Abstract
Background & Aims
The characterization of T cells infiltrating hepatocellular carcinoma (HCC) provides information on cancer immunity and also on selection of patients with precise indication of immunotherapy. The aim of the study was to characterize T-cell populations within tumor tissue and compare them with non-neoplastic liver tissue as well as circulating cells of the same patients.
Methods
The presence of unique cell populations was investigated in 36 HCC patients by multidimensional flow cytometry followed by t-distributed stochastic neighbor embedding analysis. Functional activity of tumor-infiltrating T cells was determined after activation by phorbol 12-myristate 13-acetate and ionomycin.
Results
Within the tumor there were more cells expressing CD137 and ICOS than in non-neoplastic liver tissue, possibly after recent antigenic activation. These cells contained several populations, including the following: (1) functionally impaired, proliferating CD4+ cells co-expressing Inducible T-cell costimulator (ICOS) and T cell immunoreceptor with Ig and ITIM domains (TIGIT); (2) functionally active CD8+ cells co-expressing CD38 and Programmed cell-death protein 1 (PD1); and (3) CD4-CD8 double-negative T-cell receptor αβ and γδ cells (both non–major histocompatibility complex–restricted T cells). When the identified clusters were compared with histologic classification performed on the same samples, an accumulation of activated T cells was observed in immune-inflamed HCC. The same analyses performed in 7 patients receiving nivolumab treatment showed a remarkable reduction in the functionally impaired CD4+ cells, which returned to almost normal activity over time.
Conclusions
Unique populations of activated T cells are present in HCC tissue, whose antigen specificity remains to be investigated. Some of these cell populations are functionally impaired and nivolumab treatment restores their responsiveness. The finding of ongoing immune response within the tumor shows which lymphocyte populations are impaired within the HCC and identifies the patients who might take benefit from immunotherapy.
Keywords: Cancer Immunity, Immunotherapy, tSNE Analysis
Abbreviations used in this paper: DBSCAN, density-based spatial clustering application with noise algorithm; DN, CD4-CD8 double-negative; HCC, hepatocellular carcinoma; IFN, interferon; IL, interleukin; MAIT, mucosal-associated invariant T cell; NTIL, nontumor liver-infiltrating lymphocyte; PBMC, peripheral blood mononuclear cell; PBS, phosphate-buffered saline; PMA, phorbol 12-myristate 13-acetate; TCR, T-cell receptor; TIL, tumor-infiltrating lymphocyte; TNF, tumor necrosis factor; Treg, regulatory T cell; tSNE, t-distributed stochastic neighbor embedding
Graphical abstract
Summary.
Immune profiling of hepatocellular carcinoma–infiltrating cells showed unique T-cell populations, including impaired ICOS+, TIGIT+, CD4+ T cells, activated CD8+, and non–major histocompatibility complex restricted T cells. Immunotherapy with check-point inhibitors diminished nonfunctional T cells and increased the number of responding cells.
Hepatocellular carcinoma (HCC) is the second leading cause of worldwide mortality1 and is the most common primary liver cancer, accounting for 90% of all liver cancers.2 Epidemiologic studies have shown that HCC occurs mostly in patients with chronic liver diseases including those caused by hepatitis viral infection, alcohol abuse, nonalcoholic steatohepatitis, obesity, and exposure to the fungal aflatoxin B1 toxin.2, 3
Inflammation can be an essential component of cancer development and also facilitates cell growth.4 In HCC, the chronic liver inflammation is sustained by activation of innate and adaptive immune cells that contribute to liver damage, leading to cell transformation and tumor cell invasion of liver tissue.5 Immune cells accumulating within the tumor also may exert antitumor activities, thereby their characterization may acquire an important prognostic value.6 Indeed, a recently proposed classification of tumors was based on the presence of lymphocytic infiltrate in the tumor microenvironment6: inflamed tumors are those with lymphocytic infiltrate; immune-excluded, those with lymphocytes only at tumor edges, and immune-desert, those with minimal numbers of lymphocytes near tumor tissue. Inflamed tumors usually are characterized by local production of proinflammatory cytokines and are associated with a high response rate to anticheckpoint therapy.6 However, which tumor-infiltrating lymphocytes correlate with antitumor response, or are unleashed by anticheckpoint therapy, remains unclear. Importantly, it also is unknown whether unique lymphocytes that are present in inflamed tumors are predictive of therapy response and have prognostic value.
Tumor-infiltrating lymphocytes (TILs) have been studied for their prognostic value in several human tumors,7, 8 including HCC,9 in which the presence of T cells and the absence of macrophages correlated with patient survival. These studies did not show whether unique T-cell populations preferentially infiltrate HCC.
It also remains unknown whether HCC TILs are functionally impaired and, if so, which population then becomes the antigen-nonresponder. Unresponsive T cells might represent tumor-specific lymphocytes, which are potential targets of immunomodulatory therapies. Current HCC treatment options remain unsatisfactory. The use of the immune checkpoint inhibitor nivolumab in HCC patients lead to durable objective responses, although in a small percentage of patients.10 The small efficacy might be a consequence of expression of other inhibitory receptors by HCC TILs. Therefore, the identification of the populations providing antitumor immunity might be of great help to define personalized checkpoint inhibitory therapy and increase the chance of therapy response.
In this study we performed a multidimensional analysis of TILs infiltrating HCC and compared them with T cells present in nontumor liver (non–tumor infiltrating lymphocytes [NTILs]) and peripheral blood mononuclear cells (PBMCs) of the same donors. Within HCC we found evidence of recently activated CD4+, CD8+, and CD4-CD8 double-negative (DN) unique T-cell populations. Some of these cells were exhausted effector T cells that recovered immune responsiveness upon nivolumab treatment. These phenotypic and functional characteristics of HCC TILs identify novel T-cell populations that might be the target of immunotherapy in this type of tumor.
Results
Patient and Biopsy Cohort
A total of 36 patients were studied and their clinical and pathologic characteristics are summarized in Table 1. HCC biopsy specimens were obtained from 34 patients before therapeutic treatment. Eleven HCC biopsy specimens were obtained from 7 nivolumab-treated (anti-PD1 immunotherapy) patients. The patients were predominantly male (83.3%), and half of them (47.2%) suffered from cirrhosis. From each HCC patient paired samples of peripheral blood and nontumor liver biopsy specimens also were obtained.
Table 1.
Clinical and Pathologic Characteristics of HCC Patients
| Patient | Tumor biopsy code | Cell number, 106 |
Edmondson–Steiner grade, I–IV | TIM | Intratumoral TIL, % |
Stroma TIL, % |
Time point | Sex | YOB | BCLC | Cirrhosis | Liver Disease | AFP |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | D403 | 4.5 | II | 3 | 0 | 0 | Pretreatment | M | 1944 | A | No | NAFLD | 9.5 |
| 2 | D214 | 20 | II | 2 | 0 | 30 | Pretreatment | M | 1944 | A | Yes | ALD | 7.2 |
| 3 | D316 | 3.5 | II | 3 | 0 | 0 | Pretreatment | M | 1932 | A | No | NAFLD | 1 |
| 4 | D373 | 7.5 | II | 2 | 5 | 30 | Pretreatment | F | 1938 | A | No | HEP C | 5.6 |
| 4 | D557 | 2.5 | III | 1 | 20 | 20 | Nivolumab | F | 1938 | A | No | HEP C | 5.6 |
| 5 | D143 | 2 | III | 1 | 20 | 20 | Pretreatment | F | 1954 | D | Yes | ALD | 6 |
| 6 | D330 | 30 | II | 1 | 10 | 20 | Pretreatment | M | 1938 | C | Yes | ALD | 48 |
| 6 | D448 | 1.4 | II | 1 | 10 | 30 | Nivolumab | M | 1938 | C | Yes | ALD | 48 |
| 7 | D096 | 3.5 | IV | 1 | 20 | 20 | Pretreatment | M | 1941 | A | No | ALD | 2 |
| 8 | D324 | 10 | III | 1 | 10 | 20 | Pretreatment | M | 1959 | D | Yes | ALD | 104,710 |
| 9 | D332 | 3.6 | II | 1 | 10 | 20 | Pretreatment | M | 1945 | A | No | ALD | 3.1 |
| 10 | D359 | 18 | III | 1 | 20 | NA | Pretreatment | M | 1930 | B | No | ALD and NAFLD | 5917 |
| 11 | D147 | 4.5 | III | 1 | 10 | 10 | Pretreatment | M | 1948 | A | No | HEP B | 32.5 |
| 12 | D562 | 2.5 | II | 2 | 5 | 30 | Pretreatment | M | 1943 | A | Yes | HEP C | 2.5 |
| 13 | D304 | NA | III | 3 | 5 | 0 | Pretreatment | M | 1953 | A | No | HEP C and ALD | NA |
| 14 | D588 | 2.5 | II | 3 | 0 | 0 | Pretreatment | M | 1932 | A | No | NAFLD | 338 |
| 15 | D565 | 5 | IV | 1 | 20 | 5 | Pretreatment | M | 1974 | C | No | NAFLD | 645 |
| 16 | D274 | 1.6 | III | 1 | 10 | 10 | Pretreatment | M | 1936 | A | Yes | NAFLD | 1.3 |
| 17 | D029 | 2 | II | 2 | 0 | 40 | Pretreatment | F | 1943 | B | Yes | ALD | 7.9 |
| 18 | D113 | 0.8 | III | 1 | 10 | 30 | Pretreatment | M | 1937 | A | No | ALD and NAFLD | 49.8 |
| 18 | D386 | 6 | III | 1 | 30 | 20 | Pretreatment | M | 1937 | A | No | ALD and NAFLD | 49.8 |
| 18 | D388 | 1.2 | III | 1 | 30 | 20 | Pretreatment | M | 1937 | A | No | ALD and NAFLD | 49.8 |
| 19 | D336 | 18 | II | 3 | 5 | 0 | Pretreatment | M | 1936 | C | Yes | ALD | 6.6 |
| 19 | D572 | 2 | II | 1 | 10 | 10 | Nivolumab | M | 1936 | C | Yes | ALD | 6.6 |
| 19 | D573 | 2.5 | II | 3 | 0 | 0 | Nivolumab | M | 1936 | C | Yes | ALD | 6.6 |
| 20 | D268 | 30 | II | 1 | 20 | NA | Pretreatment | M | 1946 | B | Yes | ALD and NAFLD | 10.2 |
| 21 | D240 | 60 | II | 3 | 5 | 5 | Pretreatment | M | 1944 | B | Yes | ALD and NAFLD | 5.6 |
| 22 | D228 | 60 | II | 1 | 10 | 20 | Pretreatment | M | 1936 | A | Yes | ALD | 3.6 |
| 23 | D483 | 1.2 | II | 1 | 20 | 20 | Pretreatment | M | 1934 | A | No | ALD and NAFLD | 2.7 |
| 24 | D176 | 40 | II | 3 | 0 | 0 | Pretreatment | M | 1937 | C | No | NAFLD | 0.9 |
| 25 | D020 | NA | III | 1 | 30 | 5 | Pretreatment | F | 1945 | C | Yes | ALD | 1745 |
| 26 | D302 | 150 | III | 1 | 30 | 20 | Pretreatment | F | 1949 | C | Yes | ALD | 400,000 |
| 27 | D158 | 18 | II | 2 | 0 | 30 | Pretreatment | F | 1939 | B | Yes | NAFLD | 4.6 |
| 28 | D156 | 60 | II | NA | 5 | NA | Pretreatment | M | 1949 | A | Yes | HEP B | 2 |
| 29 | D017 | NA | III | NA | 5 | NA | Pretreatment | M | 1933 | A | No | ALD | 231 |
| 30 | D045 | 1.5 | III | 3 | 0 | 5 | Pretreatment | M | 1947 | A | Yes | HEP C | 7852 |
| 31 | D135 | 1.9 | IV | 1 | 10 | 30 | Pretreatment | M | 1959 | C | Yes | HEP C | 11.4 |
| 32 | D200 | 21 | II | 3 | 0 | 0 | Pretreatment | M | 1940 | B | No | ALD and NAFLD | 1615 |
| 32 | D201 | 30 | III | 1 | 20 | NA | Pretreatment | M | 1940 | B | No | ALD and NAFLD | 1615 |
| 32 | D202 | 20 | II | 3 | 0 | NA | Pretreatment | M | 1940 | B | No | ALD and NAFLD | 1615 |
| 32 | D318 | 4 | II | 3 | 0 | 5 | Pretreatment | M | 1940 | B | No | ALD and NAFLD | 1615 |
| 32 | D319 | 2.7 | II | 3 | 0 | 0 | Pretreatment | M | 1940 | B | No | ALD and NAFLD | 1615 |
| 32 | D505 | NA | III | NA | 0 | NA | Nivolumab | M | 1940 | B | No | ALD and NAFLD | 1615 |
| 33 | D503 | NA | III | 1 | 20 | 20 | Pretreatment | M | 1941 | B | No | NASH | 3995 |
| 34 | C948 | NA | III | 1 | 20 | 20 | Pretreatment | M | 1958 | C | Yes | HEP C and ALD | 12,054 |
| 34 | C949 | NA | III | 1 | 30 | 30 | Pretreatment | M | 1958 | C | Yes | HEP C and ALD | 12,054 |
| 34 | D001 | 1.7 | III | 1 | 30 | 30 | Nivolumab | M | 1958 | C | Yes | HEP C and ALD | 12,054 |
| 35 | D470 | NA | III | 1 | 20 | 10 | Nivolumab | M | 1958 | C | No | HEP C and NASH | 3.9 |
| 36 | D152 | 60 | II | 1 | 20 | 0 | Nivolumab | M | 1943 | B | No | HEP C | 1057 |
| 36 | D153 | 60 | I | 2 | 5 | 10 | Nivolumab | M | 1943 | B | No | HEP C | 1057 |
| 36 | D340 | 6 | II | 1 | 20 | 0 | Nivolumab | M | 1943 | B | No | HEP C | 1057 |
| 36 | D341 | 7 | I | 2 | 0 | 10 | Nivolumab | M | 1943 | B | No | HEP C | 1057 |
NOTE. For the tumor-immune microenvironment, 1 indicates immune-inflamed, 2 indicates immune-excluded, and 3 indicates immune-desert.
