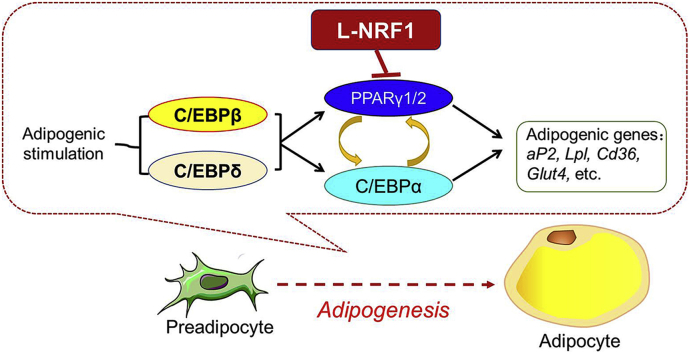
Abstract
Nuclear factor erythroid 2-related factor 1 (NRF1), a ubiquitously expressed CNC-bZIP transcription factor, plays a critical role in white adipocyte (WAC) biology, whereas the underlying mechanisms remain unknown. The mouse Nrf1 gene is transcribed in a number of alternatively spliced forms, resulting in two long protein isoforms (L-NRF1) containing 741 and 742 amino acids (aa) and multiple short isoforms (S-NRF1). Our previous study found that adipocyte-specific knockout of Nrf1 [Nrf1(f)-KO] in mice disturbs the expression of lipolytic genes in adipocytes, leading to adipocyte hypertrophy followed by inflammation, pyroptosis and insulin resistance. In the present study, we found that the stromal vascular fraction (SVF) cells isolated from white adipose tissues (WAT) of Nrf1(f)-KO mice display augmented adipogenesis showing elevated mRNA and protein expression of adipogenic markers and lipid accumulation. In 3T3-L1 cells, stable knockdown (KD) of all or long isoforms of Nrf1 (termed as A-Nrf1-KD and L-Nrf1-KD, respectively) using lentiviral shRNAs resulted in enhanced and accelerated adipogenic differentiation. Conversely, overexpression of L-NRF1-741, but not any of the S-NRF1, substantially attenuated adipogenesis in 3T3-L1 cells. These findings indicate that L-NRF1 might serve as a critical negative regulator of adipogenesis. Mechanistic investigation revealed that L-NRF1 may negatively regulates the transcription of peroxisome proliferator-activated receptor γ (PPARγ), in particular the master regulator of adipogenesis PPARγ2. Taken all together, the findings in the present study provide further evidence for a novel role of NRF1 beyond its participation in cellular antioxidant response and suggest that L-NRF1 is a negative regulator of PPARγ2 expression and thereby can suppress adipogenesis.
Keywords: NRF1, Adipogenesis, PPARγ, White adipocytes
Graphical abstract
Highlights
-
•
SVF cells isolated from WAT of Nrf1(f)-KO mice displayed augmented adipogenesis.
-
•
Stable silencing of L-Nrf1 in 3T3-L1 cells resulted in enhanced and accelerated adipogenesis.
-
•
Overexpression of L-NRF1-741, but not S-NRF1s, attenuated adipogenesis in 3T3-L1 cells.
-
•
L-NRF1 suppressed adipogenesis via downregulating PPARγ2 expression.
Abbreviations
- ADPSIN
Adiposin
- aP2
adipocyte Protein 2
- ARE
Antioxidant response element
- BAC
Brown adipocyte
- BAT
Brown adipose tissue
- CD36
Cluster of differentiation 36
- C/EBPs
CCAAT enhancer-binding proteins
- CHOP
C/EBP homologous protein
- CREB
cAMP response element-binding protein
- CREB3L4
cAMP responsive element binding protein 3 like 4
- Cont
Control
- DMIRI
Cocktail of dexamethasone 3-isobutyl-1-methylxanthine, insulin, rosiglitazone and indomethacin
- EBF1
Early B-cell factor 1
- FBS
Fetal bovine serum
- GCLC
Glutamate–cysteine ligase catalytic subunit
- GCLM
Glutamate-cysteine ligase regulatory subunit
- Glut4
Glucose transporter type 4
- HO-1
Hemeoxygenase 1
- KD
Knockdown
- KLF5 & 9
Krüppel-like factor 5 and 9
- Lpl
Lipoprotein lipase
- LRP16
Leukemia-related protein 16
- MAP4K4/NIK
Mitogen-activated protein kinase kinase kinase kinase 4/Nck-interacting kinase
- NFI
Nuclear factor I
- NQO1
NAD(P)H quinone oxidoreductase1
- NRF1
Nuclear factor-E2-related factor 1
- NRF2
Nuclear factor-E2-related factor 2
- OE
Overexpression
- ORO staining
Oil-red O staining
- Pgc1α/β
Peroxisome proliferator-activated receptor γ coactivator-α/β
- PPARγ
Peroxisome proliferator-activated receptor γ
- Rosi
rosiglitazone
- SCR
Scramble
- SREBP1
Sterol regulatory element-binding protein-1
- STAT5
Signal transducer and activator of transcription 5
- WAT
White adipose tissue; ZFP423, Zinc finger protein 423
1. Introduction
White adipose tissue (WAT), which contains adipocytes, preadipocytes, vascular cells and inflammatory and immune cells, is a dynamic and active organ that plays critical roles in regulating energy homeostasis [1]. Adipocytes, specialized for storing energy as triglycerol, are derived from mesenchymal stem cells through adipogenesis, a process of cell differentiation by which adipogenic precursor cells, so called preadipocytes, become adipocytes [2,3]. Adipogenesis is regulated by a complex network of transcription factors that coordinate expression of hundreds of proteins responsible for committing the mature adipocyte phenotype [2,4,5]. While it was firmly established that peroxisome proliferator-activated receptor γ (PPARγ) and CCAAT/enhancer-binding protein α (C/EBPα) are the terminal transcription factors responsible for adipogenic differentiation and the maintenance of the adipocyte phenotype, the signaling cascades underlying adipogenesis, especially the early events, are not fully understood.
PPARγ proteins, the principle regulator of adipogenesis, exist in two forms: PPARγ1 and PPARγ2. Both isoforms are produced by a combination of different promoter usage and alternative splicing [6]. The transcription of Pparγ is primarily under regulation of C/EBPs, a group of leucine zipper transcription factors, including C/EBPα, C/EBPβ and C/EBPδ [2,4,[7], [8], [9]]. In addition, multiple transcription factors, including early B-cell factor 1 (EBF1), Krüppel-like factor (KLF) 5 and 9, sterol regulatory element-binding protein-1 (SREBP1), zinc finger protein 423 (ZFP423), signal transducer and activator of transcription 5 (STAT5) and nuclear factor I (NFI), also play critical roles in PPARγ expression during adipogenesis [2]. Furthermore, a number of epigenetic factors have been found to modulate PPARγ expression and adipogenesis [2,6]. In contrast, only few negative regulators of PPARγ expression, such as GATA-2/3, C/EBP homologous protein (CHOP), hypoxia-inducible factor 1 (HIF1) and KLF2, have been reported [7,10]. Given the importance of PPARγ in the highly orchestrated adipogenic process, identifying novel regulators of PPARγ expression in preadipocytes is critically important.
Nuclear factor erythroid 2-related factor 1 (NRF1, also known as NFE2L1/LCRF1/TCF11) belongs to the Cap ‘n’ Collar basic-region leucine zipper (CNC-bZIP) transcription factor family, which also includes NRF2, a master regulator of the antioxidant response [11,12]. NRF1 is ubiquitously expressed in a wide range of tissues including adipose tissues [13,14]. In addition to oxidant defense, multiple physiological roles for NRF1 have been revealed, including embryonic development [15,16], proteasome stability in the brain, liver and brown adipose tissue (BAT) [14,[17], [18], [19]], lipolysis in WAT [13,20], osteoblastogenesis [21,22] and lipid metabolism in the liver [18,23,24]. As with the human analog, the mouse Nrf1 gene contains ten exons (Fig. S1A) and is transcribed in a number of alternatively spliced forms, resulting in two long protein isoforms (L-NRF1) containing 741 and 742 amino acids (aa) and multiple short isoforms (S-NRF1) with 313, 453, 572 and 583 aa, respectively (Fig. S1B) [11,25,26]. In addition, posttranslational modifications, including glycosylation and proteolytic processing, play important roles in the transactivation and stabilization of various isoforms of NRF1. Our previous studies in human HaCaT keratinocytes and MIN6 pancreatic β cells found that L-NRF1 is involved in arsenite-induced antioxidant response and protection against the cytotoxicity of arsenite [25,27]. In contrast, the S-NRF1-453, which migrates on SDS page generating a 65 kDa band, was found to be a negative regulator of L-NRF1-mediated antioxidant response [28].
To investigate the physiological function of NRF1 in brown adipose tissue (BAT), Hotamisligil's group generated brown adipocyte (BAC)-specific Nrf1-knockout mice by crossing mice carrying floxed alleles of Nfe2l1 (Nrf1-Flox) and mice bearing an uncoupling protein 1 (Ucp1) promoter-driven Cre recombinase and found that deletion of Nrf1 in BAC results in endoplasmic reticulum (ER) stress, inflammation, diminished mitochondrial function and whitening of BAT [14]. More recently, we developed a line of adipocyte-specific Nrf1-knockout [Nrf1(f)-KO] mice by crossing Nrf1-Flox and adiponectin-Cre mice and found that Nrf1(f)-KO mice exhibit a dramatically reduced subcutaneous adipose tissue mass, insulin resistance, adipocyte hypertrophy and severe adipose inflammation [13]. These new findings implicated the crucial roles of NRF1 in adipocyte biology that were previously underestimated. However, the function of NRF1 in preadipocytes have not yet been explored. In the present study, we used stromal vascular fraction (SVF) cells isolated from WAT of Nrf1(f)-KO mice and 3T3-L1 preadipocytes with altered levels of various isoforms of NRF1 to study the roles of NRF1 in adipogenesis. We found that deletion of either long or all isoforms of NRF1 resulted in augmented expression of PPARγ and adipogenesis. Conversely, overexpression of L-NRF1-741, but not S-NRF1s, in 3T3-L1 cells led to attenuated adipogenic differentiation. Overall, these data provide novel evidence that L-NRF1 is a distinct negative regulator of adipogenesis.
