Abstract
African animal trypanosomiases are caused by trypanosomes cyclically or mechanically transmitted by tsetse and other biting flies. Although molecular tools have been developed to identify drug-resistant trypanosomes in mammals, little or no investigation on drug-resistance has been undertaken on trypanosomes harbored by tsetse flies. Moreover, no data on mechanical vectors of African trypanosomes is available in most endemic areas of Cameroon. This study was designed to update our knowledge on the cyclical and mechanical vectors of African trypanosomes, and using molecular tools to identify different trypanosome species as well as diminazene aceturate resistant trypanosomes in tsetse flies trapped at Yoko in the Centre region of Cameroon.
For this study, traps were used to catch tsetse and mechanical vectors of African trypanosomes. The flies trapped were counted and identified by sex and species. DNA was extracted from tsetse and species-specific primers were used to identify different trypanosome species. PCR-RFLP was used to detect diminazene aceturate resistant strains of Trypanosoma congolense.
In all, 454 flies comprising 168 (37%) Tabanus spp., 71 (15.6%) Stomoxys spp. and 215 (47.4%) tsetse fly (i.e. 107 (49.8%) Glossina fusca congolensis, 71 (33%) Glossina fusca fusca and 37 (17.2%) Glossina palpalis palpalis) were trapped. Trypanosome infections were identified in 12.6% (27/215) of tsetse flies: 13 in G. f. congolensis, 6 in G. p. palpalis and 5 in G. f. fusca. From 24 T. congolense positive samples, PCR-RFLP was successful on 37.5% of the samples. Four samples (16.2%) harbored T. congolense strains that were resistant to diminazene aceturate while the remaining samples had drug-sensitive strains.
These results show for the first time the applicability of molecular tools for the identification of drug-resistant trypanosomes in tsetse. They revealed the existence of diminazene aceturate resistant strains of T. congolense in the tsetse-infested area of Yoko in the Centre region of Cameroon. Detection of drug-resistant trypanosomes in tsetse may enable scientists to map with accuracy specific areas where these parasites are transmitted. With such mapping, control strategies against African trypanosomiases could be improved by adapting control measures according to drug resistance distribution.
Keywords: Animal African trypanosomiasis, Tsetse fly, Trypanosomes, Drug resistance, PCR-RFLP
Graphical abstract

1. Introduction
African Animal Trypanosomiases (AAT) also known as “nagana” are parasitic diseases caused by flagellated protozoa of the genus Trypanosoma that can be transmitted cyclically by tsetse flies or mechanically by other biting flies such as Tabanus spp. and Stomoxys spp. (Steverding, 2008). As vector-borne diseases, AAT constitutes one of the most important livestock diseases in Africa because it causes acute anemia, weight loss, decrease milk production and other signs that may lead to death of cattle in endemic regions (Reis et al., 2019). In tsetse-infested areas, >50 million cattle and 230 million small ruminants are at risk of AAT (Vreysen et al., 2013). Trypanosome infections induce the loss of 10 to 50% of cattle, 2 to 10% of agricultural production, 5 to 30% of meat and 10 to 40% of milk production (Shaw, 2004). AAT remains the main constraint for livestock and agricultural development in many Sub-Saharan countries (Diall et al., 2017). It compounds poverty levels by constraining about 34% of all livestock farmers to subsist with <1.24 USD per day (Kabayo, 2002; Perry and Sones, 2007).
Many strategies including vector control, as well as the diagnosis and treatment of infected animals, have been developed to fight against AAT and its cyclical tsetse fly vector. Currently, only three trypanocides including isometamidium chloride (ISM), diminazene aceturate (DA) and ethidium bromide (Barrett et al., 2004) are available for the treatment of trypanosome infections in animals. In Africa, nearly 35 million doses of trypanocides are annually used (Holmes, 2013). This represents a figure suitable to treat about one-third of the cattle at risk (Swallow, 2000). Including of trypanocides sold informally in the African market may substantially increase the total number of doses sold annually, which may be as high as 70 million doses. Despite this demand, the budget required for the development of new drug and the low anticipated profit from the sale of trypanocides in developing countries are not enough to encourage pharmaceutical companies to investment in this field (Connor, 1992). Moreover, the inappropriate use of trypanocides has led to the emergence of resistant trypanosomes in some affected countries (Talaki et al., 2013).
The first reports on drug resistance in trypanosomes date back to the 1960s (Jones-Davies, 1967). In recent decades, drug resistance in African trypanosomes has been reported in 21 African countries (Talaki et al., 2013; Jamal et al., 2005; Mamoudou et al., 2008; Mamoudou et al., 2006; Sinyangwe et al., 2004; Odeniran et al., 2019a). However, the extent of drug resistance remains poorly understood because the in-vivo and in-vitro culture methods commonly used to determine drug resistance are laborious, difficult to implement in endemic countries and are limited by their long duration (Clausen et al., 2000). In addition, some trypanosome strains could not adapt in culture media (Clausen et al., 2000). It is in this light that molecular methods have been developed to improve the identification of drug-resistant trypanosomes (Delespaux et al., 2006; Nerima et al., 2007; Vitouley et al., 2011). Up till now, little or no studies had been tried to identify drug-resistant trypanosomes in tsetse flies, and no approach has so far been developed to highlight the presence of chemo-resistant trypanosomes in vectors. Nonetheless, the identification of drug-resistant trypanosomes in tsetse flies may constitute one of the best approaches to map drug resistance for the ultimate goal of boosting control strategies against AAT. To better understand the spread of drug resistance in AAT, it is becoming important not only to identify trypanosomes infesting animal hosts and tsetse fly vectors, but especially the tsetse flies harboring trypanosomes which are resistant to trypanocides.
This study aimed to update our knowledge on the cyclical and mechanical vectors of African trypanosomes, and to use molecular tools and methods in order to identify different trypanosome species as well as detect diminazene aceturate resistant trypanosomes in tsetse flies trapped at Yoko in the Centre region of Cameroon. Such identifications could enable rational management of existing trypanocides with the final goal of limiting the spread of drug resistance and improving peasant economy.
