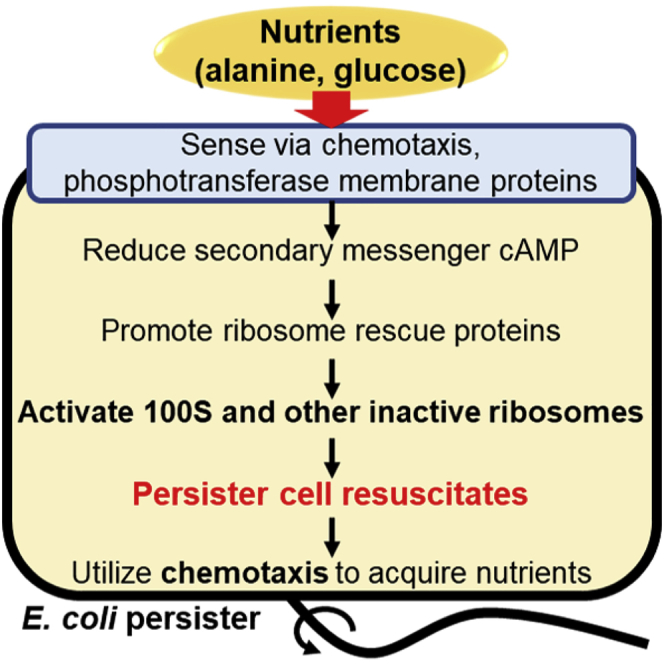
Summary
Persistence, the stress-tolerant state, is arguably the most vital phenotype since nearly all cells experience nutrient stress, which causes a sub-population to become dormant. However, how persister cells wake to reconstitute infections is not understood well. Here, using single-cell observations, we determined that Escherichia coli persister cells resuscitate primarily when presented with specific carbon sources, rather than spontaneously. In addition, we found that the mechanism of persister cell waking is through sensing nutrients by chemotaxis and phosphotransferase membrane proteins. Furthermore, nutrient transport reduces the level of secondary messenger cAMP through enzyme IIA; this reduction in cAMP levels leads to ribosome resuscitation and rescue. Resuscitating cells also immediately commence chemotaxis toward nutrients, although flagellar motion is not required for waking. Hence, persister cells wake by perceiving nutrients via membrane receptors that relay the signal to ribosomes via the secondary messenger cAMP, and persisters wake and utilize chemotaxis to acquire nutrients.
Subject Areas: Biological Sciences, Microbiology, Molecular Microbiology, Cell Biology
Graphical Abstract
Highlights
-
•
Persister cells wake primarily by sensing nutrients rather than spontaneously
-
•
Persisters wake using chemotaxis sensors and phosphotransferase membrane proteins
-
•
Persisters wake by reducing cAMP and by activating stalled/hibernating ribosomes
-
•
Persister cells undergo chemotaxis as they resuscitate
Biological Sciences; Microbiology; Molecular Microbiology; Cell Biology
Introduction
The persister cell phenotype was first noted in 1942 (Hobby et al., 1942) when penicillin did not completely sterilize a Staphylococcus aureus culture (1% of the population remained intact). The surviving subpopulation was deemed “persister cells” in 1944 (Bigger, 1944). Both groups determined that persisters are dormant (Bigger, 1944, Hobby et al., 1942), which has been corroborated (Kwan et al., 2013, Shah et al., 2006), and further research has demonstrated persister cells are not mutants (Chowdhury et al., 2016b, Kwan et al., 2015a) but instead acquire their antibiotic tolerance through this dormancy.
The persister cell phenotype is ubiquitous and has been well described in many bacteria such as Escherichia coli (Fisher et al., 2017), Pseudomonas aeruginosa (Fisher et al., 2017), and S. aureus (Fisher et al., 2017) and in Archaea (Megaw and Gilmore, 2017). Critically, the persister state arises not only after antibiotic stress but nutrient stress also creates persister cells (Bernier et al., 2013, Maisonneuve and Gerdes, 2014, Martins et al., 2018); in fact, the classic viable but not culturable state appears to be the same as the persister state (Kim et al., 2018a), so persisters form everywhere as all bacterial cells eventually face nutrient stress (Song and Wood, 2018). Hence, it may be argued that the persister state is one of the most fundamental bacterial phenotypes.
It is controversial how persister cells form. It has been argued that they form from a reduction in metabolism due to activation of a toxin of a toxin/antitoxin system. Evidence of this is that the deletion of several toxins of toxin/antitoxin systems such as MqsR (Kim and Wood, 2010, Luidalepp et al., 2011), TisB (Dörr et al., 2010), and YafQ (Harrison et al., 2009) leads to a reduction in persistence. Similarly, production of toxins unrelated to toxin/antitoxin systems can also increase persistence (Chowdhury et al., 2016a). However, recent studies have not found a connection between toxin/antitoxin systems and persistence (Goormaghtigh et al., 2018, Pontes and Groisman, 2019, Svenningsen et al., 2019). As an alternative model, we have suggested persister cells form from the inactivation of ribosomes through dimerization as a result of elevated guanosine pentaphosphate/tetraphosphate (Song and Wood, 2019).
How cells resuscitate is better understood than how they form. We have found persister cells resuscitate as soon as instantaneously in rich medium (Kim et al., 2018b) and wake based on their ribosome content (Kim et al., 2018b). For example, persister cells with 4-fold fewer ribosomes are delayed by several hours in their resuscitation while ribosome levels increase (Kim et al., 2018b). Others have suggested, but not shown, that cells may be resuscitated by reversing the effects of toxins of toxin/antitoxin systems (Cheverton et al., 2016). Hence, it is still not clear what pathway is involved in persister cell waking in regard to nutrient sensing.
To study persister cells without introducing traits of more prevalent cell phenotypes (e.g., slow-growing, tolerant stationary cells), their concentration needs to be increased so they are the dominant phenotype. Previously, we showed how to create a high percentage of E. coli persister cells (up to 70%) via rifampicin-pretreatment to stop transcription, carbonyl cyanide m-chlorophenylhydrazone (CCCP) to stop ATP production, or tetracycline to stop translation (Kwan et al., 2013); these cells were shown to be bona fide persister cells via eight different assays (multi-drug tolerance, immediate change from persistence to non-persistence in the presence of nutrients, dormancy based on lack of cell division in the absence of nutrients, dormancy via metabolic staining and cell sorting, no change in MIC compared with exponential cells, no resistance phenotype, similar morphology to ampicillin-induced persisters, and similar resuscitation as ampicillin-induced persisters) (Kim et al., 2018b). At least six other research groups have used our methods to make persister cells (Cui et al., 2018, Grassi et al., 2017, Narayanaswamy et al., 2018, Pu et al., 2019, Sulaiman et al., 2018, Tkhilaishvili et al., 2018); for example, Cui et al. Cui et al., 2018 used rifampin and tetracycline to induce persistence for screening the E. coli Keio library and found several DNA repair mutants (e.g., recA, recC, ruvA, and uvrD) with reduced persistence, Grassi et. al. (Grassi et al., 2017) used CCCP to generate P. aeruginosa and S. aureus persister cells to determine that antibiotic-tolerant phenotypes depend on different classes of antibiotics, and Narayanaswamy et al. (Narayanaswamy et al., 2018) used CCCP to generate P. aeruginosa persister cells and found poly (acetyl, arginyl) glucosamine is effective for killing them.
In the present study, we found that rather than waking spontaneously, persister cells wake primarily as a result of sensing nutrients. We then elucidated the waking pathway and found persister cells wake via chemotaxis and transport pathways that serve to reduce cAMP, activate ribosomes, and commence chemotaxis.
Results
Overview
To create a large enough population of persister cells so that we could investigate resuscitation at the single-cell level, we pre-treated exponential cells with rifampicin to stop transcription since this method converts the rare persister phenotype into the dominant cell state; i.e., we increased the number of persister cells by 105-fold, which enables us to study more readily persister cells at the single-cell level (Kwan et al., 2013). The persister cells induced by rifampicin are isolated from any remaining non-persister cells by ampicillin treatment since the non-persisters cells lyse so that the remaining population consists solely of only persister cells (Kwan et al., 2013). Please see the Transparent Methods section for details.
To study these persister cells, we utilized initially agar pates. We reasoned that, if specific persister cells wake faster, larger colonies would form on M9 agar plates since we found previously that once they wake, persister cells grow at the same rate as exponentially growing cells (Kim et al., 2018b). However, with this initial assay, colony size differences may also be influenced by differences in growth; hence, all of the significant resuscitation results found from the agar plates were confirmed by microscopic, single-cell experiments. In addition, all of the strains used in this work were tested and shown to produce roughly the same number of persister cells (so there is no defect in the formation of persister cells) and shown to grow similar to the wild-type.
