the C-terminal domain primarily functions for allosteric modulation of the signaling domain. In this scenario, the association of ADP with the regulatory domain affects the binding of cdG to the output domain. In a simple model, the outcome from these interactions leads to the attainment of an actively signaling PilB molecule which is cdG-bound without any adenine nucleotide (Figure 6). The non-phosphorylatable mutations in TtPilF [29] may influence the binding of cdG and/or domain–domain interactions that impact the attainment of this actively signaling conformation by TtPilF.
Figure 6. Model for the regulation of PilB signaling activity by cdG and ADP.
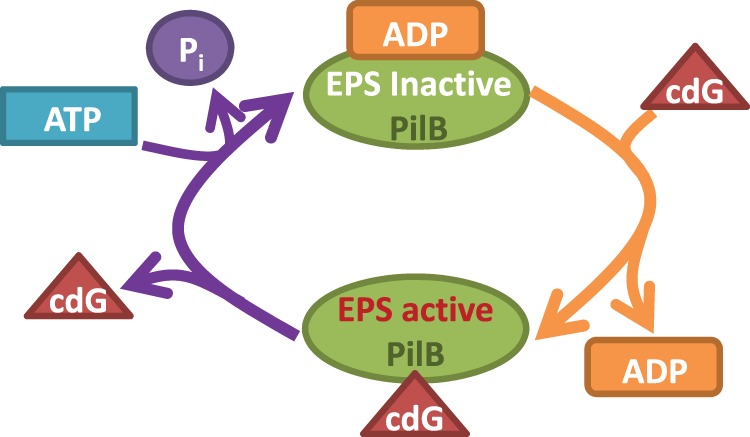
PilB is inactive in EPS signaling when it is associated with ADP. In this ADP-bound state, however, PilB has high affinity for cdG and it has a high probability to bind cdG. The binding of cdG, in turn, results in a decrease in the affinity of PilB for ADP and the release of ADP leads to the PilB–cdG complex to its active EPS-signaling conformation. The dissociation of cdG inactivates PilB in EPS signaling and may simultaneously facilitates the ATP hydrolysis cycle by PilB. The left side of this model is functionally analogous to the conversion of a G-protein from a GDP-bound and inactive signaling state to GTP-bound and active state which is catalyzed by GEF.
The binding stoichiometries of PilB with ADP and ATPγS are consistent with the rotary mechanisms of catalysis for this ATPase [12,23,25,26]. The ATPase domains of PilB are hexameric with a two-fold symmetry in crystal structures, and the two protomers at the opposite corners of the hexameric ring assume the same conformation[12,23,25]. These three pairs of protomers are proposed to represent distinct steps in ATP hydrolysis propagated through this ring ATPase by rotations. One pair is proposed to be in an ATP-bound and hydrolysis ready state, another in an immediate post-ATP-hydrolysis state with the last pair in an ADP-bound state poised for ADP/ATP exchange. Our results indicated that the catalytically active CtPilB and PilBΔN both bound approximately two ADPs per hexamer (Table 1) in its activity buffer. Similarly, two ATPγS molecules are likely bound to each PilB hexamer (Table 1) with the additional occupancy accounted for by the contaminating ADP in the commercially available ATPγS which can contain up to 10% of ADP. The variations in apparent stoichiometry by CtPilB (N = 2.8) and PilBΔN (N = 2.4) may be accounted for by their differing affinities for ADP under our experimental conditions. That is, the full-length protein, which displays a higher affinity for ADP, may have a slightly higher number of binding sites occupied by ADP than PilBΔN under conditions where ADP is not saturating. The binding stoichiometry observed here can, therefore, be explained by the proposed rotary mechanisms of catalysis for PilB [12,23,25,26].
