Abstract
Androgen receptor (AR) antagonists, such as enzalutamide, have had a major impact on the treatment of metastatic castration-resistant prostate cancer (CRPC). However, even with the advent of AR antagonist therapies, patients continue to develop resistance, and new strategies to combat continued AR signalling are needed. Here, we develop AR degraders using PROteolysis TArgeting Chimeric (PROTAC) technology in order to determine whether depletion of AR protein can overcome mechanisms of resistance commonly associated with current AR-targeting therapies. ARD-61 is the most potent of the AR degraders and effectively induces on-target AR degradation with a mechanism consistent with the PROTAC design. Compared to clinically-approved AR antagonists, administration of ARD-61 in vitro and in vivo results in more potent anti-proliferative, pro-apoptotic effects and attenuation of downstream AR target gene expression in prostate cancer cells. Importantly, we demonstrate that ARD-61 functions in enzalutamide-resistant model systems, characterized by diverse proposed mechanisms of resistance that include AR amplification/overexpression, AR mutation, and expression of AR splice variants, such as AR-V7. While AR degraders are unable to bind and degrade AR-V7, they continue to inhibit tumor cell growth in models overexpressing AR-V7. To further explore this, we developed several isogenic prostate cell line models in which AR-V7 is highly expressed, which also failed to influence the cell inhibitory effects of AR degraders, suggesting that AR-V7 is not a functional resistance mechanism for AR antagonism. These data provide compelling evidence that full-length AR remains a prominent oncogenic driver of prostate cancers which have developed resistance to AR antagonists and highlight the clinical potential of AR degraders for treatment of CRPC.
Introduction
Androgen receptor (AR) signaling is critical for prostate development and homeostasis as well as the initiation and progression of prostate cancer, including in the castration- and enzalutamide-resistant states [1], [2]. Indeed, the clinical development of second-generation AR antagonists, including enzalutamide, has confirmed that AR remains a key oncogene in castration-resistant prostate cancer (CRPC) [3], [4]. Furthermore, response to enzalutamide is temporary and incremental, and prostate cancer cells that develop resistance to AR-targeted therapy usually maintain AR expression [5], [6]. This suggests that development of new therapies that can target the remaining AR activity may provide benefit to CRPC patients that have developed resistance to current therapies.
With the hypothesis that AR protein is still active even during castration- and enzalutamide-resistant states, we employed the PROteolysis TArgeting Chimera (PROTAC) strategy to build compounds that targeted AR through proteasomal degradation. In the PROTAC approach, a chimeric molecule is designed that contains a small-molecule ligand that binds to the target protein, a second small-molecule ligand that binds to an E3 ubiquitin ligase complex, and a chemically stable linker which tethers the two ligands together [7], [8]. We have previously used this method to create potent PROTAC degraders effective at inhibiting tumor growth through targeting other oncogenic molecules, such as BET [9], [10]. Recently, we described the initial synthesis strategy for AR PROTAC degraders [11] and perform further compound optimization in the current study. Using our lead compound (ARD-61), we determined the ability of AR degraders to inhibit tumor growth in numerous prostate cancer models, including those demonstrating characteristic mechanisms of enzalutamide-resistance, such as expression of AR splice variants (e.g., AR-V7) [5]. Importantly, we show that AR degraders are effective at inhibiting growth of enzalutamide-resistant prostate cancer cells as well as those expressing AR-V7, despite no loss of AR-V7 expression. To our knowledge, this is the first study to illustrate that full-length AR protein is often essential during resistance to AR antagonists and remains an attractive target despite its full antagonism, upregulation, or high splice variant expression.
Results
Generation of ARD-61, a potent PROTAC AR degrader
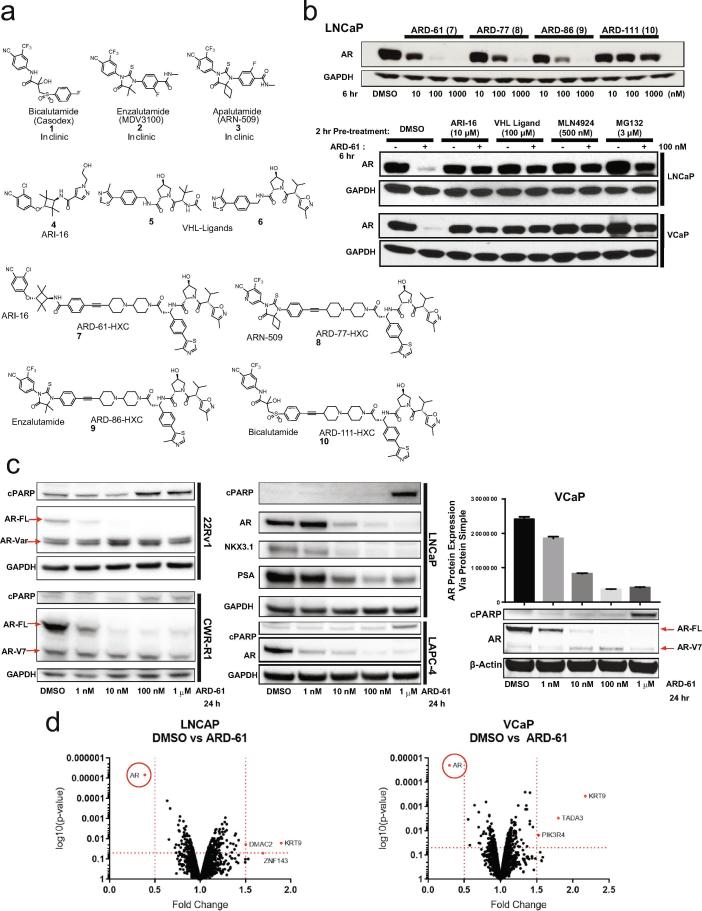
We developed four optimized functional (chimeric) molecules by synthesizing either ARI-16 (a previously described AR antagonist [11], [12]) or the FDA-approved AR antagonists bicalutamide, enzalutamide, or apalutamide to bind the target protein (AR), linked to a small-molecule ligand that would bind to an E3 ubiquitin ligase (VHL) complex and target AR to the proteasome (Fig. 1a). We assessed and compared the specificity and efficacy of our four chimeric molecules--ARD-61, ARD-77, ARD-86, and ARD-111—in targeting and degrading AR in different prostate cancer cell lines (Fig. 1b-c). ARD-61, containing ARI-16 as the AR antagonist, outperformed the other three chimeric compounds in terms of AR degradation potency (Fig. 1b) and exhibited dose- (DC50, 8.0 nM in LNCaP cells, Supplementary Fig. S1a-b) and time-dependent activity (Supplementary Fig. S1c-d, S2a); as expected, ARI-16 alone did not promote degradation (Supplementary Fig. S1c-d). Pre-treatment of cells with AR antagonist (ARI-16), VHL ligand, NEDD8 activating E1 enzyme inhibitor (MLN4924), or proteasome inhibitor (MG132) prevented AR degradation, confirming that the mechanism of degradation was consistent with the PROTAC design (Fig. 1b, Supplementary Fig. S2a).