Persister Cells Wake by Recognizing Ala
To elucidate insights about waking using defined medium, we hypothesized that a single amino acid could wake E. coli persister cells since L-alanine revives spores of Bacillus subtilis (Mutlu et al., 2018). Initially, four groups of five amino acids were tested (#1: Ala, Arg, Cys, Phe, Ser; #2: Gly, His, Thr, Val, Tyr; #3: Asn, Ile, Lys, Pro, Trp; and #4: Asp, Glu, Gln, Leu, Met) by plating E. coli persister cells on M9 agar plates with the amino acid combinations as the sole carbon source and checking for growth after 4 days (Figure S1). Larger colonies were obtained from group #1 compared with groups #3 and #4, indicating faster waking, and persister cells did not wake for group #2 (Figure S1). As expected, there was no persister cell growth with M9 agar that lacks amino acids (negative control). For the best three groups, each amino acid was tested separately for persister cell waking, and clearly substantially more persister cells were revived with Ala (Figure S2).
To confirm that the macroscopic colonies seen on M9 agar plates reflected faster waking, single E. coli persister cells were observed on M9 5X Ala agarose gel pads using light microscopy. We found that 18 ± 1% of persister cells began to wake, as evidenced by dividing or elongating within 6 h (Figure 1, average with standard deviations shown in Table S1). As a negative control, single-cell waking was monitored using Asn (5X) from group #3. Unlike with Ala, only 2 ± 2% divided after 6 h (Figure 1 and Table S1). Hence, persister cells with Ala wake nine times more frequently than those with Asn. As an additional negative control, waking on agarose gel pads that lack amino acids (and any other carbon source) was investigated, and we found no persister cells wake after 6 h (Figure 1). This lack of persister cell waking without nutrients also confirms that the persister cells used are bona fide dormant cells since, in contrast, exponential cells can wake in 6 h on agarose pads that lack nutrients (12 ± 4%, Table S1).
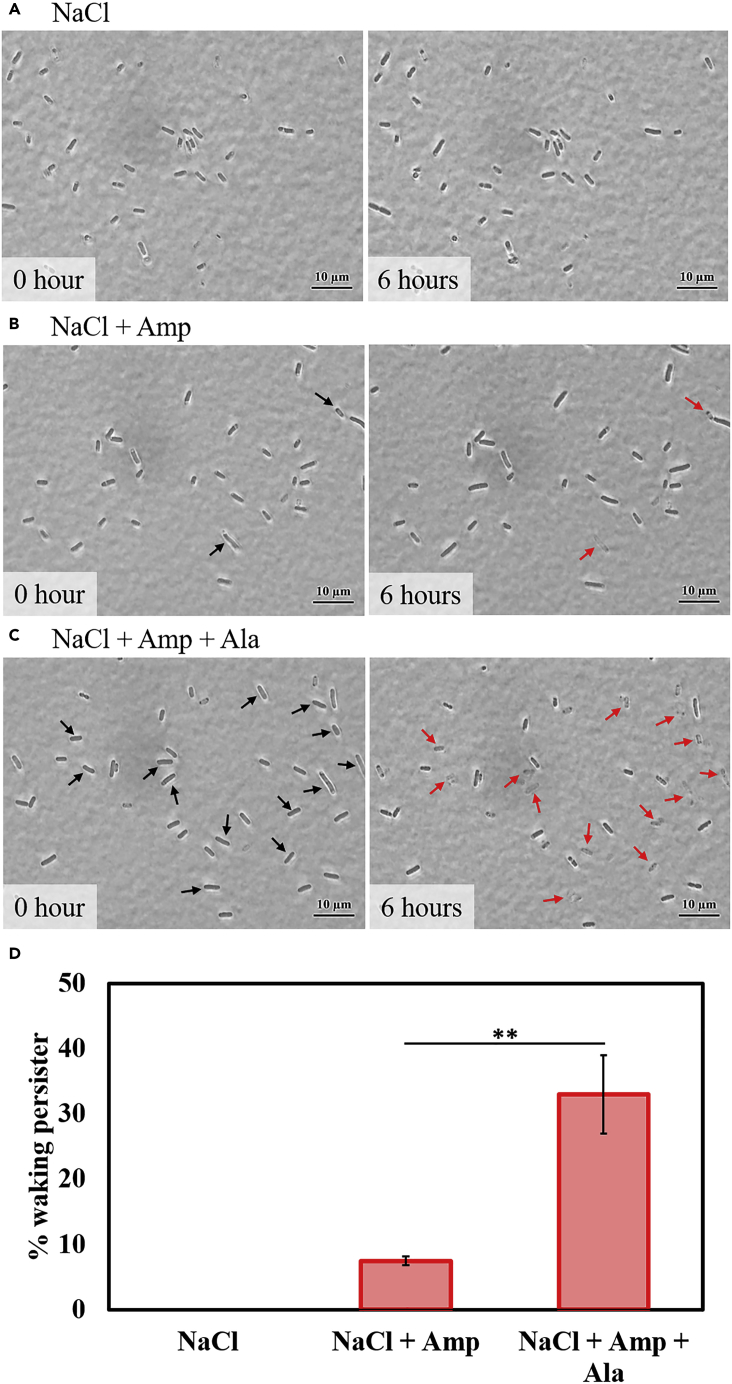
Figure 1.
Single Persister Cells Wake with Ala
E. coli BW25113 persister cells were incubated at 37°C on M9 agarose gel pads supplemented with 5X Ala (A), 5X Asn (B), or no amino acids (C), and images were captured at 0 and 6 h. Black arrows indicate cells that resuscitate. Scale bar indicates 10 μm. Representative results from two independent cultures are shown. Tabulated cell numbers are shown in Table S1.
Persister Cells Wake Primarily by Sensing Nutrients rather Than Spontaneously
Along with waking as a response to a change in environmental conditions (e.g., the addition of nutrients [Kim et al., 2018b]), it has been reported that persister cells wake spontaneously (Balaban et al., 2004); hence, we investigated whether persister cells wake in the absence of Ala. To test this, single persister cells were observed on NaCl, NaCl + ampicillin, and NaCl + ampicillin + Ala agarose gel pads. The rationale is that if the persisters wake, ampicillin will lyse the cells (Uehara et al., 2009), which is seen as a disappearing single cell. We used NaCl buffer rather than M9 buffer so that the effect of ampicillin and Ala would be clearer.
We found that persister cells wake primarily in the presence of Ala since there was 4.4-fold more waking followed by lysis with Ala + ampicillin in 6 h compared with gel pads that lacked Ala but contained ampicillin (Figure 2, statistics in Table S2). As expected, for the negative control (NaCl only), there was no cell death seen in 6 h (Figure 2A). Therefore, most persister cells do not resuscitate spontaneously but instead sense the carbon source through some pathway. Furthermore, these results suggest that the cells do not determine whether the antibiotic is present or not.
Figure 2.
Single Persister Cells Wake Primarily by Sensing Nutrients Rather than Spontaneously
(A–C) E. coli BW25113 persister cells were incubated at 37°C on (A) NaCl, (B) NaCl + ampicillin (Amp), and (C) NaCl + Amp + Ala agarose gel pads for 6 h. Cells first resuscitate (black arrows) and then lyse in the presence of ampicillin (red arrows). Scale bar indicates 10 μm. Representative results from two independent cultures are shown.
(D) Persister cell waking (%) after 6-h incubation. Tabulated cell numbers are shown in Table S2, and error bars indicate standard deviations. Student's t test was used to compare two groups (** indicates a p value <0.0003).
Alanine Is a Waking Signal
Since persister cells wake in the presence of Ala, we investigated whether Ala acts as a true signal rather than as a carbon and energy source by utilizing an isogenic dadA mutation that abolishes growth on alanine by inactivating D-amino acid dehydrogenase (Franklin and Venables, 1976, Wild and Klopotowski, 1981). We confirmed the dadA mutation abolishes growth on Ala (Figure S3) and found 4% of the single dadA cells wake with Ala, whereas no cells wake without Ala (Figure S4, average with standard deviations shown in Table S3); hence, Ala wakes persister cells in the absence of growth. Moreover, we tested whether Ala wakes persister cells in the presence of the additional carbon and energy source pyruvate (final concentration 0.24 wt%) when Ala is not used for growth (i.e., with the dadA mutant). We found pyruvate does not wake single dadA persister cells, whereas Ala wakes dadA persister cells, in the presence of pyruvate, to the same extent as in the absence of pyruvate (Figure S4 and Table S3). Therefore, Ala is a true signal for waking persister cells.