At the enzyme level, the N-terminal truncated CtPilB is more active as an ATPase than its full-length counterpart (Figure 5). This is in contrast with N-terminal truncated TtPilF variants which showed no significant change in activity from the wild type [35]. This perhaps relates to differences in the specific points of truncations and to the architecture of the proteins in question. TtPilF has three tandem MshEN-like domains at its N-terminus while the PilB from most T4P systems has only one such domain [29,35,45,46]. In addition, we observed that two deletions that removed additional residues from CtPilB resulted in proteins with very low ATPase activity (data not shown). It is known that the crystal forms of the ATPase domain of TtPilF is inactive as an ATPase [22,23]. Somewhat unexpectedly, cdG did not affect the ATPase activity of full-length CtPilB under our experimental conditions (Supplementary Figure S5), despite its effect on ADP binding (Figure 4 and Table 1). On the other hand, this is consistent with the observations that the low ATPase activity of VcMshE and GsPilB in vitro was affected by cdG only to a very limited extent [33] or not at all [25]. The biological and biochemical relevance of these observations in vitro is not clear. It is possible that these somewhat puzzling results are due to the lack of interacting partners for PilB in vitro and their resulting physical or mechanical constraints on the freedom of movement by PilB domains in vivo. In other words, as the T4P assembly motor, PilB performs mechanical work within the T4PM using energy harvested from ATP hydrolysis. In the PilB-only system here, the protein–protein interactions that apply such physical constraints and mechanical loads in vivo are absent. This absence provides one possible explanation for the lack of any observable effect of cdG on the ATPase activity of PilB. A change in load may also explain the increase in ATPase activity of the N-terminal truncated PilB (Figure 5). That is, the removal of the MshEN domain may decrease the mass that has to be displaced continuously when a PilB hexamer hydrolyzes ATP by the proposed rotary mechanism of catalysis [23–26]. Further investigations are necessary to fully understand how ATP hydrolysis by PilB is coupled to T4P assembly and how the dual and paradoxical functions of PilB in T4P biogenesis and EPS signaling are controlled in vivo.
Acknowledgements
We would like to acknowledge Andreas Sukmana for the construction of pAS104 and for performing preliminary enzyme assays. We are grateful to Dr. Shiv Kale for granting us access to the ITC instrument and more importantly for his generous assistance with his expertize. We thank Dr. Florian Schubot for critical reading and constructive discussions of this manuscript.
Abbreviations
- cdG
cyclic-di-GMP
- EPS
exopolysaccharide
- GEFs
GDP/GTP exchange factors
- ITC
isothermal titration calorimetry
- MLG
malachite green
- MSHA
mannose-sensitive hemagglutinin
- MshEN
N-terminus of MshE
- MW
molecular weight
- SEC
size exclusion chromatography
- T4P
type IV pilus
- T4PM
type IV pilus machinery
Author Contributions
Both K.J.D. and Z.Y. designed research, analyzed data, and wrote the manuscript. K.J.D. performed research and collected data.
Funding
This work was supported by National Science Foundation grants MCB-1417726 and MCB-1919455 to Z.Y.