2. Material and methods
2.1. Study area
This study was conducted in Yoko (5°35′33″N, 12°18′57″E) (Fig. 1), which is located in the “Mbam et Nkim” district of the Centre region at about 270 km to the northeast of Yaoundé, the political capital of Cameroon. Located in the buffer zone between the evergreen southern part and the northern Sahel savanna part of Cameroon, Yoko covers a surface area of about 15,000 km2. In this locality, about 135,000 ha of forest are protected and if trees are not cut down, Yoko will be able to earn stable incomes on the carbon market at the international level. The climate is of the savanna type with one short dry season from November to February and one rainy season from March to October. The average rainfall is about 916.6 mm with an average temperature of about 22 °C. The relief is formed by a wide variety of mountain ridges to the west, as well as vast plains, valleys and steep hills that disturb the monotony of the plains. The vegetation is mixed, and comprises galleries and dense forests as well as vast expanses of shrubs and herbaceous savanna. The inhabitants practice fishing, trade, and peasant agriculture dominated by diverse subsistence crops. They also practice animal farming such as breeding of pigs, sheep, goats, cattle, donkeys and poultry. The hydrographic network is dense and dominated by the river Yoko. Yoko is limited, to the north by the towns of Bankim, Banyo, Tibati and Ngaoundal in the Adamawa region of Cameroon; to the south by Ntui and Mbandjock in the Centre region; to the west by Ngambe-Tikar and Ngoro in the Centre Region; and to the East by Bétaré-Oya and Belabo in the East region, and Nsem and Nanga Eboko in the Centre region.
Fig. 1.
Map of Cameroon showing tsetse-infested area of Yoko where the entomological survey was performed.
For this study, entomological surveys were performed in two villages: Medjan Vouni (5°22.055′N, 12°25.752′E) and Kounde (5°30.691′N, 12°14.312′E).
2.2. Collection of flies
The entomological survey was conducted during the end of the rainy season from 24th October to 1st November 2017. In the two villages (Fig. 1) where the surveys were conducted, 30 Vavoua traps (Laveissière and Grébaut, 1990) were deployed for four consecutive days in various tsetse fly favorable biotopes. The geographical coordinates of each trap were recorded using a global positioning system (GPS). Trapped flies (cyclical and mechanical vectors) were collected twice a day (from 9 to 10 am and from 3 to 4 pm) and numbered according to the trap number. Each trapped fly was morphological identified by sex and species using morphological keys as described by Pollock (1982). Tsetse flies were sorted into teneral (young flies that had never taken a blood meal) and non-teneral flies as described by Pollock (1982). Each tsetse fly was subsequently put into a 1.5 mL eppendorf micro-tube containing 95% ethanol. The micro-tubes containing whole tsetse fly were kept at room temperature in the field and later transferred to −20 °C in the laboratory until use.
2.3. DNA extraction
DNA was extracted from each whole tsetse fly using the cetyl trimethyl ammonium bromide (CTAB) method as described by Navajas et al. (1998). Briefly, the alcohol used to preserve each fly was evaporated by incubating the opened micro-tubes containing flies at 80 °C in an oven for about 1 h. Thereafter, each fly was ground with a pestle in 2% CTAB solution (CTAB 2%; 1 M Tris, pH 8; 0.5 M of EDTA pH 8; 5 M of NaCl). The disrupted tissue was incubated at 60 °C for 30 min before adding the chloroform/isoamyl alcohol mixture (24/1; V/V) to extract DNA. DNA was precipitated by adding isopropanol (V/V) followed by a centrifugation at 13,000 rpm for 15 min. DNA pellets were washed twice with 70% cold ethanol and then dried at room temperature. These pellets were each finally re-suspended in 50 μL of sterile distilled water before their storage at −20 °C until use. During each DNA extraction round, a micro-tube containing the CTAB buffer that served to crush tsetse flies was used as negative control. This micro-tube was processed together with those containing tsetse flies. The content of this tube was used as additional negative control beside the normal PCR negative control made up of all PCR reagents without DNA.
2.4. Molecular identification of different trypanosome species
Four sets of species-specific primers (Table 1) were used to identify T. brucei s.l., T. vivax, and T. congolense “forest” and “savannah” types. This identification was done by PCR as described by Simo et al. (2006). For this identification, each PCR reaction was carried out in a final volume of 15 μL containing 1.5 μL of 10× PCR reaction buffer, 1.5 mM MgCl2, 200 mM of each dNTP, 10 picomoles of each primer (Table 1), 0.3 units of Taq DNA polymerase (New England Biolabs, 5 U/μL stock solution) and 3 μL of DNA extract. For each PCR amplification reaction, the cycling phase was preceded by a denaturation step at 94 °C for 5 min. This was then followed by 40 amplification cycles. Each of these cycles was made up of a denaturation step at 94 °C for 30 s; an annealing step at 60 °C for 30 s for T. vivax, T. congolense forest and savannah, and at 58 °C for 30 s for T. brucei s.l.; and an extension step at 72 °C for 1 min. After the 40 cycles, a final extension step was performed at 72 °C for 10 min.
Table 1.
Primers used for the detection of different trypanosome species.
| Trypanosome species | Primer code | Primer sequences | References |
|---|---|---|---|
| T. congolense forest type | TCF1 TCF2 |
5’-GGACACGCCAGAAGTACTT-3’ 5’-GTTCTCGCACCAAATCCAAC-3’ |
(Masiga et al., 1992) |
| T. congolense savannah type | TCN1 TCN2 |
5’- TCGAGCGAGAACGGGCACTTTGCGA-3’ 5’-ATTAGGGACAAACAAATCCCGCACA-3’ |
(Moser et al., 1989) |
| T. brucei s.l. | TBR1 TBR2 |
5’-CGAATGAATATTAAACAATGCGCAG-3’ 5’-AGAACCATTTATTAGCTTTGTTGC-3’ |
(Moser et al., 1989) |
| T. vivax | TVW1 TVW2 |
5’-CTGAGTGCTCCATGTGCCAC-3’ 5’-CCACCAGAACACCAACCTGA-3’ |
(Masiga et al., 1992) |
After the PCR reactions, 10 μL of each amplified product was resolved on a 2% agarose gel that was subsequently stained with ethidium bromide, visualized under ultraviolet light and photographed.