Persister Cells Resuscitate through the Chemotaxis Apparatus
Since most persister cells wake by responding to the presence of Ala as a signal, we investigated the mechanism of how persister cells resuscitate by looking for faster waking when individual proteins are produced. Hence, we pooled all the ASKA plasmids (each pCA24N-derived plasmid produces one E. coli protein) and electroporated them into E. coli BW25113, formed persister cells, and selected for faster growth on M9 5X Ala agar plates containing chloramphenicol to retain the plasmid. After 3 days, some colonies could be seen, and after 8 days, different phenotypes were clearly seen (Figure S5). The faster (bigger) colonies were purified and sequenced and six proteins were identified: PsiF, PanD, YmgF, YjcF, PptA, and CheY.
For these six putative proteins that stimulate persister cell waking, single cells harboring the ASKA plasmid were observed on M9 5X Ala agarose gel pads and compared with persister cells with an empty plasmid (E. coli BW25113/pCA24N) (Figures S6–S8 and Table S4). Note that, owing to the metabolic burden of the plasmid, the cells wake slower so the microscopic observations were extended to 18 h and protein production was not optimized. We found that single cells that produce PanD (23-fold) and YmgF (18-fold) wake at a higher frequency with Ala, compared with cells containing the empty plasmid (Table S4). Remarkably, all the single persister cells that produced the chemotaxis response regulator CheY resuscitated (33-fold enhancement, Figure 3 and Table S4), and 82% of the single persister cells that produced response regulator CheA resuscitated (27-fold enhancement) (Figures 3 and S9 and Table S5). In addition, single cells of deletion mutants ΔcheY and ΔcheA are completely inhibited in their waking on M9 Ala (Figure S10). Critically, the CheY- and CheA-related strains do not have defects in either persister cell formation or growth (Table S6). Together, these results demonstrate the importance of the chemotaxis system in nutrient recognition and waking in persister cells.
Figure 3.
Single Persister Cell Waking on Ala after Producing Chemotaxis-Related, cAMP-Related, and Ribosome-Resuscitation-Related Proteins
(A–E) E. coli BW25113 persister cells containing (A) pCA24N, (B) pCA24N_cheY, (C) pCA24N_trg, (D) pCA24N_cpdA, and (E) pCA24N_cpdA with 3 mM atropine were incubated at 37°C on M9 5X Ala agarose gel pads for 18 h. Black arrows indicate cells that resuscitate. Scale bar indicates 10 μm. Representative results from two independent cultures are shown. Before forming persister cells, cultures were grown without IPTG with 30 μg/mL Cm.
(F) Persister cell waking (%) after incubating for 18 h. Tabulated cell numbers are shown in Table S5, and error bars indicate standard deviations. Student's t tests were used to compare two groups (* indicates a p value <0.05 and ** indicates a p value <0.01). Additional microscope images are shown in Figures S9 and S16.
To investigate further the link between chemotaxis and persister cell waking, we tested all five of the methyl-accepting chemotaxis proteins (Tsr, Tar, Trg, Tap, and Aer) for waking on agar plates by using isogenic mutants and found inactivating Tar and Trg decreased waking compared with the wild type (Figure S11). Corroborating these results, persister cells that produce Tar and Trg woke faster than those with the empty plasmid and faster than cells producing Aer, Tsr, and Tap (Figure S12).
Single cells producing Tar (9-fold) and Trg (8-fold) also woke with a higher frequency than cells containing the empty plasmid, and single cells producing Tsr woke the same as those with the empty plasmid (Figures 3 and S9 and Table S5). Hence, persister cells wake on alanine via the chemotaxis system utilizing methyl-accepting chemotaxis proteins Tar and Trg and response regulators CheY and CheA.
Persister Cell Waking Does Not Require Flagellar Rotation
Since the chemotaxis system involves the flagellum, we investigated its role in persister cell waking. Using agar plates, there was no effect of the flagellum as tested by inactivating the flagella motor MotA (Figure S13), and single persister cells with inactivated flagella motor MotB did not wake significantly differently than wild-type persisters (Figure S14 and Table S7). Corroborating these results, inactivating the hook protein FlgE (Brown et al., 2012) had no effect on persister waking (Figure S15), so flagellum movement is not required for persister cells waking. Note the flagella system has been linked previously to persister cell formation (Shan et al., 2015) but not to resuscitation.
In contrast, inactivating FhlD and FliN reduced the frequency of single-cell waking by 3-fold (Figure S14 and Table S7). In addition, producing FliN in single cells increased the waking frequency by 4.3-fold (Figures 3 and S16 and Table S5). Hence, the master transcriptional regulator of flagellar proteins is involved in persister cell waking.
Persister Cells Wake by Lowering cAMP Levels
Having determined that persister cells wake by utilizing the chemotaxis apparatus, we explored how the external nutrient signal was propagated beyond the chemotaxis and flagellum system. Since cells can sense their environment (e.g., surfaces) by cAMP (Lee et al., 2018), and cAMP has been linked to persistence (Kwan et al., 2015b), we reasoned that persister cell waking may involve this secondary message.
To test the importance of cAMP, we produced the only adenylate cyclase in E. coli, CyaA (Tuckerman et al., 2009), via BW25113/pCA24N-cyaA and found using agar plates that cells with elevated cAMP have reduced waking compared with cells with the empty plasmid (Figure S17). Consistently, cells producing the cAMP-specific phosphodiesterase CpdA (Imamura et al., 1996) via BW25113/pCA24N-cpdA have increased waking on M9-Ala agar plates; we showed previously that CpdA can eliminate cAMP by reducing its concentration 323-fold (Kwan et al., 2015b). Corroborating these results, cells lacking CpdA (BW25113 ΔcpdA) have reduced waking on M9-Ala plates (Figure S17).
Verifying these agar plate results, single cells producing adenylate phosphodiesterase CpdA before persister formation had a 15-fold greater (0.5 mM IPTG) and 8-fold greater (0 mM IPTG) frequency of waking (Figures 3 and S9 and Table S5) by reducing cAMP levels. In addition, producing adenylate cyclase CyaA reduced the frequency of waking by 30% compared with the empty plasmid (Figures 3 and S16 and Table S5). In a consistent manner, increasing cAMP levels by inactivating CpdA completely eliminated single-cell waking (Figure S18 and Table S8) and decreasing cAMP levels by inactivating CyaA led to 4.3-fold greater frequency of waking (78%) (Figure S18 and Table S8).
As additional proof of the impact of cAMP, we used the cAMP inhibitor atropine (Huynh et al., 2012) and found single cells wake with atropine (3 mM) at twice the frequency (Figure S19). Consistently, adding exogenous cAMP (2 mM) reduced the frequency of single-cell waking 3-fold (Figure S19). Moreover, combining atropine with production of CpdA to reduce cAMP increased the frequency of waking 4-fold such that 92% of the cells woke in 6 h (Figure 3 and Table S5). Therefore, reducing cAMP levels increases persister cell waking dramatically.
Persister Cells Wake by Resuscitating and Rescuing Ribosomes
Since reducing levels of cAMP led to faster resuscitation, we explored further the mechanism for their waking. We hypothesized that, since elevated cAMP levels increase ribosome hibernation during starvation (Shimada et al., 2013), lowering cAMP levels should lead to ribosome resuscitation; hence, we tested the impact of changing the concentration of ribosome resuscitation factor HflX (Gohara and Yap, 2018); HflX also rescues ribosomes from mRNA (Zhang et al., 2015). We utilized ribosome rescue factor ArfB (Abo and Chadani, 2014) as a negative control since it is positively regulated by cAMP (Raghavan et al., 2011). In addition, since trans-translation is used to rescue ribosomes from mRNAs that lack a stop codon (Abo and Chadani, 2014), we also investigated the impact of SsrA on persister cell waking, using the strain we constructed (Wang et al., 2009).
For M9 Ala agar plates, we found cells producing HflX woke faster, compared with cells with the empty plasmid, whereas cells producing ArfB did not wake, indicating that ArfB reduces waking dramatically (Figure S20). Similarly, for single cells, we found that inactivating HflX completely inhibited waking (Figure S21 and Table S9). In contrast, inactivating ArfB in single cells had no effect (Figure S21 and Table S9). Corroborating these results, producing HflX in single cells increased the frequency of waking dramatically (7-fold) (Figure S16 and Table S5).
For ribosome rescue, using agar plates, we found that inactivating SsrA led to significantly slower waking (Figure S20). Similarly, for single cells, inactivating SsrA almost completely inhibited waking (frequency of waking reduced to less than 1%) (Figure S21 and Table S9). Therefore, resuscitating ribosomes via HflX and SsrA is vital for persister cell resuscitation. Previously, SsrA has been shown to have a modest role (2- to 8-fold) in the formation of persister cells (Shi et al., 2013).