AFP, α-fetoprotein; ALD, alcoholic liver disease; BCLC, Barcelona Clinic Liver Cancer; HEP B, hepatitis B; HEP C, hepatitis C; NA, not applicable; NAFLD, nonalcoholic fatty liver disease; NASH, nonalcoholic steatohepatitis; TIM, tumor-immune microenvironment; YOB, year of birth.
CD4+ T Cells Are Enriched in Tumor Tissue
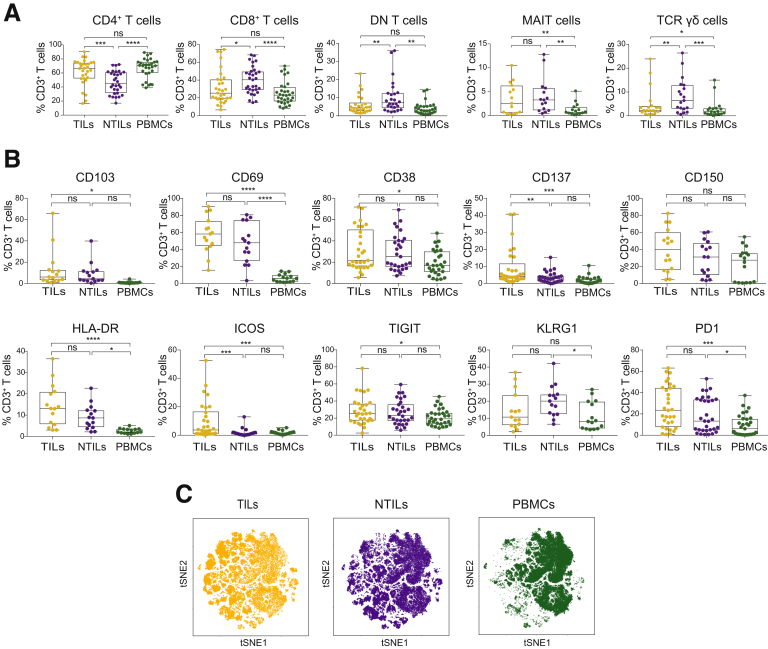
Comparative analyses were performed on samples obtained from TILs, NTILs, and PBMCs. The frequency of CD4+ T cells in TILs was significantly higher compared with NTILs (66.6% ± 3.36% vs 45.25% ± 2.9% of CD3+ T cells; P = .0001). On the contrary, CD8+ and DN T-cell frequencies were lower in TILs compared with NTILs (25.3% ± 3.41 vs 40.8% ± 2.77 for CD8+ T cells; P = .02; and 4.44% ± 1 vs 7.24% ± 1.7 for DN T cells; P = .0018). Within the DN T cells, T-cell receptor (TCR) γδ cells were reduced in the tumor compared with nontumor liver (2.85% ± 1.53 vs 7.15% ± 1.71; P = .01), whereas mucosal-associated invariant T (MAIT) cells did not show major differences in TILs vs NTILs (2.57% ± 0.8 vs 3.28% ± 0.96, respectively) (Figure 1A). The percentages of CD4+, CD8+, DN, and TCR γδ T cells were not different between TILs and PBMCs. The frequencies of MAIT cells in TILs and NTILs were higher than in the PBMCs (2.13% ± 0.5 and 0.73% ± 0.22, respectively; P = .0007), confirming the preferential localization of MAIT cells into the liver.11, 12 Taken together, these data showed that CD4+ T cells are enriched whereas CD8+, DN, and TCR γδ T cells are decreased in HCC compared with nontumor liver.
Figure 1.
Frequency and phenotype of CD3+T-cell subsets in HCC patients. (A) Box plots showing the frequency of CD4, CD8, DN, MAIT, and TCR γδ cells in TILs vs NTILs and PBMCs. Each dot represents an individual sample. (B) Box plots showing the phenotype of CD3+ cells. The percentage of cells expressing the indicated markers is indicated for each sample. (C) tSNE map of total CD3+ cells in TILs (yellow dots), NTILs (purple dots), and PBMCs (green dots). *P ≤ .05, **P ≤ .01, ***P ≤ .001, and ****P ≤ .0001, 2-tailed paired test for multiple comparison (Tukey test). ns, not significant.
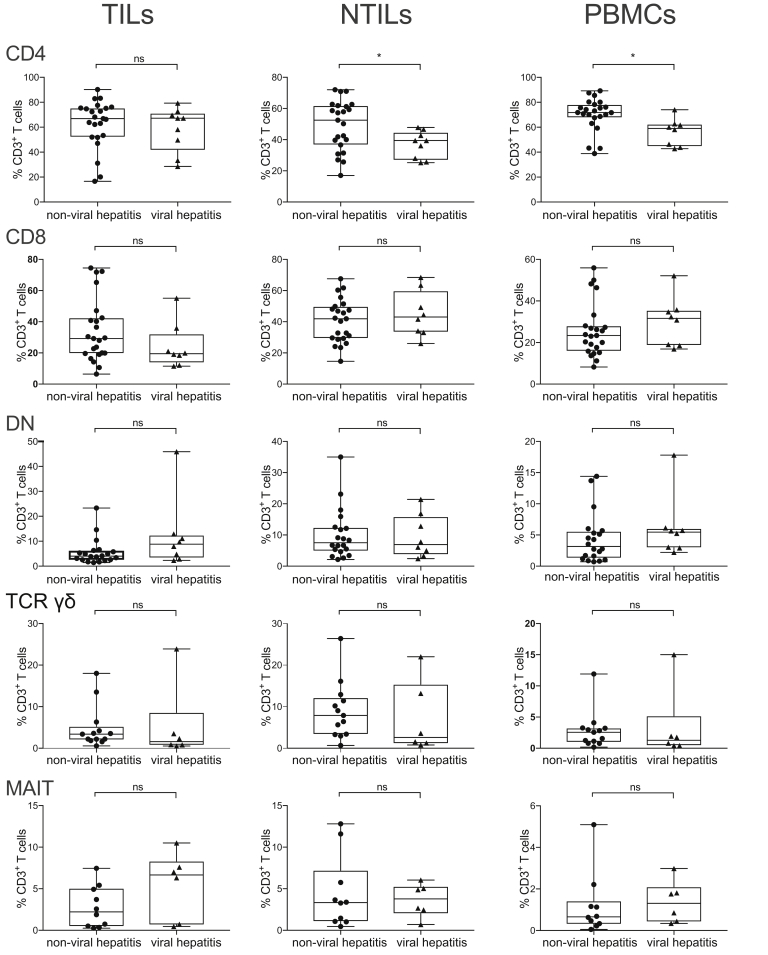
The underlying liver disease (viral hepatitis vs alcoholic hepatitis) did not show direct influence on the immune infiltrate, with the exception of a slightly larger number of CD4 cells in NTILs and PBMCs (Figure 2).
Figure 2.
Frequency and phenotype of CD3+T-cell subsets in viral and nonviral hepatitis.Box plots showing the frequency of CD4, CD8, DN, MAIT, and TCR γδ cells in TILs, NTILs, and PBMCs. Each dot represents an individual sample. *P ≤ .05, Wilcoxon signed-rank test, adjusted with the Benjamini and Hochberg method. ns, not significant.
CD137 and ICOS Expressing T Cells Are Enriched in the Tumor
Next, a multiparametric flow cytometry analysis was performed to characterize TIL phenotype and functional state. The expression of activation markers (CD103, CD69, CD38, CD137, CD150, HLA-DR, and ICOS) and inhibitory receptors (TIGIT, KLRG-1, and PD1) was compared on T cells isolated from TILs, NTILs, and PBMC (see list of analyzed molecules in Table 2). Increased expression of CD137 and ICOS were found on TILs vs NTILs (P = .005 and P = .0002, respectively) and vs PBMCs (P = .0001 and P = .0001, respectively) (Figure 1B). Of note, these markers were similarly expressed by NTILs and PBMCs, indicating that their expression was tumor-associated and not a consequence of liver homing. The increased CD137 and ICOS expression indicated the presence of recently activated T cells in the tumor.13 To assess whether these markers might show the emergence of unique T cells infiltrating HCC tissue, we analyzed all markers using the t-distributed stochastic neighbor embedding (tSNE) algorithm that reduces all data to 2 dimensions (Figure 1C). Unique populations appeared in TILs whereas the major cell clusters in NTILs and PBMCs were very similar to each other. NTILs also showed small clusters different from PBMCs, which probably reflect unique liver-resident T-cell populations.
Table 2.
Cell Surface Immunostaining Panel 1
| Marker | Dye | Host species | Reactivity | Clone | Description | Catalog no |
|---|---|---|---|---|---|---|
| CD3 | PE/Cy7 | Mouse | Human | UCHT1 | Co-receptor: CD3 T cells | 300420 |
| CD4 | Brilliant Violet 711 | Mouse | Human | OKT4 | Co-receptor: CD4 T cells | 317440 |
| CD8 | BUV 496 | Mouse | Human | RPA-T8 | Co-receptor: CD8 T cells | 564804 |
| TCR γδ | Brilliant Violet 421 | Mouse | Human | B1 | T-cell receptor: TCR γδ | 331218 |
| hMR1-5OP-RU Tetramer | PE | Human | MR1-Ag tetramer: MAIT cells | 31704 R | ||
| CD19 | PE/Cy5 | Mouse | Human | J3-119 | Co-receptor: B cells | A07771 |
| CD14 | PE/Cy5 | Mouse | Human | RMO52 | Co-receptor: monocytes | A07765 |
| CD137 | PE/Dazzle 594 | Mouse | Human | 4B4-1 | Costimulatory receptor | 309826 |
| CD278 (ICOS) | Brilliant Violet 510 | Mouse | Human | C398.4A | Costimulatory receptor | 313525 |
| CD150 (SLAM) | FITC | Mouse | Human | A12 (7D4) | Costimulatory receptor | 306306 |
| CD38 | Brilliant Violet 650 | Mouse | Human | HB-7 | cADP ribose hydrolase | 356620 |
| HLA-DR | Alexa Fluor 700 | Mouse | Human | L243 | Ag presenting molecule | 307626 |
| CD69 | APC/Cy7 | Mouse | Human | FN50 | Adhesion molecule | 310914 |
| CD103 | APC | Mouse | Human | Ber-ACT8 | Adhesion molecule | 350216 |
| TIGIT (VSTM3) | PerCP-eFluor 710 | Mouse | Human | MBSA43 | Inhibitory receptor | 46-9500-42 |
| PD1 | Brilliant Violet 785 | Mouse | Human | EH12.2H7 | Inhibitory receptor | 329930 |
| KLRG1 | Brilliant Violet 605 | Mouse | Human | 2F1/KLRG1 | Inhibitory receptor | 138419 |
Ag, antigen; cADP, cyclic adenosine diphosphate; FITC, fluorescein isothiocyanate; PE, phycoerythrin.
These data clearly indicated that HCC TILs are different form liver-resident T cells and might have immunologic relevance.