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Supplementary Material
References
- 1.Giltner C.L., Nguyen Y. and Burrows L.L. (2012) Type IV pilin proteins: versatile molecular modules. Microbiol. Mol. Biol. Rev. 76, 740–772 10.1128/MMBR.00035-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Craig L., Forest K.T. and Maier B. (2019) Type IV pili: dynamics, biophysics and functional consequences. Nat. Rev. Microbiol. 17, 429–440 10.1038/s41579-019-0195-4 [DOI] [PubMed] [Google Scholar]
- 3.Shi W. and Sun H. (2002) Type IV pilus-dependent motility and its possible role in bacterial pathogenesis. Infect. Immun. 70, 1–4 10.1128/IAI.70.1.1-4.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ellison C.K., Dalia T.N., Vidal Ceballos A., Wang J.C.-Y., Biais N., Brun Y.V. et al. (2018) Retraction of DNA-bound type IV competence pili initiates DNA uptake during natural transformation in Vibrio cholerae. Nat. Microbiol. 3, 773–780 10.1038/s41564-018-0174-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maier B. and Wong G.C.L. (2015) How bacteria use Type IV pili machinery on surfaces. Trends Microbiol. 23, 775–788 10.1016/j.tim.2015.09.002 [DOI] [PubMed] [Google Scholar]
- 6.Craig L., Pique M.E. and Tainer J.A. (2004) Type IV pilus structure and bacterial pathogenicity. Nat. Rev. Microbiol. 2, 363–378 10.1038/nrmicro885 [DOI] [PubMed] [Google Scholar]
- 7.Pelicic V. (2008) Type IV pili: e pluribus unum? Mol. Microbiol. 68, 827–837 10.1111/j.1365-2958.2008.06197.x [DOI] [PubMed] [Google Scholar]
- 8.Jakovljevic V., Leonardy S., Hoppert M. and Sogaard-Andersen L. (2008) Pilb and PilT are ATPases acting antagonistically in type IV pilus function in Myxococcus xanthus. J. Bacteriol. 190, 2411–2421 10.1128/JB.01793-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Salzer R., Joos F. and Averhoff B. (2014) Type IV pilus biogenesis, twitching motility, and DNA uptake in Thermus thermophilus: discrete roles of antagonistic ATPases PilF, PilT1, and PilT2. Appl. Environ. Microbiol. 80, 644–652 10.1128/AEM.03218-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wall D. and Kaiser D. (1999) Type IV pili and cell motility. Mol. Microbiol. 32, 1–10 10.1046/j.1365-2958.1999.01339.x [DOI] [PubMed] [Google Scholar]
- 11.Talà L., Fineberg A., Kukura P. and Persat A. (2019) Pseudomonas aeruginosa orchestrates twitching motility by sequential control of type IV pili movements. Nat. Microbiol. 4, 774–780 10.1038/s41564-019-0378-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Merz A.J., So M. and Sheetz M.P. (2000) Pilus retraction powers bacterial twitching motility. Nature 407, 98–102 10.1038/35024105 [DOI] [PubMed] [Google Scholar]
- 13.Skerker J.M. and Berg H.C. (2001) Direct observation of extension and retraction of type IV pili. Proc. Natl Acad. Sci. U.S.A. 98, 6901–6904 10.1073/pnas.121171698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clausen M., Jakovljevic V., Sogaard-Andersen L. and Maier B. (2009) High-force generation is a conserved property of type IV pilus systems. J. Bacteriol. 191, 4633–4638 10.1128/JB.00396-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Skotnicka D., Petters T., Heering J., Hoppert M., Kaever V. and Søgaard-Andersen L. (2016) Cyclic Di-GMP regulates type IV pilus-dependent motility in Myxococcus xanthus. J. Bacteriol. 198, 77–90 10.1128/JB.00281-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jain R., Sliusarenko O. and Kazmierczak B.I. (2017) Interaction of the cyclic-di-GMP binding protein FimX and the Type 4 pilus assembly ATPase promotes pilus assembly. PLoS Pathog. 13, e1006594 10.1371/journal.ppat.1006594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chang Y.-W., Rettberg L.A., Treuner-Lange A., Iwasa J., Søgaard-Andersen L. and Jensen G.J. (2016) Architecture of the type IVa pilus machine. Science 351, aad2001 10.1126/science.aad2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mattick J.S. (2002) Type IV pili and twitching motility. Annu. Rev. Microbiol. 56, 289–314 10.1146/annurev.micro.56.012302.160938 [DOI] [PubMed] [Google Scholar]
