Abstract
Background & Aims
Consumption of a low-fiber, high-fat, Western-style diet (WSD) induces adiposity and adipose inflammation characterized by increases in the M1:M2 macrophage ratio and proinflammatory cytokine expression, both of which contribute to WSD-induced metabolic syndrome. WSD-induced adipose inflammation might result from endoplasmic reticulum stress in lipid-overloaded adipocytes and/or dissemination of gut bacterial products, resulting in activation of innate immune signaling. Hence, we aimed to investigate the role of the gut microbiota, and its detection by innate immune signaling pathways, in WSD-induced adipose inflammation.
Methods
Mice were fed grain-based chow or a WSD for 8 weeks, assessed metabolically, and intestinal and adipose tissue were analyzed by flow cytometry and quantitative reverse transcription polymerase chain reaction. Microbiota was ablated via antibiotics and use of gnotobiotic mice that completely lacked microbiota (germ-free mice) or had a low-complexity microbiota (altered Schaedler flora). Innate immune signaling was ablated by genetic deletion of Toll-like receptor signaling adaptor myeloid differentiation primary response 88.
Results
Ablation of microbiota via antibiotic, germ-free, or altered Schaedler flora approaches did not significantly impact WSD-induced adiposity, yet dramatically reduced WSD-induced adipose inflammation as assessed by macrophage populations and cytokine expression. Microbiota ablation also prevented colonic neutrophil and CD103- dendritic cell infiltration. Such reduced indices of inflammation correlated with protection against WSD-induced dysglycemia, hypercholesterolemia, and liver dysfunction. Genetic deletion of myeloid differentiation primary response 88 also prevented WSD-induced adipose inflammation.
Conclusions
These results indicate that adipose inflammation, and some aspects of metabolic syndrome, are not purely a consequence of diet-induced adiposity per se but, rather, may require disturbance of intestine-microbiota interactions and subsequent activation of innate immunity.
Keywords: Metabolic Syndrome, Microbiota, Germ-Free, Antibiotics, Altered Schaedler Flora, High-Fat Diet, MyD88
Abbreviations used in this paper: ALT, alanine aminotransferase; ASF, altered Schaedler flora; AST, aspartate aminotransferase; CXCL1, chemokine (C-X-C motif) ligand 1; DC, dendritic cells; FBS, fetal bovine serum; GF, germ-free; HBSS, Hank’s balanced salt solution; IL, interleukin; KO, knockout; LPS, lipopolysaccharide; MyD88, myeloid differentiation primary response 88; PP, Peyer’s patches; SVC, stromal vascular cells; TLR, Toll-like receptor; TNF-α, tumor necrosis factor α; WSD, Western-style diet
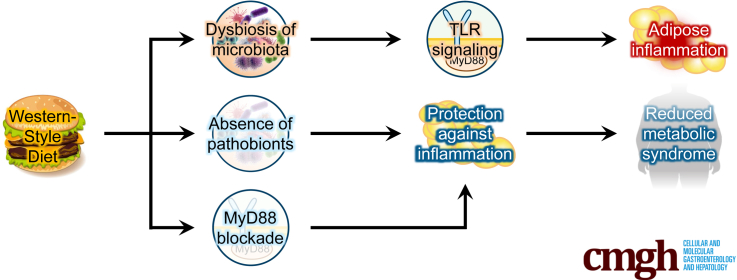
Graphical abstract
Summary.
Lack of pathobiont bacteria, achieved via use of gnotobiotic mice or antibiotics, protected against diet-induced adipose inflammation despite showing similar increases in adiposity. Despite not protecting against diet-induced adiposity, protection against adipose inflammation resulted in protection against metabolic syndrome.
Metabolic syndrome is the collective term for the interrelated metabolic abnormalities associated with obesity. Features of metabolic syndrome, which include insulin resistance, dysglycemia, hypercholesterolemia, hypertension, and hepatic steatosis, promote a variety of dangerous and costly chronic diseases that are among humanity’s most pressing public health problems. Although numerous genetic factors influence susceptibility to the development of metabolic syndrome, that the increased incidence of this disorder has occurred amidst stark societal changes in food production and dietary habits has led to the general presumption that diet is a major determinant of metabolic syndrome. In accordance, prolonged feeding of mice with a diet designed to mimic a Western-style diet (WSD), recapitulates many features of metabolic syndrome. Although some studies refer to WSD as a high-fat diet, which it is, several studies have shown that WSD differs from grain-based rodent chow in numerous ways that can impact microbiota and metabolism,1, 2, 3 including a WSD’s lack of fiber.4,5 Hence, we use the term WSD to highlight that this diet promotes metabolic syndrome for various reasons.
Metabolic syndrome increasingly is appreciated to be associated with, and driven by, chronic low-grade inflammation, especially in visceral adipose tissue. In particular, metabolic syndrome, in human beings and mice fed a WSD, is associated with a change in macrophage phenotypes in visceral adipose tissue, characterized by an increase in classic proinflammatory M1 macrophages and a decrease in anti-inflammatory M2 macrophages.6, 7, 8, 9, 10 Such changes correlate with, and drive increased adipose expression of, proinflammatory cytokines and decreased expression of anti-inflammatory cytokines. Such adipose inflammation has been proposed to result from accumulated lipids within adipocytes causing endoplasmic reticulum stress that activates intrinsic signaling pathways that result in activation of nuclear factor κ-light-chain-enhancer of activated B cells–mediated proinflammatory gene expression.11 In addition, it alternatively has been proposed that gut microbiota components might play a central role in adipose inflammation. Cani et al12 found that a WSD increased dissemination of gut bacterial–derived lipopolysaccharide (LPS) and that direct low-dose systemic administration of LPS led to increases in proinflammatory cytokines in adipose tissue gene expression and insulin resistance. This suggests that LPS, and perhaps other gut bacterial–derived activators of innate immune signaling, might be key drivers of the adipose inflammation and insulin resistance that results from a WSD.12 The general notion that gut microbiota plays a key role in driving a WSD-induced metabolic syndrome also is supported by reports that germ-free (GF) mice are protected against various features of metabolic syndrome, including adiposity and dysglycemia,13,14 although the extent of such microbiota dependence varies markedly among the several published studies in this area.1 Moreover, such studies did not examine adipose inflammation but rather attributed the metabolic impacts of microbiota ablation to inflammation-independent mechanisms, such as calorie extraction. Furthermore, a general caveat of studies relying on GF mice is that these mice have a variety of immunologic abnormalities, which might influence responses to inflammatory challenges in general.15 Hence, the central goal of this study was to examine how ablation of gut microbiota influenced WSD-induced adipose inflammation, especially in terms of the macrophage polarization that is thought to play a critical role in driving and maintaining the inflammatory state. We used 3 approaches to ablate the microbiota: antibiotics, GF mice, and gnotobiotic mice with a minimal microbiota, namely the 8-species consortium known as the altered Schaedler flora (ASF), which is sufficient to confer fairly normal gut physiology.16 In addition, we examined how such microbiota ablation impacted changes in immune cell populations in the colon, where the majority of microbiota is localized. We observed that WSD induced microbiota-dependent changes in immune cell populations in the colon and adipose tissue that were associated with adipose proinflammatory gene expression and some features of metabolic syndrome, albeit not adiposity per se.
Results
Characterization of Changes in Tissue Leukocytes in Response to Diet-Induced Obesity
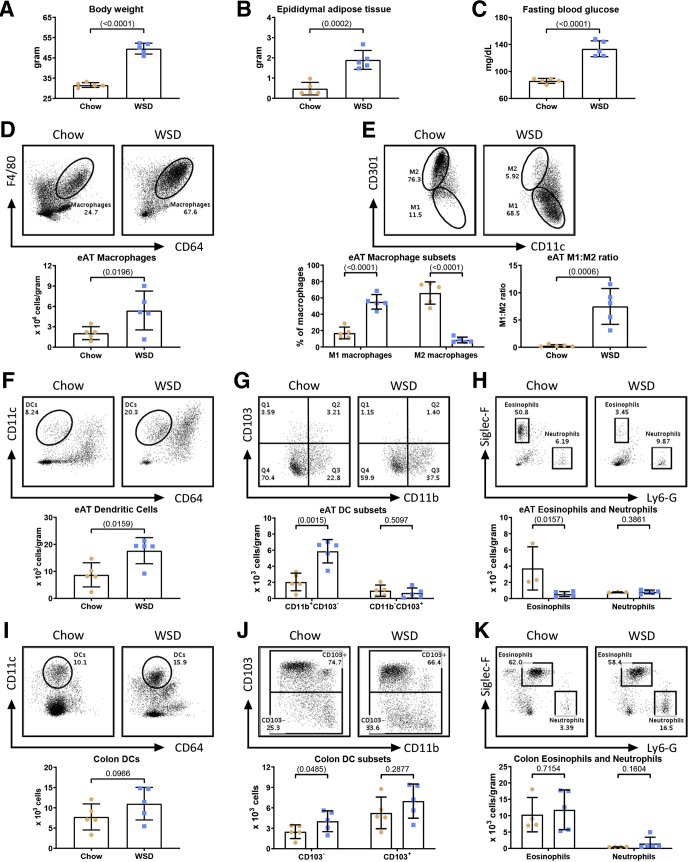
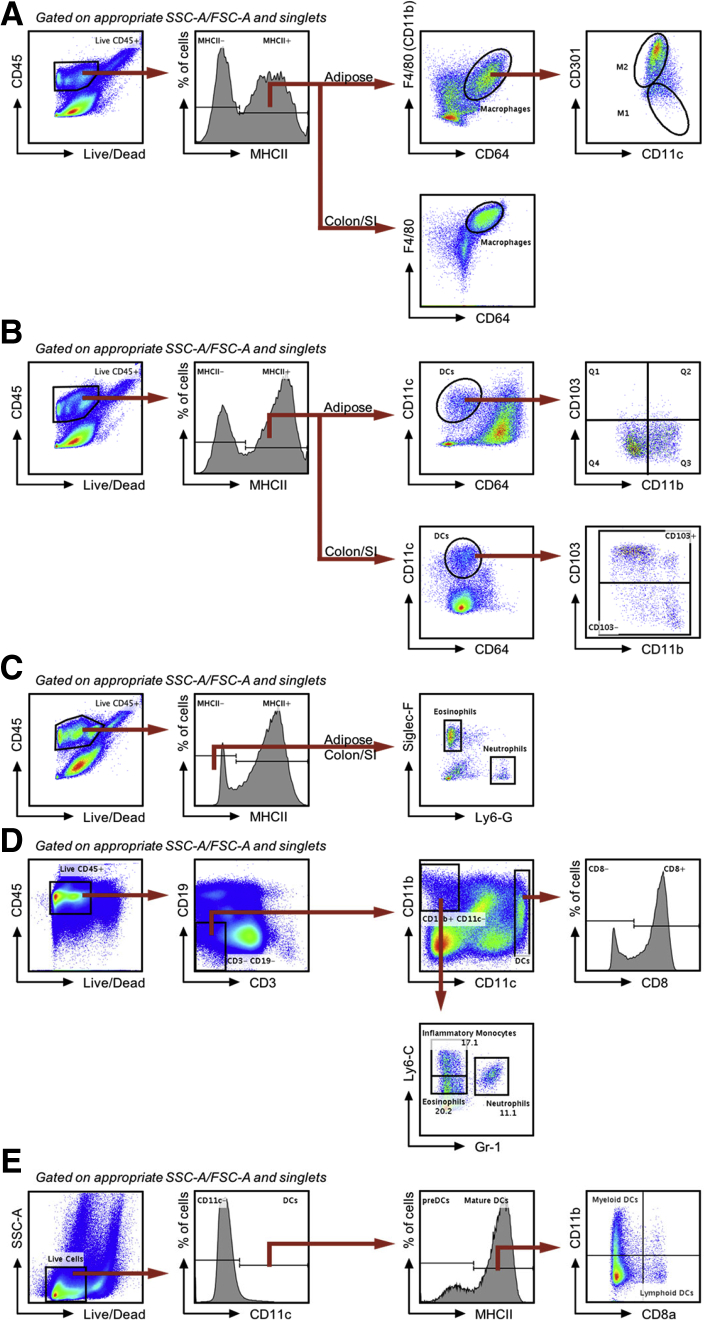