2.5. Molecular detection of diminazene aceturate resistant T. congolense in tsetse flies
The detection of T. congolense strains that are resistant to diminazene aceturate was performed by PCR-RFLP as described by Vitouley et al. (2011). For this detection, a DNA fragment of the adenosine transporter gene (TbNT10) (Munday et al., 2013) of T. congolense was first of all amplified by PCR using specific primers. The PCR product was then resolved and subsequently purified from a 2% agarose gel, then digested with DpnII endonuclease, and the products finally resolved on agarose gel and photographed.
2.5.1. Amplification of TbNT10 gene of T. congolense
The T. congolense adenosine transporter gene (TbNT10) fragment was amplified with the gene-specific primers Ade2F (5’-ATAA>/>TCAAAGCTGCCATGGATGAAG-3′) and Ade2R (5’-GATGACTAACAATATGCGGGCAAAG-3′) as described by Delespaux et al. (2006). The amplification reactions were carried out in a final volume of 50 μL containing 5 μL of 10 X PCR buffer (10 μM of Tris-HCl pH 8.3, 50 mM of KCl), 1.5 mM MgCl2, 10 mM of each dNTP, 0.5 units of Taq DNA polymerase, 10 picomoles of each primer and 10 μL of DNA extract from each T. congolense positive sample. For each PCR reaction, a negative control using nuclease free water in place of DNA template was added. Positive controls with DNA extracts coming from in vivo confirmed drug-resistant or sensitive T. congolense isolates were included. The amplification program contained an initial denaturation step at 95 °C for 5 min, followed by 40 amplification cycles. Each of these cycles was made up of a denaturation step at 95 °C for 30 s, an annealing step at 50 °C for 50 s and an extension step at 68 °C for 1 min. After the 40 cycles, a final extension step was performed at 68 °C for 5 min.
At the end of PCR reactions, the amplicons were resolved on a 2% agarose gel, then stained with ethidium bromide and visualized on ultraviolet light and photographed. Samples showing an amplified DNA fragment of 648 base pairs (648 bp) corresponding to the TbNT10 gene fragment were selected, the amplified fragment purified and digested with DpnII.
2.5.2. Purification of T. congolense TbNT10 fragment
This was done using the Monarch DNA gel extraction kit following the manufacturers' instructions. Briefly, the portion of agarose gel with the DNA fragment of 648 bp was cut under UV light, weighed and put into a 2 mL micro-tube. Thereafter, four volumes of gel dissolving buffer (400 μL of the buffer for 100 mg of agarose gel) was added to the micro-tube and incubated in a water bath at 55 °C for 10 min. The solution obtained was transferred to a mini-column and centrifuged at 13000 rpm for one minute. Each mini-column was washed twice with 200 μL each of washing solution. The DNA was eluted into a 1.5 mL micro-tube by addition of 20 μL of elution buffer followed by incubation at room temperature for one minute, and centrifugation at 13000 rpm for one minute.
After this purification, 5 μL of the purified DNA fragment were verified by electrophoresis on 2% agorose gel before digestion with DpnII.
2.5.3. Digestion of TbNT10 fragment of T. congolense
The purified TbNT10 DNA fragment was digested in a final volume of 20 μL containing 10 μL of DNA, 2 μL of 10× DpnII reaction buffer pH 7.9 (100 mM NaCl, 50 mM Tris-HCl, 10 mM MgCl2, 100 μg/BSA), 7 μL of nuclease free water, and 10 units (one micro-liter) of DpnII. The digestion was performed at 37 °C for 24 h as recommended. The digestion products were resolved by electrophoresis on 2% agarose gel at 100 V for 90 min; the gel was visualized on ultraviolet light and photographed.
2.6. Ethical statement
This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Animals, of the Molecular Parasitology and Entomology Unit of the Department of Biochemistry, in the Faculty of Science of the University of Dschang.
3. Results
3.1. Entomological data
During this study, the 30 Vavoua traps captured 454 flies comprising 215 (47.5%) tsetse flies, the cyclical vector of African trypanosomes, and 239 (52.5%) mechanical vectors made up of 168 (37%) Tabanus spp. and 71 (15.6%) Stomoxys spp. The overall apparent density of flies per day and per trap (ADP) was 1.79 for tsetse flies, 1.4 for Tabanus spp and 0.6 for Stomoxys spp.
Regarding the tsetse flies, 4.6% (10/215) were from Kounde and 95.4% (205/215) from Medjan Vouni village. The ADPs of the tsetse flies were 1.83 at Medjan Vouni and 1.25 at Kounde. One hundred and forty one (65.6%) tsetse flies were female while 74 (34.4%) were male; thus giving a sex ratio of 1.91 in favor of females. The number of female flies was significantly higher (p < .0001; X2 = 41.758) than that of males. Of the 215 tsetse flies that were captured, 49.8% (107/215) belonged to G. f. congolensis, 33% (71/215) to G. f. fusca and 17.2% (37/215) to G. p. palpalis.
3.2. Molecular identification of trypanosomes in tsetse flies
Of the 215 tsetse flies that were trapped, 12.6% (27/215) were infected by at least one trypanosome species. Trypanosoma congolense savannah “type” showed the highest infection rate of 6.5% (14/215) followed by T. congolense forest “type” with 4.7% (10/215) and T. vivax with 1.4% (3/215). No infection due to T. brucei s.l. was recorded in this study. Between male and female tsetse flies, no significant difference (P = .154) was observed in the trypanosome infection rates although 12.8% of females were infected against 8.1% of males.
