Abstract
Background
Cervical cancer (CC) is the 4th most common malignancy and cancer-related fatality in women worldwide. Circular RNA is a newly recognized noncoding RNA form. More reports have confirmed their important regulatory functions in diverse physiological and cancer processes. However, the function and mechanisms of circ-HIPK3 in CC remain unknown.
Methods
circ-HIPK3 expression status was verified between 45 paired CC and normal adjacent tissues from 45 CC patients, also the 6 CC cell lines and a normal human cervical epithelial End1/E6E7 cell line by qRT-PCR. Effects of circ-HIPK3 silence on CC cell phenotypes were estimated. Circular RNA interactome was applied to forecast binding site between circ-HIPK3 and miRNAs. Pearson correlation analysis was used to confirm the relationship between genes. Point mutation, RNA pull-down, luciferase assay and rescue experiments were applied for molecular mechanism exploration.
Results
circ-HIPK3 expression was significantly elevated in CC cells and tissues. circ-HIPK3 silence repressed growth and metastasis, while induced apoptosis in CC cells. circ-HIPK3 sponged miRNA-338-3p (miR-338-3p); miR-338-3p to up-regulate hypoxia-inducible factor-1α (HIF-1α) and CC progress. MiR-338-3p silence or HIF-1α over-expression rescued circ-HIPK3 knockdown caused inhibition of CC malignant characteristics.
Conclusion
circ-HIPK3 acts as a competing endogenous RNA of miR-338-3p to promote cell growth and metastasis in CC, via regulating HIF-1α mediated EMT. Therefore, targeting circ-HIPK3/miR-338-3p/HIF-1α axis may be a novel therapeutic strategy for CC.
Keywords: cervical cancer, circ-HIPK3, miR-338-3p, HIF-1α, EMT, metastasis
Introduction
Even though human papilloma virus (HPV) vaccine is an excellent technique to protect female from cervical cancer (CC), CC remains the 4th most common malignancy with about 450,000 new patients diagnosed each year.1–4 In the meantime, regardless of the advances in diagnosis and treatment, such as the main strategy of surgery combined with chemotherapy and radiotherapy for prolonged patient survival, the CC still remains a quite poor prognosis, and the second most frequent cancer-associated death in women,5 with a 5-year overall survival under 40% in most countries, due to the cancer metastasis and recurrence.6 The molecular mechanism of CC has not been fully revealed. Therefore, it is critical to further explore the molecular mechanism of CC oncogenesis, develop new biomarkers for the diagnosis and prognosis of CC patients, and discover the therapeutic targets for CC treatment.
As we all have known, the epithelial-mesenchymal transition (EMT) plays a crucial role in cancer metastasis by losing the epithelial characteristics and gaining the invasive and metastatic abilities.7,8 Thus, identifying factors to inhibit EMT should support the successful clinical managements.
In the recent decades, non-coding RNAs (ncRNAs), including microRNA (miRNA) and long non-coding RNA (lncRNA), have been found to be deregulated in oncogenesis.9,10
Circular RNA (circRNA) is a novel identified and special form of ncRNAs, which is distinguished by a covalently closed structure without 5′ caps, 3′ tails and protein coding capacity. CircRNAs also display cell or tissue-specific expressions, and are preserved throughout species as a result of their resistance to RNase R.11–13 CircRNAs are really stable and mainly in the cytoplasm than the linear RNAs.14,15 CircRNAs consist of miRNA response elements (MREs) that can be applied to explore the miRNA-specific antagonists. Growing evidences signify that circRNAs interrelate with RNA binding proteins (RBPs) and serve as miRNA sponges to control the target gene expression.16
Recent studies have revealed that circRNAs are involved in the CC development by diverse mechanisms, of which miRNA sponging is the most important one.17–20
Therefore, it is necessary to discover the deregulated circRNAs, find out the novel molecular mechanisms and therapeutic targets for CC treatment.