Enrichment of ICOS+ TIGIT+ CD4+ T Cells in HCC Vs Nontumor Liver and Peripheral Blood
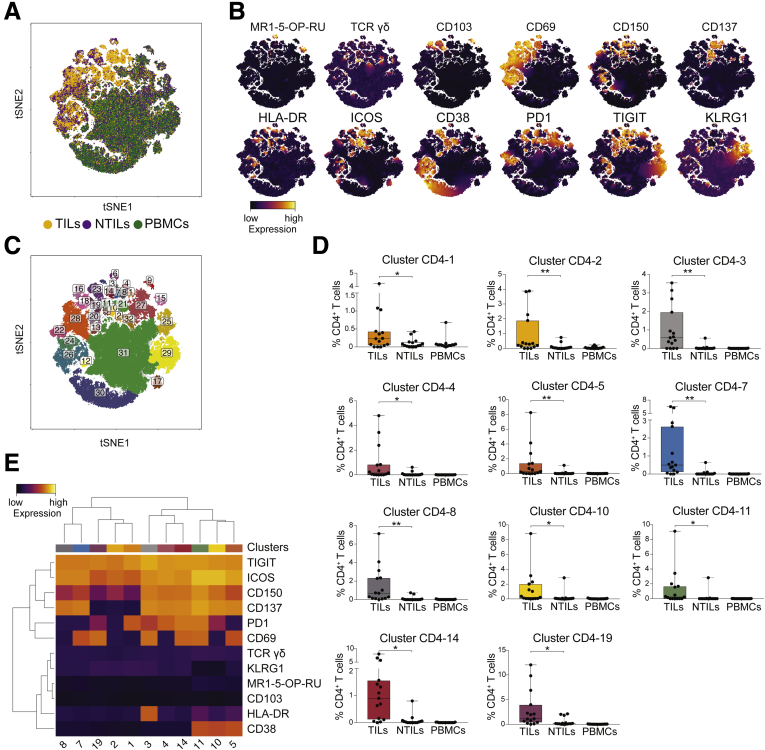
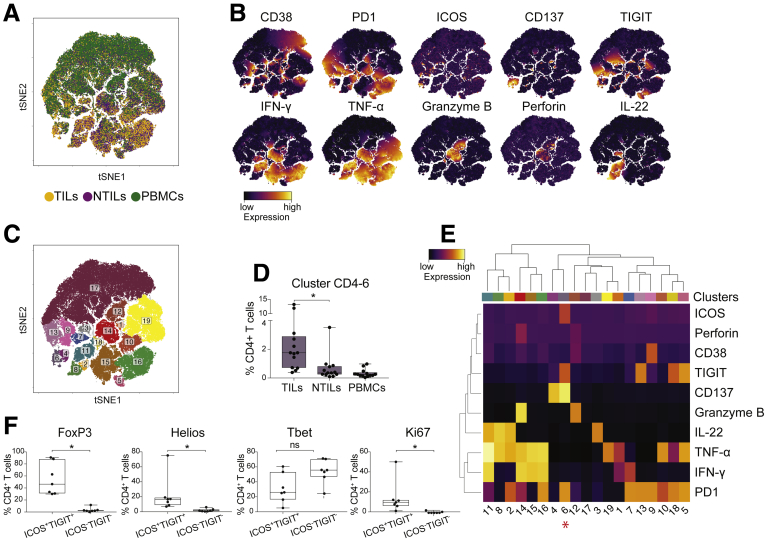
To better characterize HCC-associated clusters, tSNE analysis was performed on individual CD4+, CD8+, and DN T-cell subpopulations. The distribution of CD4+ T cells in tumor liver (yellow), in nontumor liver (purple), and in PBMCs (green) from 15 HCC patients is shown in Figure 3A. High expression of multiple activation markers (CD69, ICOS, CD150, CD137, HLA-DR, and ICOS) and inhibitory receptors (PD1 and TIGIT) mostly contributed to different clustering of CD4+ TILs (Figure 3B). As expected, small numbers of MAIT and TCR γδ cells were present in the CD4+ compartment and did not show significant differences between TILs and NTILs. Further analyses of CD4+ T cells performed using the density-based spatial clustering application with noise algorithm (DBSCAN) algorithm14 showed 11 clusters (CD4-1, -2, -3, -4, -5, -7, -8, -10, -11, -14, and -19) significantly enriched in HCC vs NTILs and PBMCs (Figure 3C and D). These clusters shared high levels of TIGIT and ICOS and differed in the expression of other activation markers and inhibitory receptors (Figure 3E). All clusters, except cluster CD4-1, CD4-2, and CD4-19, expressed high levels of CD137, a marker associated with recent T-cell activation.13 Furthermore, most CD137+ activated cells also expressed PD1, probably indicating that these cells had undergone multiple rounds of activation within HCC.15, 16 The expression of CD69 suggested that they were tissue-resident cells within the tumor,17 although this marker also may indicate previous antigen stimulation. Collectively, these data showed that CD4+ T cells co-expressing ICOS and TIGIT were preferentially accumulated in the tumor compared with the nontumor liver and PBMCs.
Figure 3.
ICOS+TIGIT+CD4+T cells are enriched in TILs. (A) tSNE map of CD4+ T cells in TILs (yellow dots), NTILs (purple dots), and PBMCs (green dots) of the first HCC cohort (n = 15). (B) tSNE maps showing the expression level and the distribution of each indicated marker. The color scale indicates the relative expression levels. (C) tSNE map of CD4+ T cells with each cluster colored differently. (D) Box plots showing the frequency of CD4+ T cells in each significantly enriched cluster in TILs vs NTILs. (E) Heatmap showing the normalized expression of the markers for selected clusters. The color scale indicates the relative expression levels. *P ≤ .05 and **P ≤ .001, Wilcoxon signed-rank test, adjusted with the Benjamini and Hochberg method.
ICOS+ TIGIT+ CD4+ TILs Proliferate in HCC But Are Functionally Impaired
The effector functions of CD4+ and CD8+ HCC TILs, were investigated in a second cohort of patients that also served as validation of the first HCC cohort. Matched TILs, NTILs, and PBMCs of 13 HCC patients were stimulated with phorbol 12-myristate 13-acetate (PMA) and ionomycin and analyzed by 13-parameter flow cytometry for their expression of activation markers and effector molecules (see list of analyzed molecules in Table 3). The analysis of interferon-γ (IFN-γ), tumor necrosis factor-α (TNF-α), interleukin (IL)22, granzyme B, and perforin showed 19 clusters in CD4+ T cells (Figure 4A and B). These results also confirmed that in this second cohort of patients, ICOS+ CD137+ TIGIT+ PD1+ T cells (cluster CD4-6) were enriched in HCC tissue (Figure 4C and D), thus validating the results observed in the first cohort of patients. Cluster CD4-6 showed a profound nonfunctional phenotype because these cells did not produce any of the investigated cytokines, although other CD4+ T cells did (Figure 4E). Remarkably, this T-cell subpopulation, compared with the ICOS- TIGIT- CD4+ counterpart, showed high levels of the transcription factors Foxp3 (P = .02) and Helios (P = .02), but not of T-bet, and also expressed the proliferation marker Ki-67 (P = .03) within the tumor, suggesting an activated phenotype (Figure 4F), in concordance with their expression of CD137 (Figure 4E).
Table 3.
Cell Surface and Intracellular Staining Panel 2
| Marker | Dye | Host species | Reactivity | Clone | Description | Catalog no |
|---|---|---|---|---|---|---|
| CD3 | Alexa Fluor 700 | Mouse | Human | UCHT1 | Co-receptor: CD3 T cells | 300424 |
| CD4 | PE | Mouse | Human | RPA-T4 | Co-receptor: CD4 T cells | 561843 |
| CD8 | BUV 496 | Mouse | Human | RPA-T8 | Co-receptor: CD8 T cells | 564804 |
| CD19 | PE/Cy5 | Mouse | Human | J3-119 | Co-receptor: B cells | A07771 |
| CD14 | PE/Cy5 | Mouse | Human | RMO52 | Co-receptor: monocytes | A07765 |
| CD137 | PE/Dazzle | Mouse | Human | 4B4-1 | Costimulatory receptor | 309826 |
| CD278 (ICOS) | Brilliant Violet 510 | Mouse | Human | C398.4A | Costimulatory receptor | 313525 |
| CD38 | Brilliant Violet 650 | Mouse | Human | HB-7 | cADP ribose hydrolase | 356620 |
| TIGIT (VSTM3) | PerCP-eFluor 710 | Mouse | Human | MBSA43 | Inhibitory receptor | 46-9500-42 |
| PD1 | Brilliant Violet 785 | Mouse | Human | EH12.2H7 | Inhibitory receptor | 329930 |
| IL22 | PE | Mouse | Human | 2G12A41 | Proinflammatory cytokine | 366704 |
| TNF-α | PE/Cy7 | Mouse | Human | MAb11 | Proinflammatory cytokine | 502930 |
| IFN-γ | APC/Cy7 | Mouse | Human | 4S.B3 | Proinflammatory cytokine | 502530 |
| Granzyme B | FITC | Mouse | Human/mouse | GB11 | Cytotoxic molecule | 515403 |
| Perforin | Brilliant Violet 421 | Mouse | Human | B-D48 | Cytotoxic molecule | 353307 |
cADP, cyclic adenosine diphosphate; DAPI, 4′,6-diamidino-2-phenylindole; FITC, fluorescein isothiocyanate; PE, phycoerythrin.
Figure 4.
ICOS+TIGIT+CD4+T cells enriched in TILs are functionally impaired. (A) tSNE map of CD4+ T cells in TILs (yellow dots), NTILs (purple dots), and TILs (green dots) of the second HCC cohort (n = 13). (B) tSNE maps showing the expression level and the distribution of each indicated marker. The color scale indicates the relative expression levels. (C) tSNE map with each cluster of CD4+ T cells colored differently. (D) Box plots showing the frequency of CD4+ T cells within cluster CD4-6 in TILs, NTILs, and PBMCs. (E) Heatmap showing the normalized expression of each marker in the indicated clusters. The color scale indicates the relative expression levels. * indicates cluster CD4-6, which is increased significantly in TILs vs NTILs. (F) Box plots showing the expression of indicated transcription factors on ICOS+ TIGIT+ CD4+ and ICOS- TIGIT- CD4+ TILs. *P ≤ .05 and **P ≤ .01, Wilcoxon signed-rank test, adjusted with the Benjamini and Hochberg method. ns, not significant.
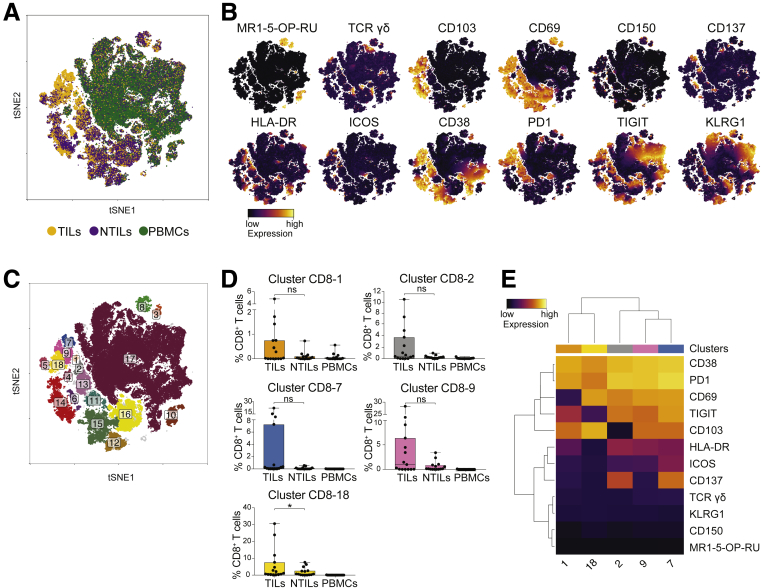
Increased Frequency of CD38+ PD1+ CD8+ T Cells in HCC
We next compared CD8+ T cells in the 3 tissues (Figure 5A). Activated CD8+ T cells preferentially accumulated in the tumor (Figure 5B), whereas CD8+ MAIT and TCR γδ cells showed similar expression in the investigated tissues (Figure 5B). Most of CD8+ TILs expressed CD38 and PD1. Although cluster CD8-18 was enriched significantly in HCC (P = .05), clusters CD8-1, -2, -7, and -9, also containing CD38+ PD1+ CD8+ T cells, were not (Figure 5C and D). Clusters CD8-1, -2, -7, and -9 also expressed the tissue-residence markers CD69 and/or CD103,18, 19 suggesting prolonged retention in HCC.20
Figure 5.
CD38+PD1+CD8+T cells are enriched in TILs. (A) tSNE map of CD8+ T cells in TILs (yellow dots), NTILs (purple dots), and PBMCs (green dots). (B) tSNE maps showing the expression level and the distribution of each indicated marker. The color scale indicates the relative expression levels. (C) tSNE map with each cluster of CD8+ T cells colored differently. (D) Box plots showing the frequency of CD8+ T cells in each cluster significantly enriched in TILs. (E) Heatmap showing the normalized expression of the markers for the indicated clusters. The color scale indicates the relative expression levels. *P ≤ .05, Wilcoxon signed-rank test, adjusted with the Benjamini and Hochberg method. ns, not significant.
The enrichment of PD1+ CD8+ T cells in HCC and the correlation of high PD1 levels with CD38 expression indicate the presence of previously activated cells, some of which could be tumor-specific within this cluster,21 as also recently reported for non–small-cell lung cancer.22
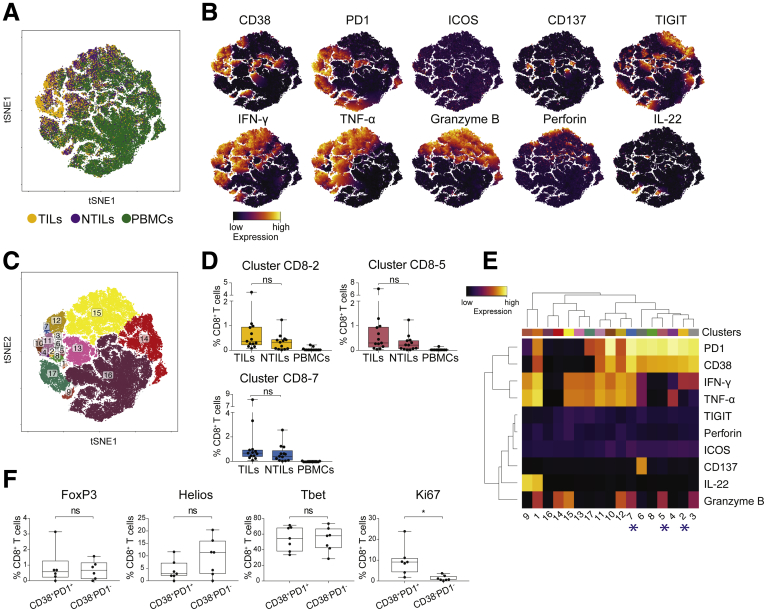
Next, we compared the functional activities of CD8+ T cells in TILs, NTILs, and PBMCs (Figure 6A and B). Seventeen clusters were identified (Figure 6C), of which 3 (clusters CD8-2, CD8-5, and CD8-7) showed accumulation in the tumor and contained CD38+ PD1+ CD8+ T cells (Figure 6D). These 3 clusters showed variable production of IFN-γ, TNF-α, and granzyme B (Figure 6E), suggesting a proinflammatory role. CD38+ PD1+ cells were significantly more proliferating (Ki-67+) than CD38- PD1- cells (P = .04) (Figure 6F), and showed similar expression of FoxP3, Helios, and T-bet transcription factors (Figure 6F). Taken together, these findings confirmed the presence within HCC of proliferating and functional CD38+ PD1+ cells, which might contribute to local inflammation and antitumor response.