2. Materials and methods
2.1. Chemicals & reagents
Dexamethasone (D1756), 3-isobutyl-1-methylxanthine (IBMX, I7018), insulin solution (human, I9278), indomethacin (I7378), sodium arsenite (71287), Oil-red O (ORO, 75087) and puromycin (P8833) were purchased from Sigma (St. Louis, MO). Rosiglitazone maleate was obtained from SmithKline Beecham Pharmaceuticals (London, UK). Culture media, fetal bovine serum (FBS), calf serum (CS) and penicillin/streptomycin were from Life Technologies (Grand Island, NY).
2.2. Animals
Nrf1(f)-KO and littermate control (Nrf1-Flox) mice used in the current study were obtained as detailed previously [13,29]. In brief, Nfe2l1(f)-KO mice were generated by crossing Nrf1-Flox mice with mice bearing an adiponectin gene promoter-driven Cre recombinase (Adipoq-Cre, 010803, Jackson Labs Technologies, Inc., Sacramento, CA) that specifically express Cre recombinase in adipocytes. All animal procedures were approved by the Animal Ethics Committee of China Medical University.
2.3. Cell culture and differentiation
3T3-L1 preadipocytes were purchased from ATCC (Manassas, VA) and cultured in high-glucose Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% CS, 50 U/ml penicillin and 50 μg/ml streptomycin (growth medium). To induce adipogenesis, the cells were cultured for 1 day after they became confluent (designated as day 0) using DMI or DMIRI protocols as detailed previously [8]. In brief, DMI protocol: 3T3-L1 preadipocytes were incubated with DMI medium containing 1 μM dexamethasone, 0.5 mM IBMX and 1 μg/ml insulin in DMEM with 10% FBS for 2 days. Then, the medium was changed to DMEM with 10% FBS and 1 μg/ml insulin (Insulin medium). The cells were maintained for an additional 3 days and the medium was refreshed every two days; DMIRI protocol: cells were differentiated by replacing growth medium with DMIRI differentiation medium containing above DMI compounds plus 1 μM rosiglitazone and 125 μM indomethacin in DMEM with 10% FBS for 3 days, then cells were maintained for additional 2 days in the same medium without additives but 10% FBS. Differentiation of preadipocytes to mature adipocytes was confirmed by ORO staining of lipid vesicles as detailed previously [8]. Representative images of ORO staining were quantified with Image J software.
The SVF cells were isolated from WAT of Nrf1(f)-KO and littermate control mice as described previously [30] and cultured in DMEM with 10% FBS, 50 U/ml penicillin and 50 μg/ml streptomycin. The DMI protocol described above was used to induce adipogenesis after the cultured SVF cells became confluent. All the cells were cultured at 37 °C in a 5% CO2 environment.
2.4. Lentiviral-based shRNA transduction
MISSION shRNA lentiviral vectors were purchased from Sigma. The packaging of lentiviral particles was performed based on the manufacturer's protocol as supplied. Lentiviral transduction of 3T3-L1 cells was performed with shRNA targeting either all or long isoforms of Nrf1 (termed as A-Nrf1 and L-Nrf1, respectively) (see Fig. S2) and compared to non-target negative control (SHC002V) as described previously [7]. The non-target shRNA control vector activates RNA-induced silencing complex and RNA interference pathway, but does not silence any mouse genes. In the current study, we define the cells transduced with the non-target shRNA control vector as Scramble cells in contrast to knockdown (KD) cells.
2.5. Lentiviral-based Nrf1 overexpression
The lentiviral transfer vectors encoding mouse NRF1-741, −583, −572 and −453 were generated by subcloning the cDNAs into a lentiviral vector pTK642, a gift from Dr. Tal Kafri, University of North Carolina at Chapel Hill. The cDNA of mouse NRF1-741 was cut from a pCMV-SPORT6-mouse Nrf1-741 plasmid (Open Biosystems, Waltham, MA). The cDNAs of mouse NRF1-583, −572 and −453 were generated by PCR amplification and cloned into XhoI and NheI sites in pTK642 vector. The vector encoding mouse NRF1-313 in lentiviral vector pLV07 (Biosettia, San Diago, CA) were generated by overlap extension PCR.
The lentiviral transfer vector DNA, psPAX2 packaging (Addgene, Watertown, MA) and pMD2.G envelope plasmid DNA (Addgene) were mixed at a respective ratio of 3:2:1. Thirty μg of the DNA mix was diluted to a final volume of 0.5 ml with distilled H2O and 125 μl of 1 M CaCl2 (Sigma). The resulting DNA solution was vigorously vortexed, during which, 0.5 ml 2× BES-buffered saline (280 mM NaCl, 50 mM HEPES, 1.5 mM Na2HPO4, pH 6.95) was added and incubated at room temperature for 30 min. Following incubation, the solution was mixed again by gentle vortexing, and then added dropwise into the HEK-293T cells (ATCC) cultured in 10-cm dish with 8 ml of DMEM without FBS. Two hours later, 1 ml FBS was added. Sixteen hours post-transfection, the medium was replaced with DMEM supplemented with 10% FBS and incubated for 36 h prior to the initial collection of viral supernatant. A second collection was made after a further 24 h. The conditioned medium from the two harvests were combined and cleared by centrifugation at 1,500 rpm for 5 min at 4 °C then passed through a 0.45 μm pore MILLEX GP filter with PES membrane (Millipore, Burlington, MA).
3T3-L1 cells were infected with the lentiviral preparations described above for 3 days, then the infected cells were selected using blasticidin (50 μg/ml) for additional 3–5 days, resulting in an approximately 70% rate of cells which were then considered stably expressing various isoforms of NRF1 due to the high efficiency of integration of lentiviral vector into mammalian genome and blasticidin resistance. The overexpression of NRF1 was confirmed using Western blot and RT-qPCR. In the current study, we define the cells transduced with negative control lentiviral vectors as Control cells (Cont) in contrast to cells overexpressing various isoforms of NRF1 (Nrf1-OE).
2.6. Quantitative real-time RT–PCR
Total mRNA was isolated with TRIzol (Life Technologies) according to manufacturer's instructions and then the potential DNA contamination was removed by running through an RNase-Free DNase Set and RNeasy Mini kit (Qiagen, Valencia, CA). The resultant DNA-free RNA was diluted in RNase-free H2O and quantified by Nanodrop (Thermo, Wilmington, DE) at 260 nm. RNA samples were stored at −80 °C until use. Total RNA was reversely transcribed with MuLV reverse transcriptase and Oligo d(T) primers (Applied Biosystems, Foster City, CA). The SYBR Green PCR Kit (Qiagen) was used for quantitative real-time RT-PCR analysis. The primers were designed using Primer Express (Applied Biosystems) and synthesized by MWG-BIOTECH Inc. (High Point, NC). The primer sequences are listed in Supplementary Table S1. Real-time fluorescence detection was carried out using an ABI PRISM 7900 HT Sequence Detector (Applied Biosystems). Analysis of data was performed as detailed previously [8], and 18s was used as loading control for normalization.
2.7. Western blot analysis
Isolation of cell fractions and Western blotting were performed as detailed previously [8]. Antibodies for NRF1 (sc-13031; 1:500), NRF2 (sc-13032; 1:500), C/EBPβ (sc-7962; 1:500) and C/EBPα (sc-61; 1:500) were obtained from Santa Cruz, Inc. (Santa Cruz, CA). Antibody for NRF1 (12936-1-AP; 1:1000) were from Proteintech Group, Inc. (Rosemont, IL). Antibodies for C/EBPδ (#2318; 1:1000), PPARγ (#2435; 1:1000), CREB (#9197; 1:1000) and p-CREB (#9198; 1:1000) were purchased from Cell Signaling Technology, Inc. (Danvers, MA). Antibody for V5 (R960-25; 1:3000) were from ThermoFisher Scientific, Inc. (Waltham, MA). Antibody for β-ACTIN (A1978; 1:2000) was purchased from Sigma. The molecular weight (MW) of each protein shown on immunoblot was estimated based on the MagicMark™ XP Western Protein Standard (Life Technologies) on 4–12% or 12% Tris-Glycine Gel (Life Technologies). Representative images of western blots were quantified with Image J software.
2.8. Pparγ2 promoter luciferase reporter assay
Serial 5′-deleted Pparγ2 promoter-driven luciferase reporters were designed as described previously [8]. The inserts with 2601, 1842, 1411, 934, 258 and 138 bp, which were designed starting from +85 bp, were amplified by PCR using mouse (C57BL/6J) genomic DNA as template and the BGLII and EcoRV-ended primers. The primer design was listed in Online Materials Table S2. These promoter sequences were digested with BGLII/EcoRV and ligated into BGLII/EcoRV-digested pGL4.10 [luc 2] vector (Promega, Madison, WI). As described in our previous study [31], the luciferase activity was measured by the Luciferase Reporter Assay System and normalized to cell viability determined by Non-Radioactive Cell-Proliferation Assay Kit (Promega).
2.9. Chromatin immunoprecipitation (ChIP) assay
ChIP analysis were performed using the SimpleChIP® Plus Sonication Chromatin IP Kit (#56383, Cell Signaling Technology) according to the manufacture's instruction. Briefly, 90% confluent 3T3-L1 cells overexpressing V5-tagged NRF1-741 were treated with DMI and 10 μM MG132 for 6 h, followed by cross-linking with 1% formaldehyde for 15 min at room temperature. Pelleted cross-linked cells were re-suspended and lysed with cell lysis buffer and nuclear lysis buffer sequentially and then followed by sonication for 6 times with 30s each time by using Q700 Sonicator (Qsonica Ilc., Newtown, CT). Ten micro gram of chromatin collected from centrifuged ultra-sonicated cell lysates was diluted at least 5 times in IP buffer. Two percent of diluted chromatin was preserved for input, and left was immunoprecipitated with antibody against V5 (R960-25, ThermoFisher) and non-specific mouse IgG overnight at 4 °C with rotation. Immune complex was recycled by magnetic beads and then washed by low and high salt buffer, respectively. Cross-linked DNA were reversed by addition of 2 μl proteinase K and 6 μl 5 M NaCl and incubated overnight at 65 °C after being eluted from magnetic beads. Enriched DNA were purified and then amplified by PCR for 34 cycles with 8 pairs of primers (Table S3) targeting continuous segments in the upstream of Pparg2 gene. PCR products were resolved with 1% agarose gels.