Persister Cells Also Wake by Recognizing Sugars
Using alanine to wake cells, it was not clear how cAMP levels are reduced; hence, we studied resuscitation with the building blocks of the biofilm matrix since we hypothesized that those materials would be readily available extracellularly. Initially, we chose four polysaccharide building blocks of the most prevalent polymers of E. coli biofilms: D-glucose from cellulose (Klemm et al., 2005), D-glucosamine and N-acetyl-D-glucosamine from poly-N-acetylglucosamine (Pokhis et al., 2015), and α-D-glucose 1-phosphate from colonic acid (Patel et al., 2012, Stevenson et al., 1996). The final concentration of each sugar was kept at 0.4%, and each polysaccharide building block was utilized as the sole carbon and energy source. We also tested mannose, sorbitol, and maltose since they are good carbon sources for E. coli.
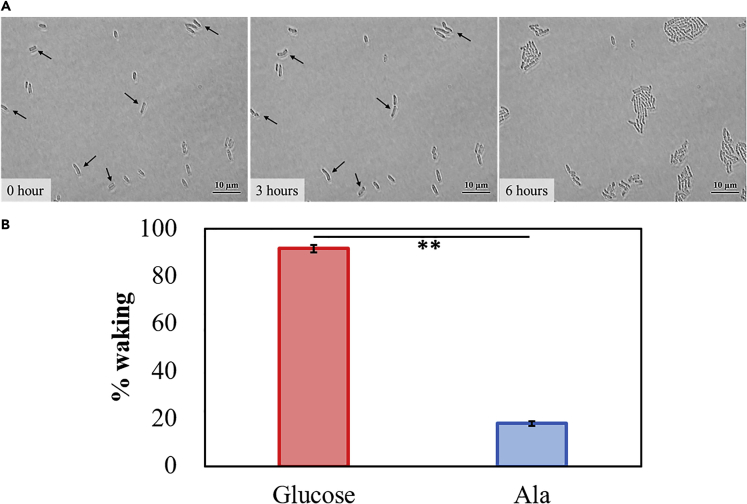
Using minimal agar plates that contain these polysaccharides, we found that persister cells wake more rapidly on biofilm-derived polymers than on Ala since colonies form 2 days faster on plates with the polysaccharide precursors (Figure S22). As expected, we found that the frequency of single-cell waking was higher with glucose compared with Ala; for example, in 6 h, 92 ± 2% cells resuscitate on glucose compared with 18 ± 1% on Ala (Figure 4 and Table S10). In addition, waking was greatest with glucose, α-D-glucose 1-phosphate, and mannose compared with glucosamine, sorbitol, and maltose as shown by the generation of colonies faster on agar plates (Figure S22); hence, resuscitation is not uniform for all sugars. Furthermore, with single cells, glucose was most effective since 22 ± 3% of the cells resuscitated in 3 h with glucose compared with 16 ± 3% with α-D-glucose 1-phosphate and 14.7 ± 0.5% with mannose (Figure S23). Since waking with glucose was most rapid, we focused on using it for persister resuscitation.
Figure 4.
Single Persister Cells Wake with Glucose
(A) E. coli BW25113 persister cells were incubated at 37°C on M9 0.4% glucose agarose gel pads and images were captured at 0, 3, and 6 h. Black arrows indicate cells that resuscitate. Scale bar indicates 10 μm. Representative results from two independent cultures are shown. Tabulated cell numbers are shown in Table S10.
(B) Comparison of single E. coli BW25113 persister cells waking on minimal medium with 0.4% glucose and 0.04% alanine for 6 h, and error bars indicate standard deviations. Student's t test was used to compare two groups (** indicates a p value <0.01).
Biofilm Persister Cells Resuscitate like Planktonic Cells
To determine if persister cell resuscitation with sugars was similar in biofilms, we generated biofilms for 48 h and examined single-cell persister resuscitation with glucose and found similar resuscitation as planktonic cells (Figure S24). Using a GFP reporter that indicates the number of active 70S ribosomes in individual persister cells (Kim et al., 2018b), we also found the biofilm persisters that resuscitate first have a higher ribosome fraction (Figure S24). Furthermore, biofilm persisters that do not have nutrients have low ribosome levels (i.e., low GFP levels) and do not resuscitate (Figure S24). Hence, biofilm persister cells resuscitate via ribosome activation like planktonic cells (Figure S24).
Glucose Waking Requires Transporters PtsG and MglB and Chemotaxis
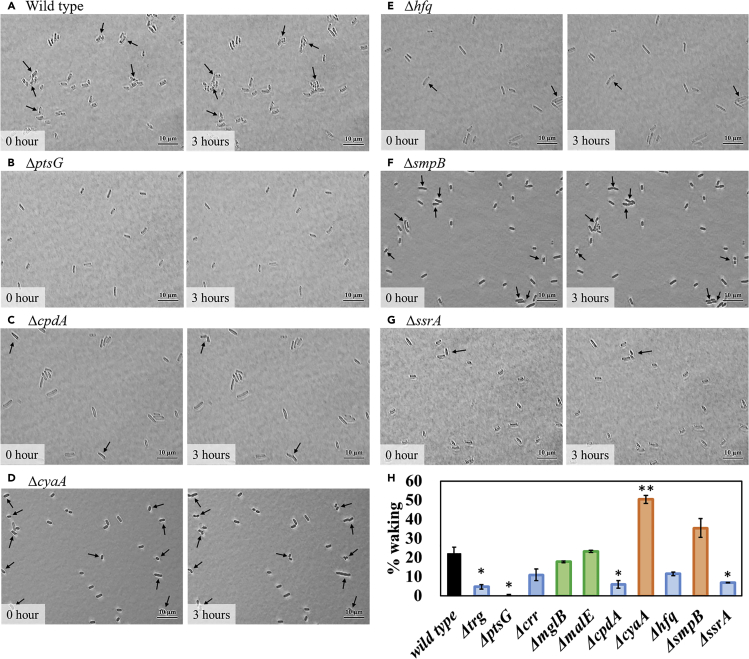
For waking on Ala, we discovered that the methyl-accepting chemotaxis proteins Trg and Tar are used for persister resuscitation. Hence, we hypothesized that the glucose phosphotransferase transport protein PtsG, which interacts indirectly with the chemotaxis system via chemoeffector CheA (Neumann et al., 2012), may be responsible for waking with glucose. Supporting this hypothesis, we found that inactivating PtsG reduced waking on agar plates (Figure S25) and nearly abolished the frequency of single-cell waking on glucose (70-fold reduction, Figure 5 and Table S11). In addition, production of PtsG from pCA24N-ptsG abolished glucose waking indicating that the stoichiometry of the PtsG proteins in the inner membrane is critical (Figures 6 and S26 and Table S12). As a negative control, the frequency of waking was tested for the maltose PTS transporter by inactivating MalE; as expected, deletion of malE had no impact on waking with glucose (Figures 5 and S27 and Table S11). Hence, persister cells wake on glucose through its phosphotransferase system.
Figure 5.
Single Persister Cell Waking on Glucose after Inactivating Proteins Related to Transport, cAMP, sRNA, and Ribosome Rescue
(A–G) E. coli BW25113 persister cells (A) and isogenic deletion mutants (B) ΔptsG, (C) ΔcpdA, (D) ΔcyaA, (E) Δhfq, (F) ΔsmpB, and (G) ΔssrA were incubated at 37°C on 0.4% glucose agarose gel pads for 3 h. Black arrows indicate cells that resuscitate. Scale bar indicates 10 μm. Representative results from two independent cultures are shown.
(H) Persister cell waking (%) after incubating for 3 h. Tabulated cell numbers are shown in Table S11, and error bars indicate standard deviations. Student's t tests were used to compare two groups (* indicates a p value <0.05 and ** indicates a p value <0.01). Additional microscope images are shown in Figure S27.