- 19.Xia J., Chen J.J., Chen Y., Qian G.L. and Liu F.Q. (2018) Type IV pilus biogenesis genes and their roles in biofilm formation in the biological control agent Lysobacter enzymogenes OH11. Appl. Microbiol. Biotechnol. 102, 833–846 10.1007/s00253-017-8619-4 [DOI] [PubMed] [Google Scholar]
- 20.Bischof L.F., Friedrich C., Harms A., Søgaard-Andersen L. and van der Does C. (2016) The type IV pilus assembly ATPase PilB of Myxococcus xanthus interacts with the inner membrane platform protein PilC and the nucleotide-binding protein PilM. J. Biol. Chem. 291, 6946–6957 10.1074/jbc.M115.701284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kruse K., Salzer R. and Averhoff B. (2019) The traffic ATPase PilF interacts with the inner membrane platform of the DNA translocator and type IV pili from Thermus thermophilus. FEBS Open Bio 9, 4–17 10.1002/2211-5463.12548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Collins R., Karuppiah V., Siebert C.A., Dajani R., Thistlethwaite A. and Derrick J.P. (2018) Structural cycle of the Thermus thermophilus PilF ATPase: the powering of type IVa pilus assembly. Sci. Rep. 8, 14022 10.1038/s41598-018-32218-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mancl J.M., Black W.P., Robinson H., Yang Z. and Schubot F.D. (2016) Crystal structure of a type IV pilus assembly ATPase: insights into the molecular mechanism of PilB from Thermus thermophilus. Structure 24, 1886–1897 10.1016/j.str.2016.08.010 [DOI] [PubMed] [Google Scholar]
- 24.McCallum M., Tammam S., Khan A., Burrows L.L. and Howell P.L. (2017) The molecular mechanism of the type IVa pilus motors. Nat. Commun. 8, 15091 10.1038/ncomms15091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Solanki V., Kapoor S. and Thakur K.G. (2018) Structural insights into the mechanism of Type IVa pilus extension and retraction ATPase motors. FEBS J. 285, 3402–3421 10.1111/febs.14619 [DOI] [PubMed] [Google Scholar]
- 26.Tsai C.-L. and Tainer J.A. (2016) The ATPase motor turns for type IV pilus assembly. Structure 24, 1857–1859 10.1016/j.str.2016.10.002 [DOI] [PubMed] [Google Scholar]
- 27.Sukmana A. and Yang Z. (2018) The type IV pilus assembly motor PilB is a robust hexameric ATPase with complex kinetics. Biochem. J. 475, 1979–1993 10.1042/BCJ20180167 [DOI] [PubMed] [Google Scholar]
- 28.Black W.P., Wang L., Jing X., Saldaña R.C., Li F., Scharf B.E. et al. (2017) The type IV pilus assembly ATPase PilB functions as a signaling protein to regulate exopolysaccharide production in Myxococcus xanthus. Sci. Rep. 7, 7263 10.1038/s41598-017-07594-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu W.-L., Liao J.-H., Lin G.-H., Lin M.-H., Chang Y.-C., Liang S.-Y. et al. (2013) Phosphoproteomic analysis reveals the effects of PilF phosphorylation on type IV pilus and biofilm formation in Thermus thermophilus HB27. Mol. Cell. Proteomics 12, 2701–2713 10.1074/mcp.M113.029330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Branda S.S., Vik S., Friedman L. and Kolter R. (2005) Biofilms: the matrix revisited. Trends Microbiol. 13, 20–26 10.1016/j.tim.2004.11.006 [DOI] [PubMed] [Google Scholar]
- 31.Jenal U., Reinders A. and Lori C. (2017) Cyclic di-GMP: second messenger extraordinaire. Nat. Rev. Microbiol. 15, 271–284 10.1038/nrmicro.2016.190 [DOI] [PubMed] [Google Scholar]