Fig. 1.
ARD-61 is a potent PROTAC AR degrader that reduces full-length AR levels in diverse prostate cancer cell lines. a) Structures of AR antagonists 1–4, VHL ligands 5–6, and synthesized PROTAC AR degraders 7–10. b) Western blot analysis of AR degraders 7–10. Degradation of AR protein can be blocked by AR antagonist ARI-16 (4), VHL ligand (5), NEDD8 activating E1 enzyme inhibitor MLN4924, and proteasome inhibitor MG132 in LNCaP and VCaP cells with a 2 hour (hr) pretreatment of these drugs and 6 hr treatment of degrader. c) Representative western blots of protein collected from whole cell lysates from a variety of AR-positive prostate cancer cell lines, with different AR mutation statuses (CWR-R1, LNCaP) and AR variant expression (CWR-R1, 22Rv1), treated with increasing concentrations of the lead compound ARD-61 (7) for 24 hours. PARP cleavage (cPARP) was utilized as a readout of apoptosis. AR target genes PSA (KLK3) and NKX3.1 shown for LNCaP. AR-FL, AR full-length; AR-V7, AR variant 7. VCaP cells treated with increasing concentrations of ARD-61 for 24 hours had total AR levels quantified using the ProteinSimple® assay (n = 3). d) Quantitative proteomic analysis of LNCaP and VCaP cells after 6 hr treatment of 100 nM ARD-61 compared to control (DMSO). AR is highlighted as the most downregulated protein in both cell lines; red dashes indicate proteins that met significance (>0.5 change compared to control; >0.05 p-value, n = 3). Tandem mass spectroscopy was performed on tandem mass tagged whole cell lysates.
In addition to wild-type, full-length AR, prostate cancer cells have been shown to express AR mutants or splice variants, with both often being cited as resistance mechanisms arising from treatment with AR targeting therapies [5], [6], [13], [14], [15], [16], [17]; one of the most well-studied variants, AR-V7, lacks the ligand-binding domain (LBD). Accordingly, in line with the PROTAC design of our compound, ARD-61 degraded full-length AR without decreasing splice variant protein levels, including AR-V7, in several cell lines expressing these variants (e.g., 22Rv1, CWR-R1, VCaP; Fig. 1c). ARD-61 also degraded AR proteins containing point mutations in the LBD (e. g., CWR-R1 – H874Y, LNCaP – T877A; Fig. 1c). Proteomics analysis confirmed the specificity of ARD-61 for AR degradation (Fig. 1d). Thus, ARD-61 demonstrated the most promise among our four chimeric compounds developed from PROTAC technology and was the focus of the remaining analyses.
Degradation of AR protein decreases AR signaling and inhibits prostate tumor growth
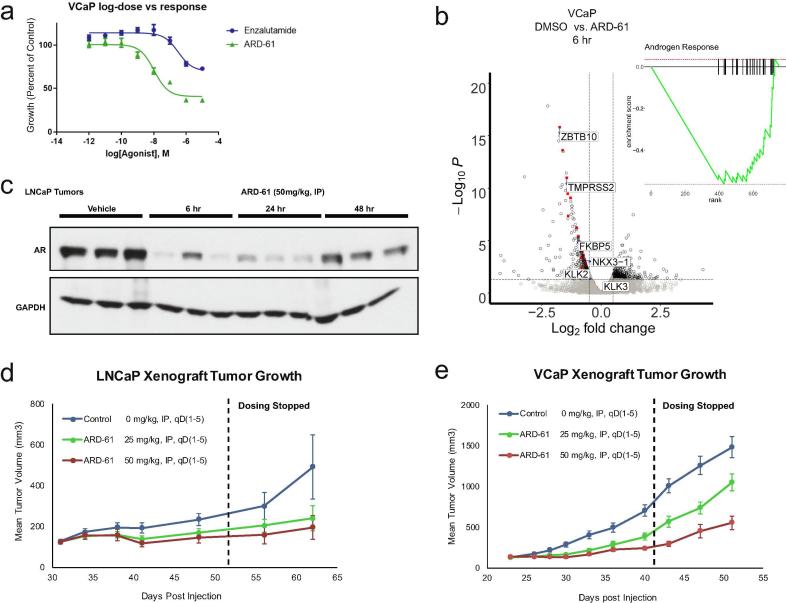
Using ARD-61, we determined whether degradation of AR protein could translate into functional effects and anti-cancer properties in vitro and in vivo. Notably, we observed that ARD-61 treatment led to PARP cleavage in all AR-dependent cell lines regardless of AR levels, AR splice variant expression, or AR mutation status (Fig. 1c); Annexin V staining confirmed induction of apoptosis by ARD-61 (Supplementary Fig. S3). Treatment of prostate cancer cells with ARD-61 resulted in dose-dependent growth inhibition to a greater degree than treatment with enzalutamide (Fig. 2a). RNA-sequencing revealed decreased AR target gene expression and signaling (Fig. 2b, Supplementary Fig. S2c-d, S9a), and common AR targets were also decreased at the protein level (Fig. 1c). These effects were mirrored in vivo using two different xenograft models, LNCaP and VCaP. Administration of ARD-61 decreased AR protein expression within 6 hours, which was sustained for up to 48 hours after a single dose without effects on AR mRNA (Fig. 2c, Supplementary Fig. S4). ARD-61 abated tumor growth in both LNCaP and VCaP models, and, notably, this inhibitory effect on tumor growth endured even after dosing stopped (Fig. 2d-e). Furthermore, ARD-61 treatment in vivo did not result in any toxicities (Supplementary Fig. S5–S7), suggesting that depletion of AR protein is a viable strategy to decrease prostate tumor growth.
Fig. 2.
ARD-61 decreases AR signaling and inhibits prostate cancer cell growth in vitro and in vivo. a) Growth curves of VCaP cells assayed by Cell Titer Glo ATP assay® treated with increasing concentrations of ARD-61 and enzalutamide for 5 days (n = 6). b) RNA-sequencing was performed on VCaP cells treated with 100 nM ARD-61 for 6 hr compared to control (DMSO, n = 3). AR target genes, in red with a few representative genes labeled, were significantly downregulated, as illustrated by gene set enrichment analysis (GSEA – inset). c) Western blot of whole tumor lysates from pharmacodynamics studies to confirm ARD-61 induces degradation of AR protein in LNCaP prostate xenograft tumor tissues in intact mice at 6, 24, and 48 hr. Efficacy analysis showed that ARD-61 at 50 and 25 mg/kg, IP, qD, 5 times a week can inhibit tumor growth (n = 10) in both LNCaP (d) and VCaP (e) models (mice intact), even after cessation of dosing (indicated by black dashed line on the graph).
