Abstract
Background
Hyperamyloidosis in the brain is known as the earliest neuropathological change and a unique etiological factor in Alzheimer's disease (AD), while progressive neurodegeneration in certain vulnerable brain regions forms the basis of clinical syndromes. It is not clear how early hyperamyloidosis is implicated in progressive neurodegeneration and what factors contribute to the selective brain vulnerability in AD.
Methods
Bioinformatics and experimental neurobiology methods were integrated to identify novel factors involved in the hyperamyloidosis-induced brain vulnerability in AD. We first examined neurodegeneration-specific gene signatures from sporadic AD patients and synaptic protein changes in young transgenic AD mice. Then, we systematically assessed the association of a top candidate gene with AD and investigated its mechanistic role in neurodegeneration.
Findings
We identified the ovary-orientated protein OCIAD1 (Ovarian-Carcinoma-Immunoreactive-Antigen-Domain-Containing-1) as a neurodegeneration-associated factor for AD. Higher levels of OCIAD1, found in vulnerable brain areas and dystrophic neurites, were correlated with disease severity. Multiple early AD pathological events, particularly Aβ/GSK-3β signaling, elevate OCIAD1, which in turn interacts with BCL-2 to impair mitochondrial function and facilitates mitochondria-associated neuronal injury. Notably, elevated OCIAD1 by Aβ increases cell susceptibility to other AD pathological challenges.
Interpretation
Our findings suggest that OCIAD1 contributes to neurodegeneration in AD by impairing mitochondria function, and subsequently leading to neuronal vulnerability, and synaptic damages.
Funding
Ting Tsung & Wei Fong Chao Foundation, John S Dunn Research Foundation, Cure Alzheimer's Fund, and NIH R01AG057635 to STCW.
Keywords: OCIAD1, Neurodegeneration, Hyperamyloidosis, Mitochondrial dysfunction, Alzheimer's disease
Abbreviations: APP, amyloid precursor protein (A4); β-tub, β-tubulin; c-CAS3, cleaved caspase-3; c-CAS9, Cleaved caspase-9; CER, cerebellum; c-PARP, cleaved PARP; CTNNB1, β-catenin. CTX, cerebral cortex; DNT, dystrophic neurite; EC, endorhinal cortex; enOD1, endogenous OCIAD1; exOD1, exogenous OCIAD1; HEK, human embryo kidney cell; HIP, hippocampus; HK1, hexokinase I. MCF, medial frontal cortex; MG, MG-132; Mid, midbrain; mit-OD1, mitochondrial OCIAD1; MMSE, Minimal Mental State Examination; MTL, mesial temporal lobe; OD1, OCIAD1; Olf, olfactory bulb; p53, tumor protein p53; PCC, posterior cingulate cortex; SFG, superior frontal gyrus; SDHB, succinate dehydrogenase complex iron sulfur subunit B; SMAC, Second mitochondria-derived activator of caspase; PUMA, p53 upregulated modulator of apoptosis; VDAC1, Voltage Dependent Anion Channel 1; VCX, primary visual cortex; Veh, vehicle
Research in context.
Evidence before this study
The pathogenesis of Alzheimer's disease (AD) is elusive and no effective therapy is available. It is known that neurodegeneration is the base of dementia and certain brain areas are more vulnerable in AD, but how hyperamyloidosis, the earliest pathological change and a unique etiological factor in AD, is implicated in neurodegeneration remains unclear. Some unknown mechanisms, other than hyperamyloidosis and taupathy, may contribute to the brain vulnerability and progressive neurodegeneration in AD. Certain early disease changes in AD (ie., hyperamyloidosis and synaptic damage) can be replicated in transgenic mice while many public research databases and bioinformatics methods enable the investigation of complex biological networks or signaling pathways in human disease. Integrating bioinformatics and biological methods to examine omics data from different species and disease stages may enhance the power of researchers to decipher complex mechanisms underlying neurodegeneration at a systematic level or identify potential etiological factors for AD.
Added value of this study
In this study, we systematically examined neurodegeneration-relevant factors in the vulnerable brain areas of patients of sporadic AD and transgenic mouse models. We integrated bioinformatics methods with neurobiological tools to assess and validate the functional role of a newly-identified factor in AD pathogenesis. We found that: (i) OCIAD1 is a factor associated with neurodegeneration and under regulation of multiple AD-relevant pathological changes, including Aβ/GSK3β and p53 signals; (ii) mis-regulated OCIAD1 interacts with BCL-2 and contributes to mitochondria dysfunction, neuronal susceptibility, and synaptic damage; and (iii) elevated OCIAD1 mediates the long-term impact of Aβ on cell vulnerability and synaptic caspase-3 activation.
Implications of all the available evidence
Our study reveals a new role of OCIAD1 in neurodegeneration, and helps better understand the pathophysiological relationship between brain vulnerability and hyperamyloidosis in AD. This methodology has the potential to help identify therapeutic targets for other neurodegenerative diseases.
Alt-text: Unlabelled box
1. Introduction
Alzheimer's disease (AD) is a complex neurological disorder with pathological hallmarks of hyperamyloidosis (senile plaques), neurofibrillary tangles containing hyperphosphorylated tau, and extensive neurodegeneration in the brain [1], [2], [3]. AD pathogenesis remains elusive and no effective therapy is available [4,5]. Neurodegeneration, including synaptic damage and neuronal loss, forms the basis of dementia in AD [1,6], and certain brain regions are more vulnerable during disease progression [7], [8], [9]. Several pathological events are implicated in this deleterious process, including hyperamyloidosis [10], tau pathology [1,9,11,12], mitochondrial dysfunction [13,14], proteostastic disturbance [15], [16], [17], and caspase-3 activation [18].
Although Aβ-driven tau pathology plays a critical role in AD [12,19,20], it is not clear how hyperamyloidosis, the earliest pathological change and a unique etiological factor in AD, is implicated in neurodegeneration [1,2,11]. Multiple evidence shows that the distribution pattern of senile plaques in the brain does not match well with that of tau tangles or neurodegeneration during disease progression [2,[10], [11], [12],21,22]. In some cases, hyperamyloidosis or tau tangle formation do not concur with neurodegeneration in the brains of human or animals [7,23,24]. Furthermore, neuronal injury initiated by amyloid beta was much less effectively rescued by suppression of tau in transgenic mice bearing mutants of human APP and tau [25]. Indeed, experimental therapies targeting β−amyloid or tau aggregates have been poorly translated from animal models to clinical trials [4,26], suggesting other mechanisms may contribute to the brain vulnerability and progressive neurodegeneration in AD.
Currently, no existing animal model of AD can mimic all aspects of AD, but certain early disease changes such as hyperamyloidosis and synaptic damage can be replicated in transgenic mice, including APP/PS1ΔE9 and 5xFAD mice [24,27,28]. Many public research databases contain large omics datasets of AD patients [29,30], and researchers have taken advantage of the conserved genomic signals in mice and humans and applied bioinformatics methods to investigate the mechanism of AD pathogenesis [31], [32], [33], [34], [35], [36], [37], [38]. Thus, integrating bioinformatics and biological methods to examine omics data [39,40], particularly from different species and disease stages, may enhance our power to decipher complex mechanisms underlying neurodegeneration at a systematic level or identify potential etiological factors for AD.
In this study, we deployed an integrative research strategy (Fig. 1a) to examine factors contributing to the progressive neurodegeneration in AD. We first analyzed early synaptic changes in young transgenic AD mice and gene expression profiles in vulnerable brain regions of AD patients. Then, we assessed and validated the association of a novel factor of interest to AD on multiple platforms. Lastly, we reported a mechanism by which misregulation of this factor is initiated by hyperamyloidosis and implicated in the neurodegeneration in AD.
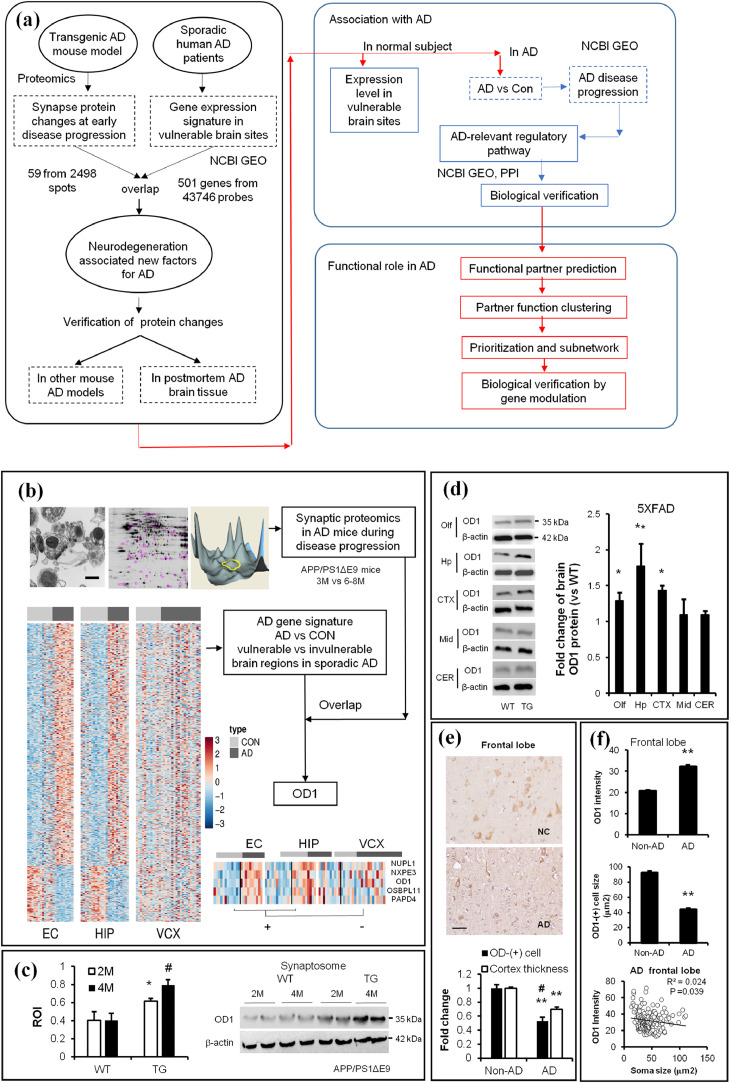
Fig. 1.
Comparative studies identify OCIAD1 as a common factor associated with neurodegeneration in both Tg AD mice and AD patients. (a) The experimental workflow implementing integrative biology to identify and evaluate the potential factor and validate its role in AD. (b) Synaptosomes isolated from mouse cortical cortex were characterized via electronic microscopy (EM) (upper left, Scale bar = 200 nm) and examined via 2D-DIGE and MS-MALDI/TOF (upper middle and right). Proteomics results overlapped with the differential gene expression between vulnerable brain regions (EC and HIP) and invulnerable region (VCX) from sporadic AD patients, as shown by the heatmap. OD1, OCIAD1; Red (upregulation), blue (down-regulation). (c-d) OD1 protein levels are changed in the synaptosome from AD mice (APP/PS1-ΔE9, c; **P < 0.01 vs. control, student t-test), and in the forebrain from 5XFAD mice (d), *P < 0.05, **P < 0.01 vs. control (n = 3, student t-test), WT, wild type; Tg, transgenic mice. 2 M, mice at age of 2 months; 4 M, mice at age of 4 months. (e) OD1-positive cells were visualized by IHC staining in the frontal lobe of normal subject (NC) and AD. The loss of OD1-positive cells was compared to the reduction in cortex thickness in the frontal lobe of AD (n = 11–30, Scale bar = 15 µm). (f) Increased OD1 intensity is correlated with a decreased soma size in OD1-(+) cells in the FTL of AD patient (n = 180 in Non-AD and 308 in AD group). Data are presented as mean±SEM, *P < 0.05, **P < 0.01 vs. Non-AD (student t-test). (See also Fig. S1).
2. Materials and methods
2.1. Experimental animal procedures
Hereditary AD transgenic (Tg) mouse models APP/PS1ΔE9 (034829-JAX) [27] and 5xFAD (034848-JAX) [28], Thy1-YFP (H-line, 003782-JAX) were obtained from Jackson Laboratory (JAX lab, Bar Harbor, Maine). Mice were bred and housed in the animal facility of Houston Methodist Research Institute. All animal experiments were approved by the Houston Methodist Research Institute Committee for Animal Care and Use (IACUC0112-0005) and in compliance with NIH guidelines. Tg mice were genotyped following a protocol of JAX lab. Heterozygous APP/PS1ΔE9 mice at 2–8, and 18 months, 5xFAD and 5xFAD::Thy1-YFP at 5-months, and age-matched littermates were used.