No trypanosome infection was found in tsetse flies from Kounde while the three different trypanosomes identified in this study were found in tsetse flies caught at Medjan Vouni. With regards to Medjan Vouni village, T. congolense savannah “type” was the most predominant species with 6.8% (14/205), followed by T. congolense forest “type” with 4.9% (10/205), and lastly T. vivax with 1.5% (3/205). A significant difference (P = .027) was observed between the infection rates of tsetse flies by the different trypanosomes in Medjan Vouni village.
Amongst the 27 tsetse flies with trypanosome infections, 11.1% (3/27) harbored mixed infections of T. congolense savannah “type” and T. congolense forest “type”, while 88.9% (24/27) had single infections of which 3 (11.1%) were T. vivax and 21 (77.8%) were T. congolense “forest” or “savannah” type.
The three different tsetse fly species were infected by different trypanosomes. Of the 24 tsetse flies with single trypanosome infections, 13 (54.2%) were found in G. f. congolensis, 6 (25%) in G. p. palpalis and 5 (20.8%) in G. f. fusca. The three mixed infections identified in the present study were found in G. p. palpalis and G. f. congolensis.
3.3. Molecular identification of diminazene aceturate resistant trypanosomes
From the 24 tsetse flies with T. congolense “savannah” or “forest” types, the adenosine transporter 1 (TbNT10) gene was successfully amplified in 38% (9/24) of them. No amplification was observed for T. vivax. After purification of the 648 bp fragment corresponding to TbNT10 gene, the digestion of the purified fragment with DpnII revealed four of the nine (44.4%) tsetse flies to be harboring profiles of diminazene aceturate resistant strains of T. congolense, while five samples (55.6%) showed profiles of diminazene aceturate sensitive strains (Fig. 2). This gives an overall prevalence of 16.7% of tsetse flies harboring T. congolense strains with resistant profile of diminazene aceturate. A T. congolense resistant strain after complete digestion of the TbNT10 gene with DpnII shows two DNA fragments (Fig. 2), while a sensitive strain shows one DNA fragment of 648 bp resulting from non-digestion of the TbNT10 gene (Fig. 2). A sample containing a mixture profile of sensitive and resistant T. congolense strains shows three DNA fragments (one for the sensitive strain and two for the resistant strain) after complete digestion of TbNT10 gene with DpnII (Fig. 2).
Fig. 2.
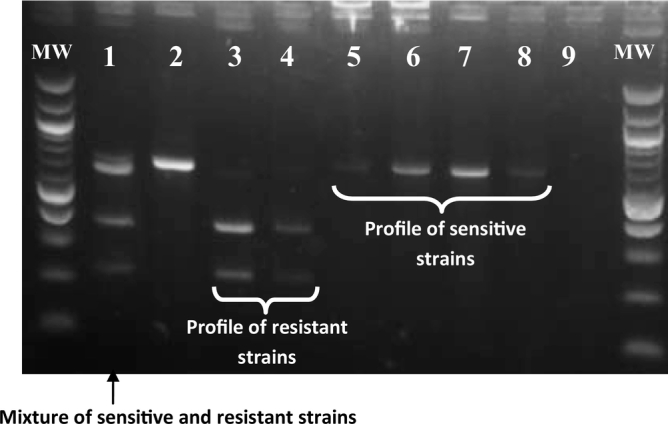
Example of an agarose gel presenting the DNA profiles resulting from the digestion of amplified DNA fragments of the TbNT10 gene of T. congolense with DpnII. Lane 1: internal positive controls made up of a DNA mixture of resistant and sensitive T. congolense strains; lanes 2, 5, 6, 7 and 8: DNA samples from diminazene aceturate sensitive strains of T. congolense (one DNA fragment illustrating non digestion of the TbNT10 gene by DpnII); lanes 3 and 4: DNA samples from diminazene aceturate resistant strains of T. congolense (two DNA fragments after the digestion with DpnII); lane 9: negative control (nuclease-free water); MW marker: 100 bp DNA ladder (New England Biolabs).
4. Discussion
In order to address the problem of African trypanosomiases within the overall objective of improving their control, considerable efforts have been undertaken at the continental level to map the distribution of tsetse fly vectors as well as the trypanosome infections (Cecchi et al., 2014). Geo-referenced data on trypanosome infections in tsetse flies are of great epidemiological importance for the control of African trypanosomiases because they enable scientists to estimate the risks of the disease from a broader perspective. In the same way, generating data on drug-resistant trypanosomes in tsetse flies will be useful for the identification of areas where these trypanosomes are transmitted. The present study falls within this continental initiative aimed at generating data on the distribution of tsetse flies and mechanical vectors of African trypanosomes, the trypanosome infection rates in tsetse flies, and especially data on drug-resistant trypanosomes.
The identification of Tabanus spp., Stomoxys spp. and three tsetse fly subspecies highlights the presence of cyclic and mechanical vectors of African trypanosomes in this area. The overall ADP of 1.79 for tsetse flies is relatively low compared to 3.1 and 3.6 recorded by Ngomtcho et al. (2017) at Gamba and Dodeo in the Adamawa region of Cameroon. These results could be explained by vector control measures undertaken in Yoko by the “pour on” of insecticides on cattle and the set up of screens by the “Special Mission for Tsetse fly Eradication” of the Ministry of Agriculture of Cameroon. Moreover, the sampling performed only during four days of the end of the wet season is an additional argument justifying the low relative density of tsetse flies since the ADP varies with seasons. The presence of three tsetse fly subspecies can be linked to the variety of biotopes that offer suitable conditions for various types of vectors. The identification of G. f. congolensis and G. f. fusca are in concordance with results of Ngomtcho et al. (2017) who suspected, on the basis of cytochrome oxidase sequences, the presence of tsetse flies belonging to G. fusca group. Our results are also in agreement with those of Tongue et al. (2015) who reported the occurrence of G. f. congolensis in northern region of Cameroon. For G. p. palpalis, its presence is in line with results of Ngomtcho et al. (2017) who reported its presence in the Adamawa region of Cameroon, and its wide distribution in the forest regions of West and Central Africa (Simo et al., 2008; Simo et al., 2012; Farikou et al., 2010; Kaba et al., 2014). For efficient vector control against AAT, our data suggest that the strategies to develop must take into account the presence of tsetse flies, as well as Tabanus spp. and Stomoxys spp that can feed on cattle (Odeniran et al., 2019b; Muzari et al., 2010).