ARD-61 remains effective in enzalutamide-resistant model systems
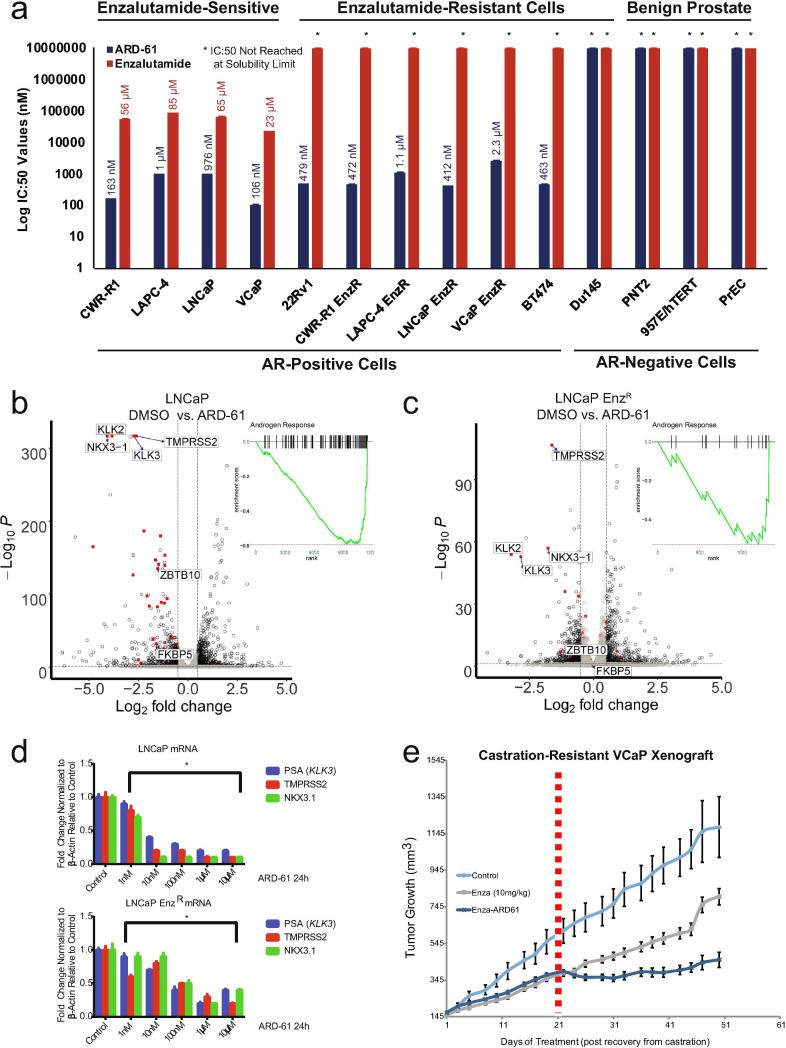
We compared the efficacy of ARD-61 across a panel of cell lines to enzalutamide, the most commonly used second-generation AR antagonist in treating advanced prostate cancer. Strikingly, ARD-61 had strong growth inhibitory effects on AR-positive prostate cancer cell lines already resistant to enzalutamide, including the AR-positive breast cancer cell line BT474, without affecting AR-negative cell lines (Fig. 3a). Specifically, enzalutamide-sensitive cell lines showed approximately 100-fold greater sensitivity to ARD-61 compared to enzalutamide, while enzalutamide-resistant cell lines showed similar sensitivity to ARD-61 (Fig. 3a). Further, ARD-61 slowed growth more efficiently and demonstrated greater PARP cleavage than AR knockdown in both enzalutamide-sensitive and -resistant cell lines (Supplementary Fig. S8). ARD-61 also decreased AR target genes similarly in both enzalutamide-sensitive (Fig. 3b,d, Supplementary Fig. S9b) and -resistant cell lines (Fig. 3c-d, Supplementary Fig. S9c), where AR signaling is decreased compared to parental [2]. Notably, in a VCaP castration-resistant xenograft model, ARD-61 treatment further improved growth inhibition in tumors previously treated with enzalutamide, and tumors treated with ARD-61 continued to grow slower than those treated with enzalutamide over time (Fig. 3e, Supplementary Fig. S2f). Together, these data suggest that AR degraders can target AR signaling that remains after direct antagonism of the receptor and demonstrate their potential in enzalutamide-resistant prostate cancer.
Fig. 3.
Enzalutamide-resistant prostate cancer models remain sensitive to ARD-61 treatment. a) Half-maximum inhibitory concentration (IC:50) values of ARD-61 and enzalutamide on the growth of AR-positive and -negative cancer cell lines, including both prostate cancer and one AR-positive breast cancer cell line, BT474 (n = 6). ARD-61 shows an effect in both enzalutamide-sensitive and -resistant cells that are AR-positive, while there is no effect in the AR-negative prostate cancer and normal cell lines. b) RNA-sequencing was performed on parental LNCaP cells treated with 100 nM ARD-61 for 24 hours compared to control. AR target genes (red), with a few labeled representative genes, are significantly downregulated, as illustrated by gene set enrichment analysis (GSEA) plot (n = 3). c) RNA-sequencing was performed on enzalutamide-resistant (EnzR) LNCaP cells with 100 nM ARD-61 for 24 hours compared to control. AR target genes (red) are downregulated from a low baseline (n = 3). GSEA of RNA-sequencing shows significant decreases of AR signaling targets normally upregulated by AR. d) Total RNA extracted from parental LNCaP and LNCaP EnzR cells treated with ARD-61 for 24 hours at concentrations ranging from 1 nM to 10 µM and analyzed by Q-RT-PCR for the mRNA transcripts of the AR target genes PSA (KLK3), TMPRSS2, and NKX3-1 (n = 3). e) Growth curves of tumor volume illustrating the effects of ARD-61 and enzalutamide on castration-resistant VCaP xenografts. Mice were injected with VCaP prostate cancer cells subcutaneously, and when the tumors reached 200 mm3 in size, the mice were castrated. Once the tumor grew back to the pre-castration size, the animals were treated with control (n = 10) or enzalutamide (10 mg/kg). After three weeks (21 days marked with dashed red line), half of the mice (n = 8) treated with enzalutamide were switched to ARD-61 (50 mg/kg).
Growth inhibition by ARD-61 involves degradation of the full-length receptor and occurs independent of AR-V7 expression
Most studies on the development of AR antagonist resistance have focused on activating mutations in the AR LBD and splice variants noted above, particularly the constitutively active AR variants like AR-V7 that lack the LBD [5], [6], [13], [14], [15], [16], [17]. While we have shown that ARD-61 will bind AR like a traditional antagonist and only degrades AR protein that contains the LBD, many cell lines with high AR splice variant expression (e.g., 22Rv1, CWR-R1, CWR-R1 EnzR, VCaP, VCaP EnzR) show sensitivity to ARD-61 (Fig. 1c, 3a, 4d) and even exhibit compensatory increases in variant expression (Fig. 1c, Supplementary Fig. S2b, S2e).