2.2. Synapse integrity and synaptic protein profile in AD mice
Synapse integrity in the mouse brain was examined by transmission electronic microscopy (TEM) on the brain tissues and the synaposomes isolated from the cerebral cortex of WT and AD Tg mice (APP/PS1-ΔE9). Mouse brain tissues were prepared as described [41] except the brain punches (around 1 mm3) were from the temporal cortex. Synaptosomes and synaptoneurosomes were prepared as described previously [42], [43] and synaptosome pellets were processed for TEM as described [41]. Examination was performed using a JEM 1010 transmission electron microscope (JEOL, Inc. Peabody, MA, USA). Digital images were obtained using AMT Imaging System (Advanced Microscopy Techniques Corp., Danvers, MA, USA). At least ten images were randomly photographed per region and sample. Synapses in coded samples were morphologically identified per unit area and calculated in a blinded way using Image J software. The mean values of synapses were used for statistics. Synapse integrity in the mouse brain was examined biochemically by western blot analysis of the presynaptic and postsynaptic protein markers (SNAP25, PSD95) in the isolated synapotosome and synaptoneurosomes (Suppl. Table 1). Equal amounts of proteins were resolved on 4–20% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE, Bio-Rad) and transferred to a polyvinylidene fluoride (PVDF) membrane using the Transblot Pack and Turbo Transfer System (Bio-Rad). The membrane was probed with primary antibodies and subsequent horseradish peroxidase (HRP)-conjugated secondary IgGs (Suppl. Table 1). Immunoblot signals were detected using ECL kit (WBKLS0500, Millipore, Billerica, MA) on Image Quant LAS4000 system (GE Healthcare, Pittsburgh, PA).
Synaptic protein profiles were determined by proteomics assay of synaptosomes. Synaptosomes were prepared from the cerebral cortex of WT and AD Tg mice (APP/PS1-ΔE9) [43] and sent to Applied Biomics (Hayward, CA) for 2D-DIGE. Differentially altered protein spots were selected and identified by mass spectrometry (MALDI-TOF/TOF) using GPS Explorer software equipped with the MASCOT. Protein candidates were verified by western blot in synaptosomes isolated from young wild type and AD Tg mice (APP/PS1-ΔE9).
2.3. Human brain vulnerability-associated gene expression
The differential gene expression data were pulled from GEO datasets (GSE5281, GSE28146, GSE53890) [29,30]. The differentially expressed genes common in two vulnerable brain regions (EC+/HIP+, AD vs. normal subjects, see abbreviates section) but not in the non-vulnerable brain region (VCX-, AD vs. normal subjects) were defined as vulnerability-relevant AD gene signatures (EC+/HIP+/VCX-) and presented as a heatmap by R program. The AD gene signature was then overlapped with the disease progression-associated proteins in transgenic AD mice to identify AD neurodegeneration-associated factors. Gene expression of OCIAD1 in vulnerable brain region was analyzed in 454 human normal subjects with microarray data from 19 brain sites in the database Oncomine [32]. OCIAD1 mRNA levels in the vulnerable brain sites of sporadic AD patients were examined in the two GEO datasets (GSE5281, GSE28146) and correlated with either disease severity (MMSE score) or NFT scores via Pearson correlation coefficient. Case information for GSE5281 are listed in [Suppl. Table 2] while human subject information for Oncomine dataset and GSE28146 can be found in publications [29,32].
2.4. Detection of OCIAD1 protein levels in the brain
OCIAD1 protein levels in the mouse brain and synaptosomes were determined by western blot. Five mouse brain regions were rapidly dissected from wild type and 5xFAD Tg mice, including the olfactory bulbs, hippocampus, cerebral cortex, midbrain, and cerebellum. Brain tissues were snap-frozen on dry ice and processed in 1xRIPA lysis buffer to obtain proteins as described previously [41,44]. OCIAD1 protein in neural cells were examined in the brain by immunostaining. For immnuohistochemistry (IHC) staining, mouse sagittal brain frozen sections (12 µm) were prepared from saline-perfusion wild type mice. Postmortem human frozen brain sections for immunohistological examination were obtained from sporadic AD patient at Houston Methodist Hospital (HMH) and processed by neuropathologists at the Department of Pathology and Genomic Medicine, following approval by the Houston Methodist Research Institute IRB. Postmortem brain samples and the neuropathologic scores are as below: NIH/AA (2012) score (A3/B3/C2), Thal phase for amyloid plaques (4A3), Braak and Braak neurofibrillary tangle stage (VB3), CERAD neuritic plaque score (Moderate C2). The control was from non-demented patients with no significant AD pathology on routine pathologic examination. OCIAD1-(+) cells were visualized by using anti-OCIAD1 antibody [45] and photographed with a microscope (Olympus X). OCIAD1-(+) cells in different mouse brain areas were examined by using a Mouse Brain Altas [46]. The total number, intensity, and soma size of OCIAD1-(+) cells in the cerebral cortex (layer I-VI) of human and wild type mouse, as well as the cortex thickness were manually examined in frontal lobe of normal subject and AD patients with Image J software. Adjacent non-stained area of same size was selected as background. OCIAD1 intensity was correlated with soma size of more than 180 cells via Pearson correlation coefficient.
For immunofluoresence staining OCIAD1 in neural cells in the mouse brain, serial sagittal sections (40 µm) were prepared with a cryostat (Leica CM 1850, Germany) and followed with autofluorescence quench. Brain sections were immersed in a block solution and followed up by a free-floating immunofluorescence staining against OCIAD1 (Suppl. Table 1), then directly mounted with anti-fade medium (Polysciences, Warrington, PA). OCIAD1 intensity in brain sections were examined via Image J while control signals were recorded from an area of the same size.
2.5. Prediction of the regulatory network of OCIAD1
All the factors that significantly regulate gene expression of OCIAD1 were collected from Gene Expression Omnibus (GEO) databases and examined in HIPPIE v2.0 [33]. Secondary interactors linking any two regulators of OCIAD1 with reliability score higher than 0.8 were sorted while the shortest path was weighed by Dijkstra's algorithm. The AD-irrelevant connections were manually filtered by query records for co-citation with keywords “node A, B, Alzheimer's” based on the NCBI PubMed. The AD-relevant pathway was then prioritized by the number of connective edges and publication record for further verification.
2.6. Modeling hyperamyloidiosis in neuronal cells
Three different cell models were used to mimic hyperamyloidosis in the brain: (i) mouse hippocampal HT22 neuronal cell line or primary cortical (CT) neuronal cells prepared from wild type mouse embryos were treated with synthesized Abeta 1–42 (5 µM, CP0012, Biomatik USA LLC., Wilmington, DA) for 16 hrs; (ii) primary neuronal cells with elevated levels of human APP were generated by transient transfection with plasmids encoding full length of human APP (wt or mutant); or (iii) primary hippocamppal neuronal cells were cultured from 5xFAD pups. Primary CT neurons were prepared from wild type mouse embryos at day E15-16 and cultured in vitro until DIV7 before treatment. For transient transfection, primary neurons were incubated for a shorter period (2 hrs) in a mixture of lipofectamine LTX/Plus reagent/plasmids (hAPPwt or APPswe, 0.5 µg per well in 12-well plate, 1.5 µg per well in 6-well plate) [44]. For preparation of primary AD hippocampal neurons, 5xFAD pups (P1) were quickly genotyped as follows: Tail tissues were placed in 50 mM NaOH solution (25 µL/each) and then heated to 95 °C for 5 min. The tubes were cooled on ice and Tris–HCl buffer (1 M) containing 10 mM EDTA (25 µL/each, pH 8.2) was added. The mixture was briefly centrifuged and the supernatant was collected for PCR using the JAX lab protocol. Hippocampus were dissected and dissociated neuronal cells were cultured in vitro until indicated time points for further treatment.
2.7. OCIAD1 level in neural cells under condition of aberrant proteostasis
OCIAD1 protein level in neuronal cells under condition of hyperamyloidosis was determined by dual immunofluorescence staining against OCIAD1 and APP [41,44]. Primary and secondary antibodies are listed in Suppl. Table 1. Cells grown on coverslips were stained and mounted with DAPI-containing anti-fade medium (Polysciences, Warrington, PA). Fluorescence signals within cells, cortical neurons over-expressing hAPPwt and hAPPswe/ind, or nearby untransfected neurons were recorded by FluoView™ FV1000 Confocal Microscope (Carl Zeiss Inc., Germany). NIH Image J was used to analyze fluorescence signals and densitometry to evaluate the protein levels and neuronal morphology. OCIAD1 mRNA level under condition of hyperamyloidiosis was determined by real-time PCR. Total RNAs were extracted from primary CT neurons (DIV14) following treatments (MG-132, 10 µM; Aβ1-42, 5 µM, 16 hrs), or from AD hippocampal neurons (DIV12) using Trizol reagent (Invitrogen) and glycogen (50 µg/ml, AM9510, Life Technologies, Carlsbad, CA). cDNAs were synthesized according to the manufacturer instructions and real-time PCR was performed by using an iScript One-Step RT-PCR kit (Bio-Rad 170–8892) on a C1000 Thermocycler (Bio-Rad). Average CT values for OCIAD1 were normalized to the values for the internal control 18S rRNA (Suppl. Table 3) and compared between experimental groups. The protein level of OCIAD1 in neural cells was determined by western blot. Primary CT neurons (DIV14) were prepared as previous described. Following pretreatment with vehicle or anisomysin (0, 20 and 40 µM, Sigma) for 2 hrs, neurons were treated by Aβ1-42 (5 µM) for 16 hrs. Primary human and murine astrocytes were purchased from Lonza (Basel, Switzerland) and cultured in 6 well plates following manufactory's instruction until 90% confluence. Before protein harvest, cells were exposed in medium with Aβ1-42 (5 µM) for 16 hrs. HT22 neuronal cells (a gift from Dr. James Simpkins, West Virginia University Health Science Center) were cultured in 6-well plates, as previously described, treated with leupeptin (10 µg/mL), NH4Cl (50 mM), chloroquine (100 µM, Sigma, St. Louis, Mo., USA), MG-132 (10 µM, M1157, AG Scientific Inc., San Diego, CA), or vehicle for 16 hrs. The cell proteins were harvested for western assays.
2.8. Modulation on GSK3beta/CTNNB1 and p53 signals in cells
To knockdown endogenous GSK3beta, HEK293 cells were treated with a mixture of RNAi-Max/DsRNAi (2–10 nM, IDT #hs.Ri.GSK3B.13.2) following manufactory's protocol. To modulate GSK3beta signals, primary CT neuronal cells were cultured until DIV12 and pre-treated with GSK3beta inhibitor LiCl (0, 0.3 at 3 mM, Sigma) for 2 hrs followed by exposure of Aβ1-42 (5 µM, 8 hrs). To modulate CTNNB1 or p53 signals, human HEK293 cells were cultured in DMEM+10% FBS and transfected transiently with a shRNAi-β-catenin plasmid to knockdown CTNNB1, or with a mixture of RNAi-Max /siRNA (10 nM) to suppress p53 or human GSK3beta (2–10 nM). Transfected cells were maintained in original culture medium for an additional 72 hrs and then treated with drugs. Cell proteins were harvested at indicated time points for western blot assay on OCIAD1 levels.
2.9. Prediction of the function of OCIAD1 via its interaction partners
The top 100 interaction partners of OCIAD1 in GeneDeck (V3.08 and V3.10) [34] and other interaction partners in Human Integrated Protein-Protein Interaction rEference (HIPPIE) and STRING databases [36] were included. Database for Annotation, Visualization and Integrated Discovery (DAVID) V6.7 was used to cluster the gene function while HIPPIE, Cytoscape, or commercial software IPA (Ingenuity, Qiagen, Venlo, Netherlands) were used to generate the interaction, identify key nodes associated with mitochondria-associated cell death, and build a sub-network among those partners presented in both functional modules.
2.10. Co-localization of OCIAD1 in the mitochondria
Dual immuofluorescence staining and laser-based confocal microscopy were used to determine the localization of OCIAD1 in the mitochondria. Mouse HT22 neuronal cells or human HEK293 cells were cultured on glass bottom dishes (35 mm, Matek, MA). HEK293 cells were transiently transfected with a DsRed-mito plasmid (1.5 µg /well) by using LTX/Plus reagent. The localization of OCIAD1 on mitochondria was determined by examination of immunofluorescence signals of OCIAD1 and SOD2, a mitochondrial protein marker [47], or immunofluorescence signals of OCIAD1 and DsRed, or by detection of OCIAD1 protein in the isolated mitochrondia from mouse brain by western blot assays. For high-throughput analysis of the co-localization of OCIAD1 on mitochondria and other colocalization signals, computerized cell segmentation and processing were applied as described previously [48,49]. Image J co-localization Plugin was also used to validate the distribution of OCIAD1 in the mitochondria.
2.11. Genetic modulation of OCIAD1 in neuronal cells
Three types of genetic modulations were applied to change OCIAD1 levels in neuronal cells. First, primary CT neurons (DIV12) were transiently transfected with a plasmid encoding mouse OCIAD1 and GFP, while CT neurons transfected with a plasmid encoding GFP only as control. Transfected cells were kept growing for 48 hrs before treatment. Second, HT22 neuronal cells were cultured in 6 well plates and infected with lentivirus particle containing a plasmid encoding OCIAD1 and GFP, or shRNAi targeting OCIAD1. Lentivirus was prepared per previous protocol [44] with minor modifications on the concentrations of lentivirus (5x, MOI) and polybrene (1 µg/ml). Infected cells were kept for an additional 72 hrs before treatment. Third, HT22 neuronal cells were cultured in 12-well plates and transiently transfected a plasmid via encoding OCIAD1 and puromycin selection marker for 48 hrs. Transfected cells were selected in medium containing puromycin (4 µM) for 2 weeks to generate single colonies, which were maintained in medium containing puromycin (1 µM).