2.10. Statistical analyses
All statistical analyses were performed using Graphpad Prism 5 (GraphPad Software, San Diego, CA), with p < 0.05 considered as significant. More specific indices of statistical significance are indicated in individual figure legends. Data are expressed as mean ± SEM. For comparison between two groups, a Student's t-test was performed. For comparison among multiple groups, two-way ANOVA with Bonferroni post hoc testing was performed.
3. Results
3.1. SVF cells isolated from WAT of Nrf1(f)-KO mice show augmented adipogenesis
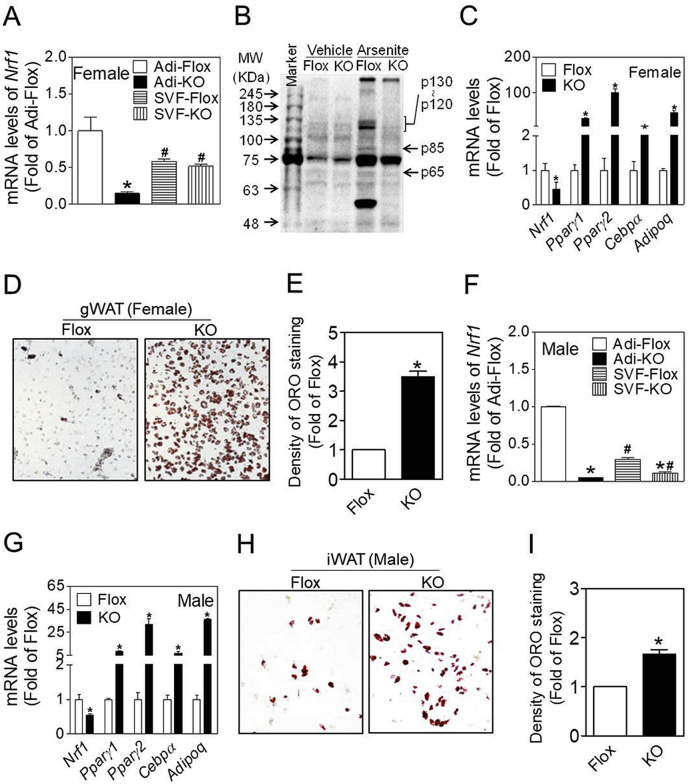
To assess that the adipocytes and SVF cells from Nrf1(f)-KO mice have reduced expression of Nrf1, we isolated and fractioned the gonadal WAT (gWAT) from female mice and inguinal WAT (iWAT) from male mice to measure the mRNA levels of Nrf1. Of note, female Nrf1(f)-KO mice had a robust adipose phenotype showing no subcutaneous adipose tissues left in adult mice [13]. Although the mRNA levels of Nrf1 in adipocyte fractions isolated from female and male Nrf1(f)-KO mice were much lower than those from Flox control mice, mRNA quantification in SVF cells from gWAT and iWAT demonstrated only a 10% and 60% reduction of Nrf1 expression, respectively (Fig. 1A and F). This meager reduction is likely due to the fact that SVF cells fractioned freshly from WAT are a mixture of fibroblasts, mesenchymal stem cells, endothelial cells, smooth muscle cells, macrophages, and others [32]. In particular, the WAT of Nrf1(f)-KO mice contains a much higher portion of macrophages than that from Flox control mice [13]. To further validate the knockout of Nrf1 in adipocytes, the protein levels of NRF1 in adipocyte fractions isolated from gWAT of Nrf1(f)-KO or Flox control mice were measured following 6 h culture in normal culture medium and arsenite-challenged condition, in which NFR1 can be activated [27]. As shown in Fig. 1B, the basal protein expression of NRF1 in adipocyte fractions, either isolated from gWAT of Nrf1(f)-KO or Flox control mice, is almost undetectable. In contrast, acute arsenite exposure in vitro dramatically increased the protein levels of NRF1 in adipocytes from Flox control mice, showing multi-bands increased on the immunoblot. As predicted, the adipocytes from gWAT of Nrf1(f)-KO mice showed no relevant bands induced by arsenite exposure. To further confirm the knockout of Nrf1 in SVF cells, the mRNA expression was also measured in the SVF cells after they were cultured in normal growth media to confluence and maintained for 5 days (Fig. 1C) or following adipogenic differentiation (Fig. 1G). In line with the key findings above, the SVF cells from Nrf1(f)-KO mice showed significantly decreased mRNA expression compared with the cells from Flox control mice.
Fig. 1.
Differentiation of SVF cells isolated from WAT of female and male mice with adipocyte-specific ablation of all isoforms of Nrf1. Animals were 16 weeks of age. Groups of n = 3 mice were used. (A and F) Relative mRNA expression of Nrf1 in adipocyte fractions and SVF of gonadal WAT (gWAT) of female mice (A) and inguinal WAT (iWAT) of male mice (F). Adi-Flox, Adi-KO, SVF-Flox and SVF-KO represent adipocyte fractions and SVF of Nrf1-Flox (Flox) and Nrf1(f)-KO (KO) mice, respectively. *p < 0.05 vs the same cell type of Flox, #p < 0.05 vs adipocytes with the same genotype. (B) NRF1 protein expression in adipocytes of gWAT of female Flox and KO mice. Adipocytes were cultured in normal growth medium with or without 5 μM sodium arsenite, a potent NRF1 inducer, for 6 h to increase expression of NRF1. (C) Basal mRNA expression of adipogenic factors in cultured SVF cells isolated from gWAT of female mice. The cells were maintained in growth medium without hormonal additives for 5 days post confluence. (D) Representative images (10 X) of ORO staining of the cells in (C). (E) Quantification of ORO staining in Fig. 1D. (G and H) mRNA expression of adipogenesis-related factors (G) and representative images (10 X) of ORO staining (H) of differentiated SVF cells isolated from iWAT of male mice. Cultured SVF cells were differentiated using DMIRI protocol (see Methods). *p < 0.05 vs Flox. (I) Quantification of ORO staining in Fig. 1H.
To determine the effect of deficiency of Nrf1 on adipogenesis, the mRNA expression of various adipogenic genes and lipid accumulation were measured in the SVF cells from WAT of Nrf1(f)-KO and Flox control mice. As shown in Fig. 1C and D, when the SVF cells from gWAT of female Nrf1(f)-KO mice were cultured to confluence in regular growth medium, the cells expressed significantly higher levels of adipogenic marker genes, including Pparγ1, Pparγ2, Cebpα and adiponectin (Adipoq) than those in Flox control cells (Fig. 1C). ORO staining of the cells showed dramatically increased levels of lipid accumulation in the cells from Nrf1(f)-KO mice compared to the cells from Flox controls (Fig. 1D and E). These findings clearly indicate that the SVF cells from gWAT of female Nrf1(f)-KO mice display a spontaneous adipogenic differentiation without DMI treatment. While the SVF cells from iWAT of male Nrf1(f)-KO mice showed no apparent spontaneous adipogenic differentiation, the SVF cells from iWAT of male Nrf1(f)-KO mice displayed dramatically increased mRNA expression of various key adipogenic genes and lipid accumulation compared to Flox control cells following a 5-day DMI protocol to induce adipogenesis (Fig. 1G–I).