- 32.Romling U., Galperin M.Y. and Gomelsky M. (2013) Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol. Mol. Biol. Rev. 77, 1–52 10.1128/MMBR.00043-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roelofs K.G., Jones C.J., Helman S.R., Shang X., Orr M.W., Goodson J.R. et al. (2015) Systematic identification of cyclic-di-GMP binding proteins in Vibrio cholerae reveals a novel class of cyclic-di-GMP-binding ATPases associated with type II secretion systems. PLoS Pathog. 11, e1005232 10.1371/journal.ppat.1005232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Y.-C., Chin K.-H., Tu Z.-L., He J., Jones C.J., Sanchez D.Z. et al. (2016) Nucleotide binding by the widespread high-affinity cyclic di-GMP receptor MshEN domain. Nat. Commun. 7, 12481 10.1038/ncomms12481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kruse K., Salzer R., Joos F. and Averhoff B. (2018) Functional dissection of the three N-terminal general secretory pathway domains and the Walker motifs of the traffic ATPase PilF from Thermus thermophilus. Extremophiles 22, 461–471 10.1007/s00792-018-1008-9 [DOI] [PubMed] [Google Scholar]
- 36.Matsuyama B.Y., Krasteva P.V. and Navarro M. (2017) Isothermal titration calorimetry to determine apparent dissociation constants (Kd) and stoichiometry of interaction (n) of C-di-GMP binding proteins. Methods Mol. Biol. 1657, 403–416 10.1007/978-1-4939-7240-1_30 [DOI] [PubMed] [Google Scholar]
- 37.Vega S., Abian O. and Velazquez-Campoy A. (2016) On the link between conformational changes, ligand binding and heat capacity. Biochim Biophys. Acta 1860, 868–878 10.1016/j.bbagen.2015.10.010 [DOI] [PubMed] [Google Scholar]
- 38.Velazquez-Campoy A., Leavitt S.A. and Freire E. (2015) Characterization of protein-protein interactions by isothermal titration calorimetry. Methods Mol. Biol. 1278, 183–204 10.1007/978-1-4939-2425-7_11 [DOI] [PubMed] [Google Scholar]
- 39.Liang Y. (2008) Applications of isothermal titration calorimetry in protein science. Acta Biochim. Biophys. Sin (Shanghai) 40, 565–576 10.1111/j.1745-7270.2008.00437.x [DOI] [PubMed] [Google Scholar]
- 40.Rosing J. and Slater E.C. (1972) The value of ΔG° for the hydrolysis of ATP. Biochim. Biophys. Acta, Bioenerg. 267, 275–290 10.1016/0005-2728(72)90116-8 [DOI] [PubMed] [Google Scholar]
- 41.Black W.P., Xu Q. and Yang Z. (2006) Type IV pili function upstream of the Dif chemotaxis pathway in Myxococcus xanthus EPS regulation. Mol. Microbiol. 61, 447–456 10.1111/j.1365-2958.2006.05230.x [DOI] [PubMed] [Google Scholar]
- 42.Syrovatkina V., Alegre K.O., Dey R. and Huang X.-Y. (2016) Regulation, signaling, and physiological functions of G-proteins. J. Mol. Biol. 428, 3850–3868 10.1016/j.jmb.2016.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vetter I.R. and Wittinghofer A. (2001) The guanine nucleotide-binding switch in three dimensions. Science 294, 1299–1304 10.1126/science.1062023 [DOI] [PubMed] [Google Scholar]
- 44.Gerya B. (2009) Regulation of G-Protein activity by GEF and GAP (https://commons.wikimedia.org/)
- 45.Collins R.F., Hassan D., Karuppiah V., Thistlethwaite A. and Derrick J.P. (2013) Structure and mechanism of the PilF DNA transformation ATPase from Thermus thermophilus. Biochem. J. 450, 417–425 10.1042/BJ20121599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chiang P., Sampaleanu L.M., Ayers M., Pahuta M., Howell P.L. and Burrows L.L. (2008) Functional role of conserved residues in the characteristic secretion NTPase motifs of the Pseudomonas aeruginosa type IV pilus motor proteins PilB, PilT and PilU. Microbiology 154, 114–126 10.1099/mic.0.2007/011320-0 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.