2.12. Neuronal apoptosis and cell viability assays
Primary neuronal cells were transfected with a plasmid encoding mouse OCIAD1 with a GFP label and sequentially treated with Aβ1-42 (10 µM, 24 hrs). Neuronal injury was determined morphologically under microscopy by observing Hoechst 33258 dye-indicated nuclear morphology. Cell injury induced by Str (1 µM, 18 hrs) in HT22-mOD1 cell line was also determined by using a LIVE/DEAD cell viability assay kit following the manual instructions. Briefly, living and dead cells were labeled by calcein-AM (2 µM) and ethidium homodimer-1 (2 µM) respectively. The dead cells and extracellular space recorded via ImageXpress (Molecular Device, Sunnyvale, CA) in nine fields per well were determined by Image J. MTT assay was examined on a Fluostar Omega multi-microplate reader (BMG Labtec) to evaluate cell injury in HT22 cells with genetic modulation on OCIAD1, which were treated with MG-132 (20 µM, 24 hrs), staurosporine (Str, 0.2, 1, 5, 25 µM, 12 or 24 hrs), CCCP (2, 10, 50, 250 µM, 24 hrs), H2O2 (1, 5 mM, 24 hrs), or glutamate (5, 25 mM, 24 hrs). Western blot assays were used to examine apoptotic activity at indicated time windows by observing key proteins that are putatively involved in extrinsic and intrinsic apoptosis. All cytotoxcity experiments were performed in triplicate. Neurite morphology within fluorescence-labeled cortical neurons over-expressing hAPP (Wt or Mt), OCIAD1, or nearby untransfected neurons was photographed via confocal microscopy. The fluorescence intensity of OCIAD1 within the cell body and the morphology of neurites within the area of a 200 µm distance to the cell body were examined by Image J with a Sholl analysis plugin. Correlation between OCIAD1 intensity and neurite morphology was determined via Pearson statistical analysis.
2.13. Detection of mitochondrial apoptotic factor release
Mitochondrial and cytosolic fractions were isolated from cultured cells with a mitochondria isolation kit (Pierce Biotechnology 89874, Rockfold, IL) following the manufacturer's instruction. CYTC, SMAC, ENDOG were examined via western blot. Mitochondria and synaptosomes were freshly isolated following protocols mentioned previously. The slur bands containing synaptosomes and mitochondria were enriched for in vitro incubation assays or snap-frozen on dry ice and stored at −80 °C for protein analysis. Mitochondria isolated from the cerebral cortex of wild type mice were added (30 µg) in 1 ml of a Tyrodes salt solution (pH 7.2) and co-incubated with CCCP (10 µM), BAX, BAX/BCL-2, BAX/OCIAD1, OCIAD1, or BAX/BCL-2/OCIAD1 (3 µg each) at 37 °C for 3 hrs as previously described [50], with BSA as the control. Human proteins (hBAX, hBCL-2), and mouse OCIAD1 protein (mOD1) were generated by removing GST tag from GST fusion proteins by using Glutathione Sepharose 4B (GE Healthcare) and thrombin (5 U in PBS, 20 °C, 18 hrs). Mitochondria or synaptosome from Tg mice (APP/PS1ΔE9) were tested using same procedures in the presence or absence of OCIAD1 (3 µg). The treated mitochondria and synaptosome were collected by centrifugation (14,000 g, 15 min), and the pellets were lysed for western blot assay on CYT C and cleaved caspase-3.
2.14. Detection of mitochondrial membrane potential
pEF1-OD1-T2A-puro vector was transfected into HT22 cells according to the aforementioned method. Cell lines stably expressing OCIAD1 (HT-OD1) and control (HT-Mock) were selected by resistance marker puromycin (4 µg/mL, 631305, Clontech Laboratories Inc. Mountain View, CA) for 2 weeks, and then verified by western blot. After treatment with Str (1 µM) or vehicle (DMSO) for 2 hrs, 4 hrs, and 6 hrs, the HT-OD1 cells and the control cells were incubated with JC-1 dye (5 μM, Life Technologies, T-3168) for 30 min at 37 °C to indicate the ΔΨm. The fluorescence signal of monomer and J-aggregate forms were recorded simultaneously by flow cytometry (BD LSR II Flow Cytometer, BD Biosciences, San Jose, CA), using argon-krypton laser line (ex488/ex568 nm) and analyzed with Cellguest Software (BD Biosciences).
2.15. Protein-protein interaction assay in vitro
Glutathione-S-transferase (GST) fusion human protein OCIAD1 (ag9977), BCL-2 (ag3508), and BAX (ag0576) were purchased from Proteintech Group (Chicago, IL), while GST fusion mouse OCIAD1 protein (mOD1) was purified from E coli BL21-DE3 cells bearing a plasmid pGEX-2T-mOD1. For GST-pulldown, GST fusion bait proteins were coupled to Glutathione sepharose B slurs (17-0756-01, GE Healthcare, PA, USA) for 1 hr at 4 °C. Mouse cortical homogenates or HEK cell lysates (200 μg) were incubated with the bait-protein-coupled beads for 2 hrs at 4 °C. Equal amounts of GST protein, beads, homeogenates, and lysates were used following the same procedures in control groups. The proteins binding with the bait-bead-complex were collected, and subjected to western blot assay. Proteins under observation included BCL-2, BCL-XL, BAX, BAD, CYTC, SMAC, PUMA, VDAC1, CAS3, CAS9, and CASP8.
2.16. Cell-based protein-protein interaction analysis
A CheckMate™ mammalian two-hybrid system kit (E2440, Promega) was used to determine the binding between OCIAD1 and its potential partners in cells. ORFs of the two test proteins were cloned into pACT-VP16 vector or pBIND-GAL4 vector, respectively, and then co-transfected into HEK293T cells according to the manufacturer's instructions. Thirty-six hours after transfection, proteins were harvested with a passive lysis buffer in a dual luciferase activity assay kit (E1910, Promega). The OCIAD1-BCL-2 interaction, OCIAD1-BAX interaction, and the effects of OCIAD1 on the BCL-2/BAX and BAX/BAX complexes were examined and positive controls were set following manual instruction while cells transfected with single gene (pOD1, pBcl-2, pBax) or dual genes (pBcl-2/pBax,or pBax/pBax) were used as internal controls in the experimental design.
2.17. Modulation of OCIAD1 on the long-term effects of Aβ on neural cell susceptibility
HT22 cells were cultured as described previously and transfected with mouse OCIAD1 siRNA (10 nM, IDT, Suppl. Table 4) for 16 hrs using a Lipofectamine RNAi MAX (Invitrogen) following the manufacturer's instructions. The transfected cells were grown for an additional 16 hrs and then treated with Aβ (1 μM) for 16 hrs. The medium containing Aβ was replaced and cells were treated with MG-132 (20 μM), or Str (1 μM) for 12 hrs to detect caspase-3 activity via western blot assay, or 24 hrs to determine cell viability.
2.18. Detection of amyloid plaque, OCIAD1 level and dystrophic neurites in AD mouse brain
For labeling amyloid plaques in the mouse brain, serial sagittal sections (40 µm) were prepared with a cryostat (Leica CM 1850, Germany) and followed with methoxy‑X04 staining after autofluorescence was quenched. Brain sections were immersed in a methoxy‑X04 solution (100 mM in 40% ethanol, pH 10, Tocris Bioscience 4920, Bristol, UK) for 10 min, rinsed briefly with tap water, and then either followed up by a free-floating immunofluorescence staining against OCIAD1 (1:500), or with mouse anti-SMI-32 (1:500, BioLegend), then directly mounted with anti-fade medium (Polysciences, Warrington, PA). OCIAD1, SMI-32 or YFP intensity within 50 µM distances around amyloid plaques in Tg mouse brain sections were examined via Image J [51] while control signals were recorded from an area of same size where no amyloid plaques and SIM-32 intensity change were found.
2.19. Statistical analysis
For biostatistics analysis, data were collected in a blinded way and normalized with control group and then analyzed using t-tests or, for multiple groups, one-way ANOVA followed by Bonferroni post hoc tests, using SPSS11.0 or Excel. Apoptosis in more than 100 primary neurons was examined and analyzed by Chi-Square test. AD gene signatures were sorted by an R program and overlapped by Excel. Correlation between OCIAD1 and MMSE was examined via Pearson correlation coefficient. Data were presented as means with standard error and statistically significant level was set as P < 0.05.
3. Results
3.1. Comparative analysis of brain omics data reveals OCIAD1 as a factor associated with AD neurodegeneration
Chronic changes in gene expression underlie synapse plasticity and the different vulnerability in brain areas during AD progression [8,29,30]. Synapse damage is an early neurodegenerative marker of AD [6] that can be replicated in transgenic (Tg) AD mice harboring mutant APP and presenilin (i.e. APP/PS1ΔE9 and 5xFAD mice) [24,27,28]. We systematically analyzed those factors associated with neurodegeneration at the early stage of AD, by examining the protein profiles of isolated synaptosomes from phenotype-verified young AD mice (APP/PS1ΔE9 at 3 months and 6–8 months, Fig. 1a). Our proteomics assay (2D-DIGE and MS-MALDI/TOF) revealed that fifty-nine of 2498 spots were associated with disease development in AD mice (Fig. 1b) when a mild synaptic loss is shown in the cerebral cortex (Fig. S1a-b).
In parallel, we compared the gene expression profiles between vulnerable brain regions (entorhinal cortex, EC; hippocampus, HIP) and non-vulnerable brain regions (primary visual cortex, VCX) of sporadic AD patients. Five hundred and one genes were specifically associated with brain vulnerability (Fig. 1b). After cross-examining our genetic and proteomic findings, three factors were associated with disease development, including the ovary-orientated protein OCIAD1, Actin Related Protein 2/3 Complex Subunit 4 (ARPC4) and NADH:Ubiquinone Oxidoreductase Subunit A1 (NDUFA11). No existing evidence links ARPC4 with neurodegeneration or AD, while the decreased level of NDUFA11 might be a result from reduced number of mitochondria in the synapse of the cerebral cortex of AD mice. Thus, the only upregulated protein, OCIAD1, was then selected for further study (Fig. 1b). The elevated OCIAD1 levels were shown in the synaptosome from young APP/PS1ΔE9 CE mice (Fig. 1c), in the forebrain of 5xFAD mice (Fig. 1d), and in the neural cells in the frontal lobe of postmortem AD patients (Fig. 1e,f).
In the brain of wild type mouse, OCIAD1-positive cells were found in the layer III-V of cerebral cortex, hippocampus, brain stem and cerebellum (Fig. S1c). In the human brain, OCIAD1 positive cells in cerebral cortex are mainly located in the layer III-V with large body size, likely pyramidal neurons (Fig. 1e). In the frontal lobe in AD, OCIAD1 positive non-neuronal cells are increased in white matter (Fig. S1d) while the loss of OCIAD1-(+) cells (47%) seems more severe in comparison to a 30% reduction in the cortex thickness (Fig. 1e). Notably, the increased OCIAD1 intensity is correlated with a reduced soma size in this OCIAD1-(+) cells in the frontal lobe of postmortem AD patients (Fig. 1f), but this correlation was not shown in the cortex of wild type mouse (Fig. S1e), suggesting an association between OCIAD1 and AD neurodegeneration.
3.2. OCIAD1 is associated with brain vulnerability and under regulation of Aβ/GSK3β signals
To elucidate the role of OCIAD1 in neurodegeneration in AD, we examined the expression profile of OCIAD1 in the brain of normal subjects using brain microarray data from Oncomine [32] and in the brain of AD patients using the GEO dataset [29,30,40] (Fig. 2a). We found a higher level of OCIAD1 in the vulnerable brain areas of both normal subjects and sporadic AD patients, including the entorhinal cortex, medial temporal cortex, anterior cingulate cortex, and hippocampus (Fig. 2b, vs. normal subjects and VCX). Notably, analysis of the differential gene expression dataset in GEO dataset revealed that increased levels of OCIAD1 in the hippocampus are correlated with disease severity (Fig. 2c), supporting that OCIAD1 level may be associated with the brain vulnerability in AD.
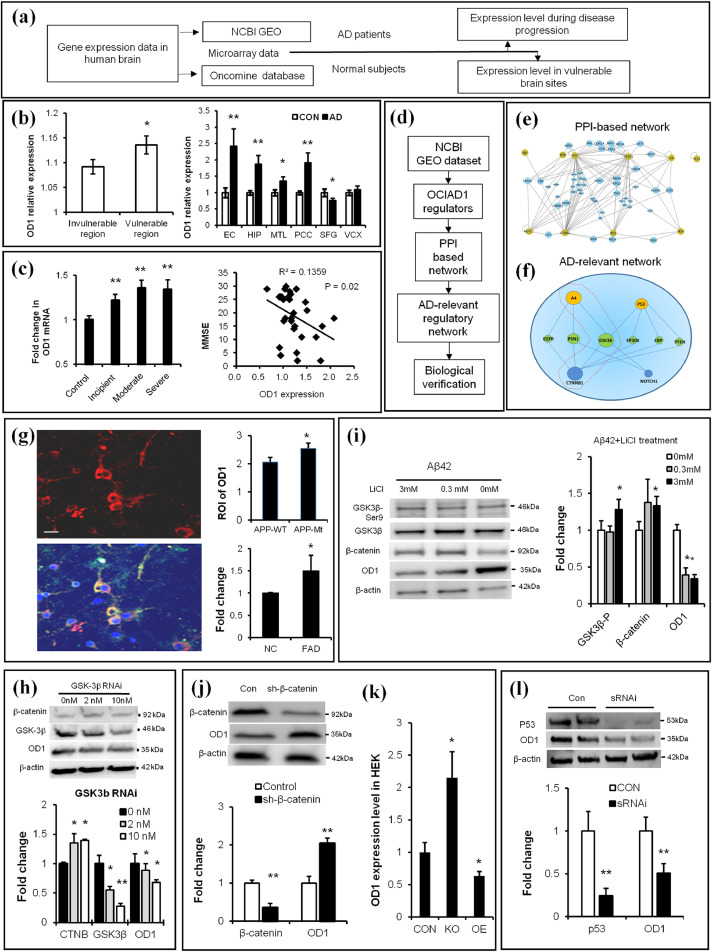
Fig. 2.