3.2. Stable knockdown of A-Nrf1 or L-Nrf1 results in increased adipogenic potency in 3T3-L1 preadipocytes
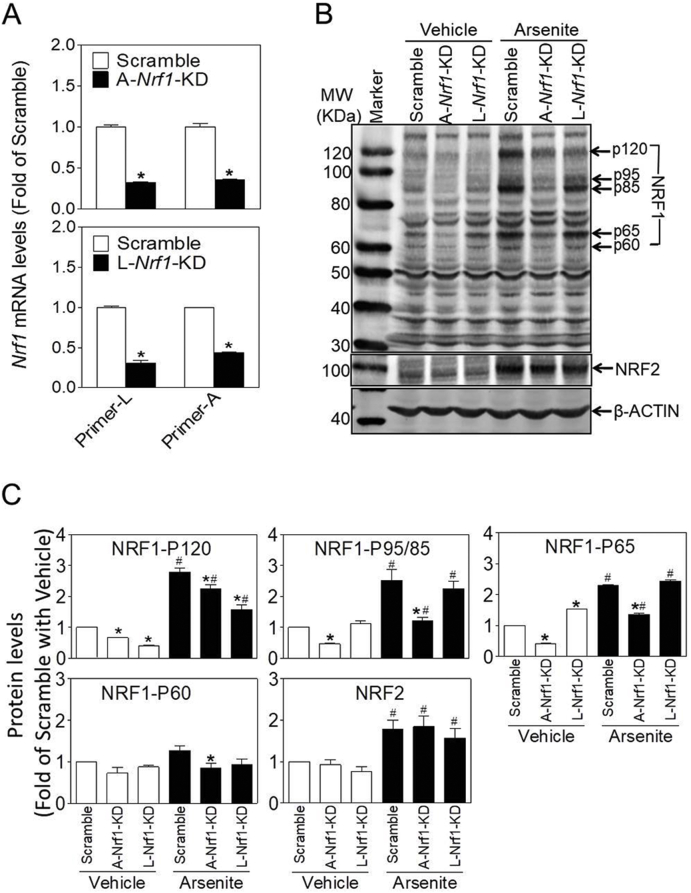
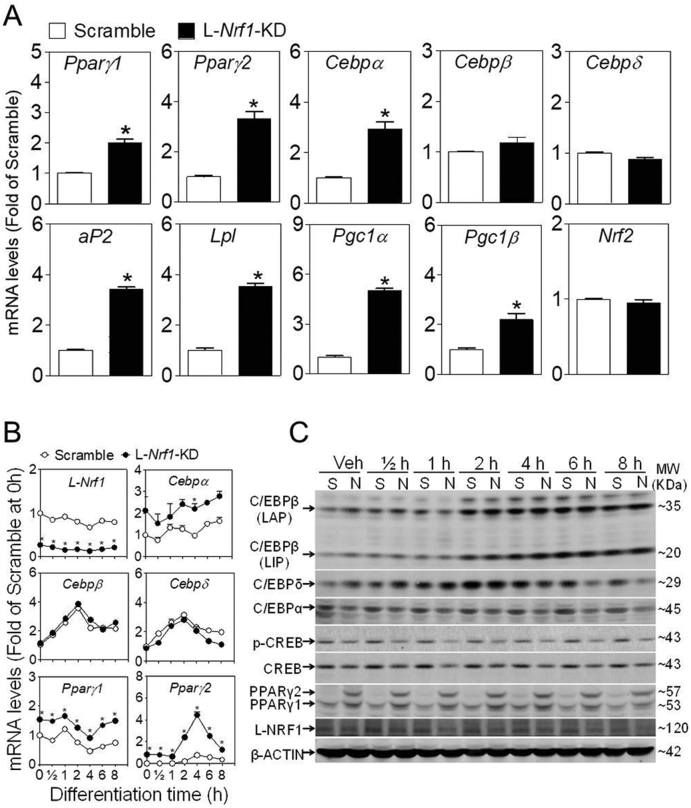
To verify the key findings in SVF cells from WAT of Nrf1(f)-KO mice and characterize the roles of various isoforms of NRF1 in adipogenesis, we used two sets of shRNA to specifically silence L-Nrf1 (termed as L-Nrf1-KD) or A-Nrf1 (termed as A-Nrf1-KD) in 3T3-L1 preadipocytes (Fig. S2). As shown in Fig. 2A, the mRNA expression of L-Nrf1 and A-Nrf1 were significantly decreased in A-Nrf1-KD (upper panel) and L-Nrf1-KD cells (lower panel). To further validate the specific silencing effect, the protein expression of NRF1 was measured under basal state (Vehicle) and arsenite-challenged condition in which NRF1 can be activated [25,27]. Western blot analysis showed that the basal protein expression of NRF1, which appears as multiple bands, including p120, p95, p85, p65 and p60 on the blot, was relatively low in Vehicle-treated cells, whereas arsenite treatment significantly induced their expression in Scramble cells (Fig. 2B and C). Compared with Scramble cells, A-Nrf1-KD cells had reduced levels of all isoforms of NRF1, in particular under arsenite-challenged condition. Remarkably, L-Nrf1-KD cells showed no diminished expression of most S-NRF1s except p120 (Fig. 2B and C), confirming the specificity of the shRNA against L-Nrf1. In agreement with previous studies [33], arsenite treatment significantly increased the expression of NRF2, although no differences were found with regard to this among Scramble, A-Nrf1-KD and L-Nrf1-KD cells. This suggests that the knockdown of Nrf1 did not affect the expression of NRF2 in 3T3-L1 cells (Fig. 2B and C).
Fig. 2.
Silencing of Nrf1 using two distinct shRNAs in 3T3-L1 cells. (A and B) Expression of mRNA (A) and protein (B) of NRF1 in 3T3-L1 cells transduced with lentiviral shRNA targeting against all or long isoforms of NRF1. A-Nrf1-KD, knockdown of all isoforms of Nrf1; L-Nrf1-KD, knockdown of L-Nrf1; Primer-L and Primer-A, the primer set detecting L-Nrf1 and A-Nrf1, respectively. Vehicle, medium; Arsenite, cells were treated with 10 μM arsenite for 6 h n = 3. *p < 0.05 vs. Scramble. (C) Quantification of protein levels in Fig. 2B. *p < 0.05 vs. Scramble under the same condition; #p < 0.05 vs. the same cell type in Vehicle group;
In line with the silencing effect, the expression of various antioxidant genes, including glutamate–cysteine ligase catalytic subunit (Gclc), glutamate-cysteine ligase regulatory subunit (Gclm) and NAD(P)H quinone oxidoreductase1 (Nqo1) but not hemeoxygenase1 (Ho1), was markedly attenuated in A-Nrf1-KD cells (Fig. S3A, upper panel), whereas only Gclc and Nqo1 were marginally decreased in L-Nrf1-KD cells (Fig. S3A, lower panel). In addition, both A-Nrf1-KD and L-Nrf1-KD cells were more sensitive than Scramble cells to the cytotoxicity induced by arsenite exposure for 24 or 48 h (Fig. S3B).
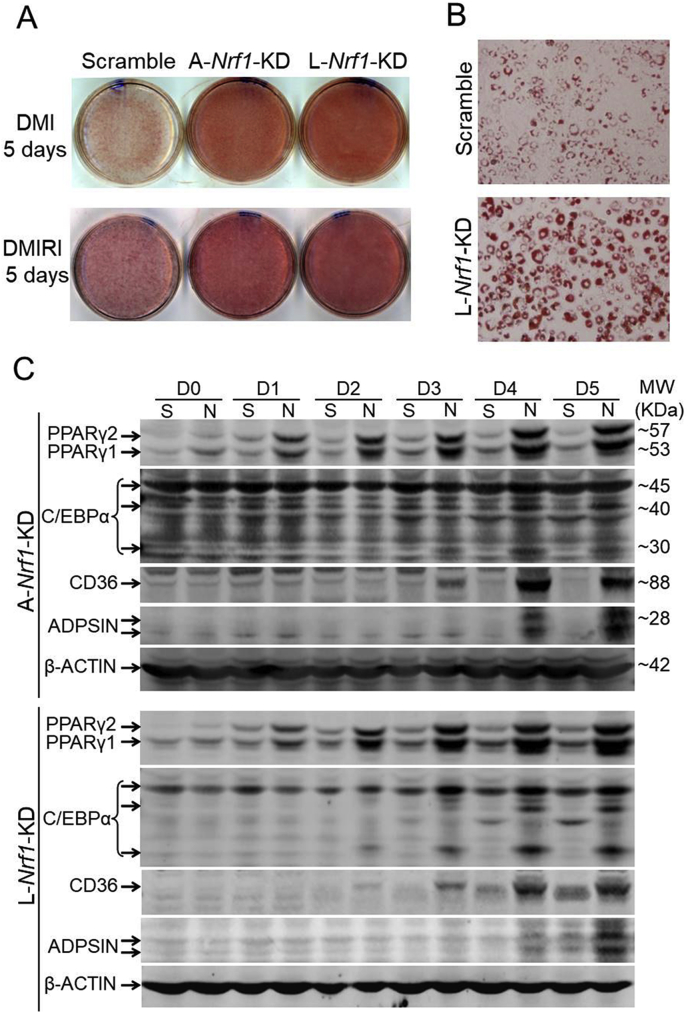
To investigate the potential role of NRF1 in adipogenesis, we cultured L-Nrf1-KD and A-Nrf1-KD 3T3-L1 cells in the medium with hormonal cocktail DMI or DMIRI to induce adipogenesis, followed by an ORO staining to monitor lipid accumulation. Compared with Scramble cells, both L-Nrf1-KD and A-Nrf1-KD cells displayed a significant increase of lipid accumulation following DMI or DMIRI treatment (Fig. 3A and B; Fig. S4), suggesting that L-NRF1, among all the isoforms, might play a dominant role in regulation of adipogenesis. To further understand the mechanisms underlying increased adipogenesis induced by L-NRF1 deficiency in 3T3-L1 cells, we examined key adipogenic markers during the differentiation process in both L-Nrf1-KD and A-Nrf1-KD cells. Western blot analyses showed elevated protein expression of PPARγ, C/EBPα, cluster of differentiation 36 (CD36) and adiposin (ADPSN) in both Nrf1 deficient cells in a time-dependent manner (Fig. 3C), indicating that L-NRF1 is likely involved in the differentiation of 3T3-L1 preadipocytes. Our remaining studies therefore mainly focused on the roles of L-NRF1 in adipogenesis.
Fig. 3.
Deficiency of A-Nrf1 or L-Nrf1 results in augmented adipogenesis in 3T3-L1 cells. (A and B) Representative images (A, 1X and B, 20X) of ORO staining of Scramble, A-Nrf1-KD and L-Nrf1-KD cells differentiated with DMI or DMIRI protocol. A representative result of 3–5 independent experiments is shown. (C) A-Nrf1-KD and L-Nrf1-KD cells show elevated protein expression of adipogenic markers during DMI-induced adipogenesis. D, Day; S, Scramble; N, A-Nrf1-KD or L-Nrf1-KD cells.
3.3. L-Nrf1-KD 3T3-L1 cells show elevated gene and/or protein expression of adipogenic factors under basal condition and during the early stage of differentiation
Among the factors involved in adipogenic differentiation, PPARγ is considered to be the master transcriptional regulator, while other critical factors such as C/EBPβ and δ, trigger the induction of PPARγ and C/EBPα at the early stage of adipogenesis [8]. To investigate the molecular details behind the enhanced adipogenesis caused by L-Nrf1 deficiency, we examined the basal mRNA expression of these adipogenic regulators in L-Nrf1-KD cells. The results showed that the basal expression of Pparγ and its downstream genes including Cebpα, adipocyte protein 2 (aP2), lipoprotein lipase (Lpl), peroxisome proliferator-activated receptor γ coactivator α/β (Pgc1α and Pgc1β), but not its upstream regulators such as Cebpβ, Cebpδ and Nrf2, were significantly elevated in L-Nrf1-KD cells under normal culture condition compared to Scramble cells (Fig. 4A), suggesting that L-Nrf1 silencing increases adipogenesis probably through upregulating the expression of Pparγ, specifically Pparγ2, the isoform that plays the dominant role in adipogenesis.
Fig. 4.