The trypanosomes identified in this study were recently reported in tsetse flies and mammals of neighboring regions to Yoko such as Bafia in the Centre region, and Dodeo and Gamba in the Adamawa region of Cameroon (Ngomtcho et al., 2017; Simo et al., 2015). Such trypanosomes have been recently identified in cattle of Yoko (Mewamba et al., Unpublished data). Our results indicate active transmission of different trypanosome species, and especially T. congolense savannah “type” which is the most pathogenic trypanosome for cattle (Bengaly et al., 2002). The high number of tsetse flies with single infections (88.9%) compared to those with mixed infections (11.1%) corroborates results reported in tsetse flies (Morlais et al., 1998a; Morlais et al., 1998b; Tchouomene-Labou et al., 2013) and domestic animals (Simo et al., 2006; Nimpaye et al., 2011; Njiokou et al., 2004) of Cameroon and other African countries (Malele et al., 2003; Masiga et al., 1996; Masiga et al., 1992). Despite the similar profiles of single and mixed infections reported across sub-Saharan African countries, the percentages of mixed infections seem to vary between countries, and even within regions of the same country. For instance, 11.1% of mixed infections were found in the present study while only 7.02% and 6.41% were reported respectively in sleeping sickness foci of Malanga in the Democratic Republic of Congo (Simo et al., 2012) and Fontem in Cameroon (Kanté Tagueu et al., 2018). The discrepancy between these results could be linked to the variety of tsetse species, the transmission dynamics in each tsetse-infested region and the technical approaches used to identify trypanosomes. In the present study, trypanosomes were identified from whole flies and in consequence, it is not possible to know if the mixed infections were from the same or different tissues (Kanté Tagueu et al., 2018). Such mixed infections highlight the probability of tsetse flies to harbor or transmit several trypanosome species. Understanding how mixed infections evolve in tsetse flies and their potential impact on the transmission dynamics of trypanosomes remain areas for future investigations.
The trypanosome infection rate of 12.6% revealed in this study is lower than 25.5% and 40% reported respectively in Bafia and Dodeo, two tsetse-infested areas neighboring to Yoko (Ngomtcho et al., 2017; Simo et al., 2015). This low infection rate could be explained by the vector control strategy deployed once a year during the dry season by the “Special Mission for Tsetse flies eradication”, and also the administration of trypanocides by farmers. Another explanation could result from the fact that only four trypanosome species were investigated in this study. In such context, we cannot rule out the presence of other species of trypanosomes like T. grayi that has been recently identified in tsetse of neighboring area of Yoko (Ngomtcho et al., 2017). Although the control strategies (such as the use of insecticides, screens, and trypanocides) implemented in Yoko have reduced the prevalence of trypanosome infections, the uncontrolled use of trypanocides without advice from veterinarians could probably lead to the development of drug-resistant trypanosomes. Collecting data on the frequency at which the trypanocides are administered as well as on the implementation of vector control strategies could enable to better understand the current epidemiological situation for the final goal of improving the control of AAT in this locality.
Investigations of point mutations on the adenosine transporter gene that have been associated to the resistant or susceptibility profiles of diminazene aceturate in T. congolense enabled to amplify this gene in 37.5% (9/24) of samples that were positive for T. congolense. Remarkably, four samples for which TbNT10 genes were amplified revealed, after digestion with DpnII, DNA profiles identical to those of diminazene aceturate resistant strains of T. congolense (Vitouley et al., 2011). These molecular tools could therefore be used for the identification of trypanosomes showing drug-resistant/susceptible profiles in mammals as well as in tsetse flies. Our results revealed for the first time the presence of T. congolense strains that show diminazene aceturate resistant profiles in the tsetse-infested area of Yoko in the Centre region of Cameroon. They are in agreement with results found in domestic animals of this locality (Mewamba et al., unpublished data) and also with those of Mamoudou et al. (2008) who reported such drug resistance in animals of the Adamawa region of Cameroon which is neighboring to Yoko. These findings have important epidemiological implications for the control of AAT. The approach developed in this study could enable to avoid culture and isolation steps that are susceptible to induce selection of some parasite strains that are not able to growth in rodents or in culture media. These strains cannot therefore be isolated for in vitro drug sensitivity tests. If such un-isolable strains are resistant ones, wrong epidemiological data on drug resistance will be generated. The identification of drug-resistant trypanosomes in mammals gives indications on the circulation of resistant parasites without telling us where the animals contracted the resistant parasites. In most tsetse-infested areas where cattle and farmers are generally subjected to transhumance, investigations on drug resistance in animals provide evidence for the circulation of resistant trypanosomes in the areas visited by cattle. These data are not enough to provide information on villages where the transmission of the drug-resistant strains occurred. The approach developed in this study could complement such information because tsetse fly movement is restricted compared to transhumance phenomena. Given the geographical position of Yoko and the restricted movement of tsetse flies, the tsetse flies infested by trypanosomes showing resistance profiles to diminazene aceturate have more likely acquired locally these infections. Data generated from tsetse flies therefore enable us to identify with accuracy where the transmission of drug-resistant trypanosomes occurred. With such information, instead of generalizing the occurrence of drug resistance in one region or country or sub-region, the identification of drug-resistant trypanosomes in tsetse flies could enable scientists to identify villages or areas presenting a high risk for a particular drug resistance. In regions where trypanosomes are resistant to one trypanocide, the resistant drug must be replaced by the one that has shown no resistance and the implementation of vector control will enable to stop the spread of resistant strains. If the two main trypanocides commonly used in AAT control are both resistant, intensive vector control measures must be implemented in this area. These control measures will have important economic implications since the treatment costs will be reduced and more likely the costs of meat and milk. These intensive vector control measures could revert the situation and the resistant trypanocide could subsequently be used after some years.