Recently, a novel circRNA, circ-HIPK3 was reported to promote the proliferation and invasiveness of glioma cells through sponging miR-124-3p to up-regulate the STAT3 expression.21 Abnormal expression of miR-338-3p has been reported to be extremely related with the development and prognosis of multiple cancers.22–27 However, the relationship between circ-HIPK3 and miR-338-3p, also their function and clinical application in CC remain unclear.
In our current study, we intended to investigate the function, molecular mechanisms and clinical implications of the circ-HIPK3 in CC. Our results demonstrated that circ-HIPK3 was up-regulated in CC cells and tissues to promote the cell proliferation and EMT. More significantly, we found that circ-HIPK3 functioned as a sponge of miRNA-338-3p (miR-338-3p), thus to up-regulate the HIF-1α expression, and consequently promote the tumorigenesis of CC. This study provided new verifications that may facilitate the advance of effective therapeutic strategies against CC in the clinical practices.
Methods
Patient Tissues
CC and paired adjacent normal cervical tissues from 45 CC patients were collected from patients experiencing surgery at the department of obstetrics and gynecology, the First People’s Hospital of Lianyungang. All tissues were snap-frozen in liquid nitrogen at the time of collection and stored at −80°C until use.
Consent to Participate and Ethics Approval
All samples were collected with the written informed consents from the patients before the study was started. All experiments were approved by the Ethics Committee of the First People’s Hospital of Lianyungang according to the Declaration of Helsinki.
Reagents
Macoy’s 5A medium and fetal bovine serum (FBS) were from Gibco (Rockford, MD, USA). Dual-Luciferase Reporter Assay System was from Promega (Madison, WI, USA). miRNeasy Mini Kit, miScript II RT Kit and miScript SYBR Green PCR Kit were from Qiagen (Duesseldorf, Germany). PrimeScrip™ RT Master Mix, RNAiso Plus and SYBR Green Premix Ex Taq™ II were from TaKaRa (Dalian, China). Matrigel was from BD (New Jersey, USA). CCK-8 assay kit was from Dojindo Corp (Kyushu, Japan). RIPA lysis buffer was from Beyotime (Shanghai, China). Lipofectamine 3000 and SuperSignal West Dura Extended Duration Substrate were from Invitrogen/Thermo Fisher Scientific (Waltham, USA). APC-Annexin V and propidium iodide were from Sigma (St. Louis, USA).
Cell Lines and Culture
Six CC cell lines (HeLa, CaSki, SiHa, C-33A, C-4I, SW756) and a normal human cervical epithelial End1/E6E7 cell line were bought from the Committee on Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). The cells were cultured in Macoy’s 5A medium-containing fetal bovine serum (FBS, 10%), penicillin (100 IU/mL) and streptomycin (100 mg/mL) at 37°C in a humidified 5% CO2 incubator.
Lentiviral Vector, siRNA, Mimics and Inhibitors
The lentivirus-mediated HIF-1α over-expression and no-load lentivirus vectors were provided by Hanbio (Shanghai, China). CC cells were infected with the lentivirus according to the instructions of the manufacturer, and stable cells were selected with puromycin.
siRNA targeting circ-HIPK3 (si-circ-HIPK3) and the control siRNA (si-NC) were got from RiboBio (Guangzhou, China); miR-338-3p inhibitor (inh-miR-338-3p) and the control siRNA (si-NC) were purchased from GenePharma (Shanghai, China), all of them were transfected into the CC cells using Lipofectamine 3000 following the manufacturer’s instruction.
Target Gene Identification
The potential sponged miRNAs by the circ-HIPK3 were predicted via the website tool Circular RNA Interactome (https://circinteractome.nia.nih.gov/). The target mRNAs of miR-338-3p were determined based on the literature.
Quantitative Real-Time PCR (qRT-PCR)
RNAiso Plus was used to isolate the total RNA. NanoDrop ND-1000 (Thermo Fisher Scientific, Wilmington, DE) was employed to estimate the RNA purity and concentration. PrimeScrip™ RT Master Mix was applied for reverse transcription, and SYBR Green Premix Ex Taq™ II was applied for cDNA amplification according to the manufacturer’s instructions.