L-Nrf1-KD 3T3-L1 cells exhibits increased mRNA and protein expression of PPARγ under basal condition and during the early stage of adipogenic differentiation. The mRNA expression of adipogenic factors in Scramble and L-Nrf1-KD cells maintained in normal growth medium (A) and post DMI treatment for indicated time (B). The mRNA levels were measured by RT-qPCR. *p < 0.05 vs. Scramble at the same time. (C) The protein expression of C/EBPs and PPARγ1/2 in Scramble and L-Nrf1-KD cells treated with DMI for indicated time. Whole cell lysates (100 μg protein) were separated on 4–12% Tris-Glycine gels and detected using the antibodies against C/EBPs, CREBs, PPARγ1/2, NRF1 and β-ACTIN. S, Scramble; N, L-Nrf1-KD cells; Veh, Vehicle; h, hour; DMI, D, Dexamethasone; M, 3-isobutyl-1-methylxanthine (IBMX); I, Insulin. A representative result of 3-5 independent experiments is shown.
To further illustrate the effect of L-Nrf1 deficiency on adipogenesis, the expression of various key adipogenic regulators was assessed during the early stage of differentiation of 3T3-L1 cells. Both the mRNA and protein expression of C/EBPβ and δ showed a dramatic induction and reached peaks at 2–4 h post DMI treatment in both Scramble and L-Nrf1-KD cells, whereas no evident difference was observed between the cells (Fig. 4B and C), indicating that deficiency of L-Nrf1 does not affect their expression. In contrast, the expression of PPARγ, in particular PPARγ2, was remarkably increased in L-Nrf1-KD cells compared to Scramble cells under basal and DMI-treated conditions. This indicates that L-Nrf1 might play a crucial negative regulatory role in PPARγ expression and its knockdown released this inhibition. Of note, the protein levels of C/EBPα, cAMP response element-binding protein (CREB) and p-CREB in L-Nrf1-KD cells showed a slightly decrease compared to Scramble cells, further highlighting that L-Nrf1 deficiency-induced upregulation of PPARγ accounted for the enhanced adipogenesis in 3T3-L1 cells.
3.4. Overexpression of L-NRF1, but not S-NRF1, suppresses DMI-induced adipogenesis in 3T3-L1 cells
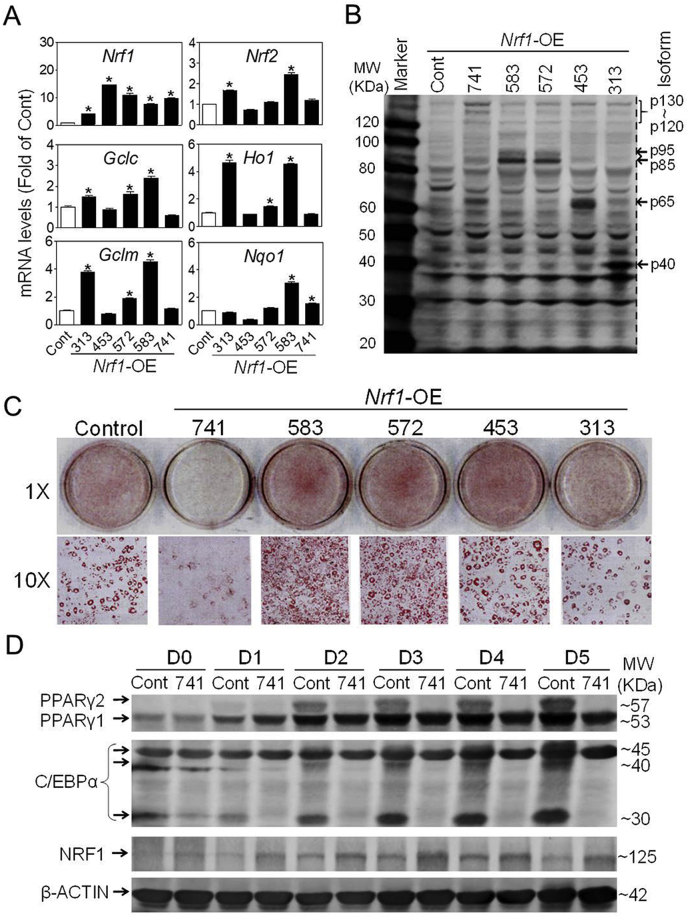
To further characterize the roles of various isoforms of NRF1 in adipogenesis, we generated multiple lines of 3T3-L1 cells, in which individual isoform of NRF1, including 313, 453, 572, 583 and 741 isoforms, was stably overexpressed (Nrf1-OE) using lentiviral vectors. As illustrated in Fig. 5, the Nrf1-OE cells showed dramatically elevated expression of various isoforms of NRF1 at mRNA (Fig. 5A) and protein (Fig. 5B; Fig. S5) levels. Remarkably, overexpression of isoform 313, 572 and 583, but not 453 and 741, resulted in elevated mRNA expression of various antioxidant genes, such as Gclc, Gclm, Ho1 and Nqo1 (Fig. 5A). In addition, overexpression of NRF1-313 or −583, but not other isoforms, in 3T3-L1 cells stably expressing an ARE-reporter [34] resulted in significantly elevated ARE activity compared to Control cells (Fig. S6). These results clearly indicate that the Nrf1-OE cells were successfully developed.
Fig. 5.
Effects of overexpression of various isoforms of NRF1 on adipogenesis in 3T3-L1 cells. (A) The mRNA expression of Nrf1, Nrf2, Gclc, Gclm, Nqo1 and Ho1 in various Nrf1-OE cells. Cont, Control; 313, 453, 572, 583 and 741 refer to the cells overexpressing indicated isoforms of Nrf1, respectively. n = 3. *p < 0.05 vs. Cont. (B) Representative image of protein expression of various isoforms of NRF1 in Nrf1-OE cells. (C) Oil-red O staining of Control and Nrf1-OE cells treated with DMI protocol for 5 days. (D) The protein expression of adipogenic factors in Nrf1-OE cells treated with DMI for indicated days. Cont, Control; 741, cells overexpressing NRF1-741; D, Day. A representative result of 3-5 independent experiments is shown. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
As expected, overexpression of L-NRF1-741 substantially suppressed DMI-induced adipogenic differentiation compared to Control group in 3T3-L1 preadipocytes (Fig. 5C; Fig. S7). In contrast, overexpression of NRF1-572 and −583 led to enhanced adipogenesis, whereas NRF1-313 and −453 overexpression caused no observable changes in the rate of lipid accumulation (Fig. 5C; Fig. S7), indicating that different isoforms of NRF1 play distinct regulatory roles in the differentiation process. To further validate the inhibitory effect of L-NRF1-741 overexpression on adipogenesis, we evaluated protein profile of key adipogenic markers during adipogenesis. The results showed that PPARγ, PPARγ2 in particular, and C/EBPα levels were significantly lower in L-NRF1-741-overexpressed cells compared to Control during DMI-induced adipogenesis (Fig. 5D). Of note, L-NRF1 continued to increase along the process of differentiation (Fig. 5D).
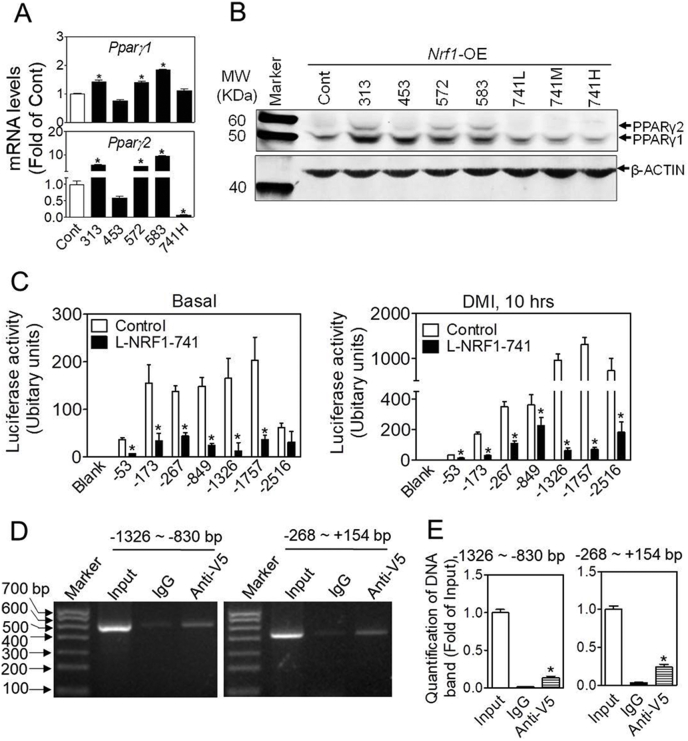
3.5. Overexpression of L-NRF1 suppresses PPARγ expression in 3T3-L1 cells
To further characterize the molecular details underlying the adipogenic phenotype induced by overexpression of various isoforms of NRF1, we measured the basal mRNA expression of adipogenic factors in Nrf1-OE cells. As shown in Fig. 6A and Fig. S8, overexpression of L-NRF1-741 significantly reduced mRNA expression of Pparγ2, Cebpα, adipsin and glucose transporter type 4 (Glut4), which is consistent with the findings in L-Nrf1-KD cells where Nrf1 deficiency resulted in upregulation of these genes (Fig. 4A), solidifying the premise that L-NRF1 is a negative regulator of Pparγ2. In contrast, overexpression of NRF1-313, −572, −583, but not −453, substantially increased the mRNA expression of Pparγ1/2, Cebpα, Cebpδ, aP2, Cd36 and Glut4, suggesting that L-NRF1 and S-NRF1 might play opposite regulatory roles in adipogenesis. In addition, Western blotting results further confirmed that S-NRF1s and L-Nrf1-741 play contrary roles in the expression of PPARγ under basal condition (Fig. 6B).
Fig. 6.