In this study, only diminazene aceturate resistance was investigated in T. congolense. Consequently, we don't know if isomethamidium chloride resistant strains circulate in this study area. Regarding T. vivax where no amplification was obtained for the adenosine transporter gene, its resistance/susceptibility profile for diminazene aceturate and isometamidium chloride is still not yet understood. For a general overview of drug resistance in this area, additional studies on drug resistance against other trypanocides are required not only in T. congolense, but also on other trypanosomes circulating in this area. Such extensive investigations will provide the real epidemiological picture of drug resistance needed in order to achieve the overall goal of improving AAT control and thus the economic situation of peasant farmers.
5. Conclusion
This study revealed the presence of Tabanus spp. and Stomoxys spp as well as three different tsetse flies which are mechanical and cyclical vectors of African trypanosomes. It revealed also different species of trypanosomes in tsetse flies trapped at Yoko. When applied to tsetse flies, the molecular tools used to identify profiles of drug-resistant trypanosomes in animal hosts revealed, in tsetse flies of Yoko, T. congolense with molecular profiles corresponding to that of diminazene aceturate resistant strains. The identification of drug-resistant trypanosomes in tsetse flies could help to map with accuracy the areas where their transmission occurs. Such specific geo-localization of transmission sites will make it possible to refine control strategies to fight against AAT.
Authors' contributions
Conceived and designed the experiments: GS, CT, PS, SR. Performed the experiments: EMKM, EMM, Performed entomological survey OF, RMNK. Analysed the data: EMKM, EMM, GS. Wrote and revised the paper: GS, SR, CT. All authors read and approved the final manuscript.
Declaration of competing interest
The authors declare that they have no competing interests. The funding agencies played no role in the design or implementation of the study, the analysis or interpretation of the data, or the preparation and submission of the manuscript.
Acknowledgement
This work was supported by the “JEAI EpiReTryp” grant of the “Institut de Recherche pour le Développement”, the UMR INTERTRYP of Montpellier and the University of Dschang. We thank the reviewers for their useful comments that enabled to improve the quality of this manuscript.
References
- Barrett M.P., Maudlin I., Holmes P.H., Miles M.A. The Trypanosomiasis. CABI Publishing; 2004. Future prospects in chemotherapy for trypanosomiasis; pp. 445–458. [Google Scholar]
- Bengaly Z., Sidibe I., Ganaba R., Desquesnes M., Boly H., Sawadogo L. Comparative pathogenicity of three genetically distinct types of Trypanosoma congolense in cattle: clinical observations and haematological changes. Vet. Parasitol. 2002;108:1–19. doi: 10.1016/s0304-4017(02)00164-4. [DOI] [PubMed] [Google Scholar]
- Cecchi G., Paone M., Feldmann U., Vreysen M.J., Diall O., Mattioli R.C. Assembling a geospatial database of tsetse-transmitted animal trypanosomosis for Africa. Parasit. Vectors. 2014;7:39. doi: 10.1186/1756-3305-7-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen P.H., Pellmann C., Scheer A., Tietjen U., Schares G., Bauer B. Application of in vitro methods for the detection of drug resistance in trypanosome field isolates. ICPTV Newsletter. 2000;2:9–12. [Google Scholar]
- Connor R.J. 1992. The Diagnosis, Treatment and Prevention of Animal Trypanosomiasis Under Field Conditions in Programme for the Control of African Animal Trypanosomiasis and Related Development: Ecological and Technical Aspects. FAO Animal Production and Health Paper No. 100. Food and Agriculture Organisation of the United Nations (FAO) (Rome, Italy) [Google Scholar]
- Delespaux V., Ayral F., Geysen D., Geerts S. PCR-RFLP using Ssu-rDNA amplification: applicability for the diagnostic of mixed infections with different trypanosome species in cattle. Vet. Parasitol. 2006;117:185–193. doi: 10.1016/j.vetpar.2003.08.004. [DOI] [PubMed] [Google Scholar]
- Diall O., Cecchi G., Wanda G., Argilés-Herrero R., Vreysen M.J.B., Cattoli G. Developing a progressive control pathway for African animal trypanosomosis. Trends Parasitol. 2017;33:499–509. doi: 10.1016/j.pt.2017.02.005. [DOI] [PubMed] [Google Scholar]
- Farikou O., Njiokou F., Mbida Mbida J.A., Njitchouang G.R., Djeunga H.N., Asonganyi T. Tripartite interactions between tsetse flies, Sodalis glossinidius and trypanosomes: an epidemiological approach in two historical human African trypanosomiasis foci in Cameroon. Infect. Genet. Evol. 2010;10:115–221. doi: 10.1016/j.meegid.2009.10.008. [DOI] [PubMed] [Google Scholar]
- Holmes P. Tsetse-transmitted trypanosomes – their biology, disease impact and control. J. Invertebr. Pathol. 2013;112:S11–S14. doi: 10.1016/j.jip.2012.07.014. [DOI] [PubMed] [Google Scholar]
- Jamal S., Sigauque I., Macuamule C., Neves L., Penzhorn B.L., Marcotty T. The susceptibility of Trypanosoma congolense isolated in Zambezia Province, Mozambique, to isometamidium chloride, diminazene aceturate and homidium chloride, Onderstepoort. J. Vet. Res. 2005;72:333–338. doi: 10.4102/ojvr.v72i4.190. [DOI] [PubMed] [Google Scholar]