Figure 6.
CD38+PD1+CD8+T cells enriched in TILs show different functionality. (A) tSNE map of CD8+ T cells in TILs (yellow dots), NTILs (purple dots), and PBMCs (green dots). (B) tSNE maps show the expression level and the distribution of each indicated marker. Color scale indicates the relative expression levels. (C) tSNE map with each cluster of CD8+ T cells colored differently. (D) Box plots show the frequency of the CD38+ PD1+ CD8+ T-cell clusters increased in TILs. (E) Heatmap shows the normalized expression of each marker in the indicated clusters. Color scale indicates the relative expression levels. The * denote the clusters increased in TILs vs NTILs. (F) Box plots show the expression of the indicated transcription factor and proliferation marker in CD38+ PD1+ CD8+ and CD38- PD1- CD8+ TILs. Wilcoxon signed-rank test, adjusted with the Benjamini and Hochberg method. ns, not significant.
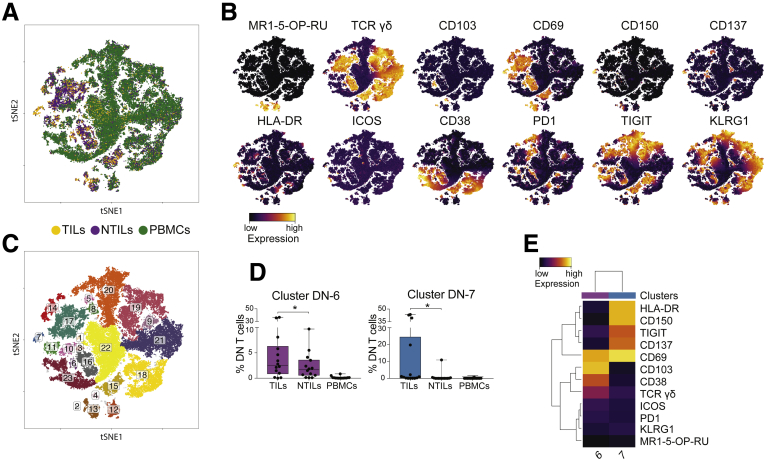
Two Unique Populations of DN T Cells Are Present Within HCC
CD4- CD8- DN T cells contain different populations of innate-like T cells, including TCR γδ and MAIT cells, and are more abundant in the liver than in PBMCs.12, 23 Multidimensional analysis of 10 markers (Figure 7A and B) showed 2 DN clusters significantly enriched in HCC biopsy specimens (Figure 7C and D). Cluster DN-6 contained activated CD38+ TCR γδ cells co-expressing the tissue-resident markers CD69 and CD10317 and cluster DN-7 composed by TCR αβ non-MAIT cells expressing the activation markers CD69, CD137, CD150, and HLA-DR, and the inhibitory TIGIT molecule (Figure 7E). Because CD137 is a marker of recent T-cell activation,13 these findings suggest that a population of DN TCR αβ cells might be activated within HCC.
Figure 7.
Unique clusters of DN T cells are enriched in TILs. (A) tSNE map of DN T cells in TILs (yellow dots), NTILs (purple dots), and PBMCs (green dots). (B) tSNE maps show the expression level and the distribution of each indicated marker. Color scale indicates the relative expression levels. (C) tSNE map with each cluster of DN T cells colored differently. (D) Box plots show the frequency of cells in each cluster significantly enriched in TILs. (E) Heatmap shows the normalized expression of the markers for the indicated clusters. Color scale indicates the relative expression levels. *P ≤ .05, Wilcoxon signed-rank test, adjusted with the Benjamini and Hochberg method.
Comparison of TIL Phenotype Correlates With Tumor Immunohistologic Profile
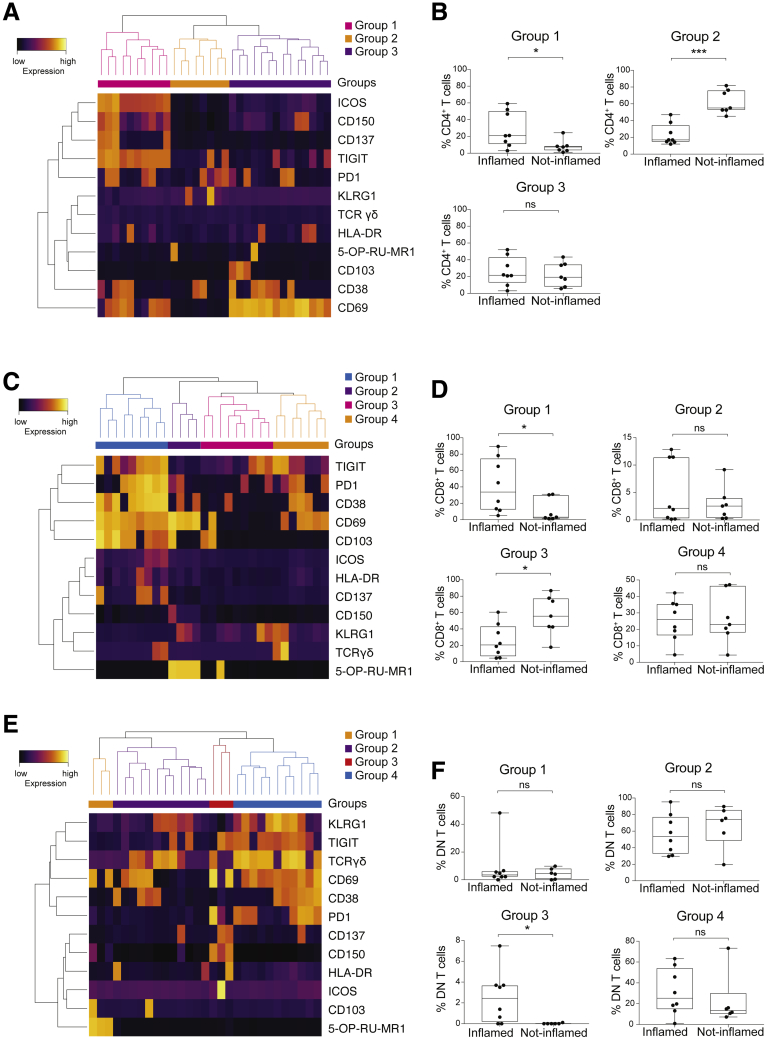
An important classification of tumors relies on the presence of mononuclear cell infiltrate (immune-inflamed, immune-excluded, and immune-desert) within tumor tissue.6 To investigate potential correlations between the presence of immune infiltrate and unique cell clusters, each biopsy also was classified by H&E staining according to the immune infiltrate after recently proposed guidelines24 (Figure 8). Nonsupervised hierarchical clustering identified 3 groups of CD4+ TILs, 4 groups of CD8+ TILs, and 4 groups of DN TILs (Figure 9A, C, and E, respectively). Next, we assessed the abundance of each group in inflamed vs noninflamed tumors (Figure 9B, D, and F). Within CD4+ TILs, group 1 cells were accumulated significantly in inflamed tumors and were highly activated (Figure 9B). All cells co-expressed ICOS and TIGIT and, with some variability, other activation markers and inhibitory receptors. Group 2 cells lacked the CD69 marker and were enriched significantly in noninflamed tumors. Finally, group 3 included liver-resident T cells equally represented in inflamed and noninflamed tumors (Figure 9A and B).
Figure 8.
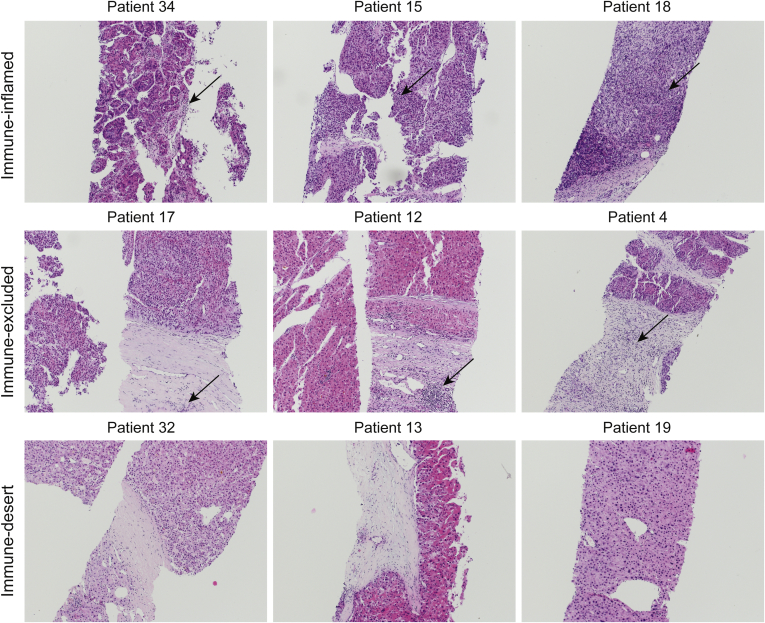
Representative histology of HCC tumors from 9 patients. (A) Immune-inflamed HCC with immune cells closely intermingled with tumor cells. (B) Immune-excluded HCC, showing ≥10% of the tumor stroma populated by immune cells located in the immediate vicinity of tumor cells. (C) Immune-desert HCC, showing less than 10% of the tumor stroma populated by lymphocytes, without dense immune cell infiltrates, and no contact of immune cells with tumor cells (original magnification, ×100, H&E stain). Arrows indicate the presence of lymphocytic infiltrates.
Figure 9.
TIL clusters correlate with tumor microenvironment. (A) Heatmap showing the normalized expression of the indicated markers for each CD4+ TIL cluster. The color scale indicates the relative expression levels. (B) Box plots showing the frequency of CD4+ TIL groups in inflamed tumors (immune-inflamed tumors) vs noninflamed tumors (immune-excluded and immune-desert tumors). (C) Heatmap showing the normalized expression of the markers for each CD8+ TIL cluster. The color scale indicates the relative expression levels. (D) Box plots showing the frequency of CD8+ TIL groups in inflamed tumors (immune-inflamed tumors) vs noninflamed tumors (immune-excluded and immune-desert tumors). (E) Heatmap showing the normalized expression of the markers for each DN TIL cluster. The color scale indicates the relative expression levels. (F) Box plots showing the frequency of DN TIL groups in inflamed tumors (immune-inflamed tumors) vs noninflamed tumors (immune-excluded and immune-desert tumors).
Within CD8+ TILs, 4 groups were identified. Group 1 cells contained resident memory CD8+ TILs, co-expressed CD38, PD1, and TIGIT, together with both tissue-residence markers CD69 and CD103. Group 1 CD8+ TILs were enriched significantly in inflamed tumors, possibly as a result of local activation. Group 2 TILs identified liver-resident MAIT cells and group 4 identified liver-resident CD8+ TILs. Both groups were not activated and were distributed equally among inflamed and noninflamed tumors. Finally, group 3 TILs contained CD8+ T cells with a nonactivated and non–tissue-residence phenotype, and were enriched in noninflamed tumors (Figure 9C and D).
In DN TILs, group 1 contained MAIT cells, and group 2 contained both TCR αβ and γδ cells characterized by low expression of activation and tissue-residence markers. In contrast, group 3 contained activated CD150+ DN cells and was enriched significantly in inflamed tumors. Finally, group 4 contained tissue-resident DN cells (Figure 9E and F). These findings showed that in HCC immune-inflamed tumors there are significantly increased populations of CD4+, CD8+, and DN TILs, which co-express unique markers of activation and tissue residence.