Overexpression of L-NRF1-741 suppresses the expression of PPARγ in 3T3-L1 cells. (A) The mRNA expression of Pparγ in control and various Nrf1-OE cells under normal culture condition. Cont (Control), 313, 453, 572, 583, and 741 refer to the cells overexpressing indicated isoforms of NRF1. *p < 0.05 vs. Control. n = 3-5. (B) Representative image of Western blot detecting PPARγ1/2 in control and various Nrf1-OE cells under basal condition. 741L/M/H refer to the cells overexpressing L-NRF1-741 using low (L), medium (M), and high (H) concentrations of lentiviral vector. n = 3. (C) The activity of Pparγ2 promoter-reporter assay. Control and NRF1-741 cells were transfected with the Pparγ2 promoter-luciferase constructs. After 2 days, the transfected cells were treated with basal growth medium (left panel) and DMI differentiation media (right panel) for 10 h. Blank, no insert of reporter; −53, −173, −267, −849, −1326, −1757 and −2516 represent the reporters driven by the 5′-truncated Pparγ2 promoter. n = 3–6; *p < 0.05 vs. Control expressing the same reporter. (D) Representative images of ChIP analysis for the interaction between NRF1-741 and promoter region of Pparg2 gene. 3T3-L1 cells overexpressing V5-tagged NRF1-741 were treated with DMI and 10 μM MG132 for 6 h when reaching 90% confluence, followed by ChIP assay. Input, enriched DNA by mouse IgG and V5 antibody were amplified by PCR with 8 pairss of primers designed to flank the continuous 3,000 bp range in the upstream of Pparγ2 gene. Non-immunoprecipitated chromatin (2%) was used as input control. PCR products were resolved with 1% DNA gel. n = 3. (E) Quantification of DNA bands in Fig. D. n = 3; *p < 0.05 vs. 2% input. (E) Quantification of DNA bands in Fig. D. n = 3; *p < 0.05 vs. 2% input.
To investigate the potential participation of NRF1 in the transcriptional regulation of Pparγ2, we analyzed the promoter sequence of Pparγ2 and identified multiple consensus ARE and ARE-like sites between −2516 bp and +1 bp region (Fig. S9). To further determine whether L-NRF1 is directly involved in the transcriptional regulation of Pparγ2, we developed a series of 5′-truncated Pparγ2 promoter-driven luciferase reporters and measured the activities in 3T3-L1 cells under basal and DMI-treated conditions. As shown in Fig. 6C, overexpression of L-NRF1-741 in 3T3-L1 cells significantly attenuated the activities of the luciferase reporters driven by −53, −173, −267, −849, −1326, −1757 and −2516 bp promoters under both basal (left panel) and/or DMI-treated conditions (right panel) in contrast to Control cells.
To study the physical interaction between long isoform NRF1 and promoter sequence of Pparγ2 gene, we performed ChIP analysis in 3T3-L1 cells overexpressing V5-tagged NRF1-741 (Fig. S10). As shown in Fig. 6D and E, following 6 h culture in the medium containing DMI and 10 μM MG132, a direct binding of NRF1-741 to DNA fragments range from −1326 to −830 bp and −268 to +154 bp in the upstream of Pparγ2 gene were observed, indicating that NRF1-741 might bind to Pparγ2 promoter to regulate its transcription.
4. Discussion
The impairment in WAT formation and/or dysfunction of WAC are associated with various metabolic disorders [3]. Our recent study showed that Adipoq-Cre-derived adipocyte-specific knockout of Nrf1 in mice results in disturbed expression of lipolytic genes in adipocytes, leading to adipocyte hypertrophy followed by severe inflammation, pyroptosis and insulin resistance, implicating an important role of NRF1 in adipocyte biology [13]. Here, we focused on the roles of NRF1 in the process of adipogenesis by which preadipocytes differentiate into mature adipocytes. We found that the SVF cells isolated from iWAT of male Nrf1(f)-KO mice, in which the expression and function of all isoforms of NRF1 were diminished in adiponectin-expressing cells, promoted adipogenesis induced by hormonal cocktails. Remarkably, the SVF cells isolated from gWAT of female Nrf1(f)-KO mice, which exhibit a more severe adipose phenotype than male mice [13], showed a spontaneous adipogenic differentiation even without DMI treatment. In 3T3-L1 cells, silencing of either all or long isoforms of NRF1 resulted in enhanced and accelerated adipogenic differentiation induced by hormonal cocktails. Conversely, overexpression of L-NRF1-741, but not S-NRF1s, substantially attenuated adipogenesis in 3T3-L1 cells. These findings clearly revealed that L-NRF1 might serve as a critical negative regulator of adipogenesis. Mechanistic studies suggest that L-NRF1 might dominate over S-NRF1s and negatively regulates the expression of PPARγ2 under basal and hormonal cocktail-treated conditions. Taken all together, the findings in the present study demonstrate a novel biological role of NRF1 beyond its participation in cellular antioxidant responses and protein homeostasis and suggest that L-NRF1 is a negative regulator of PPARγ expression and thereby can suppress adipogenesis.
Adipogenesis can be commonly initiated in vitro by treating preadipocytes with hormonal cocktails, such as DMI or DMIRI. Subsequently the early responders C/EBPβ and C/EBPδ might be upregulated during the early-stage of differentiation and eventually trigger the activation of PPARγ which forms a positive feedback loop with C/EBPα [8,35,36]. Among this adipogenic network, the transcription and activation of PPARγ have been proven to be the central events and are essential for adipogenesis [2,37]. Therefore, identifying novel factors that can directly or indirectly regulate PPARγ expression might provide valuable insight into overall adipogenic mechanisms. Previous studies including our own identified numerous positive regulators of PPARγ, such as C/EBPs and NRF2 [7,8]. More recently, cAMP responsive element binding protein 3 like 4 (CREB3L4), leukemia-related protein 16 (LRP16), mitogen-activated protein kinase kinase kinase kinase 4/Nck-interacting kinase (MAP4K4/NIK), and several microRNAs have been found to regulate the protein stability of C/EBPβ [38], activate mTOR signaling [39], control the transcription of PPARγ [40], and target the 3′-untranslated region of PPARγ [41,42], respectively. However, only few negative regulators, including GATA-2/3, CHOP, HIF1 and KLF2, have been reported [43,44]. In the current study, we found that both A-Nrf1-KD and L-Nrf1-KD 3T3-L1 cells exhibit attenuated expression of PPARγ at mRNA and protein levels, even under basal culture condition. Conversely, overexpression of L-NRF1-741 in 3T3-L1 cells substantially attenuated the expression of PPARγ and adipogenesis, but had no effect on the early-stage adipogenic factors. These findings clearly indicate that L-NRF1 might function as a negative regulator on the transcription of Pparγ and thus adipogenesis. Given the dominant roles that L-NRF1 plays in response to various stresses [14,17,18,27,45], we expected that L-NRF1 might play a crucial regulatory role in adipogenesis under various stressed conditions.
C/EBPβ and C/EBPδ have been recognized as key transcription factors regulating Pparγ expression at the early stage of adipogenesis [2,5]. Identifying the factors that regulate C/EBPβ and C/EBPδ activity as well as cooperate with other C/EBPs in an adipogenic-specific manner should provide additional insight into the mechanisms regulating adipogenesis. However, we did not find any significant effects of Nrf1 silencing, either A-Nrf1-KD or L-Nrf1-KD, on C/EBPβ expression during the early stage of adipogenesis. In our previous study, we have demonstrated that NRF2, another transcription factor of CNC-bZIP family, is involved in adipogenesis in 3T3-L1 cells via regulating the transcription of Cebpβ and Pparγ [7,8]. In contrast, the results in the current study strongly suggest a negative role of L-NRF1 in regulation of adipogenesis. This is in contrast to the positive effects of NRF2 on adipocyte differentiation, implying multiple distinct functions of members of the CNC-bZIP family in adipocyte biology. Moreover, we did not observe the crosstalk between NRF1 and NRF2 through determining the expression of NRF2 in Nrf1-KD cells (Fig. 2B) and ARE activity in NRF1-741 overexpression cells (Fig. S6B), indicating that NRF2 may not be involved in the repression of NRF1 on adipogenesis. Although both NRF1 and NRF2 belong to the CNC-bZIP family, exhibit similar antioxidant function, and even share the same binding sites, the precise role of NRF1 in adipogenesis is poorly defined, probably due to the complexity of metabolic interactions and the multiple isoforms of NRF1. NRF1 and C/EBPs belong to bZIP proteins which need binding partners for their transcriptional activity [11,[46], [47], [48], [49]]. Interestingly, L-NRF1 has been reported to bind with C/EBPβ to regulate odontoblast differentiation [21]. In the current study, we found that L-NRF1-741 might directly bind to the promoter region of Pparγ2 gene to repress its transcription. However, the exact mechanism for the suppression and potential binding partners of L-NRF1 in the process still need further investigation.
It has been reported that excess levels of ROS inhibit adipogenesis via activating CHOP, a negative regulator of C/EBPβ in the early stage of adipogenesis, or promoting HIF1, a repressor of Pparγ promoter activity [7,10]. However, a mild level of ROS may contribute to adipogenesis through enhancing C/EBPβ binding activity to activate Pparγ2 expression or increasing insulin sensitivity in normal adipocytes [[50], [51], [52]]. In the current study, we found that deficiency of either all or long isoforms of NRF1 in 3T3-L1 preadipocytes results in attenuated expression of various antioxidant genes compared to Scramble cells, implying that the loss of NRF1 could affect adipogenesis through altering the redox signaling that is involved in preadipocyte differentiation. However, whether NRF1-mediated antioxidant response affects intracellular ROS levels and thus adipogenesis and the mechanism behind the effect remain inconclusive.
Recent study showed that NRF1 could serve as a critical regulator for maintaining thermogenic function in BAT by increasing proteasomal activity [14], and may also play critical roles in proteasomal functions in mouse liver tissue [18], and in multiple human and mouse cell lines [[53], [54], [55], [56]]. Although we did not evaluate the roles of NRF1 in proteasome homeostasis in preadipocytes, we suspect that NRF1-dependent proteasome activity might contribute to regulate PPARγ expression. However, this possible mechanism needs further analysis.