- Jones-Davies W.J. A Berenil resistant strain of Trypanosoma vivax in cattle. Vet. Rec. 1967;81:567–568. doi: 10.1136/vr.81.22.567. [DOI] [PubMed] [Google Scholar]
- Kaba D., Zacarie T., Mpondi A.M., Njiokou F., Bosson-Vanga H., Kröber T. Standardising visual control devices for tsetse flies: central and west African species Glossina palpalis palpalis, PLoS Negl. Trop. Dis. 2014;8:e2601. doi: 10.1371/journal.pntd.0002601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabayo J.P. Aiming to eliminate tsetse from Africa. Trends Parasitol. 2002;18:473–475. doi: 10.1016/s1471-4922(02)02371-1. [DOI] [PubMed] [Google Scholar]
- Kanté Tagueu S., Farikou O., Njiokou F., Simo G. Prevalence of Sodalis glossinidius and different trypanosome species in Glossina palpalis palpalis caught in the Fontem sleeping sickness focus of the southern Cameroon. Parasite. 2018;25:44. doi: 10.1051/parasite/2018044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laveissière C., Grébaut P. Recherches sur les pièges à glossines (Diptera: Glossinidae). Mise au point d'un modèle économique: le piège Vavoua. Trop. Med. Parasitol. 1990;41:185–192. [PubMed] [Google Scholar]
- Malele I., Graske L., Knight C., Ferris V., Njiru Z., Zamilton P. The use of specific and genetic primers to identify trypanosomes infectious of wild tsetse flies in Tanzania by PCR. Infect. Genet. Evol. 2003;3:271–279. doi: 10.1016/s1567-1348(03)00090-x. [DOI] [PubMed] [Google Scholar]
- Mamoudou A., Zoli N., Mbahin C., Tanenbe B., Clausen P.H., Marcotty T. Prevalence and incidence of bovine trypanosomosis on the Adamaoua plateau in Cameroon 10 years after the tsetse eradication campaign. Vet. Parasitol. 2006;142:16–22. doi: 10.1016/j.vetpar.2006.06.033. [DOI] [PubMed] [Google Scholar]
- Mamoudou A., Delespaux V., Chepnda V., Hachimou Z., Andrikaye J.P., Zoli A. Assessment of the occurrence of trypanocidal drug resistance in trypanosomes of naturally infected cattle in the Adamaoua region of Cameroon using the standard mouse test and molecular tools. Acta Trop. 2008;106:115–118. doi: 10.1016/j.actatropica.2008.02.003. [DOI] [PubMed] [Google Scholar]
- Masiga D.K., Smyth A.J., Ayes P., Bromidge T.J., Gibson W.C. Sensitive detection of trypanosomes in tsetse flies by DNA amplification. Int. J. Parasitol. 1992;22:909–918. doi: 10.1016/0020-7519(92)90047-o. [DOI] [PubMed] [Google Scholar]
- Masiga D.K., McNamara J.J., Laveissière C., Truc P., Gibson W.C. A high prevalence of mixed trypanosome infections in tsetse flies in Sinfra, Côte d'Ivoire, detected by DNA amplification. Parasitol. 1996;112:75–80. doi: 10.1017/s0031182000065094. [DOI] [PubMed] [Google Scholar]
- Morlais I., Grebaut P., Bodo J.M., Djoha S., Cuny G. Characterization of trypanosome infections by polymerase chain reaction (PCR) amplification in wild tsetse flies in Cameroon. Parasitol. 1998;116:547–554. doi: 10.1017/s0031182098002625. [DOI] [PubMed] [Google Scholar]
- Morlais L., Grebaut P., Bodo J.M., Djoha S., Cuny G., Herder S. Detection and identification of trypanosomes by polymerase chain reaction in wild tsetse flies in Cameroon. Acta Trop. 1998;70:109–117. doi: 10.1016/s0001-706x(98)00014-x. [DOI] [PubMed] [Google Scholar]
- Moser D.R., Cook G.A., Ochs D.E., Bailey C.P., McKane M.R., Donelson J.E. Detection of Trypanosoma congolense and Trypanosoma brucei subspecies by DNA amplification using the polymerase chain reaction. Parasitol. 1989;99:57–66. doi: 10.1017/s0031182000061023. [DOI] [PubMed] [Google Scholar]
- Munday J.C., Rojas López K.E., Eze A.A., Delespaux V., Van Den Abbeele J., Rowan T., Barrett M.P., Morrison L.J., de Koning H.P. Functional expression of TcoAT1 reveals it to be a P1-type nucleoside transporter with no capacity for diminazene uptake. Int. J. Parasitol. Drugs Drug Resist. 2013;3:69–76. doi: 10.1016/j.ijpddr.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzari M.O., Burgess G.W., Skerratt L.F., Jones R.E., Duran T.L. Host preferences of tabanid flies based on identification of blood meals by ELISA. Vet. Parasitol. 2010;174:191–198. doi: 10.1016/j.vetpar.2010.08.040. [DOI] [PubMed] [Google Scholar]
- Navajas M., Lagnel J., Gutierrez J., Bourset P. Species wide homogeneity of nuclear ribosomal ITS2 sequences in the spider mite Tetranychus urticae contrasts with extensive mitochondrial COI polymorphism. Heredity. 1998;80:742–752. doi: 10.1046/j.1365-2540.1998.00349.x. [DOI] [PubMed] [Google Scholar]
- Nerima B., Matovu E., Lubega G.W., Enyaru J.C.K. Detection of mutant P2 adenosine transporter (TbAT1) gene in Trypanosoma brucei gambiense isolates from northwest Uganda using allele-specific polymerase chain reaction. Trop. Med. Int. Health. 2007;12:1361–1368. doi: 10.1111/j.1365-3156.2007.01918.x. [DOI] [PubMed] [Google Scholar]