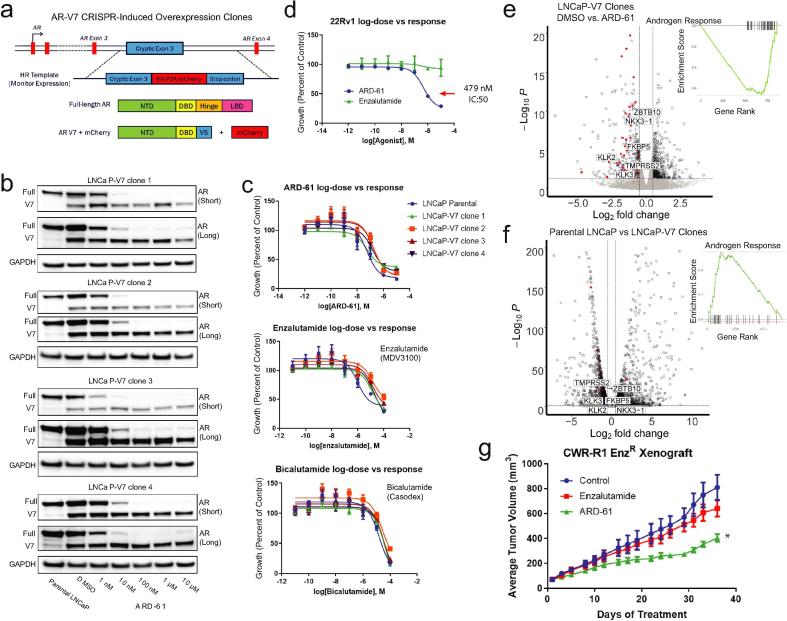
To explore this, we used CRISPR/Cas9 to engineer a variety of LNCaP-derived cell line clones with high AR-V7 expression (Fig. 4a) to test the hypothesis that high levels of AR-V7 may lead to resistance to ARD-61. We tested ARD-61 against four single cell-derived CRISPR-induced AR-V7 overexpressing clones and found that ARD-61 degraded full-length AR in all four clones but did not affect AR-V7 expression (Fig. 4b). Furthermore, we assayed the clones for their sensitivity to ARD-61 as well as to the AR antagonists enzalutamide and bicalutamide. Compared to the parental LNCaP cell line, we observed no significant difference in their sensitivities to any of the drugs, including ARD-61 (Fig. 4c), which is striking considering that 22Rv1 cells express high levels of truncated AR variants and are enzalutamide-resistant, but sensitive to ARD-61 (Fig. 4d). RNA-sequencing of the clones showed predicted decreases in canonical AR signaling when treated with ARD-61 (Fig. 4e, Supplementary Fig. S9d); however, when compared to parental LNCaP cells, AR-V7 overexpressed clones showed slight increases in AR signaling despite being similarly sensitive to ARD-61 (Fig. 4f, Supplementary Fig. S9e).
Fig. 4.
The growth inhibitory effects of ARD-61 occur through degradation of full-length AR and are independent of AR-V7 expression. a) Schematic of the creation of CRISPR-induced overexpression clones in LNCaP cells. b) Representative western blots of protein collected from whole cell lysates from four representative cell lines derived from individual single cell clones with CRISPR-induced AR-V7 expression treated with increasing concentrations of ARD-61 for 24 hours (short and long western blot exposures). c) Growth curves of LNCaP-V7 clones vs parental cells treated with ARD-61 (top panel), enzalutamide (middle panel), or bicalutamide (bottom panel) (n = 6). d) Growth curves of AR-V7 expressing 22Rv1 cells treated with ARD-61 compared to enzalutamide, assayed by Cell Titer Glo ATP® (n = 6). e) RNA-sequencing was performed on LNCaP AR-V7 overexpressing cells treated with 100 nM ARD-61 for 24 hours compared to control. AR target genes, in red with a few representative genes labeled, are significantly downregulated, and illustrated by gene set enrichment analysis (GSEA) plot (inset). f) With similar analysis to (e), RNA-sequencing of LNCaP parental cells compared to LNCaP AR-V7 overexpressing clones shows an increase in AR pathway gene expression (GSEA plot; inset). g) Growth curves of tumor volume illustrating the effects of ARD-61 and enzalutamide on enzalutamide-resistant CWR-R1 EnzRin vivo. Castrated mice were injected subcutaneously with CWR-R1 EnzR cells, and when the tumors reached 100 mm3 in size, the mice were treated with control (n = 10), enzalutamide (50 mg/kg, n = 10), or ARD-61 (50 mg/kg, n = 10) qd, 5 times a week throughout the duration of the experiment.
Moreover, we found that ARD-61 is highly effective in vivo by using the aggressive, metastatic, and castration- and enzalutamide-resistant CWR-R1 EnzR xenograft model that expresses high levels of AR-V7 [2] (Fig. 4g). Taken together, these data reveal that while AR-V7 may enhance AR signaling, full-length AR is required for the growth and survival of castration- and enzalutamide-resistant prostate cancer. Thus, full-length AR remains an attractive clinical target for AR degraders in treating advanced disease.
Discussion
Collectively, these data show the necessity for full-length AR in all stages of prostate cancer. Enzalutamide-resistant cell lines require full-length AR for sustained survival, despite continued AR antagonism, and cells with AR variant overexpression, shown previously to promote resistance to AR-targeted therapies [5], [6], [13], [14], [15], [16], [17], still require AR protein with an intact LBD to maintain growth. While AR splice variants have been shown to correlate with resistance to AR-targeted therapies (castration, AR antagonists, and androgen synthesis inhibitors [18]), their expression also highly correlates with full-length AR expression and amplification [19], which our data suggests is likely mediating the more aggressive phenotype seen during resistance.
Moreover, while most studies have shown that AR variants likely function to promote signaling and cellular growth through full-length AR [20], it has been argued by some groups that it may form functional homodimers with a unique cistrome [21], [22]. One such recent study by Cato et al. used an endogenous system and an AR-V7 specific antibody for ChIP-seq to suggest that AR-V7 functions as a transcriptional repressor for tumor suppressor genes [21]. Our data suggests that these genes are more likely not relevant in late-stage disease and resistance to AR-targeted therapies, given that in our endogenous CRISPR overexpression system, we found no difference in sensitivity to AR-targeted therapies and AR degradation (Fig. 4). Similarly, cell lines expressing endogenous AR variants are still sensitive to ARD-61.
Importantly, our data are consistent with the previously published work on resistance to AR-targeted therapies illustrating that AR is generally expressed and active after treatment [2], [23]. AR is often amplified and overexpressed in these contexts, which frequently correlates with AR variant expression that can be an excellent biomarker of aggressive disease, but AR variant expression likely plays a secondary role in mediating the phenotype that is mostly driven by full-length AR [24]. Our data illustrate the importance of continued targeting of AR in advanced prostate cancer and the potential clinical promise of the PROTAC class of drugs. In addition to ARD-61, other AR degraders have recently been under investigation, including the enzalutamide-based ARCC-4 PROTAC which showed enhanced activity compared to enzalutamide in in vitro studies [25], [26]. Ultimately, through their clinical translation, we anticipate the development of ARD-61 and other PROTAC AR degraders to be therapeutic advances for patients with metastatic CRPC via elimination of full-length AR protein.