The central goal of this study was to test the hypothesis that intestinal microbiota plays a central role in driving high-fat WSD-induced alterations in populations of leukocytes, especially those in visceral adipose tissue that mediate chronic inflammation and promote metabolic syndrome. Hence, we first sought to broadly define changes in leukocyte populations induced by an obesogenic diet. C57BL/6J mice (6-week-old males) were maintained on standard grain-based rodent chow or administered WSD for 8 weeks, at which time they were euthanized and tissues were harvested for analysis. Our focus on end point assays was designed to enable the subsequent gnotobiotic approaches described later. In accordance with previous studies, administration of a WSD resulted in a marked increase in body weight (Figure 1A), adiposity (Figure 1B), and fasting blood glucose levels (Figure 1C). Such indices of WSD-induced metabolic syndrome are associated with infiltration of, and relative population changes in, adipose tissue macrophages. Hence, we first analyzed epididymal white adipose tissue by flow cytometry using the gating procedure outlined in Figure 2A. Data were collected until 100,000 events were reached in the live/cluster of differentiation 45 (CD45+) gate. For data analysis, cells were gated for appropriate side scatter area (SSC-A)/forward scatter area (FSC-A) and singlets. Live CD45+ major histocompatibility complex class II (MHCII)+ cells were identified as macrophages if they were CD11b+ (or F4/80+) and CD64+. From these macrophages, M1 macrophages were identified as CD301-, and M2 macrophages were identified as CD301+ (Figure 2A). In accordance with previous studies, a WSD led to an increase in the number of macrophages per gram of adipose tissue (Figure 1D), as well as an increase in the proportion of proinflammatory M1 macrophages, and a decrease in anti-inflammatory M2 macrophages, thus resulting in a 25-fold increase in the M1:M2 ratio in response to WSD (Figure 1E). These changes were not observed after 4 weeks of WSD feeding and thus we restricted our studies to the 8-week time point, at which we also observed a moderate increase in the overall number of adipose dendritic cells (DCs) per gram of fat (Figure 1F), driven by an increase in the CD11b+CD103- subset of DCs during obesogenic conditions, although CD11b-CD103+ DCs were not appreciably impacted (Figure 1G; gating procedure is outlined in Figure 2B). Live CD45+MHCII- cells of the adipose tissue were identified as eosinophils if they were Sialic acid-binding immunoglobulin-type lectins F (Siglec-F+) and lymphocyte antigen 6 complex, locus G (Ly6-G-); concurrently, neutrophils were identified as Siglec-F-Ly6-G+ (Figure 2C). In accordance with previous studies, we also observed a decrease in the adipose eosinophils in response to a WSD (Figure 1H).17, 18, 19 Finally, in contrast to some studies, we did not observe an increase in adipose neutrophils in response to WSD feeding (Figure 1H).20, 21, 22
Figure 1.
Characterization of changes in tissue leukocytes in response to diet-induced obesity. Four-week-old male C57BL/6J mice were purchased from The Jackson Laboratory and housed for 2 weeks to favor microbiota stabilization. Subsequently, half of the mice were switched to a Western-style, high-fat diet (60% kcal from fat) or continued on a standard grain-based chow as a control. After 8 weeks on a WSD, mice were killed and biometric measurements are shown for (A) final body weight and (B) epididymal adipose weight. (C) In addition, a 15-hour fasting blood glucose level was measured before euthanasia. Epididymal adipose tissue was analyzed by flow cytometry to quantify the following: (D) macrophages, (E) M1:M2 macrophage ratio, (F) DCs, (G) CD11b+CD103- and CD11b-CD103+ DC subsets, and (H) eosinophils and neutrophils. Colon tissue was used for flow cytometry to quantify as cells per organ: (I) DCs, (J) CD103+/- dendritic subsets, and (K) eosinophils and neutrophils. Data are the means ± SD (N=5). Statistical significance was determined using the t test. P < .05, brackets indicate significance. eAT, epididymal adipose tissue.
Figure 2.
Gating scheme for myeloid cells in adipose, small intestine, colon tissue, spleen, and PPs. Data were collected until 100,000 events were reached in the Live/CD45+ gate. All cells were gated for appropriate side scatter area/forward scatter area and singlets. (A) Gating scheme of macrophages from epididymal adipose and intestinal tissue. Live CD45+MHCII+ cells were identified as macrophages if they were F4/80+ (or CD11b+) and CD64+. Of epididymal adipose tissue macrophages, M1 macrophages were identified as CD301-, M2 macrophages were identified as CD301+. (B) Gating scheme of DCs from epididymal adipose and intestinal tissue. Live CD45+MHCII+ cells were identified as DCs if they were CD11c+ but CD64-. DC subsets were identified as either CD11b+CD103- (Q3) or CD11b-CD103+ (Q1) in the adipose or CD103+/- in the small intestine and colon. (C) Gating scheme of eosinophils and neutrophils from epididymal adipose and intestinal tissue. Live CD45+MHCII- cells were identified as eosinophils if they were Siglec-F+Ly6-G- or neutrophils if they were Siglec-F-Ly6-G+, these populations also both are CD11b+. (D) Gating scheme of myeloid cells from splenic tissue. Live CD45+MHCII+ cells were gated to exclude B cells (CD19+) and T cells (CD3+). Of this CD45+MHCII+CD19-CD3- population, CD11chigh cells were classified as DCs, which then were classified as either CD8+/-, while CD11b+CD11c- cells were categorized as either eosinophils (Ly6-CloGr-1-), neutrophils (Gr-1+), or inflammatory monocytes (Ly6-ChighGr-1-). (E) Gating scheme of DCs from PPs. Dendritic cells were identified as live CD11c+ and subdivided as either preDCs or mature DCs based on MHCII expression. Mature MHCII+ DCs were categorized as either myeloid DCs (CD8a-CD11b+) or lymphoid DCs (CD8a+CD11b-).
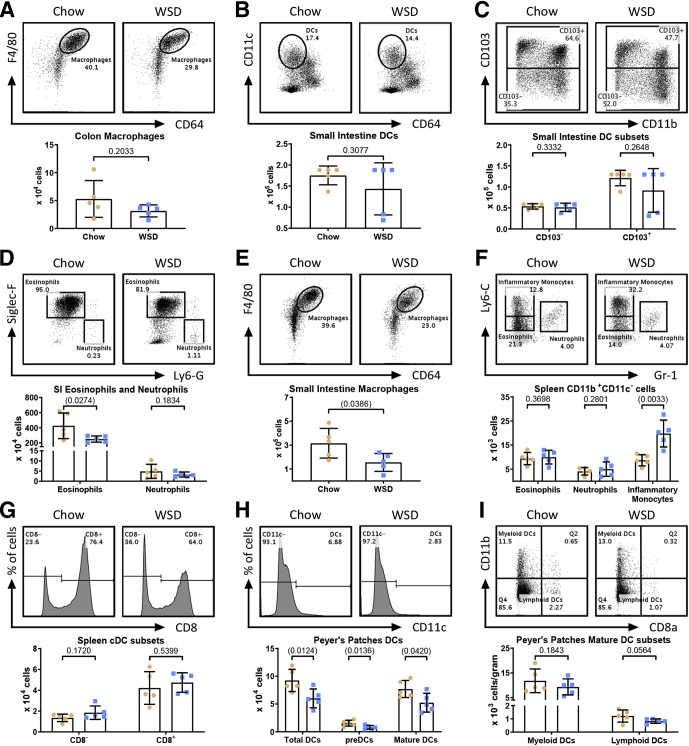
We recently reported that a WSD induces dramatic morphologic changes in the colon. Although such impacts appear to be mediated by impacts on epithelial cell proliferation, and primarily the WSD’s lack of fiber than its fat content per se, they nonetheless are thought to promote WSD-induced low-grade inflammation and metabolic syndrome.5,23 Thus, we next analyzed the extent to which a WSD might impact leukocyte populations in the small intestine and/or colon after 8 weeks of WSD consumption. Live CD45+MHCII+ cells were identified as DCs if they were CD11c+ and CD64-, and further categorized as either CD103- or CD103+ (Figure 2B). We observed a modest increase in total colonic DCs that did not reach statistical significance (Figure 1I), driven by an increase CD103- DCs and to a lesser extent by CD103+ DCs (Figure 1J). We did not observe changes in either colonic eosinophil or macrophage populations (Figures 1K and 3A). We also noted a modest trend toward increased colonic neutrophils that seemed worthy of monitoring in subsequent experiments (Figure 1K). A WSD induced only modest changes, in fact reductions, in some leukocyte populations in the small intestine and Peyer’s patches (PPs) (Figure 3), while the only observed change in the spleen was a moderate increase in inflammatory monocytes (Figure 3F; gating procedures are detailed in Figure 2). Altogether, these results indicated that the previously observed changes in adipose macrophages are among the most prominent changes in leukocytes induced by this WSD and identified less pronounced changes in colonic leukocytes that might contribute to the WSD-induced low-grade inflammation that promotes metabolic syndrome.
Figure 3.
An obesogenic diet alters myeloid cell populations in the small intestine, and to a lesser extent in the spleen and PPs. (A) After 8 weeks on chow (orange circles) or a WSD (blue squares), mice (N=5) were killed and colon tissue was analyzed for macrophages. Small intestinal tissue was analyzed for the following: (B) DCs, (C) CD103+/- DCs, (D) eosinophils and neutrophils, and (E) macrophages. Splenic tissue was analyzed for (F) eosinophils, neutrophils, inflammatory monocytes, and (G) CD8+/- classical dendritic cells (cDCs). (H–I) PPs were analyzed for total DCs and DC subsets. Data are the means ± SD. Statistical significance was determined using the t test. P < .05, brackets indicate significance.