To date, NRF1 has been found in a broad range of tissues and extensively investigated for its roles in antioxidant response, inflammation, apoptosis, metabolic homeostasis, mitochondrial function, proteasome stability, cellular differentiation and lipid metabolism [11,12,57,58]. However, its characteristics and functions in preadipocytes and WAC have not been fully explored. Our findings provided a novel insight into the regulatory roles of various isoforms of NRF1 in adipose biology and antioxidant response system. The current study identified L-NRF1 as a novel negative regulator of PPARγ expression and thus plays a critical role in adipogenesis. Given that adipogenesis is clearly complex and multifactorial, the underlying mechanisms by which NRF1 plays a role in the process need further study.
Author contributions
Conception and design of the study: Jingbo Pi, Yuanyuan Xu, Peng Xue.
Acquisition of the data: Peng Xue, Yongyong Hou, Zhuo Zuo, Huihui Wang, Zhendi Wang, Suping Ren.
Construction of plasmids and viruses: Jian Dong, Peng Xue, Jingqi Fu.
Interpretation of the data and drafting of the manuscript: Jingbo Pi, Peng Xue, Yongyong Hou, Yuanyuan Xu.
Revision of the manuscript: Melvin E. Andersen, Qiang Zhang.
All authors approved the final manuscript.
Declaration of competing interest
The authors have no conflicts of interest to disclose.
Acknowledgement
This work was supported by National Natural Science Foundation of China 81573106 (J.P.), 81830099 (J.P.), 81402661 (Y.H.), 81573187 (Y.X.), Liaoning Pandeng Scholar (J.P.), Liaoning Key Research and Development Guidance Plan 2019JH8/10300012 (J.P.) and Liaoning Revitalization Talents Program XLYC1807225 (Y.X.).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2019.101414.
Contributor Information
Yuanyuan Xu, Email: yyxu@cmu.edu.cn.
Jingbo Pi, Email: jbpi@cmu.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Rosen E.D., Spiegelman B.M. What we talk about when we talk about fat. Cell. 2014;156:20–44. doi: 10.1016/j.cell.2013.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee J.E., Schmidt H., Lai B., Ge K. Transcriptional and epigenomic regulation of adipogenesis. Mol. Cell. Biol. 2019;39 doi: 10.1128/MCB.00601-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ghaben A.L., Scherer P.E. Adipogenesis and metabolic health. Nat. Rev. Mol. Cell Biol. 2019;20:242–258. doi: 10.1038/s41580-018-0093-z. [DOI] [PubMed] [Google Scholar]
- 4.Mota de Sa P., Richard A.J., Hang H., Stephens J.M. Transcriptional regulation of adipogenesis. Comp. Physiol. 2017;7:635–674. doi: 10.1002/cphy.c160022. [DOI] [PubMed] [Google Scholar]
- 5.Rosen E.D., Walkey C.J., Puigserver P., Spiegelman B.M. Transcriptional regulation of adipogenesis. Genes Dev. 2000;14:1293–1307. [PubMed] [Google Scholar]
- 6.Lee J.E., Ge K. Transcriptional and epigenetic regulation of PPARgamma expression during adipogenesis. Cell Biosci. 2014;4:29. doi: 10.1186/2045-3701-4-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pi J., Leung L., Xue P., Wang W., Hou Y., Liu D., Yehuda-Shnaidman E., Lee C., Lau J., Kurtz T.W., Chan J.Y. Deficiency in the nuclear factor E2-related factor-2 transcription factor results in impaired adipogenesis and protects against diet-induced obesity. J. Biol. Chem. 2010;285:9292–9300. doi: 10.1074/jbc.M109.093955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hou Y., Xue P., Bai Y., Liu D., Woods C.G., Yarborough K., Fu J., Zhang Q., Sun G., Collins S., Chan J.Y., Yamamoto M., Andersen M.E., Pi J. Nuclear factor erythroid-derived factor 2-related factor 2 regulates transcription of CCAAT/enhancer-binding protein beta during adipogenesis. Free Radic. Biol. Med. 2012;52:462–472. doi: 10.1016/j.freeradbiomed.2011.10.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gross B., Pawlak M., Lefebvre P., Staels B. PPARs in obesity-induced T2DM, dyslipidaemia and NAFLD. Nat. Rev. Endocrinol. 2017;13:36–49. doi: 10.1038/nrendo.2016.135. [DOI] [PubMed] [Google Scholar]
- 10.Gummersbach C., Hemmrich K., Kroncke K.D., Suschek C.V., Fehsel K., Pallua N. New aspects of adipogenesis: radicals and oxidative stress. Differentiation. 2009;77:115–120. doi: 10.1016/j.diff.2008.09.009. [DOI] [PubMed] [Google Scholar]
- 11.Kim H.M., Han J.W., Chan J.Y. Nuclear factor erythroid-2 like 1 (NFE2L1): structure, function and regulation. Gene. 2016;584:17–25. doi: 10.1016/j.gene.2016.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Y., Xiang Y. Molecular and cellular basis for the unique functioning of Nrf1, an indispensable transcription factor for maintaining cell homoeostasis and organ integrity. Biochem. J. 2016;473:961–1000. doi: 10.1042/BJ20151182. [DOI] [PubMed] [Google Scholar]
- 13.Hou Y., Liu Z., Zuo Z., Gao T., Fu J., Wang H., Xu Y., Liu D., Yamamoto M., Zhu B., Zhang Y., Andersen M.E., Zhang Q., Pi J. Adipocyte-specific deficiency of Nfe2l1 disrupts plasticity of white adipose tissues and metabolic homeostasis in mice. Biochem. Biophys. Res. Commun. 2018;503:264–270. doi: 10.1016/j.bbrc.2018.06.013. [DOI] [PubMed] [Google Scholar]
- 14.Bartelt A., Widenmaier S.B., Schlein C., Johann K., Goncalves R.L.S., Eguchi K., Fischer A.W., Parlakgul G., Snyder N.A., Nguyen T.B., Bruns O.T., Franke D., Bawendi M.G., Lynes M.D., Leiria L.O., Tseng Y.H., Inouye K.E., Arruda A.P., Hotamisligil G.S. Brown adipose tissue thermogenic adaptation requires Nrf1-mediated proteasomal activity. Nat. Med. 2018;24:292–303. doi: 10.1038/nm.4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chan J.Y., Kwong M., Lu R., Chang J., Wang B., Yen T.S., Kan Y.W. Targeted disruption of the ubiquitous CNC-bZIP transcription factor, Nrf-1, results in anemia and embryonic lethality in mice. EMBO J. 1998;17:1779–1787. doi: 10.1093/emboj/17.6.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leung L., Kwong M., Hou S., Lee C., Chan J.Y. Deficiency of the Nrf1 and Nrf2 transcription factors results in early embryonic lethality and severe oxidative stress. J. Biol. Chem. 2003;278:48021–48029. doi: 10.1074/jbc.M308439200. [DOI] [PubMed] [Google Scholar]
- 17.Lee C.S., Lee C., Hu T., Nguyen J.M., Zhang J., Martin M.V., Vawter M.P., Huang E.J., Chan J.Y. Loss of nuclear factor E2-related factor 1 in the brain leads to dysregulation of proteasome gene expression and neurodegeneration. Proc. Natl. Acad. Sci. U. S. A. 2011;108:8408–8413. doi: 10.1073/pnas.1019209108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee C.S., Ho D.V., Chan J.Y. Nuclear factor-erythroid 2-related factor 1 regulates expression of proteasome genes in hepatocytes and protects against endoplasmic reticulum stress and steatosis in mice. FEBS J. 2013;280:3609–3620. doi: 10.1111/febs.12350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yuan J., Zhang S., Zhang Y. Nrf1 is paved as a new strategic avenue to prevent and treat cancer, neurodegenerative and other diseases. Toxicol. Appl. Pharmacol. 2018;360:273–283. doi: 10.1016/j.taap.2018.09.037. [DOI] [PubMed] [Google Scholar]
- 20.Berg D.T., Gupta A., Richardson M.A., O'Brien L.A., Calnek D., Grinnell B.W. Negative regulation of inducible nitric-oxide synthase expression mediated through transforming growth factor-beta-dependent modulation of transcription factor TCF11. J. Biol. Chem. 2007;282:36837–36844. doi: 10.1074/jbc.M706909200. [DOI] [PubMed] [Google Scholar]
- 21.Narayanan K., Ramachandran A., Peterson M.C., Hao J., Kolsto A.B., Friedman A.D., George A. The CCAAT enhancer-binding protein (C/EBP)beta and Nrf1 interact to regulate dentin sialophosphoprotein (DSPP) gene expression during odontoblast differentiation. J. Biol. Chem. 2004;279:45423–45432. doi: 10.1074/jbc.M405031200. [DOI] [PubMed] [Google Scholar]
- 22.Park S.Y., Kim S.H., Yoon H.K., Yim C.H., Lim S.K. The role of nuclear factor-E2-related factor 1 in the oxidative stress response in MC3T3-E1 osteoblastic cells. Endocrinol. Metab. (Seoul) 2016;31:336–342. doi: 10.3803/EnM.2016.31.2.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Widenmaier S.B., Snyder N.A., Nguyen T.B., Arduini A., Lee G.Y., Arruda A.P., Saksi J., Bartelt A., Hotamisligil G.S. NRF1 is an ER membrane sensor that is central to cholesterol homeostasis. Cell. 2017;171:1094–1109. doi: 10.1016/j.cell.2017.10.003. e1015. [DOI] [PubMed] [Google Scholar]
- 24.Xu Z., Chen L., Leung L., Yen T.S., Lee C., Chan J.Y. Liver-specific inactivation of the Nrf1 gene in adult mouse leads to nonalcoholic steatohepatitis and hepatic neoplasia. Proc. Natl. Acad. Sci. U. S. A. 2005;102:4120–4125. doi: 10.1073/pnas.0500660102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cui Q., Fu J., Hu Y., Li Y., Yang B., Li L., Sun J., Chen C., Sun G., Xu Y., Zhang Q., Pi J. Deficiency of long isoforms of Nfe2l1 sensitizes MIN6 pancreatic beta cells to arsenite-induced cytotoxicity. Toxicol. Appl. Pharmacol. 2017;329:67–74. doi: 10.1016/j.taap.2017.05.013. [DOI] [PubMed] [Google Scholar]