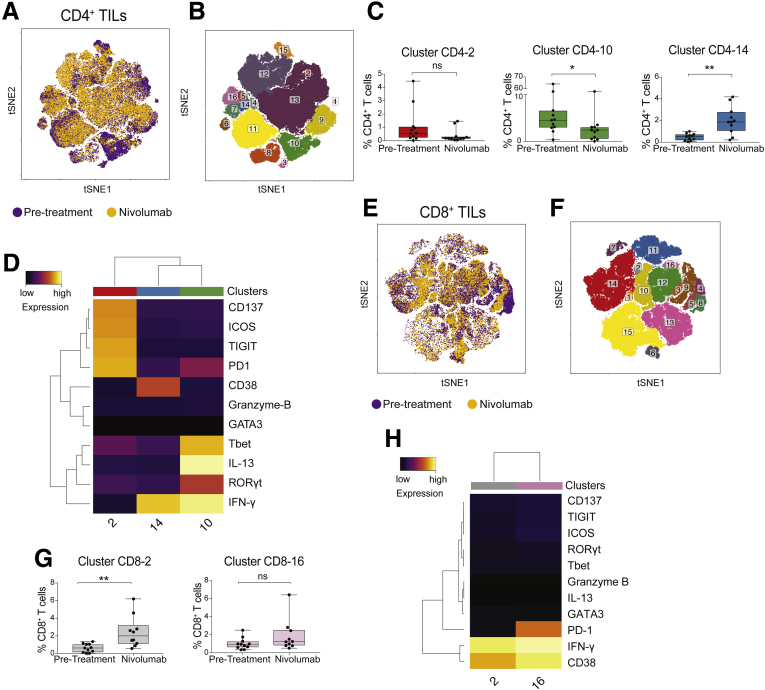
Phenotypic and Functional Changes of CD4+ and CD8+ T Cells in Tumor Biopsy Specimens Upon Nivolumab Treatment
Next, we examined the effects of the immune-checkpoint inhibitor nivolumab on TIL phenotype and function. Biopsy specimens before and during nivolumab therapy were studied in 7 HCC patients (Table 1, patients 4, 6, 19, 32, 34, 35, and 36, and Table 4 shows the list of analyzed molecules). We also found expression of CD38, ICOS, PD1 markers, and of IFN-γ, and IL13 cytokines and granzyme B in the TILs of these patients before treatment, as well as expression of the transcription factor T-bet (Figures 10A and B and 11A and B).
Table 4.
Cell Surface and Intracellular Staining Panel 4
| Marker | Dye | Host species | Reactivity | Clone | Description | Catalog no |
|---|---|---|---|---|---|---|
| CD3 | Alexa Fluor 700 | Mouse | Human | UCHT1 | Co-receptor: CD3 T cells | 300424 |
| CD4 | Brilliant Violet 711 | Mouse | Human | OKT4 | Co-receptor: CD4 T cells | 317440 |
| CD8 | BUV 496 | Mouse | Human | RPA-T8 | Co-receptor: CD8 T cells | 564804 |
| CD19 | PE/Cy5 | Mouse | Human | J3-119 | Co-receptor: B cells | A07771 |
| CD14 | PE/Cy5 | Mouse | Human | RMO52 | Co-receptor: monocytes | A07765 |
| CD137 | PE/Dazzle 594 | Mouse | Human | 4B4-1 | Costimulatory receptor | 309826 |
| CD278 (ICOS) | Brilliant Violet 510 | Mouse | Human | C398.4A | Inhibitory receptor | 313525 |
| CD38 | Brilliant Violet 650 | Mouse | Human | HB-7 | cADP ribose hydrolase | 356620 |
| PD1 | Brilliant Violet 785 | Mouse | Human | EH12.2H7 | Inhibitory receptor | 329930 |
| IFN-γ | APC/Cy7 | Mouse | Human | 4S.B3 | Proinflammatory cytokine | 502530 |
| IL13 | PE/Cy7 | Mouse | Human | JES10-5A2 | Proinflammatory cytokine | 501914 |
| Granzyme B | FITC | Mouse | Human/mouse | GB11 | Cytotoxic molecule | 515403 |
| T-bet | PE | Mouse | Human | 4B10 | Transcription factor–Th1 polarized T cell | 137236 |
| GATA-3 | Brilliant Violet 711 | Mouse | Human | 16E10A23 | Transcription factor–Th2 polarized T cell | 653814 |
| RORγt | APC | Mouse | Human/mouse | AFKJS-9 | Transcription factor–Th17 polarized T cell | 17-6988-80 |
cADP, cyclic adenosine diphosphate; FITC, fluorescein isothiocyanate; PE, phycoerythrin.
Figure 10.
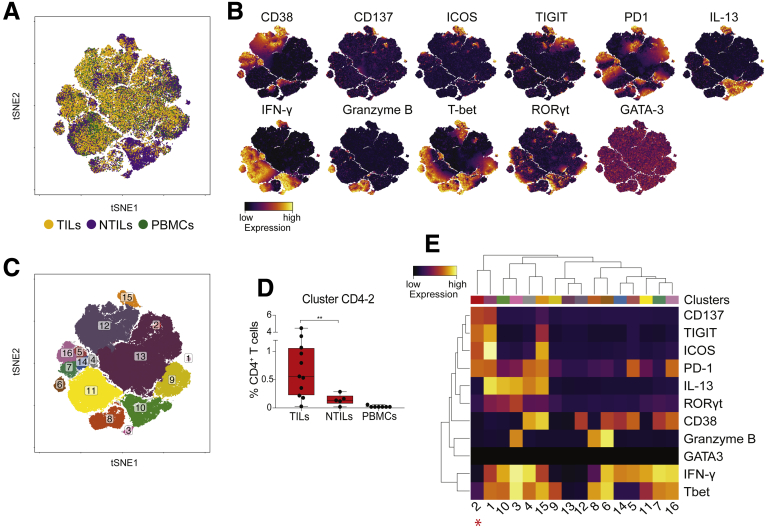
ICOS+TIGIT+PD1+CD137+CD4+T cells are enriched in patients before treatment. (A) tSNE map of CD4+ T cells from TILs (yellow dots), NTILs (purple dots), and PBMCs (green dots) of pretreatment and on-therapy HCC patients. (B) tSNE maps show the expression level and the distribution of each indicated marker. Color scale indicates the relative expression levels. (C) tSNE map colored by clusters of CD4+ T cells. (D) Box plot shows the frequency of the ICOS+ TIGIT+ PD1+ CD137+ CD4+ T-cell cluster significantly enriched in pretreatment HCC biopsy specimens. (E) Heatmap shows the normalized expression of the panel markers for the indicated clusters. Color scale indicates the relative expression levels. The red asterisk denotes the cluster 2 significantly increased in TILs vs NTILs.
Figure 11.
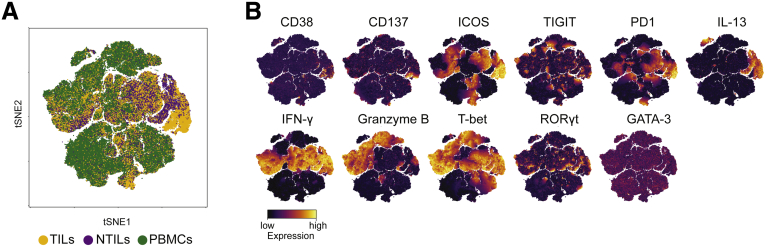
Nivolumab therapy affects CD8+T-cell phenotype. (A) tSNE map of CD8+ T cells from TILs (yellow dots), NTILs (purple dots), and PBMCs (green dots) of pretreated and on-therapy HCC patients. (B) tSNE maps showing the expression level and the distribution of each indicated marker. Color scale indicates the relative expression levels.
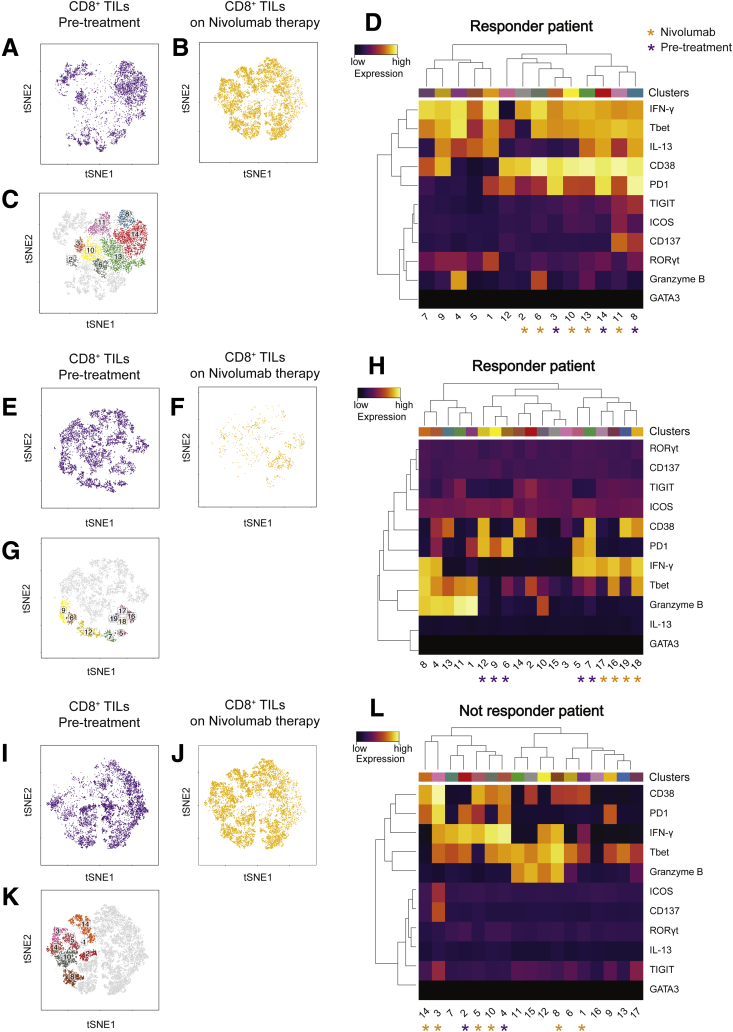
When we analyzed individual cell clusters (Figure 10C), an enrichment of activated and functionally impaired CD4+ T cells was detected in TILs before treatment (cluster CD4-2; P = .006) (Figure 10D and E). Nivolumab therapy induced changes in the composition of CD4+ and CD8+ TIL clusters (Figure 12A, B, E–G), namely a reduced percentage of cells in cluster CD4-2 (ICOS+ CD137+ TIGIT+ PD1+ CD4+ T cells), which, however, was not statistically significant (P = .06). Nivolumab therapy significantly reduced the percentage of cells in cluster CD4-10 (P = .03) that express RAR-related orphan receptor gamma T (RORγT) and T-bet and produce IFN-γ and IL13, while it increased the percentage of IFN-γ–producing CD38+ CD4+ T cells (cluster CD4-14; P = .001) (Figure 12C and D). Nivolumab therapy also promoted the enrichment of IFN-γ–producing CD38+ CD8+ T cells (cluster CD8-2; P = .01) (Figure 12H and G). Interestingly, in 2 patients with an immune-inflamed profile who responded to nivolumab, changes in patient-specific clusters of CD8+ TILs were observed (Figure 13A–H). However, therapy-induced changes also were observed in a patient who did not respond to nivolumab (Figure 13N). Because not all patients with an immune-inflamed tumor profile responded to nivolumab, our data suggest that the presence of an immune-inflamed profile is necessary but not sufficient for clinical response to anti-PD1 immunotherapy. Because only 15% to 20% of HCC patients respond to nivolumab,10 we could explain this low response rate by the high heterogeneity observed within tumor-infiltrating CD8+ T cells.
Figure 12.
Nivolumab therapy reduces the frequency of exhausted CD4+T cells in TILs and increases the frequency of activated CD4+and CD8+TILs producing IFN-γ. (A) tSNE map of CD4+ T cells infiltrating the tumor in samples before treatment (purple dots) and on-treatment with nivolumab (yellow dots). (B) tSNE map colored by clusters in CD4+ TILs. (C) Box plots showing the frequency CD4+ TIL clusters before and on nivolumab therapy. (D) Heatmap of the CD4+ TIL clusters changed after nivolumab therapy. The heatmap shows the normalized expression of the panel markers. The color scale indicates the relative expression levels. (E) tSNE map of CD8+ T cells infiltrating the tumor samples before treatment (purple dots) and on-treatment with nivolumab (yellow dots). (F) tSNE map colored by clusters of CD8+ TILs. (G) Box plots showing the frequency of CD8+ TIL clusters before and on nivolumab therapy. (H) Heatmap of the CD8+ TIL clusters changed after nivolumab therapy. The heatmap shows the normalized expression of the panel markers. The color scale indicates the relative expression levels. *P ≤ .05 and **P ≤ .001, Wilcoxon signed-rank test, adjusted with the Benjamini and Hochberg method. ns, not significant.
Figure 13.
Effect of nivolumab therapy on HCC patients. (A, E, and I) tSNE maps of CD8+ TILs from patients before nivolumab treatment. (A and E) Responder and (I) nonresponder patients. (B, F, and J) tSNE maps of CD8+ TILs from patients on nivolumab therapy. (B and F) Responder and (J) nonresponder patients. (C, G, and M) tSNE maps indicating individual clusters of CD8+ TILs in (C and G) responder and (K) nonresponder patients. (D, H, and L) Heatmaps of the normalized expression of each marker for indicated clusters. The purple asterisks indicate TIL clusters enriched in pretreatment samples and the yellow asterisks indicate TIL clusters enriched in on-therapy samples. Color scales indicate the relative expression levels of the indicated marker.
Discussion
Tumor-infiltrating immune cells greatly contribute to therapy response and disease arrest.25, 26, 27, 28, 29 Among the tumor infiltrating cell types, T lymphocytes directly participate in tumor killing and are considered favorable prognostic markers across multiple cancers.7, 8, 30, 31 However, the presence of a large number of TILs is not evidence of a favorable prognosis because the status of T-cell activation, the expression of costimulatory and co-inhibitory molecules, and the functional capacity of TILs also contribute to the prognostic outcome.32 Therefore, both TIL phenotype and functional properties are important to establish effective antitumor immunity.