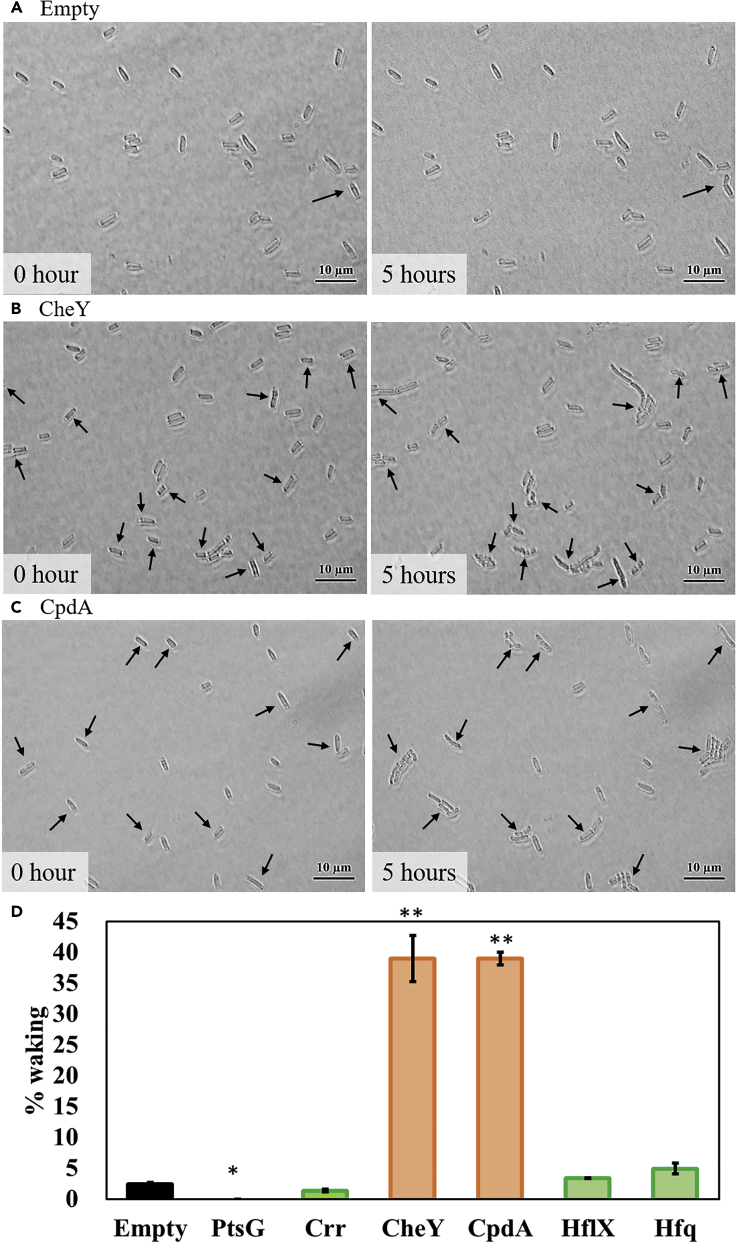
Figure 6.
Single Persister Cell Waking on Glucose after Producing Proteins Related to Transport, Chemotaxis, cAMP, Ribosome Resuscitation, and sRNA
(A–C) E. coli BW25113 persister cells containing (A) pCA24N, (B) pCA24N_cheY, and (C) pCA24N_cpdA were incubated at 37°C on M9 0.4% glucose agarose gel pads for 5 h. Black arrows indicate cells that resuscitate. Scale bar indicates 10 μm. Representative results from two independent cultures are shown. Before forming persister cells, cultures were grown without IPTG with 30 μg/mL Cm.
(D) Persister cell waking (%) after incubating for 5 h. Tabulated cell numbers are shown in Table S12, and error bars indicate standard deviations. Student's t tests were used to compare two groups (* indicates a p value <0.05 and ** indicates a p value <0.01). Other microscope images are shown in Figure S26.
Since persister waking on glucose occurs via PtsG, we tested whether the chemotaxis system was involved since the transported glucose stimulates chemotaxis (Neumann et al., 2012). By producing chemoeffector CheY, we found that the frequency of single-cell waking increased dramatically (16-fold) (Figure 6 and Table S12). Hence, persister cell waking with glucose also depends on an active chemotaxis system.
We also tested whether the resuscitation of persister cells on glucose depends on the chemotaxis ABC glucose transporter MglB and found a 1.2-fold reduction in the frequency of single-cell waking upon inactivating MglB (Figures 5 and S27 and Table S11). Corroborating this result, deleting the methyl-accepting chemotaxis protein Trg, which interacts with glucose from MglB (Neumann et al., 2012), reduced the frequency of single-cell waking by 5-fold (Figures 5 and S27 and Table S11). Hence, persister cell resuscitation on glucose requires both well-known glucose transport systems as well as the chemotaxis system.
Glucose Waking Requires Reduced cAMP
Since Ala resuscitation requires the cell to lower cAMP levels, we explored if this occurs with glucose and found again that low cAMP levels are needed since adding 2 mM cAMP, as well as inactivating phosphodiesterase CpdA (Imamura et al., 1996) to increase cAMP levels, reduces waking on glucose on agar plates (Figure S28). Corroborating these plate results, producing CpdA to lower cAMP levels increased the frequency of waking of single cells dramatically (16-fold) on glucose (Figure 6 and Table S12), and inactivating adenylate cyclase CyaA (Tuckerman et al., 2009) to eliminate cAMP increased the frequency of waking on glucose substantially (2.3-fold) (Figure 5 and Table S11). In addition, inactivating CpdA reduced the frequency of single-cell waking by 4-fold (Figure 5 and Table S11). Therefore, cAMP levels must be low for persister cells to wake with glucose.
Glucose Transport Lowers cAMP Concentrations
We hypothesized that the requisite reduction in cAMP upon resuscitation with glucose is a result of glucose transport with PtsG, since the associated phosphotransferase system enzyme EIIAGlc becomes unphosphorylated; unphosphorylated EIIAGlc inactivates adenylate cyclase CyaA and leads to low cAMP (Deutscher, 2008, Yao et al., 2011) (a well-known aspect of catabolite repression). In support of this model, we tested the crr isogenic mutant (crr encodes EIIAGlc) with single cells and found that inactivating EIIAGlc reduced the frequency of single-cell waking 2-fold (Figures 5 and S27 and Table S11). In addition, single cells producing EIIA (pCA24N-crr) had 2-fold reduced waking frequency (Figures 6 and S26 and Table S12). Therefore, reducing cAMP concentrations through EIIAGlc is important for persister cell waking.
Glucose Waking Requires Ribosome Resuscitation
Since waking on Ala requires cells to rescue ribosomes from corrupt mRNA and from dimerization via HflX and to rescue ribosomes from corrupt mRNA via SsrA, we tested whether these proteins are also necessary for persister cell resuscitation on glucose. For waking on agar plates, inactivating trans-translation (i.e., SsrA) abolished waking (Figure S29). For single-cell waking with glucose, there was only a small positive effect of producing HflX (Figures 6 and S26 and Table S12); however, inactivating trans-translation (ΔssrA) reduced waking dramatically (-3-fold) (Figure 5 and Table S11). Therefore, ribosome resuscitation through SsrA is primarily responsible for waking with glucose.
Cells Wake and Begin Chemotaxis Simultaneously
To determine if persister cells resuscitate and undergo chemotaxis simultaneously, we observed single persister cell waking inside motility agar with a glucose gradient; therefore, unlike the previous results where persister cells resuscitated on the surface of agarose gel pads, here persisters resuscitate and move planktonically inside a 0.3% agar (motility) gel. Note that the movement of the cells in these experiments is not due to fluid flow or random motion since the cell movement is always toward glucose in many repeated trials and since there is no fluid flow in this static 0.3% gel, which is a semisolid rather than a liquid.
Critically, we found that 20 ± 6% of the wild-type persister cells resuscitate and undergo chemotaxis immediately toward the glucose (Video S1). As expected, wild-type persister cells do not move without glucose. Since reducing cAMP by inactivating cyaA increases waking (Figures 5 and S18), we hypothesized that the cyaA mutation would magnify this effect of instantaneous waking; strikingly, 94 ± 2% of the cyaA persister cells moved immediately toward glucose, indicating that the reduction of cAMP primes the cells for waking and chemotaxis. Furthermore, we reasoned that inactivating EIIA would also reduce cAMP and stimulate waking and found 96 ± 6% of the crr cells woke and began chemotaxis to glucose (Video S1). Critically the crr mutant cells wake instantaneously without glucose and show random movement in the M9 motility agar that lacks glucose confirming that low cAMP levels lead to more active resuscitation. The crr mutants still formed persister cells since any non-persister cells were removed during the 3-h ampicillin treatment. Therefore, the E. coli persister cells that resuscitate immediately begin moving toward nutrients. To determine if this phenotype is general, we investigated whether P. aeruginosa PA14 persister cells undergo chemotaxis upon resuscitating and found they also immediately move toward the glucose; in contrast, P. aeruginosa PA14 persister cells that lack nutrients do not undergo chemotaxis (Video S2). Hence, persister cells wake and undergo chemotaxis to find nutrients.
The video shows the resuscitation of wild-type, ΔcyaA, and Δcrr persister cells in motility agar with a glucose gradient (0% to 5%, with the glucose at high concentrations on the left-hand side). One representative video of two independent cultures is shown.
The video shows the resuscitation of P. aeruginosa PA14 wild-type cells in motility agar with a glucose gradient (0% to 5%, with the glucose at high concentrations on the left-hand side) and without glucose. One representative video of two independent cultures is shown.
Stationary-Phase Cells Revive Differently Than Persisters
Having established how single persister cells resuscitate, we investigated how single stationary-phase cells revive for comparison. Although glucose revives both persister and stationary-phase cells, nearly all of the stationary-phase cells revive immediately (98 ± 2%), whereas only 7 ± 2% of the persister cells revive (Figure S30). Additionally, unlike with persisters, no ghost cells (dead cells with cytosolic components condensed at the poles) are seen for stationary-phase cells, and no lysing stationary-phase cells are seen due to killing as a result of residual ampicillin as seen with persisters (Figure S30). Hence, stationary phase cells are phenotypically distinct from persister cells.