Microbiota Ablation Did Not Mitigate WSD-Induced Adiposity but Ameliorated Features of Metabolic Syndrome
Investigation of the extent to which a particular phenotype requires the presence of microbiota has generally used antibiotic or GF approaches, both of which have benefits and caveats. Antibiotics reduce bacterial loads and change microbiota composition, but still result in many bacteria being present. In addition, antibiotics may have a direct impact on host enzymes and their bitter taste, when given to mice in drinking water, can result in ill health owing to dehydration. Concerns with GF mice include that they have broadly underdeveloped and dysregulated immune systems and are not able to digest, and thus harvest energy from plant-derived complex carbohydrates. Thus, a related approach is use of gnotobiotic mice colonized with a highly restricted microbiota such as 8 bacterial species known as ASF, which lack the γ-Proteobacterial pathobionts that we and others hypothesize drive low-grade inflammation and metabolic syndrome, yet have more normal immune and gut physiology.16 Hence, we used these 3 approaches to investigate the extent to which microbiota impact WSD-induced phenotypes.
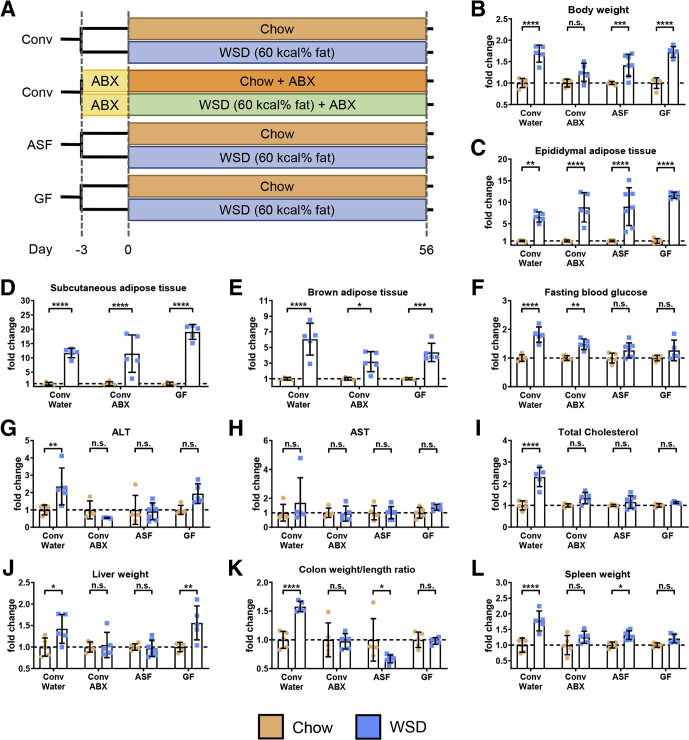
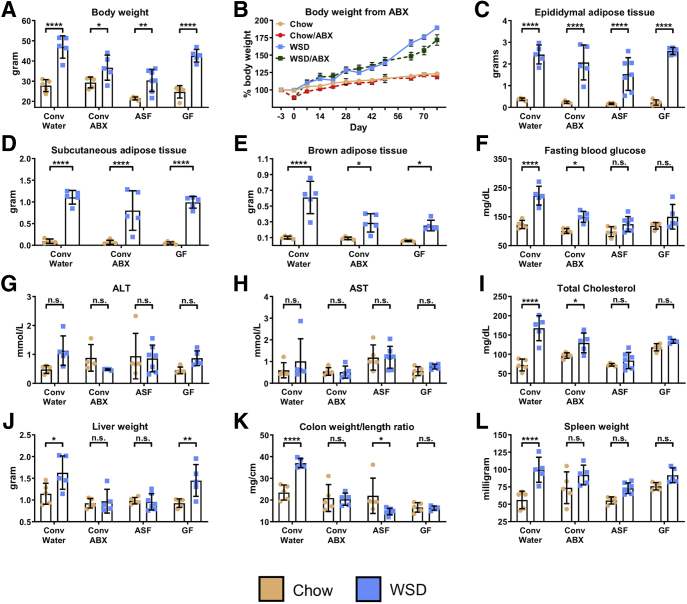
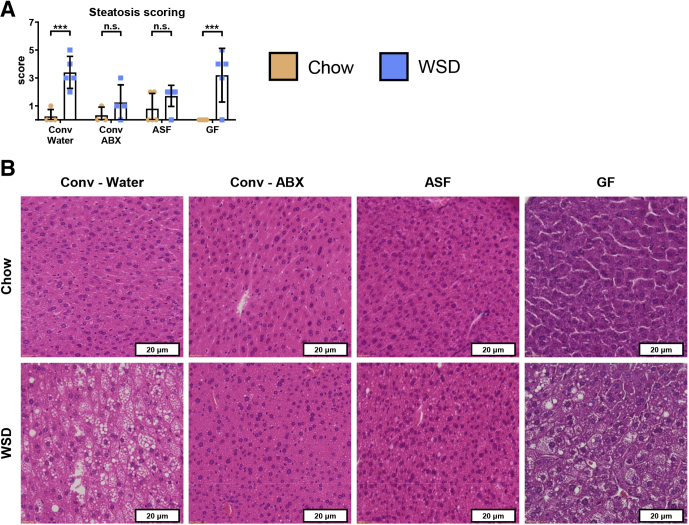
Control (i.e. conventional specific pathogen-free) and microbiota-ablated mice were maintained on standard chow (autoclaved for GF and ASF mice) or administered a WSD (irradiated for GF and ASF mice) for 8 weeks (Figure 4A), at which time they were killed, and metabolic and immunologic parameters were assayed. Absolute measures of metabolic parameters are shown in Figure 5, while relative differences between mice fed chow vs a WSD within each microbiota condition are shown in Figure 4. This visualization facilitates evaluation of the extent to which microbiota ablation impacted the metabolic effects of WSD and, at least in this case, yields similar trends as the nonnormalized data. Specifically, microbiota ablation only modestly reduced WSD-induced weight gain (Figures 4B and 5A and B) and did not reduce WSD-induced increases in epididymal fat or other adipose depots (Figure 4C–E). However, microbiota ablation protected against several indices of metabolic syndrome that otherwise result from a WSD. Specifically, the extent of WSD-induced dysglycemia was reduced (Figure 4F), as were increases in alanine aminotransferase (ALT), aspartate aminotransferase (AST), and serum cholesterol levels (Figure 4G–I), although a WSD still induced a significant increase in ALT concentration in GF mice. A similar pattern was observed for WSD-induced hepatomegaly (Figure 4J), which correlated well with WSD-induced steatosis (Figure 6) in that WSD induced changes in steatosis in conventional and GF mice but not antibiotic-treated or ASF mice. In addition, microbiota ablation prevented WSD-induced morphometric changes that reflect low-grade inflammation such as an increase in the colon weight/length ratio and spleen weight (Figure 4K and L). Thus, in contrast to some studies, including our ongoing studies that used shorter WSD exposures, ablation of microbiota did not significantly reduce the extent of adiposity that results from an 8-week exposure to WSD but ameliorated some other aspects of WSD-induced metabolic syndrome.
Figure 4.
Microbiota ablation did not mitigate WSD-induced adiposity but ameliorated features of metabolic syndrome. (A) Four-week-old male C57BL/6J conventionally raised mice were purchased from The Jackson Laboratory, 4-week-old male C57BL/6 mice possessing an ASF were obtained from the Georgia State University breeding repository, and 3- to 5-week-old male C57BL/6 GF mice were purchased from Taconic Biosciences (N=5). At day 0, animals were switched to a high-fat diet (60% kcal from fat) for 8 weeks or continued on a standard grain-based chow as a control. An additional group of conventional C57BL/6J mice were started on an antibiotic cocktail 3 days before high-fat diet administration and maintained throughout the experiment. At day 56, mice were euthanized for tissue collection and biometric measurements, data of WSD-treated mice represented here are relative to the chow-fed mice of their corresponding groups: (B) final body weight, (C) epididymal adipose tissue weight, (D) subcutaneous adipose tissue weight, (E) brown adipose tissue weight, (F) 15-hour fasting blood glucose level, (G) ALT concentration, (H) AST concentration, (I) total serum cholesterol, (J) liver weight, (K) colon weight/length ratio, and (L) spleen weight. Data are the means ± SD. Statistical significance was determined using 1-way analysis of variance corrected for multiple comparisons with a Bonferroni test. *P ≤ .05, **P ≤ .01, ***P ≤ .001, and ****P ≤ .0001. ABX, antibiotic; Conv, conventional; ns, nonsignificant.
Figure 5.
Absolute values of biometric measurements and tissue/macromolecule quantifications corresponding toFigure 4. Comparison of conventional mice with antibiotic-treated, ASF, and GF mice (n=5-7): (A) final body weight, (B) changes in weight in antibiotic-treated mice, (C) epididymal adipose tissue weight, (D) subcutaneous adipose tissue weight, (E) brown adipose tissue weight, (F) fasting blood glucose level, (G) ALT concentration, (H) AST concentration, (I) total serum cholesterol, (J) liver weight, (K) colon weight/length ratio, and (L) spleen weight. Data are the means ± SD. Statistical significance was determined using 1-way analysis of variance corrected for multiple comparisons with a Bonferroni test. *P ≤ .05, **P ≤ .01, ***P ≤ .001, and ****P ≤ .0001. ABX, antibiotic; Conv, conventional; ns, nonsignificant.
Figure 6.
Hepatic steatosis is reduced in antibiotic-treated and ASF mice on a WSD compared with conventional controls. Extended analysis of mice from Figure 2. Four-week-old male C57BL/6J conventionally raised mice were purchased from The Jackson Laboratory, 4-week-old male C57BL/6 mice possessing an ASF were obtained from the Georgia State University breeding repository, and 3- to 5-week-old male C57BL/6 GF mice were purchased from Taconic Biosciences. At day 0, animals were switched to a high-fat diet (60% kcal from fat) for 8 weeks or continued on a standard grain-based chow as a control. An additional group of conventional C57BL/6J mice were started on an antibiotic cocktail 3 days before high-fat diet administration and maintained throughout the experiment. At day 56, mice were killed and liver tissues were collected to assess (A) hepatic steatosis and (B) perform H&E staining. Data are the means ± SD. Statistical significance was determined using 1-way analysis of variance corrected for multiple comparisons with a Bonferroni test. ***P ≤ .001. ABX, antibiotic; Conv, conventional; ns, nonsignificant.