Brain OCIAD1 level is associated with AD development and under regulation of Aβ/GSK3β/p53 signals. (a) Workflow for analysis of OD1(OCIAD1) gene expression in human brain. (b-c) Higher expression level of OD1 was shown in the vulnerable brain sites in normal human subjects (left, in b) and AD patients (right, in b) while the increased level of OD1 correlates with severity of AD (c). (d-e) OD1 regulators were derived from GEO database to build PPI-based connection network (e) and generate AD-relevant regulatory network (f). (See also Table 1). (g-h) Elevated APP (g) or knockdown GSK-3β (h) change OD1 level in cells. APP-staining (red), OD1-staining (green), DAPI-staining (blue). APP mutant (APP-Mt), APP wild type (APP-Wt), wild type (NC), transgenic mice (FAD), knockdown GSK3β via sRNAi (GSK3β RNAi). (See also Fig. S2a-2c). (i) Chemical intervention of Aβ-GSK-3β signal affected levels of β-catenin and OD1 in cultured primary neurons. (j) Modulating β-catenin changed the OD1 level in HEK293 cells as shown in GEO data. (k) Knockdown (KO), over-expression (OE), knockdown β-catenin via shRNAi (sh-β-catenin). (l) Knockdown p53 reduces OD1 level in HEK293. Knockdown p53 via sRNAi (sRNAi). (See also Fig. S2d-2e). Data are presented as mean±SEM in (b–c, g–l). *P < 0.05, **P < 0.01 vs. control in (b) (student t-test, left panel, n = 32; right panel, n = 5), in c (one-way ANOVA, left panel, n = 3; Pearson Correlation Coefficient, right panel, n = 30), in h-l (student t-test, n = 3 in h-j, l, n = 6 in k),.*P < 0.05 vs. wild type in g (student t-test, n = 33 in APPWT, n = 45 in APPMt, and n = 3 in NC and FAD). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
We further examined possible connection between OCIAD1 and AD pathophysiology by analysis of the protein-protein interaction (PPI) network among OCIAD1 regulators under the context of AD (Fig. 2d). Twenty regulatory factors of OCIAD1 were found by analysis of the GEO database (Table 1), among which all 9 factors that upregulate OCIAD1 were found to be elevated in AD while the 8 factors that down-regulate OCIAD1 expression were reduced in AD as shown in literature search in the PubMed. Cross-analysis of the PPI networks of OCIAD1 in HIPPIE and the literatures in PubMed revealed that APP, an up-regulatory factor of OCIAD1, and CTNNB1 (β-catenin), a suppressing factor, interact through GSK-3β (Fig. 2e,f). In addition, cross-analysis of PPI network also revealed p53 as an upregulatory factor of OCIAD1 (Fig. 2e,f)
Table 1.
GEO-derived regulatory factors of OCIAD1.
| Regulators in GEO | Reported change in AD | Effects on OD1 |
|---|---|---|
| STAT3, p53, Myb, CD133, CXCL4/PF4, APP, SRF, Homocystein, d-serine | Elevated | Upregulation |
| CTNNB1, Notch, CREB, IRS3, IRS4, IRS1, DHT, VitE | Decreased | Suppressor |
| Estradiol, HDACs, Aging | Uncertain | Uncertain |
Note: Upregulators: STAT3, signal transducer and activator of transcription3; p53, tumor protein p53; Myb, v-myb avian myeloblastosis viral oncogene homolog; CD133, prominin 1; CXCL4/PF4,C-X-C motif chemokine 4; APP, amyloid protein precursor; and SRF, serum response factor. Downregulators: CTNNB1, β-catenin; CREB, cAMP responsive element binding protein; IRS1/3/4, insulin receptor substrate 1; and DHT, dihydrotestosterone. Uncertain: HDACs, Histone deacetylases.
Published reports suggest that aberrant Abeta-tau interaction and misregulated p53 are implicated in neurodegeneration in AD [52], while GSK-3β mediates Abeta-induced tau phosphorylation [19] and regulates CTNNB1 [53,54]. We thus tested whether Abeta/GSK3beta/CTNNB1 pathway and p53 signals regulate OCIAD1 in vitro. We mimicked hyperamyloidosis in cells by transfecting primary neurons with plasmids encoding human APP (wild type APPwt or mutant APPmt), culturing primary neurons derived from 5xFAD mouse embryos, or treating neuronal cells with synthesized Aβ1-42. The OCIAD1 levels were elevated under all these conditions in neuronal cells (Fig. 2g, Fig. S2a-b). Knocking down GSK-3β (Fig. 2h), application of GSK-3β inhibitor LiCl (Fig. 2i) or knocking down CTNNB1 (Fig. 2j) in HEK293 cells also changed the OCIAD1 protein levels, which is consistent with the GEO data from other groups (Fig. 2k).
In human brain, although OCIAD1 is also expressed in non-neuronal cells in the white matter, OCIAD1 levels were not altered in cultured astrocytes following treatment by the synthesized oligomer Aβ1-42 (Fig. S2c), suggesting a limited contribution of astrocytes in the OCIAD1 mis-regulation in AD brain. Beside Abeta/GSK3beta, other AD-relevant pathological events, such as p53 signaling (Fig. 2l) and proteostasis imbalance (Fig. S2d), also upregulates the protein level of OCIAD1. Interestingly, OCIAD1 protein is mildly increased in the brain of aged mouse (18 M vs 4 M), but its gene expression was decreased in the frontal lobe of aged human (Fig. S2e-f). These results indicate that multiple early pathological signals, such as Aβ/GSK-3β and p53, are involved in OCIAD1 misregulation in AD.
3.3. OCIAD1 mis-regulation contributes neuronal susceptibility through mitochondria-associated neurodegeneration
To determine how OCIAD1 contributes to neuronal susceptibility in AD, we predicted the function of OCIAD1 by examining its interaction partners and networks in the bioinformatics databases (GeneDeck V3.08, V3.11, HIPPIE, and STRING) [33,34,36], and by clustering its functional partners via the DAVID program [35] (Fig. 3a). We found that 28.1% of OCIAD1 partners were relevant to mitochondria, 43.1% were implicated in the cell death pathway, and many partners were involved in both (Fig. 3b, Table 2). Dual immunofluorescence staining against mitochondrial protein marker SOD2 further revealed a certain portion of OCIAD1 (49.5%) was located on the mitochondria (33%) in mouse hippocampal neuronal cells (HT22) (Fig. 3c). In human HEK293 cells, 75.4% of OCIAD1 was found on 71.2% DsRed-labeled mitochondria. Interestingly, a higher mitochondrial OCIAD1 level was also shown in AD mouse brains (APP/PS1ΔE9) at early disease stages compared to those from the WT mice (Fig. 3d).
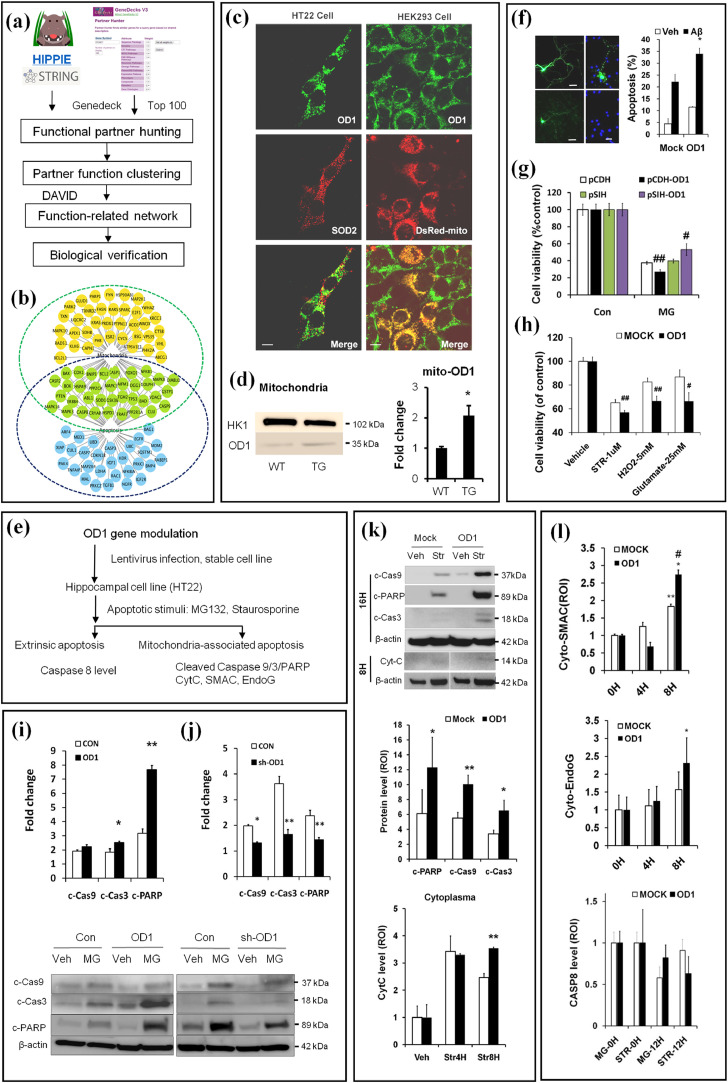
Fig. 3.
Integrative neurobiology study reveals a role of OCIAD1 in mitochondria-associated cell death. (a) Workflow for predicting OD1 (OCIAD1) function. (b) Cell death pathway and mitochondria are predicted as functional targets for OD1. Predicted functional partners of OD1 are indicated as nodes relevant to cell death pathways (blue), to mitochondria (orange), or both (green). (c-d) OD1 in the mitochondria was visualized via dual immunofluorescence staining against mitochondrial protein marks (SOD2) or Ds-Red Mito (c), and compared between the cerebral cortex of transgenic Tg AD mice (APP/PS1-ΔE9) and wild type mice (WT, d) at 4 months. Anti-OD1 (green), anti-SOD2 or DsRed (red). (e) Workflow for in vitro validation of OD1 function in mitochondria-associated cell death. (f-h) OD1 modulation affects cell injury in primary neurons induced by Aβ1-42 (f), in HT22 cells by MG132 (g), staurosporine (STR), H2O2 or glutamate (h). Cells infected with control vector encoding GFP only (CON, Mock, pCDH, pSIH), or plasmid encoding OD1 and GFP (OD1 or pCDH-OD1), shRNAi against OD1 (pSIH-OD1). (See also Fig. S3a-3d). (i) Up-regulation or down-regulation (j) of OD1 modulates the intrinsic apoptosis induced by MG-132 in HT22 cells. (k-l) Elevating OD1 in HT22 cells facilitates Str-induced apoptosis and release of cytochrome C (Cytc, k), SMAC (cyto-SMAC), EndoG (cyto-EndoG), but does not affect Caspase-8 activation (CASP8, l). Control vector (Mock), plasmid encoding OD1 (OD1). MG-12 h, MG-132 treatment for 12 hrs; STR treatment for 0 hr (0H), 4 hrs (Str4H), 8 hrs (Str8H), or 12 hrs (STR-12H). Data are presented as mean±SEM in (d-g) and (i- l)(student t-test, n = 3). * P < 0.05 vs. WT in (d) and vs.Veh in (f) #P < 0.05, ##P < 0.01 vs. Con in (g-h); *P < 0.05, **P < 0.01 vs. Con in (i-j); *P < 0.05, **P < 0.01 vs. Mock (k) or 0H (l), #P < 0.05 vs. Mock (l). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Table 2.
Clustering the predicted OCIAD1 functional partners.
| Category | Term | Count | % | P-value | Benjamini |
|---|---|---|---|---|---|
| GOTERM_BP_FAT | regulation of cell death | 69 | 43.1 | 4.50E-42 | 1.10E-38 |
| GOTERM_CC_FAT | cytosol | 65 | 40.6 | 1.70E-24 | 4.80E-22 |
| GOTERM_CC_FAT | endosome | 34 | 21.3 | 2.90E-22 | 4.10E-20 |
| GOTERM_CC_FAT | mitochondrion | 45 | 28.1 | 1.60E-13 | 1.50E-11 |
| KEGG_PATHWAY | Alzheimer's disease | 12 | 7.5 | 1.90E-03 | 5.50E-03 |
Next, we elevated OCIAD1 level in neuronal cells via gene modulation and examined cell injury following multiple challenges mimicking pathological changes in AD (Fig. 3e). Although up-regulating OCIAD1 in neuronal cells did not result in extensive cell death or proliferation suppression, a mild increase in the spontaneous apoptosis and caspase3 activation was shown in primary neuronal cells (Fig. 3f) and HT22 cells respectively (Fig. 3i). Under cell stress challenge, elevating OCIAD1 expression increased apoptosis in primary neurons challenged by Aβ aggregates (Fig. 3f) and cell injury in HT22 cells exposed to MG-132 (Fig. 3g), staurosporine (Str), H2O2, and glutamate (Fig. 3h). Elevated OCIAD1 also caused a decrease in neurite numbers (Fig. S3a-3b), enlarged extracellular space (Fig. S3c), and higher cell death following Str treatment (Fig. S3d).