Discussion
Spores are the ultimate bacterial resting state, and L-alanine revives spores of Bacillus subtilis (Mutlu et al., 2018). Hence, we hypothesized that L-alanine or another amino acid may wake E. coli persisters. By investigating all 20 amino acids, we found that L-alanine wakes E. coli persister cells much faster than other amino acids (Figures S1 and S2). To determine insights into how L-alanine wakes E. coli persisters, we pooled the ASKA clones, isolated their plasmids, electroporated them into E. coli BW25113, made persister cells, and selected for faster waking. From this selection, six proteins were identified that increased waking: PsiF, PanD, YmgF, YjcF, PptA, and CheY. From these six proteins, we found PanD and YmgF increase waking 20-fold. PanD (aspartate 1-decarboxylase) (Cronan, 1980) catalyzes the reaction of L-aspartate to β-alanine; hence, PanD produces an alanine-like compound. YmgF is related to an inner membrane division septum protein (Karimova et al., 2009); therefore, the persister cells wake and are primed for cell division. Critically, we determined that the chemotaxis response regulator CheY and the chemotaxis methyl-accepting chemotaxis proteins Tar and Trg are utilized for persister resuscitation. Surprisingly, the chemotaxis methyl-accepting protein Tsr, which is primarily responsible for Ala chemotaxis (Tajima et al., 2011), was not involved in persister waking; however, Tar is also utilized for Ala chemotaxis (Yang et al., 2015). Therefore, persister cells wake through the same nutrient sensors they utilize for chemotaxis. Once resuscitated, the cells then rapidly move toward nutrients since the chemotaxis system is primed immediately, and the lack of nutrients (and energy for protein production) is what caused the dormancy in the first place (Kwan et al., 2013).
cAMP activates ribosome hibernation as 100S dimers during starvation by activating ribosome-binding protein RMF (Shimada et al., 2013) so it makes sense for cells to reverse this process (as we find here) and reduce cAMP levels to wake in the presence of nutrients, since we identified that the activity of ribosomes dictates the speed of persister resuscitation (Kim et al., 2018b). Supporting this insight, we found that inactivation of the ribosome rescue factor HflX (heat shock-induced ribosome-dependent GTPase) completely stopped waking of single cells and production of HflX increased the frequency of waking dramatically; hflX (in the same operon as hfq) is repressed by cAMP-Crp (Lin et al., 2011). In contrast, ribosome rescue factor Arfb (peptidyl-tRNA hydrolase) had no effect because it is induced by cAMP-Crp (Raghavan et al., 2011). Critically, all bacteria have mechanisms for rescuing ribosomes since without rescue, all ribosomes would be depleted in less than one generation (Moore and Sauer, 2007), and we found that, without ribosome rescue via the trans-translation system (SsrA), persister cell waking is almost completely inhibited for Ala (Figure S21) and reduced for glucose (Figure 5). Hence, ribosome rescue during persister cell resuscitation may be a general mechanism and should provide ample ribosomes for resuscitation.
Although it is counter intuitive, in regard to the formation of persister cells, high concentrations of cAMP allow ribosomes to hibernate and prepare the cell for nutrient-depleted conditions; yet, high concentrations of cAMP actually reduce persistence (Kwan et al., 2015b) (reduce cell dormancy). This is because cells with high cAMP concentrations are more prepared for stress and have less need to become dormant; i.e., more fit cells have dramatically less persistence (Hong et al., 2012).
Overall, by studying persister cell resuscitation with alanine, we have identified that (1) persister cells wake primarily in response to nutrient stimulus (rather than spontaneously), (2) Ala is the most suitable amino acid signal for waking E. coli persister cells, (3) this nutrient signal is perceived through the chemotaxis system via CheY, Trg, and Tar, (4) second messenger cAMP concentrations are reduced for waking, and (5) ribosomes are restored for waking (Figure 7). By studying persister cell resuscitation with glucose, we confirmed that chemotaxis and ribosome rescue are important for persister waking. Critically, the phosphotransferase system that imports glucose serves to reduce cAMP concentrations by dephosphorylating EIIAGlc, which allows ribosomes to resuscitate. Notably, the same mechanism for cAMP reduction is possible for alanine resuscitation via the chemotaxis system that detects it, since CheA, which we found to be critical for resuscitation with alanine, interacts directly with unphosphorylated EIIAGlc (Neumann et al., 2012). Therefore, persister cell resuscitation is an elegant (i.e., highly regulated) response in which nutrients are perceived as environmental signals via membrane receptors; this environmental signal is propagated to ribosomes via the secondary metabolite cAMP, and persister cells commence foraging as they wake.
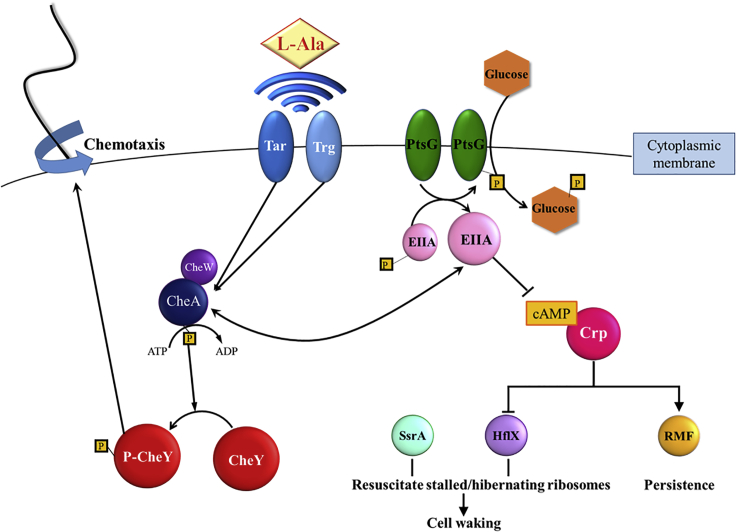
Figure 7.
Schematic of Persister Cell Waking via Alanine and Glucose
For alanine resuscitation, methyl-accepting chemotaxis proteins Tar and Trg sense the amino acid and relay this to chemotaxis response regulators CheA and CheY, which stimulate chemotaxis. For glucose resuscitation, phosphotransferase protein PtsG imports the sugar, which results in dephosphorylation of EIIA, reduction in cAMP, activation of chemotaxis, and ribosome rescue via HflX and SsrA. Spheres indicate proteins, diamonds indicate amino acids, hexagons indicate glucose,  indicates phosphate, → indicates induction, and
indicates phosphate, → indicates induction, and  indicates repression.
indicates repression.
Limitations of the Study
For our plate-based assays that were used as rapid indicator of faster cell resuscitation, it is important to note that we did not compare growth across all conditions (growth was compared for the conditions used to generate persister cells as shown in Table S6); hence, some differences in colony size may be due to differences in growth as well as differences in resuscitation. Therefore, the definitive differences in resuscitation between strains should be based on the single-cell analyses. For these single-cell microscopic experiments, the frequency of resuscitation is the fraction of the total cell population (typically 100–300 single cells for each independent culture) that grows as demonstrated by elongation or by doubling in a fixed period. Furthermore, since not all possible carbon sources were tested for persister cell resuscitation, not all mechanistic details of resuscitation are possible to be elucidated here, although recognition of nutrients via membrane receptors along with reduction in cAMP levels seem general for resuscitation since it happened for resuscitation with both a carbohydrate (glucose) and an amino acid (alanine). In addition, we note that the studies based on producing proteins via expression with pCA24N-derived constructs are not optimized for protein production.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgements
This work was supported by funds derived from the Biotechnology Endowed Professorship at the Pennsylvania State University. We thank Prof. John Parkinson and Prof. Pushkar Lele and for fruitful discussions about chemotaxis.
Author Contributions
R.Y. and S.S. performed the experiments. R.Y., S.S., M.J.B., and T.K.W. designed the experiments.
Declaration of Interests
The authors declare no competing interests.
Published: January 24, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.isci.2019.100792.
Data and Code Availability
All data are present in the manuscript.