Microbiota Ablation Reduced WSD-Induced Changes in Adipose and Colonic Leukocytes
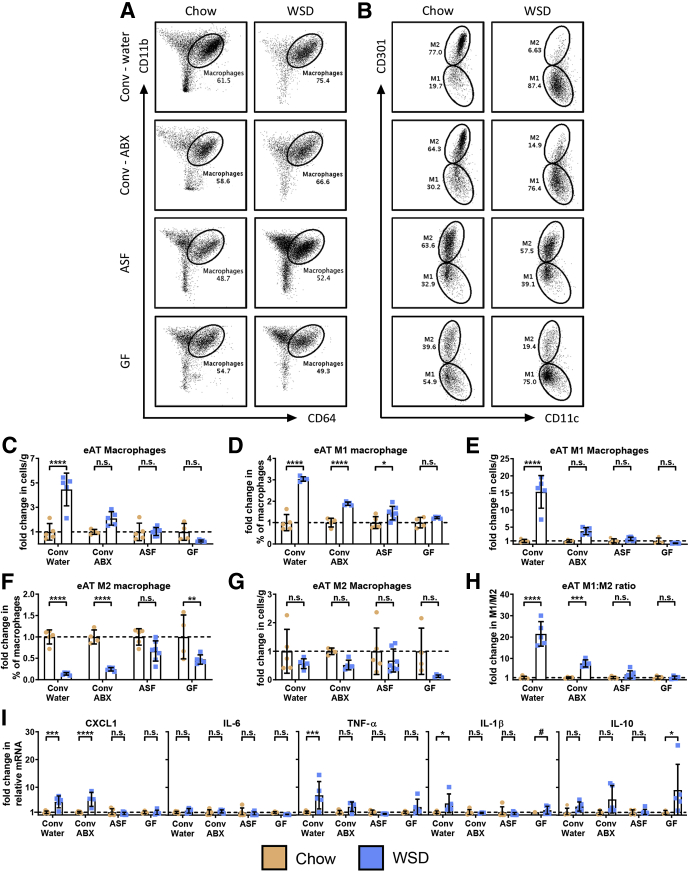
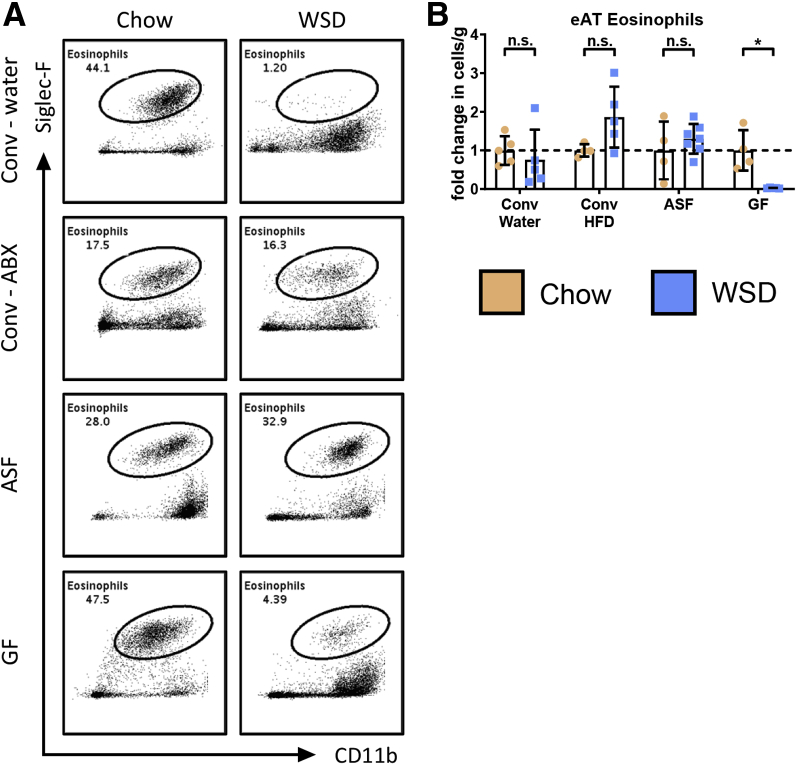
Although under the conditions used herein microbiota ablation did not impact the extent of WSD-induced adiposity, it ameliorated some aspects of metabolic syndrome, which led us to hypothesize that it might reduce the adipose inflammation that is thought to promote this disorder. In accordance with this hypothesis, ablation of microbiota by all 3 approaches, albeit more clearly for ASF and germ-free, markedly reduced the WSD-induced changes in epididymal adipose macrophages (Figure 7A–C). Specifically, microbiota ablation prevented the absolute and relative increase in M1 macrophages (Figure 7D and E) and reduced the relative, but not absolute, decrease in M2 macrophages (Figure 7F and G), thus largely preventing the increase in the M1:M2 ratio (Figure 7H). In addition, the modest trend of WSD resulting in reduced adipose eosinophils seen in conventional mice was not observed in antibiotic-treated and ASF mice (Figure 8A and B). In accordance with the notion that macrophage populations are a key determinant of the overall inflammatory tone of visceral adipose tissue, we observed that microbiota ablation largely eliminated WSD-induced increases in adipose expression of proinflammatory cytokines, including chemokine (C-X-C motif) ligand 1 (CXCL1), tumor necrosis factor α (TNF-α), and interleukin (IL)1β, while increasing expression of IL10 in antibiotic-treated and GF mice (Figure 7I).
Figure 7.
Microbiota ablation reduces macrophage infiltration and M1 macrophage polarization, reducing the M1:M2 ratio. Epididymal adipose tissue from the epididymitis of mice was analyzed using flow cytometry to quantify the following: (A) macrophages and (B) M1 and M2 macrophage subsets. Data from WSD-treated mice represented here are relative to the chow-fed mice of their corresponding groups, as follows: (C) number of macrophages per gram of adipose tissue, (D) M1 macrophages as a percentage of total macrophages, (E) M1 macrophages as the number of cells per gram of adipose tissue, (F) M2 macrophages as a percentage of total macrophages (∗∗P ≤ .01), and (G) M2 macrophages as the number of cells per gram of adipose tissue. (H) M1:M2 macrophage ratios were quantified using the percentage of M1 and M2 macrophages. (I) Epididymal adipose tissue collected at death also was analyzed for cytokines via quantitative reverse-transcription polymerase chain reaction. Data are the means ± SD (N=5-7). Statistical significance was determined using 1-way analysis of variance corrected for multiple comparisons with a Bonferroni test. *P ≤ .05, ***P ≤ .001, and ****P ≤ .0001. #Limited sample quantity prevented the use of statistical analysis. ABX, antibiotic; Conv, conventional; eAT, epididymal adipose tissue; ns, nonsignificant.
Figure 8.
Microbiota ablation modestly protects against eosinophil loss during WSD feeding. (A) Epididymal adipose tissue from the epididymitis of mice was analyzed using flow cytometry to quantify eosinophils. (B) Eosinophils of WSD-treated mice represented as the number per gram of adipose tissue is relative to chow-fed mice. Data are the means ± SD (N=5-7). Statistical significance was determined using 1-way analysis of variance corrected for multiple comparisons with a Bonferroni test. *P ≤ .05. ABX, antibiotic; Conv, conventional; eAT, epididymal adipose tissue; ns, nonsignificant.
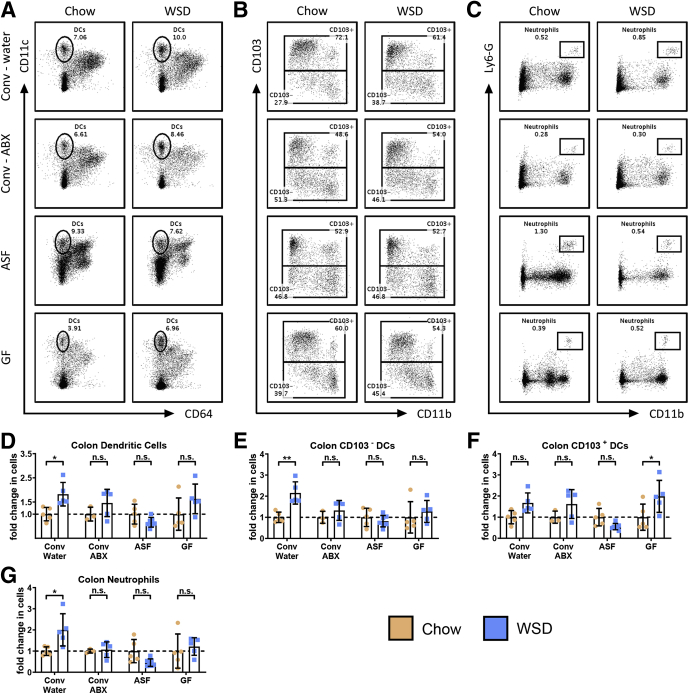
Ablation of microbiota also reduced many of the changes in leukocyte populations observed in the colon in response to WSD (Figure 9A–C). Specifically, all 3 means of reducing gut bacteria reduced the total increase in colonic DCs (Figure 9D) (P for chow vs WSD-treated conventional/antibiotic, ASF, and GF mice = .7356, .8998, and .1600, respectively) via preventing increases in CD103- DCs (Figure 9E) without impacting the trend of WSD-induced increases in CD103+ DCs (Figure 9F) (P for chow vs WSD-treated conventional/water, conventional/antibiotic, and ASF mice = .1612, .3928, and .5331, respectively). In addition, microbiota ablation prevented the modest increase in colonic neutrophils observed in response to WSD (Figure 9G), although the extent of these changes, even in conventional mice, did not result in significant changes in proinflammatory gene expression in the colon per se. Together, these results indicate that WSD-induced changes in leukocytes that mediate inflammation require the presence of a complex microbiota.
Figure 9.
Microbiota ablation reduces DC alterations and neutrophil cell infiltration in the colon. Colon tissue was analyzed using flow cytometry to quantify the following: (A) DCs, (B) CD103+/- DCs, and (C) neutrophils. Data from WSD-treated mice represented here are relative to the chow-fed mice of their corresponding groups, as follows: (D) number of DCs, (E) number of CD103- DCs, (F) number of CD103+ DCs, and (G) number of neutrophils per colon. Data are the means ± SD (N=5-7). Statistical significance was determined using 1-way analysis of variance corrected for multiple comparisons with a Bonferroni test. *P ≤ .05 and **P ≤ .01. ABX, antibiotic; Conv, conventional; ns, nonsignificant.