We then examined whether and which apoptotic pathway involves the cell injury downstream of OCIAD1 activation in vitro. As shown by the increased protein levels of c-CAS9/c-CAS3/c-PARP following treatment with MG-132 (Fig. 3i, j) or Str (Fig. 3K), and by the release of CYTC (Fig. 3k), SMAC, and ENDOG to cytoplasm (Fig. 3l), we extrapolated that the cell injury facilitated by OCIAD1 involves activation of mitochondria-associated apoptotic pathway, as no significant change was detected in total or cleaved caspase-8 (Fig. 3l), the major extrinsic apoptotic factor. These results indicate that elevated OCIAD1 can contribute to neuronal susceptibility in AD by facilitating mitochondria-associated apoptosis.
3.4. OCIAD1 impairs MMP via an interaction with BCL-2
We performed a series of experiments (Fig. 4a) to understand how OCIAD1 facilitates mitochondria-associated cell injury. We detected the mitochondrial membrane potential (MMP) in HT22 cells with elevated OCIAD1 level (HT-mOD1) by applying indicator JC-1 dye. We found that the basic MMP was 21% lower in HT-mOD1 cells than controls (Fig. 4b). After Str treatment, MMP was reduced by 42% and 51% compared to controls at 2 hrs (31%) and 6 hrs (38%) post-treatment, respectively (Fig. 4b, Fig. S4a). We then examined whether OCIAD1 elevation affects MMP depolarization-induced mitochondria-associated apoptosis. As indicated by MTT assays (left, Fig. 4c) and western blot (right in Fig. 4c, Fig. 4d), CCCP, an inducer of MMP depolarization [55], increased injury, cleaved PARP levels, and CYTC release in HT-OD1 cells, suggesting an upregulated mitochondria-associated apoptotic activity.
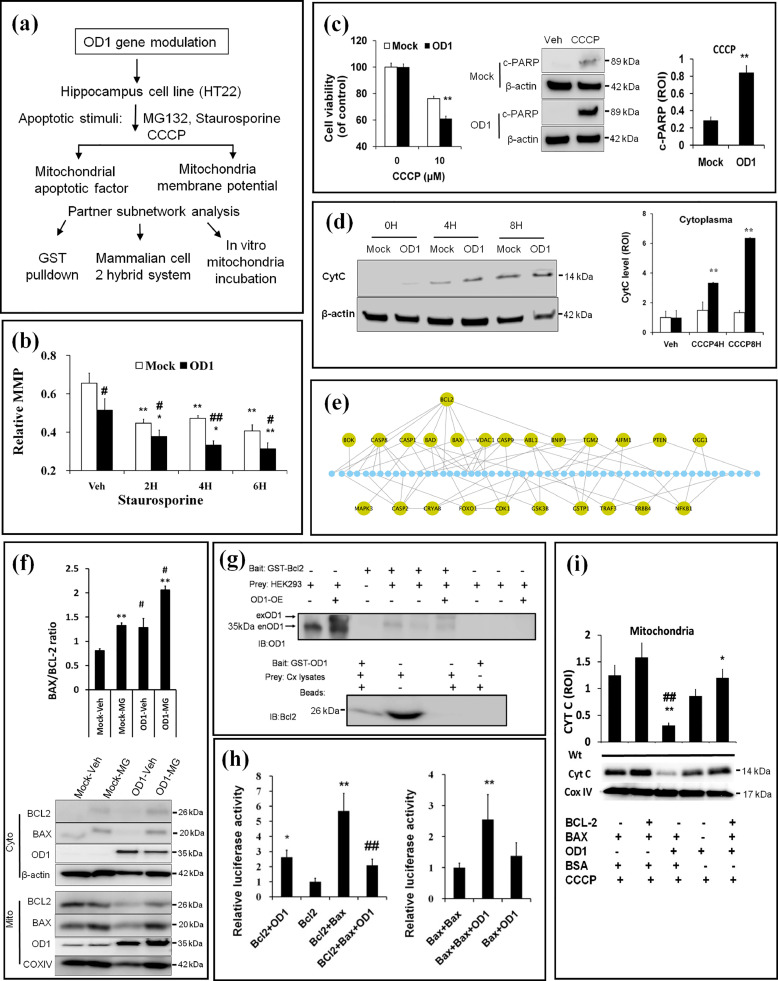
Fig. 4.
Upregulation of OCIAD1 impairs MMP via interaction with BCL-2. (a) Workflow for mechanistic study of OD1 (OCIAD1) in mitochondria-associated cell death. (b) OD1 elevation impairs MMP in HT22 cells before and after Str treatment. Cell transfected with empty vector (Mock), or plasmid encoding mouse OD1 (OD1). (See also Fig. S4a). (c-d) Effects of elevated OD1 on CCCP-induced cell injury, cleaved PARP (c), and CytC release (d) in HT22 cells. (e) PPI-based network analysis indicates BCL-2 as a key node linking OD1 to mitochondrial cell death pathway. (f) OD1 elevation interrupts the mitochondrial BCL-2/BAX ratio. Empty vector (Mock) or plasmid encoding OD1 (OD1), cytosolic fraction (Cyto), or mitochondrial fraction (Mito). (See also Fig. S4b-4d). (g) GST-pulldown assays verify BCL-2 as an interactor of OD1. (h) Dual luciferase assays show the interaction between BCL-2 and OD1, which disturbs BCL-2/BAX but facilitates BAX/BAX interaction. (i)In vitro incubation of OD1 with different combinations of BCL-2 and BAX in the isolated mitochondria from mouse cerebral cortex aggravates CytC releasing in the presence of CCCP. Data are presented as mean ± SEM in (b, f, h-i) (n = 3). * P < 0.05, ** P < 0.01 vs Veh, # P < 0.05, ## P < 0.05 vs. Mock in (b, f); ** P < 0.01 vs. Mock in (c-d). *P < 0.05, ** P < 0.01 vs. BCL-2 or vs BAX/BAX; ## P < 0.01 vs. BAX/BCL-2 in (h). * P < 0.05 and ** P < 0.01 vs. BCL-2/BAX/BSA, ## P < 0.01 vs. BAX/BSA in (i).
Next, we examined the PPI networks between the functional partners of OCIAD1 (Fig. 3b) to rank the key nodes that link MMP integrity and mitochondrial cell death pathways. BCL-2 emerged as such a key partner of OCIAD1 (Fig. 4e). In human HEK293 cells, OCIAD1 over-expression per se did not change the basal protein levels of BCL-2, BAX (Fig. 4f), Casp8 and p53. However, under condition of proteinostasis imbalance, elevated OCIAD1 reduced BCL-2 in the mitochondrial fraction to a greater extent, slightly increased OCIAD1 and BAX in the mitochondria (Fig. S4b-4d), resulting in a higher BAX/BCL-2 ratio (Fig. 4f). This suggests OCIAD1 may affect BCL-2 translocation to the mitochondria. To test this, we applied glutathione S-transferase pull down assays to examine 9 potential partners of OCIAD1 (BCL-2, BAX, BCL-XL, BAD, CYTC, SDHB, CAS3/9, CASP8) and 3 others (SMAC, PUMA, and VDAC1). The pull-down assays revealed an interaction between BCL-2 and OCIAD1 (Fig. 4g).
We then tested whether OCIAD1/BCL-2 interaction has functional meaning on BCL-2/BAX and BAX/BAX complex formation, which are crucial for maintenance of MMP in cells. By using a luciferase-based mammalian two-hybrid system, we introduced different interaction partners (BAX/BCL-2, OCIAD1/BCL-2, or BAX/BAX) in HEK293 cells and recorded an increased luciferase activity in BAX/BCL-2 group and OCIAD1/BCL-2 group, compared to control that was introduced with either BCL-2 or OCIAD1 alone (Fig. 4h). When OCIAD1 was introduced together with BAX/BCL-2 or BAX/BAX, the luciferase activity declined strikingly in BAX/BCL-2 group (5.5 vs. 2.0), but increased in BAX/BAX group (1.2 vs. 2.5), compared to control cells that were transfected with BAX/OCIAD1 (Fig. 4h), suggesting the interaction between OCIAD1-BCL-2 interferes with the dynamic relationships between BCL-2/BAX and BAX/BAX in cells.
We further evaluated the effects of OCIAD1-BCL-2 interaction on mitochondria-associated apoptosis. We isolated mitochondria from the cerebral cortex of wild type mice, incubated with a combination of proteins (OCIAD1, BCL-2, and BAX) and examined the CYTC release from the isolated mitochondria in vitro in the presence of CCCP. We found that OCIAD1 alone reduced mitochondrial CYTC level and antagonized the preserving effects of BCL-2 on BAX-induced mitochondrial CYTC reduction (Fig. 4i), suggesting OCIAD1-BCL-2 interaction can facilitate the CYTC release from mitochondria.
3.5. Elevated OCIAD1 mediates the long-term impact of Aβ on cell vulnerability and synaptic caspase-3 activation
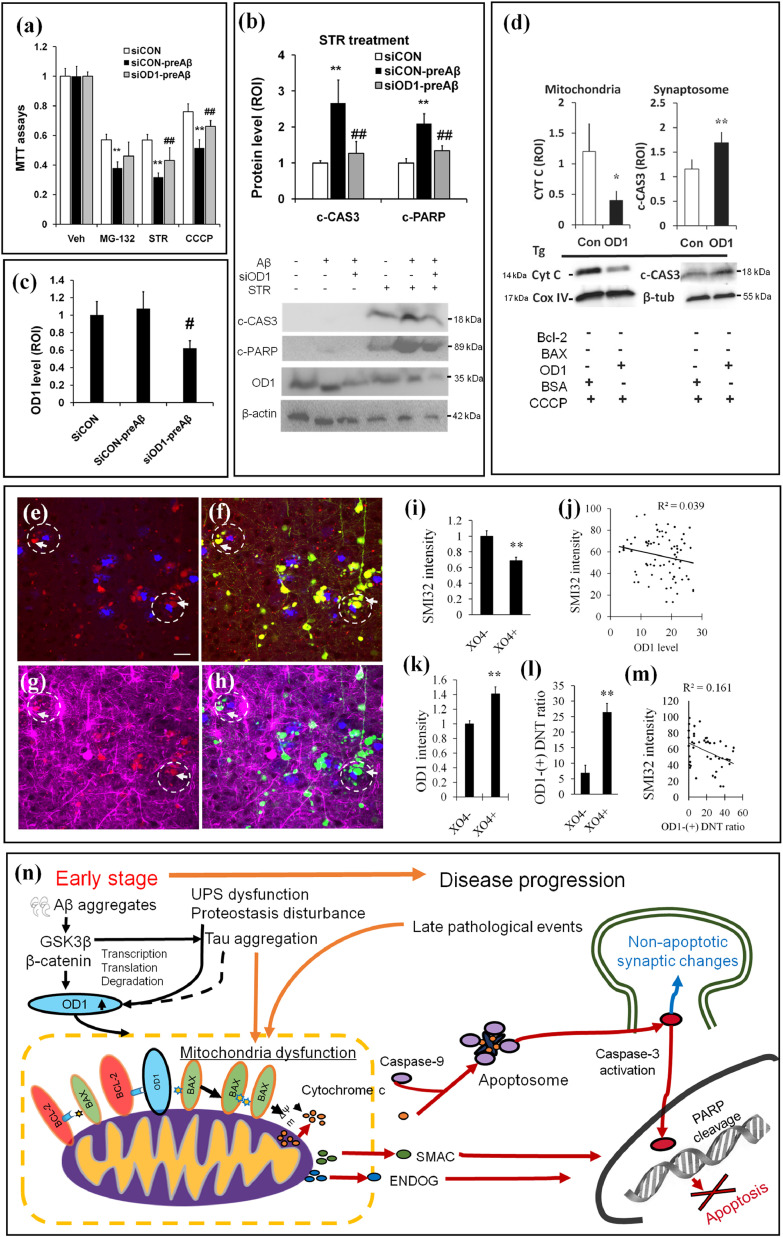
To determine whether mis-regulated OCIAD1 at early disease stages has a long-term impact on Abeta-related cell vulnerability [10], we pretreated HT22 cells with the synthesized oligomer Aβ1-42 at a nontoxic level (1 μM, 10 hrs) and examined whether OCIAD1 increases cell susceptibility to later stressors. MTT assays showed that cells pretreated with Aβ1-42 at this dosage followed by exposure to MG-132, STR, or CCCP had lower cell viability (Fig. 5a) and higher caspase-3 activation (Fig. 5b) than controls. Knockdown of OCIAD1 levels with siRNA (Fig. 5b,c) mitigated the aggravating effects of Aβ1-42 pretreatment on both cell injury (Fig. 5a) and caspase-3 activation (Fig. 5b).