Supplemental Information
References
- Abo T., Chadani Y. The fail-safe system to rescue the stalled ribosomes in Escherichia coli. Front. Microbiol. 2014;5:156. doi: 10.3389/fmicb.2014.00156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaban N.Q., Merrin J., Chait R., Kowalik L., Leibler S. Bacterial persistence as a phenotypic switch. Science. 2004;305:1622–1625. doi: 10.1126/science.1099390. [DOI] [PubMed] [Google Scholar]
- Bernier S.P., Lebeaux D., DeFrancesco A.S., Valomon A., Soubigou G., Coppee J.Y., Ghigo J.M., Beloin C. Starvation, together with the SOS response, mediates high biofilm-specific tolerance to the fluoroquinolone ofloxacin. PLoS Genet. 2013;9:e1003144. doi: 10.1371/journal.pgen.1003144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigger J.W. Treatment of staphylococcal infections with penicillin - by intermittent sterilisation. Lancet. 1944;2:497–500. [Google Scholar]
- Brown M.T., Steel B.C., Silvestrin C., Wilkinson D.A., Delalez N.J., Lumb C.N., Obara B., Armitage J.P., Berry R.M. Flagellar hook flexibility is essential for bundle formation in swimming Escherichia coli cells. J. Bacteriol. 2012;194:3495–3501. doi: 10.1128/JB.00209-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheverton A.M., Gollan B., Przydacz M., Wong C.T., Mylona A., Hare S.A., Helaine S. A salmonella toxin promotes persister formation through acetylation of tRNA. Mol. Cell. 2016;63:86–96. doi: 10.1016/j.molcel.2016.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury N., Kwan B.W., Wood T.K. Persistence increases in the absence of the alarmone guanosine tetraphosphate by reducing cell growth. Sci. Rep. 2016;6:20519. doi: 10.1038/srep20519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury N., Wood T.L., Martinez-Vazquez M., Garcia-Contreras R., Wood T.K. DNA-crosslinker cisplatin eradicates bacterial persister cells. Biotechnol. Bioeng. 2016;113:1984–1992. doi: 10.1002/bit.25963. [DOI] [PubMed] [Google Scholar]
- Cronan J.E. Beta-alanine synthesis in Escherichia-coli. J. Bacteriol. 1980;141:1291–1297. doi: 10.1128/jb.141.3.1291-1297.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui P., Niu H., Shi W., Zhang S., Zhang W., Zhang Y. Identification of genes involved in bacteriostatic antibiotic-induced persister formation. Front. Microbiol. 2018;9:413. doi: 10.3389/fmicb.2018.00413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher J. The mechanisms of carbon catabolite repression in bacteria. Curr. Opin. Microbiol. 2008;11:87–93. doi: 10.1016/j.mib.2008.02.007. [DOI] [PubMed] [Google Scholar]
- Dörr T., Vulić M., Lewis K. Ciprofloxacin causes persister formation by inducing the TisB toxin in Escherichia coli. PLoS Biol. 2010;8:e1000317. doi: 10.1371/journal.pbio.1000317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher R.A., Gollan B., Helaine S. Persistent bacterial infections and persister cells. Nat. Rev. Microbiol. 2017;15:453–464. doi: 10.1038/nrmicro.2017.42. [DOI] [PubMed] [Google Scholar]
- Franklin F.C.H., Venables W.A. Biochemical, genetic, and regulatory studies of alanine catabolism in Escherichia coil K12. Mol. Gen. Genet. 1976;149:229–237. doi: 10.1007/BF00332894. [DOI] [PubMed] [Google Scholar]
- Gohara D.W., Yap M.-N.F. Survival of the drowsiest: the hibernating 100S ribosome in bacterial stress management. Curr. Genet. 2018;64:753–760. doi: 10.1007/s00294-017-0796-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goormaghtigh F., Fraikin N., Putrinš M., Hallaert T., Hauryliuk V., Garcia-Pino A., Sjödin A., Kasvandik S., Udekwu K., Tenson T. Reassessing the role of type II toxin-antitoxin systems in formation of Escherichia coli type II persister cells. mBio. 2018;9 doi: 10.1128/mBio.00640-18. e00640–00618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grassi L., Di Luca M., Maisetta G., Rinaldi A.C., Esin S., Trampuz A., Batoni G. Generation of persister cells of Pseudomonas aeruginosa and Staphylococcus aureus by chemical treatment and evaluation of their susceptibility to membrane-targeting agents. Front. Microbiol. 2017;8:1917. doi: 10.3389/fmicb.2017.01917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison J.J., Wade W.D., Akierman S., Vacchi-Suzzi C., Stremick C.A., Turner R.J., Ceri H. The chromosomal toxin gene yafQ is a determinant of multidrug tolerance for Escherichia coli growing in a biofilm. Antimicrob. Agents Chemother. 2009;53:2253–2258. doi: 10.1128/AAC.00043-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobby G.L., Meyer K., Chaffee E. Observations on the mechanism of action of penicillin. Proc. Soc. Exp. Biol. Med. 1942;50:281–285. [Google Scholar]
- Hong S.H., Wang X., O'Connor H.F., Benedik M.J., Wood T.K. Bacterial persistence increases as environmental fitness decreases. Microb. Biotechnol. 2012;5:509–522. doi: 10.1111/j.1751-7915.2011.00327.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh T.T., McDougald D., Klebensberger J., Al Qarni B., Barraud N., Rice S.A., Kjelleberg S., Schleheck D. Glucose starvation-induced dispersal of Pseudomonas aeruginosa biofilms is cAMP and energy dependent. PLoS One. 2012;7:e42874. doi: 10.1371/journal.pone.0042874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura R., Yamanaka K., Ogura T., Hiraga S., Fujita N., Ishihama A., Niki H. Identification of the cpdA gene encoding cyclic 3ʹ,5ʹ-adenosine monophosphate phosphodiesterase in Escherichia coli. J. Biol. Chem. 1996;271:25423–25429. doi: 10.1074/jbc.271.41.25423. [DOI] [PubMed] [Google Scholar]
- Karimova G., Robichon C., Ladant D. Characterization of YmgF, a 72-residue inner membrane protein that associates with the Escherichia coli cell division machinery. J. Bacteriol. 2009;191:333–346. doi: 10.1128/JB.00331-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.-S., Chowdhury N., Yamasaki R., Wood T.K. Viable but non-culturable and persistence describe the same bacterial stress state. Environ. Microbiol. 2018;20:2038–2048. doi: 10.1111/1462-2920.14075. [DOI] [PubMed] [Google Scholar]
- Kim J.-S., Yamasaki R., Song S., Zhang W., Wood T.K. Single cell observations show persister cells wake based on ribosome content. Environ. Microbiol. 2018;20:2085–2098. doi: 10.1111/1462-2920.14093. [DOI] [PubMed] [Google Scholar]
- Kim Y., Wood T.K. Toxins Hha and CspD and small RNA regulator Hfq are involved in persister cell formation through MqsR in Escherichia coli. Biochem. Biophys. Res. Commun. 2010;391:209–213. doi: 10.1016/j.bbrc.2009.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemm D., Heublein B., Fink H.P., Bohn A. Cellulose: fascinating biopolymer and sustainable raw material. Angew. Chem. Int. Ed. 2005;44:3358–3393. doi: 10.1002/anie.200460587. [DOI] [PubMed] [Google Scholar]
- Kwan B.W., Chowdhury N., Wood T.K. Combatting bacterial infections by killing persister cells with mitomycin C. Environ. Microbiol. 2015;17:4406–4414. doi: 10.1111/1462-2920.12873. [DOI] [PubMed] [Google Scholar]
- Kwan B.W., Osbourne D.O., Hu Y., Benedik M.J., Wood T.K. Phosphodiesterase DosP increases persistence by reducing cAMP which reduces the signal indole. Biotechnol. Bioeng. 2015;112:588–600. doi: 10.1002/bit.25456. [DOI] [PubMed] [Google Scholar]
- Kwan B.W., Valenta J.A., Benedik M.J., Wood T.K. Arrested protein synthesis increases persister-like cell formation. Antimicrob. Agents Chemother. 2013;57:1468–1473. doi: 10.1128/AAC.02135-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.K., de Anda J., Baker A.E., Bennett R.R., Luo Y., Lee E.Y., Keefe J.A., Helali J.S., Ma J., Zhao K. Multigenerational memory and adaptive adhesion in early bacterial biofilm communities. Proc. Natl. Acad. Sci. U S A. 2018;115:4471–4476. doi: 10.1073/pnas.1720071115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H.H., Hsu C.C., Yang C.D., Ju Y.W., Chen Y.P., Tseng C.P. Negative effect of glucose on ompA mRNA stability: a potential role of cyclic AMP in the repression of hfq in Escherichia coli. J. Bacteriol. 2011;193:5833–5840. doi: 10.1128/JB.05359-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luidalepp H., Jõers A., Kaldalu N., Tenson T. Age of inoculum strongly influences persister frequency and can mask effects of mutations implicated in altered persistence. J. Bacteriol. 2011;193:3598–3605. doi: 10.1128/JB.00085-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maisonneuve E., Gerdes K. Molecular mechanisms underlying bacterial persisters. Cell. 2014;157:539–548. doi: 10.1016/j.cell.2014.02.050. [DOI] [PubMed] [Google Scholar]