Genetic Depletion of Toll-like Receptor–Signaling Adaptor Myeloid Differentiation Primary Response 88 Phenocopies Microbiota Ablation in Response to WSD
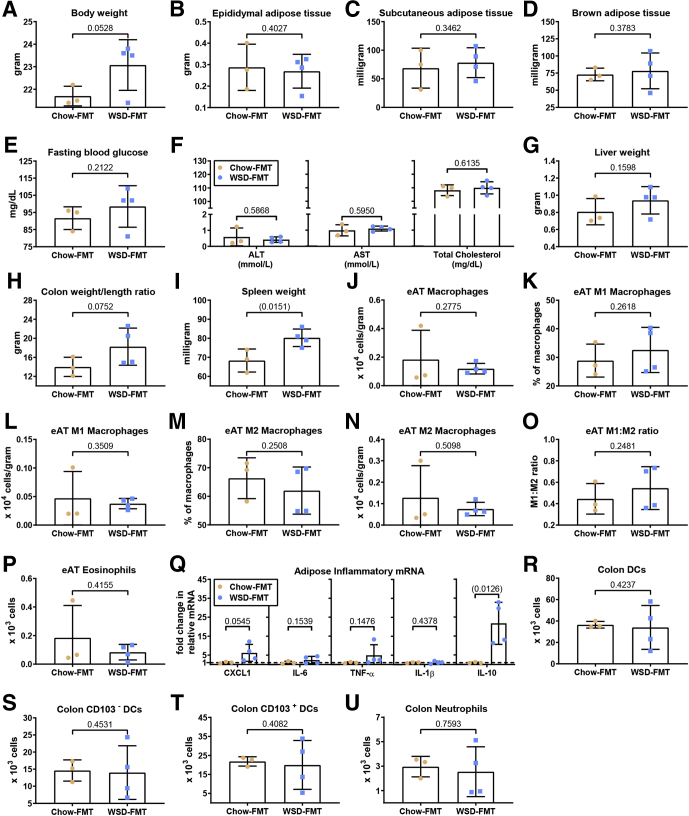
Transplant of microbiota from genetically modified mice that had developed metabolic syndrome when consuming standard rodent chow to wild-type GF mice transferred some indices of metabolic syndrome, which indicates that microbiota composition rather than the mere presence of a complex microbiota is a determinant of metabolic disease.24 Hence, we next examined the extent to which transplant of microbiota from WSD-fed wild-type mice to GF mice, maintained on rodent chow (ie, never directly exposed to WSD), might transplant adipose inflammation and/or other aspects of metabolic syndrome. Transplanted mice were killed 4 weeks after transplant, which is a time point that other transplant studies, including ours, have found sufficient to transfer some inflammatory/metabolic phenotypes,24,25 whereas longer times may result in the recipient host reshaping the transplanted microbiota. In accordance with work from Rabot et al,3 we observed that, relative to GF mice conventionalized with feces from chow-fed mice, transplant of microbiota from WSD-fed mice did not show increased adiposity or other features of metabolic syndrome (Figure 10A–I). Moreover, transplant of such WSD microbiota did not result in any differences in leukocyte populations in adipose tissue or colon (Figure 10J–U). Interestingly, WSD microbiota recipients did show modest, albeit not statistically significant, increases in CXCL1 and TNF-α in the adipose tissue, which was counterbalanced by a significant increase in IL10, arguing against the notion that the adipose tissue was in a state of low-grade inflammation (Figure 10Q). Thus, transfer of WSD-derived microbiota alone did not recapitulate adipose inflammation or metabolic syndrome, indicating that WSD-induced dysbiosis not maintained in chow-fed mice and/or that altered microbiota, by itself, is not sufficient to induce this state.
Figure 10.
Transplant of microbiota from obese mice mildly recapitulates features of obesity and myeloid cell alterations. Four-week-old male C57BL/6 GF mice were purchased from Taconic Biosciences and were administered feces from either chow-(blue boxes) or WSD-(orange circles)fed mice by oral gavage. These animals subsequently were maintained in a sterile environment for 4 weeks before terminal tissue collection and biometric quantification, data are as follows: (A) final body weight; (B) epididymal adipose tissue weight; (C) subcutaneous adipose tissue weight; (D) brown adipose tissue weight; (E) 15-hour fasting blood glucose level; (F) ALT concentration, AST concentration, and total serum cholesterol; (G) liver weight; (H) colon weight/length ratio; and (I) spleen weight. Epididymal adipose tissue was analyzed using flow cytometry to quantify the following: (J) macrophages, (K and L) M1 macrophages, (M and N) M2 macrophages, (O) M1:M2 ratio, and (P) eosinophils. (Q) Epididymal adipose tissue collected at death also was analyzed for cytokines via quantitative reverse-transcription polymerase chain reaction. Colon tissue was analyzed using flow cytometry to quantify the following: (R) DCs, (S) CD103- DCs, (T) CD103+ DCs, and (U) neutrophils. Data are the means ± SD (N=3-4). Statistical significance was determined using the t test. P < .05, brackets indicates significance. eAT, epididymal adipose tissue; FMT, fecal microbiota transplant; mRNA, messenger RNA.
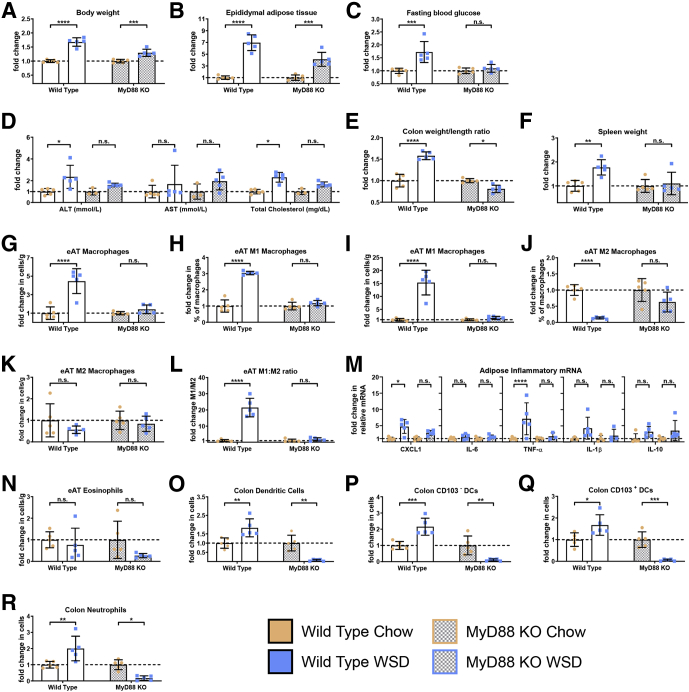
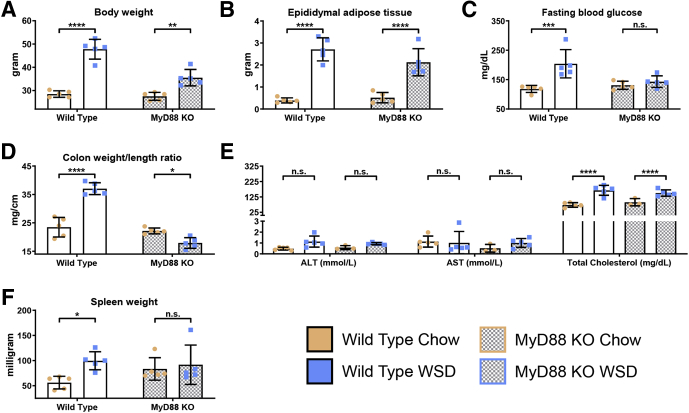
We hypothesize that inability to manage microbiota, and pathobionts in particular, as a result of WSD’s impact on the colon results in microbiota promoting inflammation via activation of innate immune signaling pathways. This hypothesis predicts that absence of the global Toll-like receptor (TLR)-signaling adaptor protein myeloid differentiation primary response 88 (MyD88), which has been observed to protect against some aspects of WSD-induced metabolic syndrome, should protect against adipose inflammation. In agreement with this notion and other studies,26 the relative and absolute extent of weight gain and adiposity induced by WSD was blunted in MyD88 knockout (KO) mice (Figures 11A and B and 12A and B). Furthermore, absence of MyD88 protected against WSD-induced indices of metabolic syndrome including dysglycemia, increased ALT levels, and hypercholesterolemia (Figures 11C and D and 12C and E). MyD88 KO mice also were protected from WSD-induced inflammation as assessed by the colon weight:length ratio and spleen weight (Figures 11E and F and 12D and F). Moreover, in response to WSD, MyD88 did not show adipose inflammation as assessed by changes in leukocyte populations or cytokine expression (Figure 11G–N). In addition, they did not show increases in colonic DC or neutrophils (Figure 11O–R). Thus, the absence of MyD88 largely phenocopied microbiota ablation, thus supporting our hypothesis that WSD-induced adipose inflammation does not result from lipid accumulation per se, but rather is promoted by microbiota or their products activating innate immune signaling pathways.
Figure 11.
Genetic depletion of TLR signaling adaptor MyD88 phenocopies microbiota ablation in response to WSD. Conventionally raised 3- to 5-week-old male C57BL/6J, wild-type and MyD88 KO mice were purchased from The Jackson Laboratory. Animals were placed on a high-fat diet (60% kcal from fat) for 8 weeks or continued on a standard grain-based chow as a control. At day 56, mice were killed for tissue collection and biometric measurements, data from WSD-treated mice represented here are relative to the chow-fed mice of their corresponding groups, as follows: (A) final body weight; (B) epididymal adipose tissue weight; (C) 15-hour fasting blood glucose level; (D) ALT concentration, AST concentration, and total cholesterol; (E) colon weight/length ratio; and (F) spleen weight. Epididymal adipose tissue from the epididymitis of mice was analyzed using flow cytometry and quantitative reverse-transcription polymerase chain reaction to quantify the following: (G) macrophages, (H and I) M1 macrophages, (J and K) M2 macrophages, (L) M1:M2 ratio, (M) inflammatory cytokines, and (N) eosinophils. Colon tissue was used to quantify the following: (O) DCs, (P) CD103- DCs, (Q) CD103+ DCs, and (R) neutrophils. Data are the means ± SD (N=5). Statistical significance was determined using 1-way analysis of variance corrected for multiple comparisons with a Bonferroni test. *P ≤ .05, **P ≤ .01, ***P ≤ .001, and ****P ≤ .0001. eAT, epididymal adipose tissue; mRNA, messenger RNA; ns, nonsignificant.
Figure 12.
Absolute values of biometric measurements and tissue/macromolecule quantifications corresponding toFigure 11: (A) body weight; (B) epididymal adipose tissue weight; (C) 15-hour fasting blood glucose level; (D) colon weight/length ratio; (E) ALT concentration, AST concentration, and total cholesterol; and (F) spleen weight. Data are the means ± SD (N=5). Statistical significance was determined using 1-way analysis of variance corrected for multiple comparisons with a Bonferroni test. *P ≤ .05, **P ≤ .01, ***P ≤ .001, and ****P ≤ .0001. ns, nonsignificant.