Fig. 5.
OCIAD1 mediates the long-term effects of Aβ aggregates on neurodegeneration in Alzheimer's disease by facilitating cell injury and neurite dysfunction.(a–c) Pretreatment with a sub-lethal level of Aβ (1 μM, 10 hrs, preAβ) aggravated the cell injury induced by sequential stressors (a, MG-132, STR or CCCP), and facilitated caspase-3 activation induced by Str (b). Knockdown OD1 (c, siOD1) alleviated cell injury and caspase-3 activation in HT22 cells (b). **P < 0.01 vs. normal control (siCON), #P < 0.05 and ##P < 0.01 vs. experimental control (siCON-preAβ). (d) CCCP-induced CYTC releasing and caspase-3 activation in isolated mitochondria or synaptosome from the cerebral cortex of Tg AD mice (APP-PS1-ΔE9) in the presence of OD1.*P < 0.05 and **P < 0.01 vs. control treated with bovine serum albumin (BSA). β-tubulin (β-tub). (e-m) OD1 signal (red), YFP-labeled neuron and neurite (yellow) and neurite density (SMI-32, magenta) were shown in the area adjacent to amyloid plaque (X04, blue) in brain sections of the 5xFAD:YFP mice (5 M). Less neurite density, higher OD1 signal and ratio of OD1-postive dystrophic neurite was indicated by arrows in the circle with dashed line. OD1: OCIAD1, DNT: dystrophic neurite. **P < 0.01 vs. control X04- (area with no amyloid plaque and neurite loss). (n) A schematic summary of the mechanistic role of OD1 in mitochondria-associated neurodegeneration in AD. Early stage pathological changes (ie. Aβ aggregates) elevate OD1 through multiple pathways, including GSK3β. Upregulated OD1 leads to mitochondrial dysfunction through binding BCL-2, disturbing BCL-2/BAX interaction and impairing MMP (ΔΨm). In the presence of late stage cell injury stress, susceptible mitochondria release apoptotic factors to initiate cell death or activate caspase-3 causing non-apoptotic synaptic changes.
As caspase 3 activation has a role in toxic Abeta-induced synapse damage [18], we next tested whether OCIAD1 is involved in this pathological event in the context of AD. By in vitro incubation of mitochondria and synaptosomes isolated from transgenic AD mice (APP-PS1-ΔE9) with CCCP, we found that OCIAD1 facilitated CYTC release and caspase-3 activation (Fig. 5d), indicating that elevated OCIAD1 by early pathological changes in AD increases cell susceptibility and plays a role in synapse damage.
We further investigated the possible role of OCIAD1 in synaptic toxicity under conditions of hyperamyloidosis in vivo. The OCIAD1 level (Fig. 5e), dystrophic neurite (Fig. 5f), and neurite density (Fig. 5g) were examined in brain sections of the 5xFAD:YFP mice. We found that the decreased neurite density (Fig. 5g, i) correlated to the increased OCIAD1 signal (Fig. 5e, j, k) in the area adjacent to X04-labeled amyloid plaque. In this area, the increased ratio of OCIAD1-postive dystrophic neurite (Fig. 5f, l) correlated to the decreased SMI-32 signals (Fig. 5h, m), supporting a role of OCIAD1 in synapse damage.
The mechanistic role of OCIAD1 in mitochondrial dysfunction and progressive neurodegeneration in AD is summarized in Fig. 5n.
4. Discussion
In this study, we integrated bioinformatics and biological methods to examine neurodegeneration-associated factors in Alzheimer's disease (AD) systematically and comparatively at two different disease stages (early and late) on two biological entities (mouse and human). We identified an ovarian-orientated protein OCIAD1 as a novel factor that is up-regulated by early disease signal Aβ/GSK3β and contributes to neurodegeneration in AD via mitochondrial dysfunction and synapse damage. These findings indicate a new possibility that OCIAD1 misregulation mediates the brain vulnerability and progressive neurodgeneration imprinted by amyloidogenesis at early disease stage, and help us understand the complex interplay among amyloid β, tau aggregates, mitochondria dysfunction, and neurodegeneration during the course of AD.
A key finding of this study is that we discovered a novel pathological role of OCIAD1 in the central nervous system through its contribution to neurodegeneration in AD. OCIAD1 was originally identified in the human ovary but is expressed ubiquitously [56], [57], [58]. In Drosophila, OCIAD1 has a vital role in regulating endocytosis and notch signaling [59], [60], two processes relevant to gamma secretase activity. OCIAD1 was found in the synapse and in the brain with relatively high expression levels [56,61,62]. Changed OCIAD1 was found in the lipid raft in AD mouse model [63] while an OCIAD family member was associated with APP metabolism via activation of gamma secretase [64]. The relationship between OCIAD1 and neurodegeneration is supported by several lines of evidence. For example, in the cerebral cortex, OCIAD1 is majorly found in neurons of large body size (likely the vulnerable pyramidal neurons) in layers III-V. This protein is up-regulated in neuronal cells in the afflicted brain of both Tg AD mice and sporadic AD patients. Higher levels of OCIAD1 are correlated with the shrunken soma size of neurons in the vulnerable brain site and disease severity in late stage of AD, as shown by gene signature analysis and immunohistochemical staining, respectively. In addition, different OCIAD1 levels in the brain of 5xFAD Tg mice and APP/PS1ΔE9 Tg mice match the phenotype difference between these two AD mouse strains, where 5xFAD mice show more striking neuronal loss [28] while APP/PS1ΔE9 mice only a mild synapse loss [27]. Moreover, protein levels of OCIAD1 are increased in the synapse and the dystrophic neurites in young AD mice, while elevation of OCIAD1 reduces neurite numbers in cells and activates caspase-3 in isolated synaptosome from AD mice when synapse damage occurs. Considering OCIAD1 is up-regulated by multiple early pathological changes (i.e., Aβ/GSK3β signal, p53), and CYTC/caspase-3 activation in the synapse has a non-apoptotic role in synapse damage [65], our findings indicate that OCIAD1 contributes to neurodegeneration in the early stage of disease in two ways: (i) it mediates synapse damage, and (ii) it imprints neuronal cells more vulnerable to subsequent AD-relevant pathophysiological challenges.
Another highlight of this study is that we reported that mis-regulated OCIAD1 leads to mitochondria dysfunction and facilitates mitochondria-associated apoptosis. Mitochondrial dysfunction has been known as a common pathogenic factor for various neurodegenerative diseases [13,14]. In AD, mitochondrial dysfunction, reduced BCL-2, up-regulated BAX, and CYTC release have been recorded [13,14,65,66]. In addition, a reduced basal MMP was found in cortical cells from AD mice [67]. Toxic Aβ accumulation, impaired MMP, increased BAX/BCL-2 ratio, cytochrome c release, and activation of CAS-9/-3 were also reported in a rat AD model [68]. Under conditions of hyperamyloidosis, many changes are linked to mitochondrial dysfunction, including mis-translocation of certain proteins to the mitochondrial membrane (ie., BAX, BCL-2, and mortalin) [65], [68], [69], aberrant interaction with appoptosin [18], p-tau [70], or LPPRC1 [71], and impaired mitochondria dynamics via Aβ/GSK-3β or p53 [52,72]. We demonstrate that OCIAD1 is located in the mitochondria and interacts with BCL-2, which was first reported by us in SFN2014 and subsequently supported by the Human Protein Atlas database (ENSG00000109180). Similar to our finding, Shetty et al. reported that OCIAD1 regulates mitochondrial function in human stem cells [73]. More recently, OCIAD1 was found in the mitochondria of mouse hematopoietic stem cells (HSC) and has a role in spontaneous apoptosis of this type of cells [74]. Therefore, our findings are consistent with the current scientific literature and match findings on mitochondrial dysfunction in AD.
Furthermore, we demonstrated a role of OCIAD1 mis-regulation in MMP impairment and the sequential release of apoptotic factors under pathological conditions of AD. These pathological changes in mitochondria result from aberrant OCIAD1-BCL2 interaction. They are not likely from the p53 pathway, because p53 levels did not change significantly following OCIAD1 over-expression in mouse HT22 neuronal cells and HEK293 cells. We found that down-regulation of p53 reduces OCIAD1 expression level, in contrast to the report that OCIAD1 regulates p53 in mouse HSC [74], suggesting that interplay between OCIAD1 and p53 may be different in stem cell and non-stem cells.
Our findings raised a possibility that OCIAD1 may mediate the pathological interactions at early disease stage among amyloid β, p-tau pathology, and neurodegeneration. The supportive evidence includes that OCIAD1 is under regulation of multiple AD-relevant pathological events [15,16], particularly early disease signal d‑serine [75] and Aβ/GSK-3β signals which trigger tau pathology in AD [19,21,53,54]. It is known that co-existence of amyloidosis and tau pathology is crucial for disease development in AD while Aβ-driven p-Tau cascade plays a role in neurodegeneration [12,19,25]. However, the distribution pattern of senile plaques does not match with either tau tangles or neurodegeneration [7,11,21,23]. Also, reducing p-Tau signal in mice bearing both human APP mutant and tau mutant did not attenuate the toxic Aβ-initiated neurodegeneration, although the tauopathy was alleviated in mice bearing human tau mutants only [17,25]. More recently, Abeta-induced vulnerability in the brain's default mode network is associated with an interaction between local Abeta aggregation and hypometabolism, but independent of tauopathy [10]. Similarly, we did not find a correlation between OCIAD1 level and either NFT pathological score or amyloid plaque distribution in postmortem brain section of AD patients, although an elevated level of OCIAD1 was found in the area near senile plaques in the brain of young AD Tg mice, as shown by our immunohistochemical staining. One possible explanation for this discrepancy is that OCIAD1 is upregulated in neuronal cells by the early local toxic Aβ signal, gradually causing these cells' demise during disease progression before, or independent of, formation of mature senile plaques and tau tangles. Indeed, besides Abeta/GSK3beta signals, we found multiple Abeta-relevant pathological events, such as UPS dysfunction and lysosome impairment, upregulates OCIAD1 at protein level. Further investigation on the interaction among GSK3beta\tau\OCIAD1, and its other regulators (e.g., β-catenin, p53) in cell models, or postmortem brain of AD patients at different disease stages would help determine whether Aβ-GSK3-OCIAD1 axis exists in vivo and its relationship to tauopathy under pathological context of AD.
The lower OCIAD1 level in the frontal lobe of AD patients is not a surprise as it is consistent with other reports [76]. Although the mechanism of lower OCIAD1 in the frontal lobe is unknown, poor correlation between mRNA level and protein product of a gene has been well reported [77]. There are several possible explanations for the discrepancy between OCIAD1 levels in the frontal lobe and other vulnerable brain regions in AD. First, the regulatory network for OCIAD1 gene expression may be different among the brain regions afflicted by normal aging or AD pathogens. For example, it has been known that aging is a key factor afflicting the frontal lobe in normal subjects while tauopathy and hyperamyloidosis affect other AD brain regions [29]. Indeed, we found OCIAD1 gene expression was suppressed in the frontal lobe of normal subjects. Second, the aged healthy controls in the dataset are at Barak stage I-II while AD patients are majorly at Barak stage V-VI when cells with higher OCIAD1 levels have already demised significantly. Indeed, we found a significant decrease in OCIAD1-(+) cells in the medial frontal cortex. Another explanation may be the biased sampling, as pyramidal neurons were dissected from the AD brain based on the cell size. We found that cells with large soma contain less OCIAD1 during neurodegeneration. A future investigation on the profiles of OCIAD1 regulatory factors among these brain sites, or a temporal examination on the protein level of OCIAD1 among these different brain sites during disease progression, may help to find whether OCIAD1 has a different role in normal aging and AD.
5. Conclusions
We systematically examined neurodegeneration-relevant factors in the vulnerable brain regions of patients of sporadic Alzheimer's disease (AD) and transgenic mouse models. In this study, both bioinformatics methods and neurobiological tools were integrated to assess and validate the functional role of the newly-identified factor OCIAD1 in AD pathogenesis. OCIAD1 is under regulation of multiple AD-relevant pathological changes, including Aβ/GSK3β signaling. Elevated OCIAD1 at early disease stage contributes to mitochondria dysfunction, neuronal vulnerability, and synaptic damage in AD. The significance of this study is that it reveals a new role of OCIAD1 in neurodegeneration and helps better understand the pathophysiological relationship between brain vulnerability and hyperamyloidosis in AD. This methodology also has the potential to help identify therapeutic targets for other neurodegenerative diseases.
Author contributions
X.L. conceived the hypothesis and methodology. X.L., H.Z., and S.T.C.W. designed the study. X.L., L.W., and T.H. conducted the dry lab works. X.L., J.C., M.C., and A.R. processed samples and conducted experiments. X.L., L.W., T.L., and H.T. collected and analyzed data. X.L., J.C., and L. W. worked on data visualization. X.L. interpreted experimental results. S.T.C.W and S.P. supervised the resources and the project. S.T.C.W acquired the funding. X.L. drafted the original article while X.L., H. Z., S.T.C.W., and W.X. reviewed & edited the article.
Funding
This study is supported by Ting Tsung & Wei Fong Chao Foundation, John S Dunn Research Foundation, Cure Alzheimer's Fund, and NIA R01AG057635 to STCW.
Declaration of Competing Interest
The authors declare no competing financial interests.