Methods
Cell lines, culture, and viability assay
R1881 was purchased from Sigma-Aldrich (St. Louis, MO), and enzalutamide (MDV3100), MLN4924, and MG132 were purchased from Selleck Chemicals (Houston, TX), and stored at −20 °C in ethanol, and −80 °C in DMSO, respectively. CWR-22Rv1 (22Rv1), BT474, DU145, and PNT2 cell lines were purchased from American Type Culture Collection (Manassas, VA) and were validated and cultured as described [2], [27]. CWR-R1, VCaP, LAPC4, LNCaP, and enzalutamide-resistant counterparts, in addition to 957E/hTERT and PrEC cells, were generously provided by Dr. Donald J. Vander Griend at the University of Illinois at Chicago and have been previously characterized and cultured as described [2], [28], [29]. For viability assays, cells were seeded in 96-well plates at 2000–10,000 cells/well (optimum density for growth) in a total volume of 100 μl media containing 10% FBS. Serially diluted compounds in 100 μl media were added to the cells 12 hours later, with six biological replicates per condition. Following 5 days of incubation, cell viability was assessed by Cell-Titer GLO (Promega, Madison, WI). The values were normalized and IC:50 was calculated using GraphPad Prism 7 software. Apoptosis and cell cycle assays were performed following the manufacturer’s instructions (Thermo-Fisher Scientific, Waltham, MA). Non-targeting and AR siRNA SMARTpools were purchased from Dharmacon (Lafayette, CO, see Supplementary Methods for sequences), and transfected using Lipofectamine RNAiMAX reagent according to manufacturer’s protocol (Thermo-Fisher Scientific). CRISPR knock-in AR-V7 overexpressing lines were generated as previously described [30] using unique guide RNA and homologous recombination template sequences (see Supplementary Methods for sequences).
Antibodies, immunoblot, and proteomic analyses
Antibodies used in the immunoblotting (IB) assays are AR (Millipore, Billerica, MA, Cat. # 06-680), cPARP (Cell Signaling Technology, Danvers, MA Cat. # 9541), GAPDH (Cell Signaling, Cat. # 3683S), Histone H3 (Cell Signaling, Cat. # 2650), NKX3.1 (Cell Signaling, D2Y1A XP, Cat. # 83700), PSA (Dako Cat. #A0562), and β-actin (Sigma-Aldrich, Cat. #A5316). All antibodies were employed at dilutions suggested by the manufacturers. Whole-cell lysates collected from cells seeded at 1 × 106 cells per well of a 6 well plate (Becton, Dickinson and Company, Franklin Lakes, New Jersey) were lysed in RIPA-PIC buffer [150 mM sodium chloride, 1.0% Igepal CA-630 (Sigma-Aldrich), 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris, pH 8.0, 1 × protease inhibitor cocktail (Roche Molecular Biochemicals; Penzberg, Germany)], scraped, and sonicated (Fisher Scientific; Hampton, NH; model FB-120 Sonic Dismembrator). Protein was quantified by BCA assay (Thermo-Fisher Scientific); 30 μg of protein were loaded per lane, separated by SDS-PAGE and transferred onto nitrocellulose membrane (GE Healthcare, Chicago, IL). The membrane was incubated for 1 hour in blocking buffer [Tris-buffered saline, 0.1% Tween (TBS-T), 5% nonfat dry milk] followed by incubation overnight at 4 °C with the primary antibody. Following a wash with TBS-T, the blot was incubated with horseradish peroxidase-conjugated secondary antibody and signals were visualized by enhanced chemiluminescence system as per manufacturer's protocol (GE Healthcare). For ProteinSimple® quantification, whole cell lysates were separated by capillary electrophoresis on the ProteinSimple® WES platform using the 12–230 kDa size separation kit and following the manufacturer’s instructions (ProteinSimple®, San Jose, CA, USA). Proteomic analyses were all performed as previously described [9], [10].
RNA isolation, quantitative real-time PCR, and RNA-seq
Total RNA was isolated from either cells grown as previously described or whole homogenized tumor xenograft tissue using miRNAeasy kit, including the optional DNAse digestion (Qiagen, Valencia, CA), and cDNA was synthesized from 1000 ng total RNA using Maxima First Strand cDNA Synthesis III Kit for RT-qPCR (Thermo Fisher Scientific). Quantitative real-time PCR was performed in triplicate using standard SYBR green reagents and protocols on a StepOnePlus Real-Time PCR system (Applied Biosystems). The target mRNA expression was quantified using the ΔΔCt method and normalized to HMBS expression. All primers were designed using Primer 3 (http://frodo.wi.mit.edu/primer3/) and synthesized by Integrated DNA Technologies (Coralville, IA). See Supplementary Methods for primer sequences. RNA-seq was performed with triplicate biological replicates using the Illumina HiSeq 2000 in paired end mode, as previously described [31]. For each gene, a rank list was generated by ordering each gene in the differential expression analysis by the DESeq2 [32] log fold change value (log2foldchange). These rank lists were used in a weighted, pre-ranked GSEA [33] analysis against MSigDBv5 [34]. Significant associations were determined for any gene set having an FWER p-value below 0.01. Genomic analyses were all performed as previously described [9], [10].
Murine prostate tumor xenograft models
Four week-old male SCID CB17 mice were obtained from a breeding colony at University of Michigan maintained by our group. Mice were anesthetized using 2% Isoflurane (inhalation) and either 2 × 106 VCaP, 1x106 LNCaP, or CWR-R1 EnzR cells suspended in 100 μl of PBS with 50% Matrigel (BD Biosciences) were implanted subcutaneously into the dorsal flank on both sides of the mice. Once the tumors reached a palpable stage (100 mm3), the animals were randomized and treated with ARD-61 or vehicle control (10% PEG400: 3% Cremophor: 87% PBS) via intraperitoneal injection respectively five times a week, or enzalutamide or vehicle [1% Carboxymethylcellulose (Sigma Aldrich), 0.25% TWEEN-80 (Sigma Aldrich), and 98.75% PBS] by oral gavage. Growth in tumor volume was recorded using digital calipers and tumor volumes were estimated using the formula (π/6) (L × W2), where L = length of tumor and W = width. Loss of body weight during the course of the study was also monitored. At the end of the studies, mice were sacrificed and tumors were extracted for the downstream analyses. For the CRPC experiment, VCaP tumor bearing mice were castrated when the tumors were approximately 200 mm3 in size. Once the tumor grew back to the pre-castration size, the animals were treated with control or enzalutamide (10 mg/kg). After three weeks, half of the mice treated with enzalutamide (10 mg/kg) were switched to ARD-61 (50 mg/kg). For CWR-R1 EnzR, castrated mice were injected subcutaneously with cells, and when the tumors reached 100 mm3 in size, the mice were treated with control, enzalutamide (50 mg/kg), or ARD-61 (50 mg/kg) qd, 5 times a week throughout the duration of the experiment. All procedures involving mice were approved by the University Committee on Use and Care of Animals (UCUCA) at the University of Michigan and conform to all regulatory standards.