Discussion
Inflammation of visceral adipose tissue is viewed as a central link between obesity and the constellation of metabolic abnormalities referred to as metabolic syndrome. Hence, there is great interest in understanding mechanisms by which adipose tissue acquires an inflammatory profile and how this profile impacts adiposity and other aspects of metabolic syndrome. Excess lipid loading can be sufficient to drive proinflammatory gene expression, which provides a plausible explanation whereby increased adiposity, by itself, can drive adipose inflammation.27 However, adipose inflammation and parameters of metabolic syndrome also can be induced by transplant of microbiota with proinflammatory features24 or direct administration of LPS,12 which indicates that microbiota is also a plausible determinant of adipose inflammation. Hence, the central goal of this study was to investigate the extent to which gut microbiota was necessary for low-grade inflammation, especially in adipose tissue, and, consequently, other aspects of metabolic syndrome. We hypothesized that adipose inflammation would depend on the presence of gut microbiota and that adipose inflammation would be critical for driving WSD-induced metabolic syndrome. We used 3 approaches of ablating microbiota, all of which yielded a generally similar pattern of results. In accordance with our hypothesis, adipose inflammation required the presence of complex pathobiont-containing microbiota. However, such adipose inflammation was not needed for increased adiposity, arguing against our hypothesis that the former provides critical positive feedback needed to promote the latter. However, lack of adipose inflammation was associated with less severe dysglycemia, which is a central defining feature of metabolic syndrome. Such reduced dysglycemia is in accordance with work from Membrez et al28 that antibiotics decreased plasma LPS and dysglycemia independent of adiposity. We also observed that a lack of WSD-induced adipose inflammation, resulting from microbiota ablation, also reduced other indices of metabolic syndrome, including increases in hepatic steatosis, AST/ALT levels, and cholesterol levels. Together, these results suggest that increased adiposity without adipose inflammation may be less likely to eventuate in severe diseases such as cardiovascular disease. Rather, obesity without inflammation may result in a state of healthy obesity, which is a state achieved by some human beings. Although considering this notion makes it tempting to suggest using antibiotics to improve the metabolic parameters of obese subjects, attempts to do so have not been effective, potentially suggesting a disconnect between the WSD model and human metabolic syndrome, and studies using this model, including ours, have focused on prevention rather than treatment.29
Although our observation that microbiota was not required for WSD-induced adiposity per se might seem to contradict some published work in this field, overall, we submit our results are in accordance with the variety of studies in this field in which some studies observed GF mice were markedly protected against WSD-induced obesity while others observed similar adiposity in GF and conventional mice.1 Some of this variance might be accounted for by the array of different approaches used. For example, Backhed et al13 compared GF mice with conventionalized mice, which are prone to obesity, relative to conventional mice, whereas studies by Rabot et al14 compared conventional and GF mice fed WSD and did not assess the impact of WSD relative to mice maintained on a control diet. However, we do not submit that such differences are sufficient to explain the observed differences in results from various studies, and we do not dispute the notion that, in some conditions, ablation of microbiota can protect against WSD-induced adiposity. Indeed, we have observed some antibiotic regimens protect against WSD-induced adiposity,4,5 while recent experiments in our laboratory continue to indicate that, in response to a 4-week WSD treatment, GF and ASF mice show reduced adiposity relative to conventional mice. However, this study focused on adipose inflammation, particularly as characterized by changes in the macrophage population, which was not observed in response to 4 weeks of WSD feeding, thus precluding this time point herein.
On the one hand, some consequences of WSD feeding occur very rapidly. Relative to mice fed grain-based chow, WSD induces a rapid (within 3 days) reduction of colonic bacterial density, marked changes in species composition, and a stark reduction in colonocyte proliferation that results in marked morphologic changes accompanied by microbiota encroachment,5 which we propose drives inflammation and dysglycemia.30 On the other hand, more traditional markers of inflammation such as increases in serum cytokine levels and leukocyte infiltration generally have been reported only in response to longer-term WSD feeding. Some metabolic consequences of WSD occur quite rapidly, with dysglycemia being present within days of exposure to WSD, although robust development of metabolic syndrome, including more severe insulin resistance, requires longer-term exposure to WSD.31 Genetic approaches that seek to block leukocyte activation indicate that inflammation is dispensable for the rapid changes, but critical for long-term insulin resistance.31 Thus, we conclude that irrespective of whether or not it impacts adiposity per se, ablation of microbiota protects against adipose inflammation, which is a key driver of metabolic syndrome.
That absence of MyD88, which is necessary for generation of proinflammatory signaling by TLRs (except TLR3) and inflammasome cytokines IL1β and IL18, thus making it effectively necessary for many aspects of nucleotide-binding oligomerization domain–like receptor signaling as well, fully mimicked the protection against adipose inflammation that resulted from microbiota ablation, which suggests a role for bacterial products in driving adipose inflammation. This result could reflect role(s) of MyD88 in multiple cell types including epithelial cells, adipocytes, and/or immune cells. In addition, that inflammation itself promotes microbiota dysbiosis32 suggests that a lack of MyD88 may ameliorate WSD-induced changes in microbiota, which also might contribute to protection of MyD88 against WSD. We anticipate future studies involving microbiota analyses and tissue-specific MyD88 KO will be able to investigate these questions. The combination of microbiota encroachment amidst reduced gut barrier function in response to WSD can be envisaged to result in bacterial products or perhaps even bacteria reaching the visceral adipose tissue, but we certainly do not exclude the possibility that adipose inflammation is driven by bacterial-induced cytokines in other tissues that then act upon adipocytes. ASF mice were protected from WSD-induced adipose inflammation, and its associated metabolic consequences, which underscores that microbiota composition, rather than the mere presence of bacteria in the gut, is a determinant of susceptibility to WSD and, moreover, supports the possibility that pathobiont bacteria might be key contributors to WSD-induced metabolic syndrome. On the other hand, in contrast to ASF and antibiotic-treated mice, GF mice showed WSD-induced liver dysfunction (hepatomegaly, steatosis, and increased ALT), which highlights that a total lack of bacteria might be detrimental, possibly as a result of immune and/or digestive abnormalities.
In conclusion, our findings indicate that the mechanism by which WSD-induced obesity results in adipose inflammation is dependent on the intestinal microbiota, possibly reflecting a deterioration in intestinal microbiota homeostasis that results in activation of innate immune signaling to protect against encroaching and/or translocating bacteria. Should such events occur in human beings, they might, in part, be a basis for why some obese persons remain relatively metabolically healthy while others develop metabolic syndrome and downstream deleterious consequences. Moreover, we speculate that irrespective of its impact on adiposity, restoration of a host microbiota homeostasis through diet, pharmacologic, and/or perhaps prebiotic/probiotic approaches might be a means to alleviate visceral adipose tissue inflammation and its metabolic consequences.
Materials and Methods
Mice and High-Fat Diet Administration
Four-week-old (wild-type and MyD88 KO) male C57BL/6 mice were purchased from Jackson Laboratory (Bar Harbor, ME) and maintained at Georgia State University (Atlanta, GA) under institutionally approved protocols (Institutional Animal Care and Use Committee A18006), housed 5 mice per cage with ALPHA-dri bedding (Shepherd Specialty Papers, Watertown, TN) and nestlets (Ancare, Bellmore, NY) and fed ad libitum. Mice were fed standard grain-based chow (LabDiet, St. Louis, MO), which is composed of relatively unrefined ingredients and given 2 weeks to allow for microbiota stabilization. Afterward, half the cohort of mice then was switched to a diet composed of 60% kcal from fat (D12492; Research Diet, New Brunswick, NJ) for 8 weeks, while the remaining half continued on the grain-based chow as a control. For antibiotic treatment, 3 days before WSD feeding mice were administered drinking water containing ampicillin (1 g/L), vancomycin (0.25 g/L), neomycin (1 g/L), and metronidazole (1 g/L). The water supply was renewed every 3 days and maintained during WSD feeding. Mice then were killed, and body weight, colon length, and weights of adipose depots (perigonadal visceral [ie, epididymal], adipose, interscapular brown adipose, and posterior subcutaneous adipose), colon, liver, and spleen were measured. Serum and organs were collected for downstream analysis.
GF and ASF Mice Experiments
C57BL/6 ASF mice obtained from the Georgia State University breeding repository and GF mice purchased from Taconic Biosciences (Rensselaer, NY) were housed in a Tecniplast (West Chester, PA) IsoCage Bioexclusion System in our GF facility. Mice were fed a modified rodent diet with 60 kcal% fat and 1.5× Vitamin Mix irradiated twice with 10–20 kGy γ-irradiation (D12492-1.5Vi; Research Diet) for 8 weeks. Samples then were collected as described previously.
Serum Panel Analyses
Mice were bled after 15-hour fasting and hemolysis-free serum was collected using serum separating tubes (BD Biosciences, San Jose, CA). Serum cholesterol, AST, and ALT levels were measured using biochemical kits from Randox Laboratories (Kearneysville, WV).
RNA Extraction and Quantitative Reverse-Transcription Polymerase Chain Reaction
Total RNA extraction was performed as previously described.33 Briefly, RNAs were extracted using TRIzol reagent (15-596-026; Invitrogen, Carlsbad, CA), with RNAs from adipose tissue purified using the RNeasy Mini Kit (74106; Qiagen, Germantown, MD). Purified RNAs were reverse-transcribed and quantitative polymerase chain reaction was performed in duplicate using an iTaq Universal SYBR Green One-Step Kit (1725151; Bio-Rad, Hercules, CA) on a CFX96 apparatus (Bio-Rad). The sense and antisense oligonucleotides used, respectively, were as follows: 36B4: 5’- TCCAGGCTTTGGGCATCA-3’ and 5’-TCTTTATCAGCTGCACATCACTCAGA-3’, CXCL1: 5’-ACTGCACCCAAACCGAAGTC-3’ and 5’-TGGGGACACCTTTTAGCATCTT-3’, IL6: 5’-GTGGCTAAGGACCAAGACCA-3’ and 5’-GGTTTGCCGAGTAGATCTCA-3’, TNF-α: 5’-CGAGTGACAAGCCTGTAGCC-3’ and 5’-CATGCCGTTGGCCAGGA-3’, IL1β: 5’-TCATGGGATGATGATAACCTGCT-3’ and 5’-CCCATACTTTAGGAAGACACGGATT-3’, and IL10: 5’-ACCTGGTAGAAGTGATGCCCCAGGCA-3’ and 5’-CTATGCAGTTGATGAAGATGTCAAA-3’. Results were normalized to the housekeeping 36B4 gene. Fold-induction was calculated using the threshold cycles (C)t method as follows: ΔΔCt = (Cttarget gene- Cthousekeeping gene)treatment - (Cttarget gene- Cthousekeeping gene)nontreatment, and the final data were derived from 2−ΔΔCT.
Fasting Blood Glucose
Mice were fasted for 15 hours during the nocturnal cycle, and baseline blood glucose levels were measured by a Nova Max plus Glucose meter (Nova Biomedical, Billerica, MA) and expressed in milligrams per deciliter.