Acknowledgements
The authors would like to thank Dr. Stanley Appel (Houston Methodist Neurological Institute), Dr. Hui Zheng (Baylor College of Medicine) and Dr. Weidong Le (Shanghai Jiao Tong University) for their insightful discussions. Also, we thank Dr. Dennis Selkoe (Brigham and Women's Hospital) for his APP plasmids; Drs. Kemi Cui and Zhen Zhao (Houston Methodist Research Institute) for their assistance in imaging preparation and analysis; Dr. Fuhai Li (Washington University at St. Louis) for suggestion on bioinformatics tools; Ying Li (Houston Methodist Research Institute) for her preliminary proteomics work; and Drs. Rebecca Danforth and James Mancuso for their reading and editing.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2019.11.030.
Contributor Information
Xuping Li, Email: xli2@houstonmethodist.org.
Stephen T.C. Wong, Email: stwong@houstonmethodist.org.
Appendix. Supplementary materials
Reference
- 1.Hyman B.T., Phelps C.H., Beach T.G., Bigio E.H., Cairns N.J., Carrillo M.C., Dickson D.W., Duyckaerts C., Frosch M.P., Masliah E., Mirra S.S., Nelson P.T., Schneider J.A., Thal D.R., Thies B., Trojanowski J.Q., Vinters H.V., Montine T.J. National institute on aging–Alzheimer's association guidelines for the neuropathologic assessment of Alzheimer's disease. Alzheimers Dement. 2012;8:1–13. doi: 10.1016/j.jalz.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jack C.R., Jr, Holtzman D.M. Biomarker modeling of Alzheimer's disease. Neuron. 2013;80:1347–1358. doi: 10.1016/j.neuron.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alzheimer's Association Alzheimer's disease facts and figures. Alzheimers Dement. 2016;12:459–509. doi: 10.1016/j.jalz.2016.03.001. [DOI] [PubMed] [Google Scholar]
- 4.Cummings J.L., Morstorf T., Zhong K. Alzheimer's disease drug-development pipeline: few candidates, frequent failures. Alzheimer's Res Ther. 2014;6:37. doi: 10.1186/alzrt269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karch C.M., Cruchaga C.A., Goate A.M. Alzheimer's disease genetics: from the bench to the clinic. Neuron. 2014;83:11–26. doi: 10.1016/j.neuron.2014.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sheng M., Sabatini B.L., Südhof T.C. Synapses and Alzheimer's disease. Cold Spring Harb Perspect Biol. 2012;4 doi: 10.1101/cshperspect.a005777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Serrano-Pozo A., Frosch M.P., Masliah E., Hyman B.T. Neuropathological alterations in Alzheimer disease. Cold Spring Harb Perspect Med. 2011;1 doi: 10.1101/cshperspect.a006189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Freer R., Sormanni P., Vecchi G., Ciryam P., Dobson C.M., Vendruscolo M. A protein homeostasis signature in healthy brains recapitulates tissue vulnerability to Alzheimer's disease. Sci Adv. 2016;2 doi: 10.1126/sciadv.1600947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fu H., Possenti A., Freer R., Nakano Y., Villegas N.C.H., Tang M., Cauhy P.V.M., Lassus B.A., Chen S., Fowler S.L., Figueroa H.Y., Huey E.D., Johnson G.V.W., Vendruscolo M., Duff K.E. A tau homeostasis signature is linked with the cellular and regional vulnerability of excitatory neurons to tau pathology. Nat Neurosci. 2019;22:47–56. doi: 10.1038/s41593-018-0298-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pascoal T.A., Mathotaarachchi S., Kang M.S., Mohaddes S., Shin M., Park A.Y., Parent M.J., Benedet A.L., Chamoun M., Therriault J., Hwang H., Cuello A.C., Misic B., Soucy J.P., Aston J.A.D., Gauthier S., Rosa-Neto P. Aβ-induced vulnerability propagates via the brain's default mode network. Nat Commun. 2019;10:2353. doi: 10.1038/s41467-019-10217-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Castellani R.J., Perry G. The complexities of the pathology-pathogenesis relationship in Alzheimer disease. Biochem Pharmacol. 2014;88:671–676. doi: 10.1016/j.bcp.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 12.Spires-Jones T.L., Hyman B.T. The intersection of amyloid beta and tau at synapses in Alzheimer's disease. Neuron. 2014;82:756–771. doi: 10.1016/j.neuron.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eckert G.P., Renner K., Eckert S.H., Eckmann J., Hagl S., Abdel-Kader R M., Kurz C., Leuner K., Muller W.E. Mitochondrial dysfunction–a pharmacological target in Alzheimer's disease. Mol Neurobio. 2012;46:136–150. doi: 10.1007/s12035-012-8271-z. [DOI] [PubMed] [Google Scholar]
- 14.Swerdlow R.H., Burns J.M., Khan S.M. The Alzheimer's disease mitochondrial cascade hypothesis: progress and perspectives. Biochim Biophys Acta. 2014;1842:1219–1231. doi: 10.1016/j.bbadis.2013.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ihara Y., Morishima-Kawashima M., Nixon R. The ubiquitin-proteasome system and the autophagic-lysosomal system in Alzheimer disease. Cold Spring Harb Perspect Med. 2012;2 doi: 10.1101/cshperspect.a006361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Polito V.A., Li H., Martini-Stoica H., Wang B., Yang L., Xu Y., Swartzlander D.B., Palmieri M., di Ronza A., Lee V.M., Sardiello M., Ballabio A., Zheng H. Selective clearance of aberrant tau proteins and rescue of neurotoxicity by transcription factor EB. EMBO Mol Med. 2014;6:1142–1160. doi: 10.15252/emmm.201303671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lasagna-Reeves C.A., de Haro M., Hao S., Park J., Rousseaux M.W., Al-Ramahi I., Jafar-Nejad P., Vilanova-Velez L., See L., De Maio A., Nitschke L., Wu Z., Troncoso J.C., Westbrook T.F., Tang J., Botas J., Zoghbi H.Y. Reduction of nuak1 decreases tau and reverses phenotypes in a tauopathy mouse model. Neuron. 2016;92:407–418. doi: 10.1016/j.neuron.2016.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang H., Zhang Y.W., Chen Y., Huang X., Zhou F., Wang W., Xian B., Zhang X., Masliah E., Chen Q., Han J.D., Bu G., Reed J.C., Liao F.F., Chen Y.G., Xu H. Appoptosin is a novel pro-apoptotic protein and mediates cell death in neurodegeneration. J Neurosci. 2012;32:15565–15576. doi: 10.1523/JNEUROSCI.3668-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choi S.H., Kim Y.H., Hebisch M., Sliwinski C., Lee S., D'Avanzo C., Chen H., Hooli B., Asselin C., Muffat J., Klee J.B., Zhang C., Wainger B.J., Peitz M., Kovacs D.M., Woolf C.J., Wagner S.L., Tanzi R.E., Kim D.Y. A three-dimensional human neural cell culture model of Alzheimer's disease. Nature. 2014;515:274–278. doi: 10.1038/nature13800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bloom G.S. Amyloid-β and tau: the trigger and bullet in Alzheimer disease pathogenesis. JAMA Neurol. 2014;71:505–508. doi: 10.1001/jamaneurol.2013.5847. [DOI] [PubMed] [Google Scholar]
- 21.Bateman R.J., Xiong C., Benzinger T.L., Fagan A.M., Goate A., Fox N.C., Marcus D.S., Cairns N.J., Xie X., BlazeyTM Holtzman DM, Santacruz A., Buckles V., Oliver A., Krista Moulder R.N., Aisen P.S., Ghetti B., Klunk W.E., McDade E., Martins R.N. Clinical and biomarker changes in dominantly inherited Alzheimer's disease. N Engl J Med. 2012;367:795–804. doi: 10.1056/NEJMoa1202753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Espuny-Camacho I., Arranz A.M., Fiers M., Snellinx A., Ando K., Munck S., Bonnefont J., Lambot L., Corthout N., Omodho L., Vanden Eynden E., Radaelli E., Tesseur I., Wray S., Ebneth A., Hardy J., Leroy K., Brion J.P., Vanderhaeghen P., De Strooper B. Hallmarks of Alzheimer's disease in stem-cell-derived human neurons transplanted into mouse brain. Neuron. 2017;93:1066–1081. doi: 10.1016/j.neuron.2017.02.001. [DOI] [PubMed] [Google Scholar]
- 23.Jack C.R., Jr, DA Bennett, Blennow K., Carrillo M.C., Feldman H.H., Frisoni G.B., Hampel H., Jagust W.J., Johnson K.A., Knopman D.S., Petersen R.C., Scheltens P., Sperling R.A., Dubois B. A/T/N: an unbiased descriptive classification scheme for Alzheimer disease biomarkers. Neurology. 2016;87:539–547. doi: 10.1212/WNL.0000000000002923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jankowsky J.L., Zheng H. Practical considerations for choosing a mouse model of Alzheimer's disease. Mol Neurodegener. 2017;12:89. doi: 10.1186/s13024-017-0231-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Busche M.A., Wegmann S., Dujardin S., Commins C., Schiantarelli J., Klickstein N., Kamath T.V., Carlson G.A., Nelken I., Hyman B.T. Tau impairs neural circuits, dominating amyloid-β effects, in Alzheimer models in vivo. Nat Neurosci. 2019;22:57–64. doi: 10.1038/s41593-018-0289-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Honig L.S., Vellas B., Woodward M., Boada M., Bullock R., Borrie M., Hager K., Andreasen N., Scarpini E., Liu-Seifert H., Case M., Dean R.A., Hake A., Sundell K., Poole Hoffmann V., Carlson C., Khanna R., Mintun M., DeMattos R., Selzler K.J. Trial of solanezumab for mild dementia due to Alzheimer's disease. N Engl J Med. 2018;378:321–330. doi: 10.1056/NEJMoa1705971. [DOI] [PubMed] [Google Scholar]
- 27.Jankowsky J.L., Fadale D.J., Anderson J., Xu G.M., Gonzales V., Jenkins N.A., Copeland N.G., Lee M.K., Younkin L.H., Wagner S.L., Younkin S.G., Borchelt D.R. Mutant presenilins specifically elevate the levels of the 42 residue beta-amyloid peptide in vivo: evidence for augmentation of a 42-specific gamma secretase. Hum Mol Genet. 2004;13:159–170. doi: 10.1093/hmg/ddh019. [DOI] [PubMed] [Google Scholar]
- 28.Oakley H., Cole S.L., Logan S., Maus E., Shao P., Craft J., Guillozet-Bongaarts A., Ohno M., Disterhoft J., Van Eldik L., Berry R., Vassar R. Intraneuronal beta-amyloid aggregates, neurodegeneration, and neuron loss in transgenic mice with five familial Alzheimer's disease mutations: potential factors in amyloid plaque formation. J Neurosci. 2006;26:10129–10140. doi: 10.1523/JNEUROSCI.1202-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liang W.S., Reiman E.M., Valla J., Dunckley T., Beach T.G., Grover A., Niedzielko T.L., Schneider L.E., Mastroeni D., Caselli R., Kukull W., Morris J.C., Hulette C.M., Schmechel D., Rogers J., Stephan D.A. Alzheimer's disease is associated with reduced expression of energy metabolism genes in posterior cingulate neurons. Proc Natl Acad Sci USA. 2008;105:4441–4446. doi: 10.1073/pnas.0709259105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blalock E.M., Buechel H.M., Popovic J., Geddes J.W., Landfield P.W. Microarray analyses of laser-captured hippocampus reveal distinct gray and white matter signatures associated with incipient Alzheimer's disease. J Chem Neuroanat. 2011;42:118–126. doi: 10.1016/j.jchemneu.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang B., Gaiteri C., Bodea L.G., Wang Z., McElwee J., Podtelezhnikov A.A., Zhang C., Xie T., Tran L., Dobrin R., Fluder E., Clurman B., Melquist S., Narayanan M., Suver C., Shah H., Mahajan M., Gillis T., Mysore J., MacDonald M.E. Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer's disease. Cell. 2013;153:707–720. doi: 10.1016/j.cell.2013.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rhodes D.R., Yu J., Shanker K., Deshpande N., Varambally R., Ghosh D., Barrette T., Pandey A., Chinnaiyan A.M. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schaefer M.H., Fontaine J.F., Vinayagam A., Porras P., Wanker E.E., Andrade-Navarro M.A. HIPPIE: integrating protein interaction networks with experiment based quality scores. PLoS ONE. 2012;7:e31826. doi: 10.1371/journal.pone.0031826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stelzer G., Inger A., Olender T., Iny-Stein T., Dalah I., Harel A., Safran M., Lancet D. GeneDecks: paralog hunting and gene-set distillation with genecards annotation. OMICS. 2009;13:477–487. doi: 10.1089/omi.2009.0069. [DOI] [PubMed] [Google Scholar]
- 35.Huangda W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using David bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 36.Szklarczyk D., Franceschini A., Wyder S., Forslund K., Heller D., Huerta-Cepas J., Simonovic M., Roth A., Santos A., Tsafou K.P., Kuhn M., Bork P., Jensen L.J., von Mering C. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gjoneska E., Pfenning A.R., Mathys H., Quon G., Kundaje A., Tsai L.H., Kellis M. Conserved epigenomic signals in mice and humans reveal immune basis of Alzheimer's disease. Nature. 2015;518:365–369. doi: 10.1038/nature14252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paban V., Loriod B., Villard C., Buee L., Blum D., Pietropaolo S., Cho Y.H., Gory-Faure S., Mansour E., Gharbi A., Alescio-Lautier B. Omics analysis of mouse brain models of human diseases. Gene. 2017;600:90–100. doi: 10.1016/j.gene.2016.11.022. [DOI] [PubMed] [Google Scholar]
- 39.Zhao H., Jin G., Cui K., Ren D., Liu T., Chen P., Wong S., Li F., Fan Y., Rodriguez A., Chang J., Wong S.T. Novel modeling of cancer cell signaling pathways enables systematic drug repositioning for distinct breast cancer metastases. Cancer Res. 2013;73:6149–6163. doi: 10.1158/0008-5472.CAN-12-4617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu T., Ren D., Zhu X., Yin Z., Jin G., Zhao Z., Robinson D., Li X., Wong K., Cui K., Zhao H., Wong S.T. Transcriptional signaling pathways inversely regulated in Alzheimer's disease and glioblastoma multiform. Sci Rep. 2013;3:3467. doi: 10.1038/srep03467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xie W., Li X., Li C., Zhu W., Jankovic J., Le W. Proteasome inhibition modeling nigral neuron degeneration in Parkinson's disease. J Neurochem. 2010;115:188–199. doi: 10.1111/j.1471-4159.2010.06914.x. [DOI] [PubMed] [Google Scholar]
- 42.Villasana L.E., Klann E., Tejada-Simon M.V. Rapid isolation of synaptoneurosomes and postsynaptic densities from adult mouse hippocampus. J Neurosci Methods. 2006;158:30–36. doi: 10.1016/j.jneumeth.2006.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dunkley P.R., Jarvie P.E., Robinson P.J. A rapid Percoll gradient procedure for preparation of synaptosomes. Nat Protoc. 2008;3:1718–1728. doi: 10.1038/nprot.2008.171. [DOI] [PubMed] [Google Scholar]
- 44.Li X.P., Xie W.J., Zhang Z., Kansara S., Jankovic J., Le W.D. A mechanistic study of proteasome inhibition-induced iron misregulation in dopamine neuron degeneration. Neurosignals. 2012;20:223–236. doi: 10.1159/000332954. [DOI] [PubMed] [Google Scholar]
- 45.Cykowski M.D., Takei H., Van Eldik L.J., Schmitt F.A., Jicha G.A., Powell S.Z., Nelson P.T. Hippocampal sclerosis but not normal aging or Alzheimer disease is associated with TDP-43 pathology in the basal forebrain of aged persons. J Neuropathol Exp Neurol. 2016;75:397–407. doi: 10.1093/jnen/nlw014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Franklin K., Paxinos G. Compact 3rd edition. Academic Press; 2008. The mouse brain in stereotaxic coordinates. ISBN: 9780080921365. [Google Scholar]
- 47.Karnati S., Lüers G., Pfreimer S., Baumgart-Vogt E. Mammalian SOD2 is exclusively located in mitochondria and not present in peroxisomes. Histochem Cell Biol. 2013;140:105–117. doi: 10.1007/s00418-013-1099-4. [DOI] [PubMed] [Google Scholar]
- 48.Zhang J, Hu J. Image segmentation based on 2D OTSU method with histogram analysis, 6. IEEE. p. 105-108.