CRediT authorship contribution statement
Steven Kregel: Conceptualization, Data Curation, Formal analysis, Investigation, Methodology, Project administration, Validation, Visualization and Writing - original draft, Writing - review & editing. Chao Wang: Data Curation, Formal analysis, Investigation, Methodology. Xin Han: Data Curation, Investigation, Methodology. Lanbo Xiao: Data Curation, Investigation. Ester Fernandez-Salas: Data Curation, Investigation. Pushpinder Bawa: Data Curation, Formal analysis, Investigation. Brooke L. McCollum: Data Curation, Investigation. Kari Wilder-Romans: Data Curation, Investigation. Ingrid J. Apel: Data Curation, Investigation. Xuhong Cao: Data Curation, Investigation, Project administration. Corey Speers: Investigation, Project administration, Resources, Supervision. Shaomeng Wang: Conceptualization, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision. Arul M. Chinnaiyan: Conceptualization, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Writing - review & editing.
Competing interests
The University of Michigan has filed a number of patent applications on AR degraders, including the AR degrader used in this research, for which S.W., C.W., and X.H. are co-inventors. These patents have been licensed by Oncopia Therapeutics, LLC for clinical development. S.W. and A.M.C. are co-founders of Oncopia Therapeutics, LLC. S.W. serves on the board of directors and A.M.C. serves on the scientific advisory board of Oncopia. Both S.W. and A.M.C. are paid consultants of Oncopia. S.W. receives a research contract from Oncopia, which did not support this research. Oncopia was not involved in the design, funding, or approval of this study. E.F.-S. previously consulted for Oncopia but is now employed at Bristol-Myers Squibb.
Acknowledgments
We thank Stephanie Ellison for scientific editing and configuration of this study and Sisi Gao for critically reading the manuscript, scientific editing, and the submission of documents. Fengyun Su, Rui Wang, and Kristin Juckette provided technical assistance. S.K. is supported by a Cancer Biology Training Grant (T32-CA09676), the Department of Defense (DoD) Prostate Cancer Research Program (PCRP) Early Investigator Research Award (W81XWH-17-1-0155), and a Prostate Cancer Foundation Young Investigator Award. S.W. is supported by NCI R01 (R01CA215758). L.X. is supported by a DoD Postdoctoral Award (W81XWH-16-1-0195). A.M.C. is an NCI Outstanding Investigator, United States (R35CA231996), Howard Hughes Medical Institute Investigator, United States, A. Alfred Taubman Scholar, and American Cancer Society Professor, and is supported by NCI R01 (R01CA200660), United States.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.neo.2019.12.003.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Supplementary Figure S1. Degradation of AR protein in LNCaP and VCaP cells by ARD-61, with enzalutamide and the corresponding AR antagonist ARI-16 included as the controls. a) LNCaP cells were treated with different concentrations of ARD-61 or the base AR antagonist ARI-16 for 24 hr. b) Calculation of DC50 (concentration at which half of cellular AR protein level was degraded) in LNCaP cells. c) LNCaP or (d) VCAP cells were treated with ARD-61 at 100 nM for different times (2-24 hrs). Enzalutamide (Enza) and ARI-16 antagonists were included as controls and showed no degradation of AR. Supplementary Figure S2. AR degradation in both the nucleus and cytoplasm leads to downstream decreases in AR targets. a) Nuclear and cytoplasmic extraction assay results via western blot in LNCaP and VCaP cells. AR antagonist (ARI-16) or VHL ligand treatment can block the observed degradation in each cellular compartment. GAPDH serves as cytoplasmic loading control, and Histone H3 serves as a nuclear loading control. b) Time course of 200 nM of ARD-61 treatment in VCaP cells illustrating AR degradation and cPARP cleavage. Gene expression analysis of AR target genes FKBP5, PSA (KLK3), and TMPRSS2 in LNCaP cells in a dose- and time-dependent manner (c and d, respectively). CT, control; Enz, enzalutamide. e) mRNA extracted from VCaP cells treated with 200 nM over various time points. f) Human FKBP5 and TMPRSS2 mRNA expression from total RNA extracted from castration-resistant VCaP xenografts (model used in Fig. 3e) treated with ARD-61 alone, enzalutamide alone (ENZA), or enzalutamide then ARD-61 (ENZA-ARD-61). Supplementary Figure S3. ARD-61 induces apoptosis in prostate cancer cell lines. Comparison of apoptosis induction effects of AR degrader and antagonists in LNCaP (a) and VCaP (b) cell lines. Cells were treated for 6 days. Supplementary Figure S4. Gene expression analysis of LNCaP xenograft tumors. Q-RT-PCR analysis of LNCaP tumors dosed intraperitoneal (IP) with ARD-61 and ARI-16 for AR (a), TMPRSS2 (b), FKBP5 (c), and PSA (KLK3) (d) for 6, 24, and 48 hours. Supplementary Figure S5. ARD-61 administration does not affect body weights of mice. Body weights of mice in the LNCaP (a) and VCaP (c) xenograft studies. Percent weight change of mice in the LNCaP (b) and VCaP (d) xenograft studies. Supplementary Figure S6. Pharmacokinetics of ARD-61 in mouse plasma and LNCaP tumor, testis, and prostate tissues. ARD-61 was dosed intraperitoneally (IP) with a single dose at 50 mg/kg. Mouse plasma (a), xenograft tumors (b), testes (c), and prostates (d) were collected at the indicated time points for PK analysis (e). Supplementary Figure S7. ARD-61 and ARI-16 pharmacokinetics for VCaP models. Pharmacokinetics of ARD-61 and ARI-16 in mouse plasma (a) and VCaP tumors (b). ARD-61 was dosed intraperitoneally (IP) with a single dose at 50 mg/kg. Mouse plasma and xenograft tumors were collected at the indicated time points for PK analysis (c). Supplementary Figure S8. Growth inhibitory effects of AR knockdown and ARD-61 in LNCaP and LNCaP-EnzR cells. AR knockdown through transfection of siAR or non-targeting (NT) control in LNCaP and LNCaP-EnzR compared to 100 nM ARD-61. a) Western blot confirmed knockdown and degradation of AR as well as PARP cleavage. Growth curves over 7 days in (b) LNCaP and (c) LNCaP-EnzR compared to 20 µM enzalutamide, assayed by Cell Titer Glo ATP®. Supplementary Figure S9. Significantly prioritized Gene Set Enrichment (GSEA) pathways by RNA-sequencing. GSEA hallmark pathways prioritized by RNA-seq for (a) VCaP treated with 100 nM ARD-61 compared to control for 6 hours, (b) LNCaP treated with 100 nM ARD-61 compared to control for 24 hours, (c) LNCaP-EnzR treated with 100 nM ARD-61 compared to control for 24 hours, (d) LNCaP AR-V7 overexpressing cells derived from CRISPR clones treated with 100 nM ARD-61 compared to control DMSO for 24 hours, (e) control LNCaP parental cells compared to LNCaP AR-V7 overexpressing cells derived from CRISPR clones, and (f) control LNCaP parental cells compared to LNCaP AR-V7 overexpressing cells derived from CRISPR clones, both treated with 100 nM ARD-61 for 24 hours.