- Martins P.M.M., Merfa M.V., Takita M.A., De Souza A.A. Persistence in phytopathogenic bacteria: do we know enough? Front. Microbiol. 2018;9:1099. doi: 10.3389/fmicb.2018.01099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megaw J., Gilmore B.F. Archaeal persisters: persister cell formation as a stress response in Haloferax volcanii. Front. Microbiol. 2017;8:1589. doi: 10.3389/fmicb.2017.01589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore S.D., Sauer R.T. The tmRNA system for translational surveillance and ribosome rescue. Annu. Rev. Biochem. 2007;76:101–124. doi: 10.1146/annurev.biochem.75.103004.142733. [DOI] [PubMed] [Google Scholar]
- Mutlu A., Trauth S., Ziesack M., Nagler K., Bergeest J.P., Rohr K., Becker N., Hofer T., Bischofs I.B. Phenotypic memory in Bacillus subtilis links dormancy entry and exit by a spore quantity-quality tradeoff. Nat. Commun. 2018;9:69. doi: 10.1038/s41467-017-02477-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanaswamy V.P., Keagy L.L., Duris K., Wiesmann W., Loughran A.J., Townsend S.M., Baker S. Novel glycopolymer eradicates antibiotic- and CCCP-induced persister cells in Pseudomonas aeruginosa. Front. Microbiol. 2018;9:1724. doi: 10.3389/fmicb.2018.01724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann S., Grosse K., Sourjik V. Chemotactic signaling via carbohydrate phosphotransferase systems in Escherichia coli. Proc. Natl. Acad. Sci. U S A. 2012;109:12159–12164. doi: 10.1073/pnas.1205307109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel K.B., Toh E., Fernandez X.B., Hanuszkiewicz A., Hardy G.G., Brun Y.V., Bernards M.A., Valvano M.A. Functional characterization of UDP-glucose:undecaprenyl-phosphate glucose-1-phosphate transferases of Escherichia coli and Caulobacter crescentus. J. Bacteriol. 2012;194:2646–2657. doi: 10.1128/JB.06052-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokhis K., Bitterlich N., Cornelli U., Cassano G. Efficacy of polyglucosamine for weight loss confirmed in a randomized double-blind, placebo-controlled clinical investigation. BMC Obes. 2015;2:25. doi: 10.1186/s40608-015-0053-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontes M.H., Groisman E.A. Slow growth determines nonheritable antibiotic resistance in Salmonella enterica. Sci. Signal. 2019;12:eaax3938. doi: 10.1126/scisignal.aax3938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu Y., Li Y., Jin X., Tian T., Ma Q., Zhao Z., Lin S.-y., Chen Z., Li B., Yao G. ATP-dependent dynamic protein aggregation regulates bacterial dormancy depth critical for antibiotic tolerance. Mol. Cell. 2019;73:143–156. doi: 10.1016/j.molcel.2018.10.022. [DOI] [PubMed] [Google Scholar]
- Raghavan R., Sage A., Ochman H. Genome-wide identification of transcription start sites yields a novel thermosensing RNA and new cyclic AMP receptor protein-regulated genes in Escherichia coli. J. Bacteriol. 2011;193:2871–2874. doi: 10.1128/JB.00398-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah D., Zhang Z., Khodursky A., Kaldalu N., Kurg K., Lewis K. Persisters: a distinct physiological state of E. coli. BMC Microbiol. 2006;6:53. doi: 10.1186/1471-2180-6-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan Y., Lazinski D., Rowe S., Camilli A., Lewis K. Genetic basis of persister tolerance to aminoglycosides in Escherichia coli. mBio. 2015;6 doi: 10.1128/mBio.00078-15. e00078–00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi W., Li J., Ji L., Zhang Y., Xie J. Trans-translation mediates tolerance to multiple antibiotics and stresses in Escherichia coli. J. Antimicrob. Chemother. 2013;68:2477–2481. doi: 10.1093/jac/dkt231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada T., Yoshida H., Ishihama A. Involvement of cyclic AMP receptor protein in regulation of the rmf gene encoding the ribosome modulation factor in Escherichia coli. J. Bacteriol. 2013;195:2212–2219. doi: 10.1128/JB.02279-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S., Wood T.K. Post-segregational killing and phage inhibition are not mediated by cell death through toxin/antitoxin systems. Front. Microbiol. 2018;9:814. doi: 10.3389/fmicb.2018.00814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S., Wood T.K. ppGpp ribosome dimerization model for bacterial persister formation and resuscitation. bioRxiv. 2019 doi: 10.1016/j.bbrc.2020.01.102. [DOI] [PubMed] [Google Scholar]
- Stevenson G., Andrianopoulos K., Hobbs M., Reeves P.R. Organization of the Escherichia coli K-12 gene cluster responsible for production of the extracellular polysaccharide colanic acid. J. Bacteriol. 1996;178:4885–4893. doi: 10.1128/jb.178.16.4885-4893.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulaiman J.E., Hao C., Lam H. Specific enrichment and proteomics analysis of Escherichia coli persisters from rifampin pretreatment. J. Proteome Res. 2018;17:3984–3996. doi: 10.1021/acs.jproteome.8b00625. [DOI] [PubMed] [Google Scholar]
- Svenningsen M.S., Veress A., Harms A., Mitarai N., Semsey S. Birth and resuscitation of (p)ppGpp induced antibiotic tolerant persister cells. Sci. Rep. 2019;9:6056. doi: 10.1038/s41598-019-42403-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima H., Imada K., Sakuma M., Hattori F., Nara T., Kamo N., Homma M., Kawagishi I. Ligand specificity determined by differentially arranged common ligand-binding residues in bacterial amino acid chemoreceptors Tsr and Tar. J. Biol. Chem. 2011;286:42200–42210. doi: 10.1074/jbc.M111.221887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tkhilaishvili T., Lombardi L., Klatt A.-B., Trampuz A., Di Luca M. Bacteriophage Sb-1 enhances antibiotic activity against biofilm, degrades exopolysaccharide matrix and targets persisters of Staphylococcus aureus. Int. J. Antimicrob. Agents. 2018;52:842–853. doi: 10.1016/j.ijantimicag.2018.09.006. [DOI] [PubMed] [Google Scholar]
- Tuckerman J.R., Gonzalez G., Sousa E.H.S., Wan X., Saito J.A., Alam M., Gilles-Gonzalez M.-A. An oxygen-sensing diguanylate cyclase and phosphodiesterase couple for c-di-GMP control. Biochemistry. 2009;48:9764–9774. doi: 10.1021/bi901409g. [DOI] [PubMed] [Google Scholar]
- Uehara T., Dinh T., Bernhardt T.G. LytM-domain factors are required for daughter cell separation and rapid ampicillin-induced lysis in Escherichia coli. J. Bacteriol. 2009;191:5094–5107. doi: 10.1128/JB.00505-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Kim Y., Wood T.K. Control and benefits of CP4-57 prophage excision in Escherichia coli biofilms. ISME J. 2009;3:1164–1179. doi: 10.1038/ismej.2009.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild J., Klopotowski T. D-amino acid dehydrogenase of Escherichia coli K12: positive selection of mutants defective in enzyme activity and localization of the structural gene. Mol. Gen. Genet. 1981;181:373–378. doi: 10.1007/BF00425614. [DOI] [PubMed] [Google Scholar]
- Yang Y.M., Pollard A., Höfler C., Poschet G., Wirtz M., Hell R., Sourjik V. Relation between chemotaxis and consumption of amino acids in bacteria. Mol. Microbiol. 2015;96:1272–1282. doi: 10.1111/mmi.13006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao R., Hirose Y., Sarkar D., Nakahigashi K., Ye Q., Shimizu K. Catabolic regulation analysis of Escherichia coli and its crp, mlc, mgsA, pgi and ptsG mutants. Microb. Cell Fact. 2011;10:67. doi: 10.1186/1475-2859-10-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Mandava C.S., Cao W., Li X., Zhang D., Li N., Zhang Y., Zhang X., Qin Y., Mi K. HflX is a ribosome-splitting factor rescuing stalled ribosomes under stress conditions. Nat. Struct. Mol. Biol. 2015;22:906. doi: 10.1038/nsmb.3103. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The video shows the resuscitation of wild-type, ΔcyaA, and Δcrr persister cells in motility agar with a glucose gradient (0% to 5%, with the glucose at high concentrations on the left-hand side). One representative video of two independent cultures is shown.
The video shows the resuscitation of P. aeruginosa PA14 wild-type cells in motility agar with a glucose gradient (0% to 5%, with the glucose at high concentrations on the left-hand side) and without glucose. One representative video of two independent cultures is shown.
Data Availability Statement
All data are present in the manuscript.