- Ngomtcho S.C.H., Weber J.S., Ngo Bum E., Gbem T.T., Kelm S., Achukwi M.D. Molecular screening of tsetse flies and cattle reveal different Trypanosoma species including T. grayi and T. theileri in northern Cameroon, Parasit. Vect. 2017;10:631. doi: 10.1186/s13071-017-2540-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimpaye H., Njiokou F., Njine T., Njitchouang G.R., Cuny G., Herder S. Trypanosoma vivax, T. congolense forest type and T. simiae: prevalence in domestic animals of sleeping sickness foci of Cameroon. Parasite. 2011;18:171–179. doi: 10.1051/parasite/2011182171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Njiokou F., Simo G., Nkinin S., Herder S. Infection rate of Trypanosoma brucei s.l., T. vivax, T. congolense "forest type" and T. simiae in small wild vertebrates in south Cameroon. Acta Trop. 2004;92:139–146. doi: 10.1016/j.actatropica.2004.04.011. [DOI] [PubMed] [Google Scholar]
- Odeniran P.O., Macleod E.T., Ademola I.O., Welburn S.C. Suspected resistance of Trypanosoma species to diminazene aceturate on a cattle farm in Nigeria. Trop. Anim. Health Prod. 2019;51:2091–2094. doi: 10.1007/s11250-019-01902-5. [DOI] [PubMed] [Google Scholar]
- Odeniran P.O., Macleod E.T., Ademola I.O., Welburn S.C. Molecular identification of bloodmeal sources and trypanosomes in Glossina spp. Tabanus spp. and Stomoxys spp. trapped on cattle farm settlements in southwest Nigeria. Med. Vet. Entomol. 2019;33:269–281. doi: 10.1111/mve.12358. [DOI] [PubMed] [Google Scholar]
- Perry B., Sones K. Poverty reduction through animal health. Science. 2007;315:333–334. doi: 10.1126/science.1138614. [DOI] [PubMed] [Google Scholar]
- Pollock J.N. Food and agriculture organization of the United Nations; Rome: 1982. Training Manual for Tsetse Control Personnel: Tsetse Biology, Systematics and Distribution; Techniques.http://www.fao.org/3/ap5178e.pdf [Google Scholar]
- Reis M.O., Souza F.R., Albuquerque A.S., Monteiro F., Oliveira L.F.D.S., Raymundo D.L., Wouters F., Wouters A.T.B., Peconick A.P., Varaschin M.S. Epizootic infection by Trypanosoma vivax in cattle from the state of Minas Gerais, Brazil. Korean J. Parasitol. 2019;57(2):191–195. doi: 10.3347/kjp.2019.57.2.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw P.M. Economics of African Trypanosomiasis. In: Maudlin I., Holmes P.H., Miles M.A., editors. The Trypanosomiases. CABI Publishing; Wallingford, UK: 2004. pp. 369–402. [Google Scholar]
- Simo G., Asonganyi T., Nkinin S.W., Njiokou F., Herder S. High prevalence of Trypanosoma brucei gambiense group 1 in pigs from the Fontem sleeping sickness focus in Cameroon. Vet. Parasitol. 2006;139:57–66. doi: 10.1016/j.vetpar.2006.02.026. [DOI] [PubMed] [Google Scholar]
- Simo G., Njiokou F., Mbida Mbida J.A., Njitchouang G.R., Herder S., Asonganyi T. Host preferences of tsetse flies from Cameroonian sleeping sickness foci: epidemiological implications. Infect. Genet. Evol. 2008;8:34–39. doi: 10.1016/j.meegid.2007.09.005. [DOI] [PubMed] [Google Scholar]
- Simo G., Silatsa B., Njiokou F., Lutumba P., Mansinsa P., Madinga J. Identification of different trypanosome species in the mid-guts of tsetse flies of the Malanga (Kimpese) sleeping sickness focus of the Democratic Republic of Congo, Parasit. Vect. 2012;5:201. doi: 10.1186/1756-3305-5-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simo G., Fongho P., Farikou O., Ndjeuto-Tchouli P.I., Tchouomene-Labou J., Njiokou F. Trypanosome infection rates in tsetse flies in the "silent" sleeping sickness focus of Bafia in the Centre region in Cameroon, Parasit. Vect. 2015;8:528. doi: 10.1186/s13071-015-1156-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinyangwe L., Delespaux V., Brandt J., Geerts S., Mubanga J., Machila N. Trypanocidal drug resistance in eastern province of Zambia. Vet. Parasitol. 2004;119:125–135. doi: 10.1016/j.vetpar.2003.11.007. [DOI] [PubMed] [Google Scholar]
- Steverding D. The history of African trypanosomiasis. Parasit. Vectors. 2008;1:1–8. doi: 10.1186/1756-3305-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swallow B.M. 2000. Impacts of Trypanosomiasis on African Agriculture. PAAT Technical and Scientific Series No. 2. Food and Agriculture Organisation of the United Nations (FAO) (Rome, Italy) [Google Scholar]
- Talaki E., Sidibe I., Akoda K., Belem A.M.G., Pangui L.J. Chimiorésistance aux trypanocides dans les élevages en Afrique subsaharienne. Rev Afr. Santé Prod. Anim. 2013;11:77–83. [Google Scholar]
- Tchouomene-Labou J., Nana-Djeunga H., Simo G., Njitchouang G.R., Cuny G., Asonganyi T. Spatial and temporal variations relevant to tsetse control in the Bipindi focus of southern Cameroon. Parasit. Vectors. 2013;6:193. doi: 10.1186/1756-3305-6-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tongue L.K., Acapovi-Yao G.L., Kaba D. Abah. E.N. Nchiwan, Updating tsetse distribution: presence of Austenina subgenus group in Northern Cameroon, J. Basic Appl. Res. Int. 2015;7:66–72. [Google Scholar]
- Vitouley H.S., Mungube E.O., Allegye-Cudjoe E., Diall O., Bocoum Z., Diarra B. Improved PCR-RFLP for the detection of diminazene resistance in Trypanosoma congolense under field conditions using filter papers for sample storage. PLoS Negl. Trop. Dis. 2011;5:1–4. doi: 10.1371/journal.pntd.0001223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vreysen M.J.B., Seck M.T., Sall B., Bouyer J. Tsetse flies: their biology and control using area wide integrated pest management approaches. J. Invertebr. Pathol. 2013;112:S15–S25. doi: 10.1016/j.jip.2012.07.026. [DOI] [PubMed] [Google Scholar]