References
- 1.Lonergan P.E., Tindall D.J. Androgen receptor signaling in prostate cancer development and progression. J Carcinog. 2011;10:20. doi: 10.4103/1477-3163.83937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kregel S., Chen J.L., Tom W., Krishnan V., Kach J., Brechka H. Acquired resistance to the second-generation androgen receptor antagonist enzalutamide in castration-resistant prostate cancer. Oncotarget. 2016;7:26259–26274. doi: 10.18632/oncotarget.8456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beer T.M., Armstrong A.J., Rathkopf D.E., Loriot Y., Sternberg C.N., Higano C.S. Enzalutamide in metastatic prostate cancer before chemotherapy. N Engl J Med. 2014;371:424–433. doi: 10.1056/NEJMoa1405095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scher H.I., Fizazi K., Saad F., Taplin M.E., Sternberg C.N., Miller K. Increased survival with enzalutamide in prostate cancer after chemotherapy. N Engl J Med. 2012;367:1187–1197. doi: 10.1056/NEJMoa1207506. [DOI] [PubMed] [Google Scholar]
- 5.Antonarakis E.S., Lu C., Wang H., Luber B., Nakazawa M., Roeser J.C. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N Engl J Med. 2014;371:1028–1038. doi: 10.1056/NEJMoa1315815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Joseph J.D., Lu N., Qian J., Sensintaffar J., Shao G., Brigham D. A clinically relevant androgen receptor mutation confers resistance to second-generation antiandrogens enzalutamide and ARN-509. Cancer Discov. 2013;3:1020–1029. doi: 10.1158/2159-8290.CD-13-0226. [DOI] [PubMed] [Google Scholar]
- 7.Ottis P., Crews C.M. Proteolysis-targeting chimeras: induced protein degradation as a therapeutic strategy. ACS Chem Biol. 2017;12:892–898. doi: 10.1021/acschembio.6b01068. [DOI] [PubMed] [Google Scholar]
- 8.Toure M., Crews C.M. Small-molecule PROTACS: new approaches to protein degradation. Angew Chem Int Ed Engl. 2016;55:1966–1973. doi: 10.1002/anie.201507978. [DOI] [PubMed] [Google Scholar]
- 9.Bai L., Zhou B., Yang C.-Y., Ji J., McEachern D., Przybranowski S. Targeted degradation of BET proteins in triple-negative breast cancer. Cancer Res. 2017;77:2476–2487. doi: 10.1158/0008-5472.CAN-16-2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kregel S., Malik R., Asangani I.A., Wilder-Romans K., Rajendiran T., Xiao L. Functional and mechanistic interrogation of BET bromodomain degraders for the treatment of metastatic castration-resistant prostate cancer. Clin Cancer Res. 2019;25:4038–4048. doi: 10.1158/1078-0432.CCR-18-3776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Han X., Wang C., Qin C., Xiang W., Fernandez-Salas E., Yang C.Y. Discovery of ARD-69 as a highly potent proteolysis targeting chimera (PROTAC) degrader of androgen receptor (AR) for the treatment of prostate cancer. J Med Chem. 2019;62:941–964. doi: 10.1021/acs.jmedchem.8b01631. [DOI] [PubMed] [Google Scholar]
- 12.Guo C., Linton A., Kephart S., Ornelas M., Pairish M., Gonzalez J. Discovery of aryloxy tetramethylcyclobutanes as novel androgen receptor antagonists. J Med Chem. 2011;54:7693–7704. doi: 10.1021/jm201059s. [DOI] [PubMed] [Google Scholar]
- 13.Efstathiou E., Titus M., Wen S., Hoang A., Karlou M., Ashe R. Molecular characterization of enzalutamide-treated bone metastatic castration-resistant prostate cancer. Eur Urol. 2014;67:53–60. doi: 10.1016/j.eururo.2014.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Korpal M., Korn J.M., Gao X., Rakiec D.P., Ruddy D.A., Doshi S. An F876L mutation in androgen receptor confers genetic and phenotypic resistance to MDV3100 (enzalutamide) Cancer Discov. 2013;3:1030–1043. doi: 10.1158/2159-8290.CD-13-0142. [DOI] [PubMed] [Google Scholar]
- 15.Li Y., Chan S.C., Brand L.J., Hwang T.H., Silverstein K.A., Dehm S.M. Androgen receptor splice variants mediate enzalutamide resistance in castration-resistant prostate cancer cell lines. Cancer Res. 2013;73:483–489. doi: 10.1158/0008-5472.CAN-12-3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu L.L., Xie N., Sun S., Plymate S., Mostaghel E., Dong X. Mechanisms of the androgen receptor splicing in prostate cancer cells. Oncogene. 2014;33:3140–3150. doi: 10.1038/onc.2013.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sobel R.E., Sadar M.D. Cell lines used in prostate cancer research: a compendium of old and new lines–part 1. J Urol. 2005;173:342–359. doi: 10.1097/01.ju.0000141580.30910.57. [DOI] [PubMed] [Google Scholar]
- 18.Scher H.I., Lu D., Schreiber N.A., Louw J., Graf R.P., Vargas H.A. Association of AR-V7 on circulating tumor cells as a treatment-specific biomarker with outcomes and survival in castration-resistant prostate cancer. JAMA Oncol. 2016;2:1441–1449. doi: 10.1001/jamaoncol.2016.1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu Z., Chen S., Sowalsky A.G., Voznesensky O.S., Mostaghel E.A., Nelson P.S. Rapid induction of androgen receptor splice variants by androgen deprivation in prostate cancer. Clin Cancer Res. 2014;20:1590–1600. doi: 10.1158/1078-0432.CCR-13-1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Watson P.A., Chen Y.F., Balbas M.D., Wongvipat J., Socci N.D., Viale A. Constitutively active androgen receptor splice variants expressed in castration-resistant prostate cancer require full-length androgen receptor. Proc Natl Acad Sci USA. 2010;107:16759–16765. doi: 10.1073/pnas.1012443107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cato L., de Tribolet-Hardy J., Lee I., Rottenberg J.T., Coleman I., Melchers D. ARv7 represses tumor-suppressor genes in castration-resistant prostate cancer. Cancer Cell. 2019;35:401–413. doi: 10.1016/j.ccell.2019.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu J., Lonergan P.E., Nacusi L.P., Wang L., Schmidt L.J., Sun Z. The cistrome and gene signature of androgen receptor splice variants in castration resistant prostate cancer cells. J Urol. 2015;193:690–698. doi: 10.1016/j.juro.2014.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robinson D., Van Allen E.M., Wu Y.M., Schultz N., Lonigro R.J., Mosquera J.M. Integrative clinical genomics of advanced prostate cancer. Cell. 2015;161:1215–1228. doi: 10.1016/j.cell.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ho Y., Dehm S.M. Androgen receptor rearrangement and splicing variants in resistance to endocrine therapies in prostate cancer. Endocrinology. 