In recent studies the TIL landscape in HCC has been investigated using different approaches.33, 34, 35, 36 Single-cell transcriptome studies were performed on small numbers of cells (<150 each in 6 HCC patients) and showed the presence of different CD8+ populations expanded from a few precursors and characterized by exhausted and regulatory T cell (Treg) signatures.33 A second study also showed the presence of a large numbers of both effector Tregs and CTLA-4+ CD8+ T cells in nonalcoholic steatohepatitis–related HCC patients, whereas large numbers of PD1+CD8+ T cells were observed in hepatitis B virus–related HCC patients.34 In a third study, Treg and CD8+ resident memory T cells were found enriched in a hepatitis B virus–related HCC microenvironment, which also was shown to contain more exhausted T cells than the microenvironment of non–viral-related HCC.36 Despite these informative studies, a comprehensive assessment of phenotypic and functional T-cell populations in HCC still is lacking. In particular, it would be very important to identify which cell population(s) mediate an effective antitumor immune response at least in a group of HCC patients, and also to show the mechanisms contributing to immune-desert or immune-inflamed HCC. These factors might be useful to predict the response to immunotherapy.37
In the present study we performed multidimensional flow cytometric analysis of T cells isolated from tumor and nontumor liver biopsy specimens and from peripheral blood of HCC patients. These investigations provide several novel insights into the immune response to HCC.
First, the tumor environment was found to be enriched in CD4+ T cells whereas CD8+ T cells and DNs were more abundant in nontumor liver tissue, in agreement with data from transcriptomics studies.33 We also found that TCR γδ cells were less abundant in the tumor microenvironment than in normal liver, whereas MAIT cells were equally present. Whether these 2 populations of innate-like T cells have a role in the immune response to HCC remains open, although these findings seem to exclude local activation and recognition of tumor cells. On the contrary, a population of tumor-infiltrating CD4+ T cells expressed CD137, a marker up-regulated in a transient manner upon T-cell antigen recognition.38 CD4+ cells also showed high expression of ICOS, an inducible costimulatory molecule expressed on previously activated T cells and T-follicular helper cells.39 The presence of cells expressing both CD137 and ICOS markers might indicate that CD4-expressing cells are activated within the HCC microenvironment and perhaps recognize locally presented antigens. This possibility is in agreement with previous studies showing that CD137+ cells isolated from ovarian cancers or melanomas are enriched in tumor-specific T cells.31, 40 The nature of the HLA class II–expressing antigen-presenting cells remains to be investigated, as does the identification of the antigens that stimulate CD4+ cells within HCC.
Interestingly, we did not find an accumulation of CD8+ or DN cells that expressed CD137, suggesting the absence of recent stimulation within HCC, or, alternatively, their unresponsive state.
A second finding was the presence of exclusive T-cell populations within HCC. The tSNE analyses showed the presence of unique CD4+, CD8+, and DN TILs, which were under-represented in NTILs and PBMCs. Within CD4+ cells, ICOS and TIGIT-co-expressing cells were tumor-enriched. TIGIT is a co-inhibitory molecule present in activated T cells, and contributes to down-modulation of T-cell responses.41 The co-expression of ICOS and TIGIT strongly suggest that CD4+ cells activated within the tumor are probably becoming poor antigen-responders.27 Most ICOS+ and TIGIT+ CD4+ cells also expressed the CD137 marker, possibly suggesting recent antigen recognition. Finally, the majority of these cells also expressed the PD1 inhibitory molecule. Although transient expression of inhibitory receptors is a characteristic of normal T-cell activation, the sustained expression of multiple immune checkpoint molecules is a hallmark of T-cell exhaustion after chronic stimulation by an antigen.27 The latter possibility may be the case of this unique CD4 population, which showed a remarkable hyporesponsiveness and did not produce cytokines upon stimulation, even with PMA + ionomycin. Not all cells co-expressing PD1 and TIGIT were functionally impaired, suggesting that only some cells were chronically stimulated within the tumor.42
Recent studies described the increased frequency of ICOS+ FoxP3+ Tregs in HCC43 and, more recently, a single-cell transcriptomics study proposed a unique HCC-infiltrating Tregs gene signature33 that overlapped with signatures in melanoma, breast, colon, and lung cancers.44, 45, 46 In accordance with these publications, our studies detected a high percentage of FoxP3- and Helios-expressing T cells within ICOS+ TIGIT+ CD4+ TILs, which could indicate a regulatory function. However, FoxP3 and ICOS markers also are expressed by recently stimulated T cells, and thus these cells might represent T cells responding to antigen within the tumor. Because of the extremely small number of this population in the biopsy specimens and lack of information on antigen specificity, it was not possible to sort these cells and test their putative inhibitory function.
In HCC biopsy specimens we also found unique clusters of CD38+, PD1+, and CD8+ TILs, which proliferated and expressed inflammatory (TNF-α and IFN-γ) cytokines and cytotoxic (granzyme B) molecules upon stimulation. These cells showed a high interpatient heterogeneity that probably contributes to the high variability in tumor immunity.
The liver is an organ containing several innate immune cell populations.47 Among these, there is enrichment of unconventional T cells, including MAIT, TCR γδ T and DN TCR αβ T cells12, 47, 48 that recognize nonpeptide ligands presented by non–major histocompatibility complex antigen-presenting molecules.49 Their antitumor activity recently has been debated.50, 51
In HCC, we found an enrichment of activated TCR γδ cells expressing CD38, CD69, and CD103 markers. The antigens and the presenting molecules stimulating γδ cells in HCC remain unknown and might provide useful information to further investigate recruitment and antitumor activity of TCR γδ cells in HCC.
A second significantly increased TIL population was CD4-CD8 DN and expressed a TCR αβ together with the activation markers CD137, HLA-DR, and CD69. These cells also up-regulated CD150+, a costimulatory molecule present in activated lymphocytes and facilitating efficient T- and B-cell responses.52 The expression of all such activation markers is a strong indication that unconventional T cells also might become stimulated within HCC. The identification of the stimulatory antigens may provide unexpected information to anti-HCC immunity and may designate these DN T cells as a novel immunotherapy target population.
Our study also provided 2 additional facts. The first is the correlation of the presence of exclusive T cells with currently adopted histologic classification. Immune inflamed tumors showed an increase of unique HCC-specific CD4+, CD8+, and DN TILs. Importantly, these cells expressed markers of activation and tissue residence, possibly suggesting activation within the tumor.
On the contrary, in immune-excluded tumors there was a large number of cells lacking activation and tissue-residence markers, probably reflecting recent blood extravasation. These findings support the clinical relevance of the histologic classification and identify novel markers to recognize nonfunctional TILs in HCC.
The second additional fact is regarding the effects of nivolumab therapy on HCC TILs. Phases 1–2 clinical studies have shown that nivolumab treatment induces a tumor reduction and an objective patient response to the therapy in 15%–20% of patients with advanced HCC.10 Based on our findings on nontreated HCC patients, we aimed to characterize the effects of anti-PD1 immunotherapy on the phenotype and function of the unique populations of HCC TILs. Although significant variations in the frequency of total T cells were not observed, important changes of HCC unique TILs were detected. The effects on circulating lymphocytes and NTILs were minimal, whereas drastic changes were observed in TILs because they diminished unresponsive ICOS+ TIGIT+ CD4+ T cells and increased activated CD4+ and CD8+ T cells expressing CD38 and producing IFN-γ. Thus, in HCC, anti-PD1 treatment promotes the reduction of unresponsive T cells and establishment of an inflamed tumor microenvironment. The increased IFN-γ release might modulate both immune and tumor cell functions,53, 54 thus positively influencing antitumor immunity. These preliminary findings could indicate that the histologic enumeration of ICOS+ TIGIT+ CD4+ TILs in inflamed HCC represents a prognostic indicator of therapy responsiveness. However, this possibility has to be considered with caution. Indeed, we are aware that the analysis described here is based on a small number of patients and that validation of its prognostic value requires ad hoc prospective studies that include more patients.
In conclusion, our detailed characterization of HCC-infiltrating TILs showed a unique populations of locally activated T cells that may provide effective antitumor immunity. These studies set the point for the identification of the tumor antigens stimulating these T cells and their possible exploitation as immunotherapeutic targets in HCC.
Materials and Methods
Patient Samples
Blood and liver needle biopsy specimens from HCC patients (n = 36) (Table 1) were obtained in the outpatient clinic of the Division of Gastroenterology and Hepatology at the University Hospital Basel (Basel, Switzerland). The use of biopsy material for this project was approved by the ethics committee of the northwestern part of Switzerland (protocol EKNZ 2014-099).
Histology
Liver biopsy specimens were fixed in 10% buffered formalin and embedded in paraffin. Sections were stained with H&E. Histopathologic evaluation was assessed by a board-certified pathologist (L.T.). Tumors were classified histologically according to the Edmondson–Steiner grading system and according to their immune-phenotype.55 In our HCC cohort, 3 patterns were described as follows: (1) immune-desert tumors, in which less than 10% of tumor stroma was populated by lymphocytes, and no dense immune cell infiltrates or contacts of the immune cells with tumor cells were detected; (2) immune-excluded tumors, in which ≥10% of tumor stroma was populated by lymphocytes located in the immediate vicinity of tumor cells; and (3) immune-inflamed, if lymphocytes were present in the tumor parenchyma in close proximity to tumor cells. This classification was not immediately applicable in every case because some tumors showed certain degrees of overlap between the different patterns described. In such cases, the tumors were classified according to the most prevalent pattern. Necrotic areas were excluded from evaluation. Macrophages and granulocytes were not considered for this classification. Our HCC cohort included prevalently biopsy specimens with an immune-inflamed phenotype.
PBMCs and Liver Infiltrating Immune Cells
PBMCs were isolated from 15 mL of blood by standard density-gradient centrifugation protocols, washed twice in phosphate-buffered saline (PBS), and cryopreserved in medium containing 90% fetal bovine serum and 10% dimethyl sulfoxide until fluorescence-activated cell sorter analysis. For isolation of liver-infiltrating lymphocytes, cylinders of liver biopsy specimens (diameter, ∼1 mm; length, 2 cm) were obtained from tumor and nontumor liver of HCC patients. The samples were washed in PBS to avoid blood contamination and mechanically smashed above a cell strainer (40-mm nylon). The recovered cells were washed twice in PBS and cryopreserved.
Antibodies
The primary monoclonal antibodies and MHC class I-related molecule (MR1) tetramers used are described in Table 5.
Table 5.
Reagents
| Marker | Dye | Host species | Reactivity | Clone | Company | Catalog no |
|---|---|---|---|---|---|---|
| CD3 | PE/Cy7 | Mouse | Human | UCHT1 | Biolegend | 300420 |
| CD3 | Alexa Fluor 700 | Mouse | Human | UCHT1 | Biolegend | 300424 |
| TCR γδ | Brilliant Violet 421 | Mouse | Human | B1 | Biolegend | 331218 |
| CD4 | Brilliant Violet 711 | Mouse | Human | OKT4 | Biolegend | 317440 |
| HLA-DR | Alexa Fluor 700 | Mouse | Human | L243 | Biolegend | 307626 |
| CD38 | Brilliant Violet 650 | Mouse | Human | HB-7 | Biolegend | 356620 |
| CD137 | PE/Dazzle 594 | Mouse | Human | 4B4-1 | Biolegend | 309826 |
| CD278 (ICOS) | Brilliant Violet 510 | Mouse | Human | C398.4A | Biolegend | 313525 |
| CD150 (SLAM) | FITC | Mouse | Human | A12 (7D4) | Biolegend | 306306 |
| CD69 | APC/Cy7 | Mouse | Human | FN50 | Biolegend | 310914 |
| CD103 | APC | Mouse | Human | Ber-ACT8 | Biolegend | 350216 |
| PD1 | Brilliant Violet 785 | Mouse | Human | EH12.2H7 | Biolegend | 329930 |
| KLRG1 | Brilliant Violet 605 | Mouse | Human | 2F1/KLRG1 | Biolegend | 138419 |
| TIGIT (VSTM3) | Brilliant Violet 605 | Mouse | Human | A15153G | Biolegend | 372706 |
| IL22 | PE | Mouse | Human | 2G12A41 | Biolegend | 366704 |
| TNF-α | PE/Cy7 | Mouse | Human | MAb11 | Biolegend | 502930 |
| IFN-γ | APC/Cy7 | Mouse | Human | 4S.B3 | Biolegend | 502530 |
| T-bet | PE | Mouse | Human | 4B10 | Biolegend | 137236 |
| Granzyme B | FITC | Mouse | Human/mouse | GB11 | Biolegend | 515403 |
| Perforin | Brilliant Violet 421 | Mouse | Human | B-D48 | Biolegend | 353307 |
| Helios | PE/Cy7 | Mouse | Human | 22F6 | Biolegend | 137236 |
| Foxp3 | Alexa Fluor 647 | Mouse | Human | 150D | Biolegend | 320014 |
| IL13 | PE/Cy7 | Mouse | Human | JES10-5A2 | Biolegend | 501914 |
| GATA-3 | Brilliant Violet 711 | Mouse | Human | 16E10A23 | Biolegend | 653814 |
| Ki-67 | Alexa 488 | Mouse | Human | Ki-67 | Biolegend | 350508 |
| CD19 | PE/Cy5 | Mouse | Human | J3-119 | Beckman Coulter (1260 Nyon, France) | A07771 |
| CD14 | PE/Cy5 | Mouse | Human | RMO52 | Beckman Coulter | A07765 |
| CD4 | PE | Mouse | Human | RPA-T4 | BD Bioscience (60322 Frankfurt am Main, Germany) | 561843 |
| CD4 | APC | Mouse | Human | RPA-T4 | BD Bioscience | 555349 |
| CD8 | BUV 496 | Mouse | Human | RPA-T8 | BD Bioscience | 564804 |
| TIGIT (VSTM3) | PerCP-eFluor 710 | Mouse | Human | MBSA43 | eBioscience | 46-9500-42 |
| ROR-γt | APC | Mouse | Human/mouse | AFKJS-9 | eBioscience | 17-6988-80 |
| hMR1-5OP-RU tetramer | PE | Human | National Institutes of Health, Bethesda, MD | 31704 R | ||
| hMR1-6-FP tetramer | PE | Human | National Institutes of Health | 31924 F |
FITC, fluorescein isothiocyanate; PE, phycoerythrin.