Fecal Microbiota Transplants
Feces from WSD- or chow-treated mice were suspended in 30% glycerol diluted in phosphate-buffered saline (1.0 mL) and stored at −80°C.25 GF C57BL/6 mice (5 weeks old) purchased from Taconic Biosciences were administered 200 μL orally of the earlier-described fecal suspension and housed in a Tecniplast IsoCage Bioexclusion System.
Adipose Tissue Preparation
Isolation and preparation of stromal vascular cells (SVCs) from the adipose tissue was performed as previously described.34 Both perigonadal fat pads were digested in 10 mL of 1 mg/mL of type IV collagenase buffer in Hank’s balanced salt solution (HBSS) (with Ca2+ and Mg2+) with 0.5% bovine serum albumin for 20 minutes at 37°C. After digestion, EDTA was added to a final concentration of 10 mmol/L and incubated at 37°C for an additional 10 minutes. SVCs were passed through a 100-μm nylon filter and a 40-μm nylon filter, respectively, before centrifugation at 500 × g for 10 minutes at 4°C to separate adipocytes from SVCs. The adipocyte containing supernatant was removed and the SVC pellet was resuspended in 0.5 mL of red blood cell lysis buffer and incubated for 5 minutes at room temperature. Then 5 mL of fluorescence-activated cell sorting buffer was added to neutralize the red blood cell lysis buffer. Cells then were pelleted by centrifugation at 500 × g for 10 minutes at 4°C before staining for leukocyte surface markers.
Intestinal Tissue Preparation
Isolation and preparation of cells from intestinal tissue was performed as previously described.35 Briefly, colons and small intestines from mice were cut into small 1.0-cm long pieces and placed into 20 mL of HBSS with 5% fetal bovine serum (FBS) and 2 mmol/L EDTA and incubated with shaking for 20 minutes at 37°C. Then the aliquot was passed through a sieve and the intestinal pieces were washed on the sieve with 10 mL HBSS with 5% FBS. Incubation and wash steps were repeated once more before the colon pieces were minced with a small razor. The minced intestines then were incubated in 20 mL HBSS with 5% FBS, 1 mg/mL collagenase type VIII, and 0.1 mg/mL DNase I with shaking for 10 minutes at 37°C. After incubation, 20 mL HBSS with 5% FBS was added to the cells before vortexing and centrifugation for 10 minutes at 1500 rpm. Cells then were resuspended in HBSS with 5% FBS and passed through a 100-μm filter and then a 40 μm cell strainer. After centrifugation, cells then were stained for leukocyte surface markers.
Splenic Tissue Preparation
After euthanasia, spleens were crushed between 2 frosted microscope slides and added to 5 mL media RPMI with 10% FBS. Cells then were filtered through a 100-μmol/L filter with an additional 5 mL RPMI with 10% FBS used to wash the filter. Cells were centrifuged at 1500 rpm for 5 minutes and cell pellets were resuspended in 5 mL red blood cell lysis buffer and incubated at 37°C for 10 minutes before centrifugation at 1500 rpm for 5 minutes. Resuspended cells were filtered through a 40-μmol/L filter before another wash step. After centrifugation, cells then were stained for leukocyte surface markers.
PP Preparation
Isolation and preparation of cells from intestinal tissue was performed as previously described.36 Briefly, removed PPs were transferred into 5 mL dissociation medium (RPMI with 1 mg/mL collagenase IV, 0.1 mg/mL DNase, and 5% FBS) and incubated for 20 minutes at 37°C with vigorous shaking. PPs were placed onto a sterile 70-μm nylon mesh cell strainer and forcibly grinded into the mesh. EDTA was added to a final concentration of 1 mmol/L to the cell suspension and incubated on a rocker for 5 minutes at room temperature. The cell suspension was passed through a second 70-μm cell strainer to remove any remaining cellular aggregates or tissue debris. Afterward, cells were centrifuged for 5 minutes at 1500 rpm and resulting cell pellets were stained for leukocyte surface markers.
Live/Dead and Leukocyte Discrimination
After isolation, all cells were washed with 300 μL cold phosphate-buffered saline twice. Cells then were resuspended in 100 μL Alexa 430 NHS ester (Thermo Fisher Scientific, Waltham, MA) at working dilution and incubated at 4°C for 20 minutes in the dark. After incubation, cells were washed with fluorescence-activated cell sorting buffer (phosphate-buffered saline with 1 mmol/L EDTA and 0.5% bovine serum albumin) before incubation with anti-mouse CD16/CD32 to block nonspecific binding of fragment, crystallizable region receptors. Staining cocktails were added to cells as outlined in Table 1. Finally, cells were washed 3 times before fixation by 1% paraformaldehyde.
Table 1.
Staining Cocktails
| Marker | Fluorophore | Dilution | Supplier |
|---|---|---|---|
| Macrophages | |||
| CD11c | PacBlue | 1:100 | eBioscience (San Diego, CA) |
| CD301 | APC | 1:50 | Biolegend (San Diego, CA) |
| CD45 | PerCP | 1:100 | Biolegend |
| CD64 | PE/Dazzle | 1:100 | Biolegend |
| F4/80 (or CD11b) | PE | 1:100 | Biolegend |
| MHCII | AF700 | 1:100 | eBioscience |
| Dendritic cells | |||
| CD103 | APC | 1:50 | Biolegend |
| CD11b | PE | 1:100 | Biolegend |
| CD11c | PacBlue | 1:100 | Biolegend |
| CD45 | PerCP | 1:100 | Biolegend |
| CD64 | PE/Dazzle | 1:100 | Biolegend |
| MHCII | AF700 | 1:100 | eBioscience |
| Eosinophils and neutrophils | |||
| CD11b | PE | 1:100 | Biolegend |
| Ly6-G | FITC | 1:100 | Biolegend |
| MHCII | AF700 | 1:100 | eBioscience |
| Siglec-F | BV421 | 1:100 | BD Biosciences |
| CD45 | PerCP | 1:100 | Biolegend |
| Splenic tissue | |||
| CD11b | Pe/Cy5 | 1:100 | Biolegend |
| CD11c | Pe/Cy7 | 1:300 | Biolegend |
| CD19 | APC | 1:100 | eBioscience |
| CD3 | PE | 1:100 | Biolegend |
| CD45 | PerCP | 1:100 | Biolegend |
| CD8 | eFluor 450 | 1:100 | eBioscience |
| Gr-1 | FITC | 1:100 | Biolegend |
| Ly6-C | BV605 | 1:100 | Biolegend |
| Peyer's patch | |||
| CD11b | Pe/Cy5 | 1:100 | Biolegend |
| CD11c | APC | 1:100 | Biolegend |
| CD8a | eFluor 450 | 1:100 | eBioscience |
| MHCII | AF700 | 1:100 | eBioscience |
Flow Cytometry Data Acquisition
Single stained controls were made using UltraComp eBeads Compensation Beads (01-2222-41; Invitrogen) using the manufacturer’s instructions. Data acquisition was performed on a BD LSRFortessa (BD Biosciences, San Jose, CA), for every sample, 100,000 events were collected in the LiveCD45+ gate and cell populations were analyzed on FlowJo 10 (BD Biosciences).
H&E Staining of Liver Tissue and Steatosis Scoring
After euthanasia, mouse liver tissue was fixed in 10% buffered formalin at room temperature overnight. Tissues then were washed in methanol 2 × 30 minutes, ethanol 2 × 15 minutes, ethanol/xylene (1:1) for 15 minutes, and xylene 2 × 15 minutes, followed by embedding in paraffin. Tissues were sectioned at 5-μm thickness and stained with H&E using standard protocols. H&E-stained slides were assigned scores of 0–5, wherein a score of 0 was assigned for no discernable steatosis, and scores of 1–5 were assigned to grade modest to severe steatosis and hepatocyte ballooning.
Statistical Analysis
Statistical significance was evaluated via the Student t test or 1-way analysis of variance with Bonferroni multiple comparisons (version 8.2.0; GraphPad Prism software). For experiments in which conditions were defined in advance as positive or negative controls that were reproducing previous results, we used t tests. In other cases, for example, testing multiple conditions without a specific predefined outcome, we used analysis of variance with post hoc. Significance determined by t test was expressed using their P values, with brackets indicating significance P ≤ .05. Significance determined by analysis of variance is noted as either P ≤ .05, P ≤ .01, P ≤ .001, P ≤ .0001, or NS.
All authors had access to the study data and reviewed and approved the final manuscript.
Footnotes
Author contributions Hao Tran, Benoit Chassaing, and Andrew T. Gewirtz were responsible for the study conception and design; Hao Tran, Benoit Chassaing, and Andrew T. Gewirtz developed the methodology; Hao Tran, Alexis Bretin, Aneseh Adeshirlarijaney, Beng San Yeoh, and Jun Zou acquired the data; Hao Tran, Benoit Chassaing, and Andrew T. Gewirtz analyzed and interpreted the data; and Hao Tran, Alexis Bretin, Aneseh Adeshirlarijaney, Beng San Yeoh, Matam Vijay-Kumar, Jun Zou, Timothy L. Denning, Benoit Chassaing, and Andrew T. Gewirtz wrote, reviewed, and/or revised the manuscript.
Conflicts of interest The authors disclose no conflicts.
Funding This work was supported by National Institute of Diabetes and Digestive and Kidney Diseases grants DK099071 and DK083890 (A.T.G.), and by a Career Development Award from the Crohn's and Colitis Foundation of America (B.C.).