2017;158:1533–1542. doi: 10.1210/en.2017-00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gustafson J.L., Neklesa T.K., Cox C.S., Roth A.G., Buckley D.L., Tae H.S. Small-molecule-mediated degradation of the androgen receptor through hydrophobic tagging. Angew Chem Int Ed Engl. 2015;54:9659–9662. doi: 10.1002/anie.201503720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Salami J., Alabi S., Willard R.R., Vitale N.J., Wang J., Dong H. Androgen receptor degradation by the proteolysis-targeting chimera ARCC-4 outperforms enzalutamide in cellular models of prostate cancer drug resistance. Commun Biol. 2018;1:100. doi: 10.1038/s42003-018-0105-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Steinkamp M.P., O'Mahony O.A., Brogley M., Rehman H., Lapensee E.W., Dhanasekaran S. Treatment-dependent androgen receptor mutations in prostate cancer exploit multiple mechanisms to evade therapy. Cancer Res. 2009;69:4434–4442. doi: 10.1158/0008-5472.CAN-08-3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Litvinov I.V., Vander Griend D.J., Xu Y., Antony L., Dalrymple S.L., Isaacs J.T. Low-calcium serum-free defined medium selects for growth of normal prostatic epithelial stem cells. Cancer Res. 2006;66:8598–8607. doi: 10.1158/0008-5472.CAN-06-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kregel S., Kiriluk K.J., Rosen A.M., Cai Y., Reyes E.E., Otto K.B. Sox2 is an androgen receptor-repressed gene that promotes castration-resistant prostate cancer. PLoS ONE. 2013;8 doi: 10.1371/journal.pone.0053701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cieslik M., Chugh R., Wu Y.M., Wu M., Brennan C., Lonigro R. The use of exome capture RNA-seq for highly degraded RNA with application to clinical cancer sequencing. Genome Res. 2015;25:1372–1381. doi: 10.1101/gr.189621.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anders S., Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liberzon A., Birger C., Thorvaldsdottir H., Ghandi M., Mesirov J.P., Tamayo P. The molecular signatures database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1. Degradation of AR protein in LNCaP and VCaP cells by ARD-61, with enzalutamide and the corresponding AR antagonist ARI-16 included as the controls. a) LNCaP cells were treated with different concentrations of ARD-61 or the base AR antagonist ARI-16 for 24 hr. b) Calculation of DC50 (concentration at which half of cellular AR protein level was degraded) in LNCaP cells. c) LNCaP or (d) VCAP cells were treated with ARD-61 at 100 nM for different times (2-24 hrs). Enzalutamide (Enza) and ARI-16 antagonists were included as controls and showed no degradation of AR. Supplementary Figure S2. AR degradation in both the nucleus and cytoplasm leads to downstream decreases in AR targets. a) Nuclear and cytoplasmic extraction assay results via western blot in LNCaP and VCaP cells. AR antagonist (ARI-16) or VHL ligand treatment can block the observed degradation in each cellular compartment. GAPDH serves as cytoplasmic loading control, and Histone H3 serves as a nuclear loading control. b) Time course of 200 nM of ARD-61 treatment in VCaP cells illustrating AR degradation and cPARP cleavage. Gene expression analysis of AR target genes FKBP5, PSA (KLK3), and TMPRSS2 in LNCaP cells in a dose- and time-dependent manner (c and d, respectively). CT, control; Enz, enzalutamide. e) mRNA extracted from VCaP cells treated with 200 nM over various time points. f) Human FKBP5 and TMPRSS2 mRNA expression from total RNA extracted from castration-resistant VCaP xenografts (model used in Fig. 3e) treated with ARD-61 alone, enzalutamide alone (ENZA), or enzalutamide then ARD-61 (ENZA-ARD-61). Supplementary Figure S3. ARD-61 induces apoptosis in prostate cancer cell lines. Comparison of apoptosis induction effects of AR degrader and antagonists in LNCaP (a) and VCaP (b) cell lines. Cells were treated for 6 days. Supplementary Figure S4. Gene expression analysis of LNCaP xenograft tumors. Q-RT-PCR analysis of LNCaP tumors dosed intraperitoneal (IP) with ARD-61 and ARI-16 for AR (a), TMPRSS2 (b), FKBP5 (c), and PSA (KLK3) (d) for 6, 24, and 48 hours. Supplementary Figure S5. ARD-61 administration does not affect body weights of mice. Body weights of mice in the LNCaP (a) and VCaP (c) xenograft studies. Percent weight change of mice in the LNCaP (b) and VCaP (d) xenograft studies. Supplementary Figure S6. Pharmacokinetics of ARD-61 in mouse plasma and LNCaP tumor, testis, and prostate tissues. ARD-61 was dosed intraperitoneally (IP) with a single dose at 50 mg/kg. Mouse plasma (a), xenograft tumors (b), testes (c), and prostates (d) were collected at the indicated time points for PK analysis (e). Supplementary Figure S7. ARD-61 and ARI-16 pharmacokinetics for VCaP models. Pharmacokinetics of ARD-61 and ARI-16 in mouse plasma (a) and VCaP tumors (b). ARD-61 was dosed intraperitoneally (IP) with a single dose at 50 mg/kg. Mouse plasma and xenograft tumors were collected at the indicated time points for PK analysis (c). Supplementary Figure S8. Growth inhibitory effects of AR knockdown and ARD-61 in LNCaP and LNCaP-EnzR cells. AR knockdown through transfection of siAR or non-targeting (NT) control in LNCaP and LNCaP-EnzR compared to 100 nM ARD-61. a) Western blot confirmed knockdown and degradation of AR as well as PARP cleavage. Growth curves over 7 days in (b) LNCaP and (c) LNCaP-EnzR compared to 20 µM enzalutamide, assayed by Cell Titer Glo ATP®. Supplementary Figure S9. Significantly prioritized Gene Set Enrichment (GSEA) pathways by RNA-sequencing. GSEA hallmark pathways prioritized by RNA-seq for (a) VCaP treated with 100 nM ARD-61 compared to control for 6 hours, (b) LNCaP treated with 100 nM ARD-61 compared to control for 24 hours, (c) LNCaP-EnzR treated with 100 nM ARD-61 compared to control for 24 hours, (d) LNCaP AR-V7 overexpressing cells derived from CRISPR clones treated with 100 nM ARD-61 compared to control DMSO for 24 hours, (e) control LNCaP parental cells compared to LNCaP AR-V7 overexpressing cells derived from CRISPR clones, and (f) control LNCaP parental cells compared to LNCaP AR-V7 overexpressing cells derived from CRISPR clones, both treated with 100 nM ARD-61 for 24 hours.