Intracellular Cytokine Induction
Liver infiltrating lymphocytes and PBMCs were thawed in PBS with 10 μg/mL DNase I (Sigma-Aldrich, St. Louis, MO) to prevent cell clumping. The cells were resuspended in RPMI 1640 medium (Gibco, Gaithersburg, MD) supplemented with 10% heat-inactivated fetal bovine serum (Bioconcept, 4123 Allschwil, CH), 100 U/mL kanamycin, 2 mmol/L stable glutamine, 1 mmol/L sodium pyruvate, and 1% minimal essential medium (MEM) nonessential amino acids (all from Bioconcept), and 10 μg/mL DNase I. The cells were rested for 1 hour at 37°C before further processing. For intracellular cytokine induction, the cells were washed and resuspended in complete medium containing PMA (25 ng/mL), calcium ionophore ionomycin (1 μg/mL), and Brefeldin A (10 μg/mL; all from Sigma-Aldrich) for 4 hours. The LIVE/DEAD Fixable Aqua Dead Cell Stain Kit (Invitrogen, Norcross, GA) was used to identify viable cells before intracellular staining.
Flow Cytometry
Liver-infiltrating lymphocytes and PBMCs were washed and resuspended in blocking buffer (PBS containing 50% human serum) for 30 minutes at room temperature.
Cells were resuspended in staining buffer (PBS containing 0.02% NaN3 and 0.5% human albumin) and stained with MR1 tetramers (National Institutes of Health Tetramer Core Facility, Emory University, Atlanta, GA) for 40 minutes at room temperature, followed by other surface stains for 20 minutes at 4°C in the dark. For intracellular staining, cells subsequently were washed in staining buffer twice before fixation and permeabilization using Biolegend (San Diego, CA) buffers for intracellular staining. In short, cells were suspended in the fixation buffer in the dark for 30 minutes, washed with permeabilization wash buffer, and stained for intracellular markers for 20 minutes at 4°C in the dark. Samples were analyzed on LSR Fortessa flow (BD Bioscience, San Jose, CA).
Flow Cytometry Data Analysis
Flow cytometry data were analyzed using FlowJo v10.1 software (TreeStar, 2060 Antwerpen, Belgium), before exporting to R, and transformed using an inverse hyperbolic sine function. Dimensionality reduction was performed on transformed data with the Barnes–Hutt implementation of tSNE,56 using a custom R script (available on request). For each set of analyses, individual cell subsets (CD4+, CD8+, and DN T cells) from each patient were randomly down-sampled to an equal number of events (5000 events). Flow cytometry data were transformed using an inverse hyperbolic sine function.
For identification of T-cell clusters, multiple runs of the DBSCAN were performed. For each cluster identified by DBSCAN, we quantified the differences between the frequency in HCC and nontumor liver samples and HCC and PBMC using the Wilcoxon signed-rank statistic test.
Statistical Analysis
The results are expressed as means ± SD. In all analyses, the Wilcoxon signed-rank test, adjusted with the Benjamini and Hochberg method and a 2-tailed paired test multicomparison (Tukey test) were used. A P value of .05 or less (95% CI) was considered statistically significant.
All authors had access to the study data and reviewed and approved the final manuscript.
Acknowledgments
The authors thank Tobias Rutishauser for help in setting a computational analysis pipeline and P. Cullen for revising the manuscript. The MR1 tetramer technology was developed jointly by Dr James McCluskey, Dr Jamie Rossjohn, and Dr David Fairlie, and the material was produced by the National Institutes of Health Tetramer Core Facility as permitted to be distributed by the University of Melbourne.
Footnotes
Author contributions Daniela Di Blasi designed and performed experiments, analyzed data, and wrote the manuscript; Tujana Boldanova and Markus H. Heim recruited patients, performed biopsies, and collected and curated clinical annotation data; Luigi Terracciano performed the histopathologic analysis; Lucia Mori discussed the data and wrote the manuscript; Markus H. Heim and Gennaro De Libero conceived and designed experiments, coordinated the study, supervised the work and the interpretation of the results, and obtained funding; and Gennaro De Libero wrote the manuscript.
Conflicts of interest The authors disclose no conflicts.
Funding This work was supported by grants SNF 310030-173240, KFS-3730-08-2015, and PMB-02-17 (G.D.L.), and by European Research Council Synergy609883 Mechanisms of Evasive Resistance in Cancer (MERiC) and SystemX (MERIC) (M.H.H.).
Contributor Information
Markus H. Heim, Email: markus.heim@unibas.ch.
Gennaro De Libero, Email: gennaro.delibero@unibas.ch.
References
- 1.Torre L.A., Siegel R.L., Ward E.M., Jemal A. Global cancer incidence and mortality rates and trends--an update. Cancer Epidemiol Biomarkers Prev. 2016;25:16–27. doi: 10.1158/1055-9965.EPI-15-0578. [DOI] [PubMed] [Google Scholar]
- 2.Marquardt J.U., Andersen J.B., Thorgeirsson S.S. Functional and genetic deconstruction of the cellular origin in liver cancer. Nat Rev Cancer. 2015;15:653–667. doi: 10.1038/nrc4017. [DOI] [PubMed] [Google Scholar]
- 3.El-Serag H.B. Hepatocellular carcinoma. N Engl J Med. 2011;365:1118–1127. doi: 10.1056/NEJMra1001683. [DOI] [PubMed] [Google Scholar]
- 4.Zamarron B.F., Chen W. Dual roles of immune cells and their factors in cancer development and progression. Int J Biol Sci. 2011;7:651–658. doi: 10.7150/ijbs.7.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Capece D., Fischietti M., Verzella D., Gaggiano A., Cicciarelli G., Tessitore A., Zazzeroni F., Alesse E. The inflammatory microenvironment in hepatocellular carcinoma: a pivotal role for tumor-associated macrophages. Biomed Res Int. 2013;2013:187–204. doi: 10.1155/2013/187204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen D.S., Mellman I. Elements of cancer immunity and the cancer-immune set point. Nature. 2017;541:321–330. doi: 10.1038/nature21349. [DOI] [PubMed] [Google Scholar]
- 7.Zheng X., Song X., Shao Y., Xu B., Chen L., Zhou Q., Hu W., Zhang D., Wu C., Tao M., Zhu Y., Jiang J. Prognostic role of tumor-infiltrating lymphocytes in gastric cancer: a meta-analysis. Oncotarget. 2017;8:57386–57398. doi: 10.18632/oncotarget.18065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu Q., Wang C., Yuan X., Feng Z., Han Z. Prognostic value of tumor-infiltrating lymphocytes for patients with head and neck squamous cell carcinoma. Transl Oncol. 2017;10:10–16. doi: 10.1016/j.tranon.2016.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Foerster F., Hess M., Gerhold-Ay A., Marquardt J.U., Becker D., Galle P.R., Schuppan D., Binder H., Bockamp E. The immune contexture of hepatocellular carcinoma predicts clinical outcome. Sci Rep. 2018;8:5351. doi: 10.1038/s41598-018-21937-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.El-Khoueiry A.B., Sangro B., Yau T., Crocenzi T.S., Kudo M., Hsu C., Kim T.Y., Choo S.P., Trojan J., Welling T.H.R., Meyer T., Kang Y.K., Yeo W., Chopra A., Anderson J., Dela Cruz C., Lang L., Neely J., Tang H., Dastani H.B., Melero I. Nivolumab in patients with advanced hepatocellular carcinoma (CheckMate 040): an open-label, non-comparative, phase 1/2 dose escalation and expansion trial. Lancet. 2017;389:2492–2502. doi: 10.1016/S0140-6736(17)31046-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lepore M., Kalinichenko A., Colone A., Paleja B., Singhal A., Tschumi A., Lee B., Poidinger M., Zolezzi F., Quagliata L., Sander P., Newell E., Bertoletti A., Terracciano L., De Libero G., Mori L. Parallel T-cell cloning and deep sequencing of human MAIT cells reveal stable oligoclonal TCRbeta repertoire. Nat Commun. 2014;5:3866. doi: 10.1038/ncomms4866. [DOI] [PubMed] [Google Scholar]
- 12.Kurioka A., Walker L.J., Klenerman P., Willberg C.B. MAIT cells: new guardians of the liver. Clin Transl Immunol. 2016;5:e98. doi: 10.1038/cti.2016.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ye Q., Song D.G., Poussin M., Yamamoto T., Best A., Li C., Coukos G., Powell D.J., Jr. CD137 accurately identifies and enriches for naturally occurring tumor-reactive T cells in tumor. Clin Cancer Res. 2014;20:44–55. doi: 10.1158/1078-0432.CCR-13-0945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hahsler MM. dbscan: density based clustering of applications with noise (DBSCAN) and related algorithms. R package version 1.0-0 2017. https://cran.r-project.org/web/packages/dbscan/vignettes/dbscan.pdf.