References
- 1.Fleissner C.K., Huebel N., Abd El-Bary M.M., Loh G., Klaus S., Blaut M. Absence of intestinal microbiota does not protect mice from diet-induced obesity. Br J Nutr. 2010;104:919–929. doi: 10.1017/S0007114510001303. [DOI] [PubMed] [Google Scholar]
- 2.Kubeck R., Bonet-Ripoll C., Hoffmann C., Walker A., Muller V.M., Schuppel V.L., Lagkouvardos I., Scholz B., Engel K.H., Daniel H., Schmitt-Kopplin P., Haller D., Clavel T., Klingenspor M. Dietary fat and gut microbiota interactions determine diet-induced obesity in mice. Mol Metab. 2016;5:1162–1174. doi: 10.1016/j.molmet.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rabot S., Membrez M., Blancher F., Berger B., Moine D., Krause L., Bibiloni R., Bruneau A., Gerard P., Siddharth J., Lauber C.L., Chou C.J. High fat diet drives obesity regardless the composition of gut microbiota in mice. Sci Rep. 2016;6:32484. doi: 10.1038/srep32484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miles J.P., Zou J., Kumar M.V., Pellizzon M., Ulman E., Ricci M., Gewirtz A.T., Chassaing B. Supplementation of low- and high-fat diets with fermentable fiber exacerbates severity of DSS-induced acute colitis. Inflamm Bowel Dis. 2017;23:1133–1143. doi: 10.1097/MIB.0000000000001155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zou J., Chassaing B., Singh V., Pellizzon M., Ricci M., Fythe M.D., Kumar M.V., Gewirtz A.T. Fiber-mediated nourishment of gut microbiota protects against diet-induced obesity by restoring IL-22-mediated colonic health. Cell Host Microbe. 2018;23:41–53 e4. doi: 10.1016/j.chom.2017.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Desvergne B. PPARdelta/beta: the lobbyist switching macrophage allegiance in favor of metabolism. Cell Metab. 2008;7:467–469. doi: 10.1016/j.cmet.2008.05.002. [DOI] [PubMed] [Google Scholar]
- 7.Hevener A.L., Olefsky J.M., Reichart D., Nguyen M.T., Bandyopadyhay G., Leung H.Y., Watt M.J., Benner C., Febbraio M.A., Nguyen A.K., Folian B., Subramaniam S., Gonzalez F.J., Glass C.K., Ricote M. Macrophage PPAR gamma is required for normal skeletal muscle and hepatic insulin sensitivity and full antidiabetic effects of thiazolidinediones. J Clin Invest. 2007;117:1658–1669. doi: 10.1172/JCI31561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Odegaard J.I., Ricardo-Gonzalez R.R., Goforth M.H., Morel C.R., Subramanian V., Mukundan L., Red Eagle A., Vats D., Brombacher F., Ferrante A.W., Chawla A. Macrophage-specific PPARgamma controls alternative activation and improves insulin resistance. Nature. 2007;447:1116–1120. doi: 10.1038/nature05894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weisberg S.P., McCann D., Desai M., Rosenbaum M., Leibel R.L., Ferrante A.W., Jr. Obesity is associated with macrophage accumulation in adipose tissue. J Clin Invest. 2003;112:1796–1808. doi: 10.1172/JCI19246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu H., Barnes G.T., Yang Q., Tan G., Yang D., Chou C.J., Sole J., Nichols A., Ross J.S., Tartaglia L.A., Chen H. Chronic inflammation in fat plays a crucial role in the development of obesity-related insulin resistance. J Clin Invest. 2003;112:1821–1830. doi: 10.1172/JCI19451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kawasaki N., Asada R., Saito A., Kanemoto S., Imaizumi K. Obesity-induced endoplasmic reticulum stress causes chronic inflammation in adipose tissue. Sci Rep. 2012;2:799. doi: 10.1038/srep00799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cani P.D., Amar J., Iglesias M.A., Poggi M., Knauf C., Bastelica D., Neyrinck A.M., Fava F., Tuohy K.M., Chabo C., Waget A., Delmee E., Cousin B., Sulpice T., Chamontin B., Ferrieres J., Tanti J.F., Gibson G.R., Casteilla L., Delzenne N.M., Alessi M.C., Burcelin R. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes. 2007;56:1761–1772. doi: 10.2337/db06-1491. [DOI] [PubMed] [Google Scholar]
- 13.Backhed F., Ding H., Wang T., Hooper L.V., Koh G.Y., Nagy A., Semenkovich C.F., Gordon J.I. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci U S A. 2004;101:15718–15723. doi: 10.1073/pnas.0407076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rabot S., Membrez M., Bruneau A., Gerard P., Harach T., Moser M., Raymond F., Mansourian R., Chou C.J. Germ-free C57BL/6J mice are resistant to high-fat-diet-induced insulin resistance and have altered cholesterol metabolism. FASEB J. 2010;24:4948–4959. doi: 10.1096/fj.10-164921. [DOI] [PubMed] [Google Scholar]
- 15.Fiebiger U., Bereswill S., Heimesaat M.M. Dissecting the interplay between intestinal microbiota and host immunity in health and disease: lessons learned from germfree and gnotobiotic animal models. Eur J Microbiol Immunol (Bp) 2016;6:253–271. doi: 10.1556/1886.2016.00036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wymore Brand M., Wannemuehler M.J., Phillips G.J., Proctor A., Overstreet A.M., Jergens A.E., Orcutt R.P., Fox J.G. The altered Schaedler flora: continued applications of a defined murine microbial community. ILAR J. 2015;56:169–178. doi: 10.1093/ilar/ilv012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bolus W.R., Peterson K.R., Hubler M.J., Kennedy A.J., Gruen M.L., Hasty A.H. Elevating adipose eosinophils in obese mice to physiologically normal levels does not rescue metabolic impairments. Mol Metab. 2018;8:86–95. doi: 10.1016/j.molmet.2017.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bolus W.R., Kennedy A.J., Hasty A.H. Obesity-induced reduction of adipose eosinophils is reversed with low-calorie dietary intervention. Physiol Rep. 2018;6 doi: 10.14814/phy2.13919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu D., Molofsky A.B., Liang H.E., Ricardo-Gonzalez R.R., Jouihan H.A., Bando J.K., Chawla A., Locksley R.M. Eosinophils sustain adipose alternatively activated macrophages associated with glucose homeostasis. Science. 2011;332:243–247. doi: 10.1126/science.1201475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elgazar-Carmon V., Rudich A., Hadad N., Levy R. Neutrophils transiently infiltrate intra-abdominal fat early in the course of high-fat feeding. J Lipid Res. 2008;49:1894–1903. doi: 10.1194/jlr.M800132-JLR200. [DOI] [PubMed] [Google Scholar]
- 21.Nagareddy P.R., Kraakman M., Masters S.L., Stirzaker R.A., Gorman D.J., Grant R.W., Dragoljevic D., Hong E.S., Abdel-Latif A., Smyth S.S., Choi S.H., Korner J., Bornfeldt K.E., Fisher E.A., Dixit V.D., Tall A.R., Goldberg I.J., Murphy A.J. Adipose tissue macrophages promote myelopoiesis and monocytosis in obesity. Cell Metab. 2014;19:821–835. doi: 10.1016/j.cmet.2014.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Talukdar S., Oh D.Y., Bandyopadhyay G., Li D., Xu J., McNelis J., Lu M., Li P., Yan Q., Zhu Y., Ofrecio J., Lin M., Brenner M.B., Olefsky J.M. Neutrophils mediate insulin resistance in mice fed a high-fat diet through secreted elastase. Nat Med. 2012;18:1407–1412. doi: 10.1038/nm.2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chassaing B., Miles-Brown J., Pellizzon M., Ulman E., Ricci M., Zhang L., Patterson A.D., Vijay-Kumar M., Gewirtz A.T. Lack of soluble fiber drives diet-induced adiposity in mice. Am J Physiol Gastrointest Liver Physiol. 2015;309:G528–G541. doi: 10.1152/ajpgi.00172.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vijay-Kumar M., Aitken J.D., Carvalho F.A., Cullender T.C., Mwangi S., Srinivasan S., Sitaraman S.V., Knight R., Ley R.E., Gewirtz A.T. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science. 2010;328:228–231. doi: 10.1126/science.1179721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chassaing B., Koren O., Goodrich J.K., Poole A.C., Srinivasan S., Ley R.E., Gewirtz A.T. Dietary emulsifiers impact the mouse gut microbiota promoting colitis and metabolic syndrome. Nature. 2015;519:92–96. doi: 10.1038/nature14232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Caesar R., Tremaroli V., Kovatcheva-Datchary P., Cani P.D., Backhed F. Crosstalk between gut microbiota and dietary lipids aggravates WAT inflammation through TLR signaling. Cell Metab. 2015;22:658–668. doi: 10.1016/j.cmet.2015.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ertunc M.E., Hotamisligil G.S. Lipid signaling and lipotoxicity in metaflammation: indications for metabolic disease pathogenesis and treatment. J Lipid Res. 2016;57:2099–2114. doi: 10.1194/jlr.R066514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Membrez M., Blancher F., Jaquet M., Bibiloni R., Cani P.D., Burcelin R.G., Corthesy I., Mace K., Chou C.J. Gut microbiota modulation with norfloxacin and ampicillin enhances glucose tolerance in mice. FASEB J. 2008;22:2416–2426. doi: 10.1096/fj.07-102723. [DOI] [PubMed] [Google Scholar]
- 29.Reijnders D., Goossens G.H., Hermes G.D., Neis E.P., van der Beek C.M., Most J., Holst J.J., Lenaerts K., Kootte R.S., Nieuwdorp M., Groen A.K., Olde Damink S.W., Boekschoten M.V., Smidt H., Zoetendal E.G., Dejong C.H., Blaak E.E. Effects of gut microbiota manipulation by antibiotics on host metabolism in obese humans: a randomized double-blind placebo-controlled trial. Cell Metab. 2016;24:63–74. doi: 10.1016/j.cmet.2016.06.016. [DOI] [PubMed] [Google Scholar]
- 30.Chassaing B., Raja S.M., Lewis J.D., Srinivasan S., Gewirtz A.T. Colonic microbiota encroachment correlates with dysglycemia in humans. Cell Mol Gastroenterol Hepatol. 2017;4:205–221. doi: 10.1016/j.jcmgh.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee Y.S., Li P., Huh J.Y., Hwang I.J., Lu M., Kim J.I., Ham M., Talukdar S., Chen A., Lu W.J., Bandyopadhyay G.K., Schwendener R., Olefsky J., Kim J.B. Inflammation is necessary for long-term but not short-term high-fat diet-induced insulin resistance. Diabetes. 2011;60:2474–2483. doi: 10.2337/db11-0194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Winter S.E., Lopez C.A., Baumler A.J. The dynamics of gut-associated microbial communities during inflammation. EMBO Rep. 2013;14:319–327. doi: 10.1038/embor.2013.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bretin A., Carriere J., Dalmasso G., Bergougnoux A., B'Chir W., Maurin A.C., Muller S., Seibold F., Barnich N., Bruhat A., Darfeuille-Michaud A., Nguyen H.T. Activation of the EIF2AK4-EIF2A/eIF2alpha-ATF4 pathway triggers autophagy response to Crohn disease-associated adherent-invasive Escherichia coli infection. Autophagy. 2016;12:770–783. doi: 10.1080/15548627.2016.1156823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cho K.W., Morris D.L., Lumeng C.N. Flow cytometry analyses of adipose tissue macrophages. Methods Enzymol. 2014;537:297–314. doi: 10.1016/B978-0-12-411619-1.00016-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harusato A., Geem D., Denning T.L. Macrophage isolation from the mouse small and large intestine. Methods Mol Biol. 2016;1422:171–180. doi: 10.1007/978-1-4939-3603-8_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.De Jesus M., Ahlawat S., Mantis N.J. Isolating and immunostaining lymphocytes and dendritic cells from murine Peyer's patches. J Vis Exp. 2013;73 doi: 10.3791/50167. [DOI] [PMC free article] [PubMed] [Google Scholar]