- 26.Zheng H., Fu J., Xue P., Zhao R., Dong J., Liu D., Yamamoto M., Tong Q., Teng W., Qu W., Zhang Q., Andersen M.E., Pi J. CNC-bZIP protein nrf1-dependent regulation of glucose-stimulated insulin secretion. Antioxidants Redox Signal. 2015;22:819–831. doi: 10.1089/ars.2014.6017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao R., Hou Y., Xue P., Woods C.G., Fu J., Feng B., Guan D., Sun G., Chan J.Y., Waalkes M.P., Andersen M.E., Pi J. Long isoforms of NRF1 contribute to arsenic-induced antioxidant response in human keratinocytes. Environ. Health Perspect. 2011;119:56–62. doi: 10.1289/ehp.1002304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang W., Kwok A.M., Chan J.Y. The p65 isoform of Nrf1 is a dominant negative inhibitor of ARE-mediated transcription. J. Biol. Chem. 2007;282:24670–24678. doi: 10.1074/jbc.M700159200. [DOI] [PubMed] [Google Scholar]
- 29.Ohtsuji M., Katsuoka F., Kobayashi A., Aburatani H., Hayes J.D., Yamamoto M. Nrf1 and Nrf2 play distinct roles in activation of antioxidant response element-dependent genes. J. Biol. Chem. 2008;283:33554–33562. doi: 10.1074/jbc.M804597200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hou Y., Xue P., Woods C.G., Wang X., Fu J., Yarborough K., Qu W., Zhang Q., Andersen M.E., Pi J. Association between arsenic suppression of adipogenesis and induction of CHOP10 via the endoplasmic reticulum stress response. Environ. Health Perspect. 2013;121:237–243. doi: 10.1289/ehp.1205731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Woods C.G., Fu J., Xue P., Hou Y., Pluta L.J., Yang L., Zhang Q., Thomas R.S., Andersen M.E., Pi J. Dose-dependent transitions in Nrf2-mediated adaptive response and related stress responses to hypochlorous acid in mouse macrophages. Toxicol. Appl. Pharmacol. 2009;238:27–36. doi: 10.1016/j.taap.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramakrishnan V.M., Boyd N.L. The adipose stromal vascular fraction as a complex cellular source for tissue engineering applications. Tissue Eng. B Rev. 2018;24:289–299. doi: 10.1089/ten.teb.2017.0061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xue P., Hou Y., Zhang Q., Woods C.G., Yarborough K., Liu H., Sun G., Andersen M.E., Pi J. Prolonged inorganic arsenite exposure suppresses insulin-stimulated AKT S473 phosphorylation and glucose uptake in 3T3-L1 adipocytes: involvement of the adaptive antioxidant response. Biochem. Biophys. Res. Commun. 2011;407:360–365. doi: 10.1016/j.bbrc.2011.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen Y., Xue P., Hou Y., Zhang H., Zheng H., Zhou T., Qu W., Teng W., Zhang Q., Andersen M.E., Pi J. Isoniazid suppresses antioxidant response element activities and impairs adipogenesis in mouse and human preadipocytes. Toxicol. Appl. Pharmacol. 2013;273:435–441. doi: 10.1016/j.taap.2013.10.005. [DOI] [PubMed] [Google Scholar]
- 35.Lowe C.E., O'Rahilly S., Rochford J.J. Adipogenesis at a glance. J. Cell Sci. 2011;124:2681–2686. doi: 10.1242/jcs.079699. [DOI] [PubMed] [Google Scholar]
- 36.Spiegelman B.M., Flier J.S. Adipogenesis and obesity: rounding out the big picture. Cell. 1996;87:377–389. doi: 10.1016/s0092-8674(00)81359-8. [DOI] [PubMed] [Google Scholar]
- 37.Lefterova M.I., Haakonsson A.K., Lazar M.A., Mandrup S. PPARgamma and the global map of adipogenesis and beyond. Trends Endocrinol. Metab. 2014;25:293–302. doi: 10.1016/j.tem.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim T.H., Jo S.H., Choi H., Park J.M., Kim M.Y., Nojima H., Kim J.W., Ahn Y.H. Identification of Creb3l4 as an essential negative regulator of adipogenesis. Cell Death Dis. 2014;5 doi: 10.1038/cddis.2014.490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zang L., Xue B., Lu Z., Li X., Yang G., Guo Q., Ba J., Zou X., Dou J., Lu J., Pan C., Mu Y. Identification of LRP16 as a negative regulator of insulin action and adipogenesis in 3T3-L1 adipocytes. Horm. Metab. Res. 2013;45:349–358. doi: 10.1055/s-0032-1331215. [DOI] [PubMed] [Google Scholar]
- 40.Tang X., Guilherme A., Chakladar A., Powelka A.M., Konda S., Virbasius J.V., Nicoloro S.M., Straubhaar J., Czech M.P. An RNA interference-based screen identifies MAP4K4/NIK as a negative regulator of PPARgamma, adipogenesis, and insulin-responsive hexose transport. Proc. Natl. Acad. Sci. U. S. A. 2006;103:2087–2092. doi: 10.1073/pnas.0507660103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Engin A.B. MicroRNA and adipogenesis. Adv. Exp. Med. Biol. 2017;960:489–509. doi: 10.1007/978-3-319-48382-5_21. [DOI] [PubMed] [Google Scholar]
- 42.McGregor R.A., Choi M.S. microRNAs in the regulation of adipogenesis and obesity. Curr. Mol. Med. 2011;11:304–316. doi: 10.2174/156652411795677990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sarjeant K., Stephens J.M. Adipogenesis. Cold Spring Harb. Perspect. Biol. 2012;4:a008417. doi: 10.1101/cshperspect.a008417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yun Z., Maecker H.L., Johnson R.S., Giaccia A.J. Inhibition of PPAR gamma 2 gene expression by the HIF-1-regulated gene DEC1/Stra13: a mechanism for regulation of adipogenesis by hypoxia. Dev. Cell. 2002;2:331–341. doi: 10.1016/s1534-5807(02)00131-4. [DOI] [PubMed] [Google Scholar]
- 45.Sha Z., Goldberg A.L. Proteasome-mediated processing of Nrf1 is essential for coordinate induction of all proteasome subunits and p97. Curr. Biol. 2014;24:1573–1583. doi: 10.1016/j.cub.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Johnsen O., Murphy P., Prydz H., Kolsto A.B. Interaction of the CNC-bZIP factor TCF11/LCR-F1/Nrf1 with MafG: binding-site selection and regulation of transcription. Nucleic Acids Res. 1998;26:512–520. doi: 10.1093/nar/26.2.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Murphy P., Kolsto A. Expression of the bZIP transcription factor TCF11 and its potential dimerization partners during development. Mech. Dev. 2000;97:141–148. doi: 10.1016/s0925-4773(00)00413-5. [DOI] [PubMed] [Google Scholar]
- 48.Ramji D.P., Foka P. CCAAT/enhancer-binding proteins: structure, function and regulation. Biochem. J. 2002;365:561–575. doi: 10.1042/BJ20020508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Amoutzias G.D., Veron A.S., Weiner J., 3rd, Robinson-Rechavi M., Bornberg-Bauer E., Oliver S.G., Robertson D.L. One billion years of bZIP transcription factor evolution: conservation and change in dimerization and DNA-binding site specificity. Mol. Biol. Evol. 2007;24:827–835. doi: 10.1093/molbev/msl211. [DOI] [PubMed] [Google Scholar]
- 50.Lee H., Lee Y.J., Choi H., Ko E.H., Kim J.W. Reactive oxygen species facilitate adipocyte differentiation by accelerating mitotic clonal expansion. J. Biol. Chem. 2009;284:10601–10609. doi: 10.1074/jbc.M808742200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Loh K., Deng H., Fukushima A., Cai X., Boivin B., Galic S., Bruce C., Shields B.J., Skiba B., Ooms L.M., Stepto N., Wu B., Mitchell C.A., Tonks N.K., Watt M.J., Febbraio M.A., Crack P.J., Andrikopoulos S., Tiganis T. Reactive oxygen species enhance insulin sensitivity. Cell Metabol. 2009;10:260–272. doi: 10.1016/j.cmet.2009.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Castro J.P., Grune T., Speckmann B. The two faces of reactive oxygen species (ROS) in adipocyte function and dysfunction. Biol. Chem. 2016;397:709–724. doi: 10.1515/hsz-2015-0305. [DOI] [PubMed] [Google Scholar]
- 53.Vangala J.R., Radhakrishnan S.K. Nrf1-mediated transcriptional regulation of the proteasome requires a functional TIP60 complex. J. Biol. Chem. 2019;294:2036–2045. doi: 10.1074/jbc.RA118.006290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Koizumi S., Hamazaki J., Murata S. Transcriptional regulation of the 26S proteasome by Nrf1. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2018;94:325–336. doi: 10.2183/pjab.94.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee D., Ryu K.Y. Effect of cellular ubiquitin levels on the regulation of oxidative stress response and proteasome function via Nrf1. Biochem. Biophys. Res. Commun. 2017;485:234–240. doi: 10.1016/j.bbrc.2017.02.105. [DOI] [PubMed] [Google Scholar]
- 56.Radhakrishnan S.K., Lee C.S., Young P., Beskow A., Chan J.Y., Deshaies R.J. Transcription factor Nrf1 mediates the proteasome recovery pathway after proteasome inhibition in mammalian cells. Mol. Cell. 2010;38:17–28. doi: 10.1016/j.molcel.2010.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bugno M., Daniel M., Chepelev N.L., Willmore W.G. Changing gears in Nrf1 research, from mechanisms of regulation to its role in disease and prevention. Biochim. Biophys. Acta. 2015;1849:1260–1276. doi: 10.1016/j.bbagrm.2015.08.001. [DOI] [PubMed] [Google Scholar]
- 58.Biswas M., Chan J.Y. Role of Nrf1 in antioxidant response element-mediated gene expression and beyond. Toxicol. Appl. Pharmacol. 2010;244:16–20. doi: 10.1016/j.taap.2009.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.