- 15.Chemnitz J.M., Parry R.V., Nichols K.E., June C.H., Riley J.L. SHP-1 and SHP-2 associate with immunoreceptor tyrosine-based switch motif of programmed death 1 upon primary human T cell stimulation, but only receptor ligation prevents T cell activation. J Immunol. 2004;173:945–954. doi: 10.4049/jimmunol.173.2.945. [DOI] [PubMed] [Google Scholar]
- 16.Petrovas C., Casazza J.P., Brenchley J.M., Price D.A., Gostick E., Adams W.C., Precopio M.L., Schacker T., Roederer M., Douek D.C., Koup R.A. PD-1 is a regulator of virus-specific CD8+ T cell survival in HIV infection. J Exp Med. 2006;203:2281–2292. doi: 10.1084/jem.20061496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kumar B.V., Ma W., Miron M., Granot T., Guyer R.S., Carpenter D.J., Senda T., Sun X., Ho S.H., Lerner H., Friedman A.L., Shen Y., Farber D.L. Human tissue-resident memory T cells are defined by core transcriptional and functional signatures in lymphoid and mucosal sites. Cell Rep. 2017;20:2921–2934. doi: 10.1016/j.celrep.2017.08.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lukas Yani S., Keller M., Melzer F.L., Weinberger B., Pangrazzi L., Sopper S., Trieb K., Lobina M., Orru V., Fiorillo E., Cucca F., Grubeck-Loebenstein B. CD8(+)HLADR(+) regulatory T cells change with aging: they increase in number, but lose checkpoint inhibitory molecules and suppressive function. Front Immunol. 2018;9:1201. doi: 10.3389/fimmu.2018.01201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ganesan A.P., Clarke J., Wood O., Garrido-Martin E.M., Chee S.J., Mellows T., Samaniego-Castruita D., Singh D., Seumois G., Alzetani A., Woo E., Friedmann P.S., King E.V., Thomas G.J., Sanchez-Elsner T., Vijayanand P., Ottensmeier C.H. Tissue-resident memory features are linked to the magnitude of cytotoxic T cell responses in human lung cancer. Nat Immunol. 2017;18:940–950. doi: 10.1038/ni.3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park S.L., Mackay L.K., Gebhardt T. Distinct recirculation potential of CD69(+)CD103(-) and CD103(+) thymic memory CD8(+) T cells. Immunol Cell Biol. 2016;94:975–980. doi: 10.1038/icb.2016.60. [DOI] [PubMed] [Google Scholar]
- 21.Kim R., Coppola D., Wang E., Chang Y.D., Kim Y., Anaya D., Kim D.W. Prognostic value of CD8CD45RO tumor infiltrating lymphocytes in patients with extrahepatic cholangiocarcinoma. Oncotarget. 2018;9:23366–23372. doi: 10.18632/oncotarget.25163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thommen D.S., Koelzer V.H., Herzig P., Roller A., Trefny M., Dimeloe S., Kiialainen A., Hanhart J., Schill C., Hess C., Savic Prince S., Wiese M., Lardinois D., Ho P.C., Klein C., Karanikas V., Mertz K.D., Schumacher T.N., Zippelius A. A transcriptionally and functionally distinct PD-1(+) CD8(+) T cell pool with predictive potential in non-small-cell lung cancer treated with PD-1 blockade. Nat Med. 2018;24:994–1004. doi: 10.1038/s41591-018-0057-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang X., Tian Z. Gammadelta T cells in liver diseases. Front Med. 2018;12:262–268. doi: 10.1007/s11684-017-0584-x. [DOI] [PubMed] [Google Scholar]
- 24.Hendry S., Salgado R., Gevaert T., Russell P.A., John T., Thapa B., Christie M., van de Vijver K., Estrada M.V., Gonzalez-Ericsson P.I., Sanders M., Solomon B., Solinas C., Van den Eynden G., Allory Y., Preusser M., Hainfellner J., Pruneri G., Vingiani A., Demaria S., Symmans F., Nuciforo P., Comerma L., Thompson E.A., Lakhani S., Kim S.R., Schnitt S., Colpaert C., Sotiriou C., Scherer S.J., Ignatiadis M., Badve S., Pierce R.H., Viale G., Sirtaine N., Penault-Llorca F., Sugie T., Fineberg S., Paik S., Srinivasan A., Richardson A., Wang Y., Chmielik E., Brock J., Johnson D.B., Balko J., Wienert S., Bossuyt V., Michiels S., Ternes N., Burchardi N., Luen S.J., Savas P., Klauschen F., Watson P.H., Nelson B.H., Criscitiello C., O'Toole S., Larsimont D., de Wind R., Curigliano G., Andre F., Lacroix-Triki M., van de Vijver M., Rojo F., Floris G., Bedri S., Sparano J., Rimm D., Nielsen T., Kos Z., Hewitt S., Singh B., Farshid G., Loibl S., Allison K.H., Tung N., Adams S., Willard-Gallo K., Horlings H.M., Gandhi L., Moreira A., Hirsch F., Dieci M.V., Urbanowicz M., Brcic I., Korski K., Gaire F., Koeppen H., Lo A., Giltnane J., Rebelatto M.C., Steele K.E., Zha J., Emancipator K., Juco J.W., Denkert C., Reis-Filho J., Loi S., Fox S.B. Assessing tumor-infiltrating lymphocytes in solid tumors: a practical review for pathologists and proposal for a standardized method from the International Immuno-Oncology Biomarkers Working Group: part 2: TILs in melanoma, gastrointestinal tract carcinomas, non-small cell lung carcinoma and mesothelioma, endometrial and ovarian carcinomas, squamous cell carcinoma of the head and neck, genitourinary carcinomas, and primary brain tumors. Adv Anat Pathol. 2017;24:311–335. doi: 10.1097/PAP.0000000000000161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Denkert C., von Minckwitz G., Darb-Esfahani S., Lederer B., Heppner B.I., Weber K.E., Budczies J., Huober J., Klauschen F., Furlanetto J., Schmitt W.D., Blohmer J.U., Karn T., Pfitzner B.M., Kummel S., Engels K., Schneeweiss A., Hartmann A., Noske A., Fasching P.A., Jackisch C., van Mackelenbergh M., Sinn P., Schem C., Hanusch C., Untch M., Loibl S. Tumour-infiltrating lymphocytes and prognosis in different subtypes of breast cancer: a pooled analysis of 3771 patients treated with neoadjuvant therapy. Lancet Oncol. 2018;19:40–50. doi: 10.1016/S1470-2045(17)30904-X. [DOI] [PubMed] [Google Scholar]
- 26.Lee N., Zakka L.R., Mihm M.C., Jr., Schatton T. Tumour-infiltrating lymphocytes in melanoma prognosis and cancer immunotherapy. Pathology. 2016;48:177–187. doi: 10.1016/j.pathol.2015.12.006. [DOI] [PubMed] [Google Scholar]
- 27.Thommen D.S., Schumacher T.N. T cell dysfunction in cancer. Cancer Cell. 2018;33:547–562. doi: 10.1016/j.ccell.2018.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barnes T.A., Amir E. HYPE or HOPE: the prognostic value of infiltrating immune cells in cancer. Br J Cancer. 2018;118:e5. doi: 10.1038/bjc.2017.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pages F., Galon J., Dieu-Nosjean M.C., Tartour E., Sautes-Fridman C., Fridman W.H. Immune infiltration in human tumors: a prognostic factor that should not be ignored. Oncogene. 2010;29:1093–1102. doi: 10.1038/onc.2009.416. [DOI] [PubMed] [Google Scholar]
- 30.Hwang W.T., Adams S.F., Tahirovic E., Hagemann I.S., Coukos G. Prognostic significance of tumor-infiltrating T cells in ovarian cancer: a meta-analysis. Gynecol Oncol. 2012;124:192–198. doi: 10.1016/j.ygyno.2011.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adams S., Gray R.J., Demaria S., Goldstein L., Perez E.A., Shulman L.N., Martino S., Wang M., Jones V.E., Saphner T.J., Wolff A.C., Wood W.C., Davidson N.E., Sledge G.W., Sparano J.A., Badve S.S. Prognostic value of tumor-infiltrating lymphocytes in triple-negative breast cancers from two phase III randomized adjuvant breast cancer trials: ECOG 2197 and ECOG 1199. J Clin Oncol. 2014;32:2959–2966. doi: 10.1200/JCO.2013.55.0491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu L., Bai Y., Wang Z. Elevated T cell activation score is associated with improved survival of breast cancer. Breast Cancer Res Treat. 2017;164:689–696. doi: 10.1007/s10549-017-4281-x. [DOI] [PubMed] [Google Scholar]
- 33.Zheng C., Zheng L., Yoo J.K., Guo H., Zhang Y., Guo X., Kang B., Hu R., Huang J.Y., Zhang Q., Liu Z., Dong M., Hu X., Ouyang W., Peng J., Zhang Z. Landscape of infiltrating T cells in liver cancer revealed by single-cell sequencing. Cell. 2017;169:1342–1356 e1316. doi: 10.1016/j.cell.2017.05.035. [DOI] [PubMed] [Google Scholar]
- 34.Inada Y., Mizukoshi E., Seike T., Tamai T., Iida N., Kitahara M., Yamashita T., Arai K., Terashima T., Fushimi K., Yamashita T., Honda M., Kaneko S. Characteristics of immune response to tumor-associated antigens and immune cell profile in patients with hepatocellular carcinoma. Hepatology. 2019;69:653–665. doi: 10.1002/hep.30212. [DOI] [PubMed] [Google Scholar]
- 35.Ou X., Guan J., Chen J.S., Ying J.C., Liu X.P., Tian P.K., Liu J.K., Nie L.P., Zhao Y., Yu G.Y. LAP(+)CD4(+) T cells are elevated among the peripheral blood mononuclear cells and tumor tissue of patients with hepatocellular carcinoma. Exp Ther Med. 2018;16:788–796. doi: 10.3892/etm.2018.6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lim C.J., Lee Y.H., Pan L., Lai L., Chua C., Wasser M., Lim T.K.H., Yeong J., Toh H.C., Lee S.Y., Chan C.Y., Goh B.K., Chung A., Heikenwalder M., Ng I.O., Chow P., Albani S., Chew V. Multidimensional analyses reveal distinct immune microenvironment in hepatitis B virus-related hepatocellular carcinoma. Gut. 2019;68:916–927. doi: 10.1136/gutjnl-2018-316510. [DOI] [PubMed] [Google Scholar]
- 37.Osorio J.C., Harding J.J. Understanding and quantifying the immune microenvironment in hepatocellular carcinoma. Transl Gastroenterol Hepatol. 2018;3:107. doi: 10.21037/tgh.2018.12.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wolfl M., Kuball J., Ho W.Y., Nguyen H., Manley T.J., Bleakley M., Greenberg P.D. Activation-induced expression of CD137 permits detection, isolation, and expansion of the full repertoire of CD8+ T cells responding to antigen without requiring knowledge of epitope specificities. Blood. 2007;110:201–210. doi: 10.1182/blood-2006-11-056168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Simpson T.R., Quezada S.A., Allison J.P. Regulation of CD4 T cell activation and effector function by inducible costimulator (ICOS) Curr Opin Immunol. 2010;22:326–332. doi: 10.1016/j.coi.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 40.Zhu Y., Chen L. CD137 as a biomarker for tumor-reactive T cells: finding gold in the desert. Clin Cancer Res. 2014;20:3–5. doi: 10.1158/1078-0432.CCR-13-2573. [DOI] [PubMed] [Google Scholar]
- 41.Pauken K.E., Wherry E.J. TIGIT and CD226: tipping the balance between costimulatory and coinhibitory molecules to augment the cancer immunotherapy toolkit. Cancer Cell. 2014;26:785–787. doi: 10.1016/j.ccell.2014.11.016. [DOI] [PubMed] [Google Scholar]
- 42.Wherry E.J. T cell exhaustion. Nat Immunol. 2011;12:492–499. doi: 10.1038/ni.2035. [DOI] [PubMed] [Google Scholar]
- 43.Tu J.F., Ding Y.H., Ying X.H., Wu F.Z., Zhou X.M., Zhang D.K., Zou H., Ji J.S. Regulatory T cells, especially ICOS(+) FOXP3(+) regulatory T cells, are increased in the hepatocellular carcinoma microenvironment and predict reduced survival. Sci Rep. 2016;6:35056. doi: 10.1038/srep35056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.De Simone M., Arrigoni A., Rossetti G., Gruarin P., Ranzani V., Politano C., Bonnal R.J.P., Provasi E., Sarnicola M.L., Panzeri I., Moro M., Crosti M., Mazzara S., Vaira V., Bosari S., Palleschi A., Santambrogio L., Bovo G., Zucchini N., Totis M., Gianotti L., Cesana G., Perego R.A., Maroni N., Pisani Ceretti A., Opocher E., De Francesco R., Geginat J., Stunnenberg H.G., Abrignani S., Pagani M. Transcriptional landscape of human tissue lymphocytes unveils uniqueness of tumor-infiltrating T regulatory cells. Immunity. 2016;45:1135–1147. doi: 10.1016/j.immuni.2016.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Plitas G., Konopacki C., Wu K., Bos P.D., Morrow M., Putintseva E.V., Chudakov D.M., Rudensky A.Y. Regulatory T cells exhibit distinct features in human breast cancer. Immunity. 2016;45:1122–1134. doi: 10.1016/j.immuni.2016.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tanchot C., Terme M., Pere H., Tran T., Benhamouda N., Strioga M., Banissi C., Galluzzi L., Kroemer G., Tartour E. Tumor-infiltrating regulatory T cells: phenotype, role, mechanism of expansion in situ and clinical significance. Cancer Microenviron. 2013;6:147–157. doi: 10.1007/s12307-012-0122-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gao B., Jeong W.I., Tian Z. Liver: An organ with predominant innate immunity. Hepatology. 2008;47:729–736. doi: 10.1002/hep.22034. [DOI] [PubMed] [Google Scholar]
- 48.Mehal W.Z., Azzaroli F., Crispe I.N. Immunology of the healthy liver: old questions and new insights. Gastroenterology. 2001;120:250–260. doi: 10.1053/gast.2001.20947. [DOI] [PubMed] [Google Scholar]
- 49.Mori L., Lepore M., De Libero G. The Immunology of CD1- and MR1-restricted T cells. Annu Rev Immunol. 2016;34:479–510. doi: 10.1146/annurev-immunol-032414-112008. [DOI] [PubMed] [Google Scholar]
- 50.Godfrey D.I., Le Nours J., Andrews D.M., Uldrich A.P., Rossjohn J. Unconventional T cell targets for cancer immunotherapy. Immunity. 2018;48:453–473. doi: 10.1016/j.immuni.2018.03.009. [DOI] [PubMed] [Google Scholar]
- 51.Silva-Santos B., Serre K., Norell H. Gammadelta T cells in cancer. Nat Rev Immunol. 2015;15:683–691. doi: 10.1038/nri3904. [DOI] [PubMed] [Google Scholar]
- 52.Ma C.S., Nichols K.E., Tangye S.G. Regulation of cellular and humoral immune responses by the SLAM and SAP families of molecules. Annu Rev Immunol. 2007;25:337–379. doi: 10.1146/annurev.immunol.25.022106.141651. [DOI] [PubMed] [Google Scholar]
- 53.Ayers M., Lunceford J., Nebozhyn M., Murphy E., Loboda A., Kaufman D.R., Albright A., Cheng J.D., Kang S.P., Shankaran V., Piha-Paul S.A., Yearley J., Seiwert T.Y., Ribas A., McClanahan T.K. IFN-gamma-related mRNA profile predicts clinical response to PD-1 blockade. J Clin Invest. 2017;127:2930–2940. doi: 10.1172/JCI91190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Patel S.J., Sanjana N.E., Kishton R.J., Eidizadeh A., Vodnala S.K., Cam M., Gartner J.J., Jia L., Steinberg S.M., Yamamoto T.N., Merchant A.S., Mehta G.U., Chichura A., Shalem O., Tran E., Eil R., Sukumar M., Guijarro E.P., Day C.P., Robbins P., Feldman S., Merlino G., Zhang F., Restifo N.P. Identification of essential genes for cancer immunotherapy. Nature. 2017;548:537–542. doi: 10.1038/nature23477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Edmondson H.A., Steiner P.E. Primary carcinoma of the liver: a study of 100 cases among 48,900 necropsies. Cancer. 1954;7:462–503. doi: 10.1002/1097-0142(195405)7:3<462::aid-cncr2820070308>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 56.van der Maaten L. Accelerating t-SNE using tree-based algorithms. J Machine Learning Res. 2014;15:3221–3245. [Google Scholar]