- 49.He T, Xue Z, Wong ST. A three-dimensional medical image segmentation app using graphic theory, Proceedings of the IEEE-EMBS international conference on in biomedical and health informatics (BHI): 268–71. IEEE.
- 50.SerulleY Sugimori M, Llinás R.R. Imaging synaptosomal calcium concentration microdomains and vesicle fusion by using total internal reflection fluorescent microscopy. Proc Natl Acad Sci USA. 2007;104:1697–1702. doi: 10.1073/pnas.0610741104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Frautschy S.A., Yang F., Irrizarry M., Hyman B., Saido T.C., Hsiao K., Cole G.M. Microglial response to amyloid plaques in APPsw transgenic mice. Am J Pathol. 1998;152:307–317. [PMC free article] [PubMed] [Google Scholar]
- 52.Ohyagi Y., Asahara H., Chui D.H., Tsuruta Y., Sakae N., Miyoshi K., Yamada T., Kikuchi H., Taniwaki T., Murai H., Ikezoe K., Furuya H., Kawarabayashi T., Shoji M., Checler F., Iwaki T., Makifuchi T., Takeda K., Kira J., Tabira T. Intracellular Abeta42 activates p53 promoter: a pathway to neurodegeneration in Alzheimer's disease. FASEB J. 2005;19:255–257. doi: 10.1096/fj.04-2637fje. [DOI] [PubMed] [Google Scholar]
- 53.He P., Shen Y. Interruption of beta-catenin signaling reduces neurogenesis in Alzheimer's disease. J Neurosci. 2009;29:6545–6557. doi: 10.1523/JNEUROSCI.0421-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Llorens-Martín M., Jurado J., Hernández F., Avila J. GSK-3β, a pivotal kinase in Alzheimer disease. Front Mol Neurosci. 2014;7:46. doi: 10.3389/fnmol.2014.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.de Graaf A.O., van den Heuvel L.P., Dijkman H.B., de Abreu R.A., Birkenkamp K.U., de Witte T., van der Reijden B.A., Smeitink J.A., Jansen J.H. Bcl-2 prevents loss of mitochondria in CCCP-induced apoptosis. Exp Cell Res. 2004;299:533–540. doi: 10.1016/j.yexcr.2004.06.024. [DOI] [PubMed] [Google Scholar]
- 56.Luo L.Y., Soosaipillai A., Diamandis E.P. Molecular cloning of a novel human gene on chromosome 4p11 by immunoscreening of an ovarian carcinoma cDNA library. Biochem Biophys Res Commun. 2001;280:401–406. doi: 10.1006/bbrc.2000.4126. [DOI] [PubMed] [Google Scholar]
- 57.Wang C., Michener C.M., Belinson J.L., Vaziri S., Ganapathi R., Sengupta S. Role of the 18:1 lysophosphatidic acid-ovarian cancer immunoreactive antigen domain containing 1 (OCIAD1)-integrin axis in generating late-stage ovarian cancer. Mol Cancer Ther. 2010;9:1709–1718. doi: 10.1158/1535-7163.MCT-09-1024. [DOI] [PubMed] [Google Scholar]
- 58.Yang A.H., Chen J.Y., Lee C.H., Chen J.Y. Expression of NCAM and OCIAD1 in well-differentiated thyroid carcinoma: correlation with the risk of distant metastasis. J Clin Pathol. 2012;65:206–212. doi: 10.1136/jclinpath-2011-200416. [DOI] [PubMed] [Google Scholar]
- 59.Morel E., Chamoun Z., Lasiecka Z.M., Chan R.B., Williamson R.L., Vetanovetz C., Dall'Armi C., Simoes S., Point Du Jour K.S., McCabe B.D., Small S.A., Di Paolo G. Phosphatidylinositol-3-phosphate regulates sorting and processing of amyloid precursor protein through the endosomal system. Nat Commun. 2013;4:2250. doi: 10.1038/ncomms3250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Khadilkar R.J., Rodrigues D., Mote R.D., Sinha A.R., Kulkarni V., Magadi S.S., Inamdar M.S. ARF1-GTP regulates ASRIJ to provide endocytic control of Drosophila blood cell homeostasis. Proc Natl Acad Sci USA. 2014;111:4898–4903. doi: 10.1073/pnas.1303559111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mukhopadhyay A., Das D., Inamdar M.S. Embryonic stem cell and tissue specific expression of a novel conserved gene, asrij. Dev Dyn. 2003;227:578–586. doi: 10.1002/dvdy.10332. [DOI] [PubMed] [Google Scholar]
- 62.Wilhelm B.G., Mandad S., Truckenbrodt S., Kröhnert K., Schäfer C., Rammner B., Koo S.J., Claßen G.A., Krauss M., Haucke V., Urlaub H., Rizzoli S.O. Composition of isolated synaptic boutons reveals the amounts of vesicle trafficking proteins. Science. 2014;344:1023–1028. doi: 10.1126/science.1252884. [DOI] [PubMed] [Google Scholar]
- 63.Chadwick W., Brenneman R., Martin B., Maudsley S. Complex and multidimensional lipid raft alterations in a murine model of Alzheimer's disease. Int J Alzheimers Dis. 2010:60479. doi: 10.4061/2010/604792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Han J., Jung S., Jang J., Kam T.I., Choi H., Kim B.J., Nah J., Jo D.G., Nakagawa T., Nishimura M., Jung Y.K. OCIAD2 activates γ-secretase to enhance amyloid β production by interacting with nicastrin. Cell Mol Life Sci. 2014;71:2561–2576. doi: 10.1007/s00018-013-1515-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hyman B.T., Yuan J. Apoptotic and non-apoptotic roles of caspases in neuronal physiology and pathophysiology. Nat Rev Neurosci. 2012;13:395–406. doi: 10.1038/nrn3228. [DOI] [PubMed] [Google Scholar]
- 66.Vaux D.L. Apoptogenic factors released from mitochondria. Biochim Biophys Acta. 2011;1813:546–550. doi: 10.1016/j.bbamcr.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 67.Rhein V., Song X., Wiesner A., Ittner L.M., Baysang G., Meier F., Ozmen L., Bluethmann H., Dröse S., Brandt U., Savaskan E., Czech C., Götz J., Eckert A. Amyloid-beta and tau synergistically impair the oxidative phosphorylation system in triple transgenic Alzheimer's disease mice. Proc Natl Acad Sci USA. 2009;106:20057–20062. doi: 10.1073/pnas.0905529106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ding H, Wang H, Zhao Y, Sun D, Zhai X. Protective Effects of Baicalin on Aβ₁₋₄₂-Induced Learning and Memory Deficit, Oxidative Stress, and Apoptosis in Rat. Cell Mol Neurobiol. 2015;35:623–632. doi: 10.1007/s10571-015-0156-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Park S.J., Shin J.H., Jeong J.I., Song J.H., Jo Y.K., Kim E.S., Lee E.H., Hwang J.J., Lee E.K., Chung S.J., Koh J.Y., Jo D.G., Cho D.H. Down-regulation of mortalin exacerbates Aβ-mediated mitochondrial fragmentation and dysfunction. J Biol Chem. 2014;289:2195–2204. doi: 10.1074/jbc.M113.492587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Manczak M., Reddy P.H. Abnormal interaction between the mitochondrial fission protein DRP1 and hyperphosphorylated tau in Alzheimer's disease neurons: implications for mitochondrial dysfunction and neuronal damage. Hum Mol Genet. 2012;21:2538–2547. doi: 10.1093/hmg/dds072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hosp F., Vossfeldt H., Heinig M., Vasiljevic D., Arumughan A., Wyler E., Genetic and environmental Risk for Alzheimer's Disease GERAD1 Consortium. Landthaler M., Hubner N., Wanker EE., Lannfelt L., Ingelsson M., Lalowski M., Voigt A., Selbach M. Quantitative interaction proteomics of neurodegenerative disease proteins. Cell Rep. 2015;11:1134–1146. doi: 10.1016/j.celrep.2015.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yan J., Liu X.H., Han M.Z., Wang Y.M., Sun X.L., Yu N., Li T., Su B., Chen Z.Y. Blockage of GSK3β-mediated Drp1 phosphorylation provides neuroprotection in neuronal and mouse models of Alzheimer's disease. Neurobiol Aging. 2015;36:211–227. doi: 10.1016/j.neurobiolaging.2014.08.005. [DOI] [PubMed] [Google Scholar]
- 73.Shetty D.K., Kalamkar K.P., Inamdar M.S. OCIAD1 controls electron transport chain complex i activity to regulate energy metabolism in human pluripotent stem cells. Stem Cell Rep. 2018;11:128–141. doi: 10.1016/j.stemcr.2018.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sinha S., Dwivedi T.R., Yengkhom R., Bheemsetty V.A., Abe T., Kiyonari H., VijayRaghavan K., Inamdar M.S. Asrij/OCIAD1 suppresses CSN5-mediated p53 degradation and maintains mouse hematopoietic stem cell quiescence. Blood. 2019;133:2385–2400. doi: 10.1182/blood.2019000530. [DOI] [PubMed] [Google Scholar]
- 75.Madeira C., Lourenco M.V., Vargas-Lopes C., Suemoto C.K., Brandão C.O., Reis T., Leite R.E., Laks J., Jacob-Filho W., Pasqualucci C.A., Grinberg L.T., Ferreira S.T., Panizzutti R. d-serine levels in Alzheimer's disease: implications for novel biomarker development. Transl Psychiatry. 2015;5:e561. doi: 10.1038/tp.2015.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Canchi S., Raao B., Masliah D., Rosenthal S.B., Sasik R., Fisch K.M., De Jager P.L., Bennett D.A., Rissman R.A. Integrating gene and protein expression reveals perturbed functional networks in Alzheimer's disease. Cell Rep. 2019;28:1103–1116. doi: 10.1016/j.celrep.2019.06.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liu Y., Beyer A., Aebersold R. On the dependency of cellular protein levels on mRNA abundance. Cell. 2016;165:535–550. doi: 10.1016/j.cell.2016.03.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.