According to the manufacturer’s instructions, a miRNeasy Mini Kit was used to extract miRNAs, a miScript II RT Kit was applied for reverse transcription and a miScript SYBR Green PCR Kit was applied for cDNA amplification. A qRT-PCR was carried out on an AB7300 thermo recycler (Applied Biosystems, Carlsbad, CA) with the TaqMan Universal PCR Master Mix. GAPDH served as the internal control for mRNA and circRNA. U6 served as the internal control for miRNA. Relative gene expression was calculated with the 2−ΔΔCt method. The primers are as follows:
circ-HIPK3: Forward: 5ʹ-CGTGGCGAAACAGATTGCAT-3ʹ
Reverse: 5ʹ-GCTGTCACAACTGTCACTGG-3ʹ
miR-338-3p: Forward: 5ʹ-ATCCAGTGCGTGTCGTGG-3ʹ
Reverse: 5ʹ- TGCTTCCAGCATCAGTGAT-3ʹ
HIF-1α: Forward: 5ʹ-GAACGTCGAAAAGAAAAGTCTCG −3ʹ
Reverse: 5ʹ- CCTTATCAAGATGCGAACTCACA −3ʹ
GAPDH: Forward: 5ʹ-TCCACCACCAACTCCTTACC-3ʹ
Reverse: 5ʹ-GCCATGGACTGTGGTCATGAG-3ʹ
U6: Forward: 5ʹ- TCCTTCGCCACCACATATAC-3ʹ
Reverse: 5ʹ- AGGGCCCATCCTAATCTTCT-3ʹ
Luciferase Assays
CC cells (5 ×104/well) were seeded in 24-well plates at 24hrs before transfection. Then, cells were co-transfected with circ-HIPK3 WT or mutated reporter and inh-miR-338-3p or si-NC using Lipofectamine 3000 according to the manufacturer’s instructions, luciferase activities were evaluated using the Dual-Luciferase Reporter Assay System 48 hrs later.
Cell Proliferation Assay
Cell proliferation assay was carried out with the CCK-8 kit following the manufacturer’s instruction. In brief, CC cells were seeded into a 96-well plate (2000 cells/well) and incubated for the indicated times, then 10 μL of CCK-8 reagent was directly loaded into the culture medium and incubated for another 2 hrs in dark at 37°C. The optional density (OD) at a wavelength of 450 nm was evaluated with a 680 Microplate Reader (Bio-Rad, Hercules).
Colony Formation Assay
CC cells were seeded in a 6-well plate (1000 cells/well) and cultured at 37°C for 7 days in the fresh medium. On the 7th day, cells were fixed for 10 min with 4% paraformaldehyde and stained for 5 min with 0.5% crystal violet solution. Then, the colony number was calculated with ImageJ and images were photographed under a light microscope (Olympus, Japan).
Cell Apoptosis Assay
CC cells to be tested were washed with pre-cold PBS, re-suspended in binding buffer and fixed, then incubated with APC-Annexin V and propidium iodide for 15 min in the dark at room temperature. Then, the fluorescence intensity was determined with a BD FACSCalibur flow cytometer. Cell Quest software (Becton Dickinson, USA) was used to calculate the percentage of cell apoptosis.
Cell Migration and Invasion Assay
The cell migration and invasive abilities were evaluated using the 24-well transwells (Corning Costar, 8.0 μm pore size). The transwell filter on the upper chamber was coated with (for the invasion assay) or without (for the migration assay) Matrigel. Briefly, 200μL of serum-free medium containing 3 × 105 cells was loaded into the upper chamber, the lower chamber was supplied with 500 μL of medium containing 10% FBS. After 48 hrs incubation, the non-migrated and non-invaded cells on the upper surface of the upper chamber were removed with a cotton swab, while the cells migrated or invaded to the opposite side were fixed with 4% paraformaldehyde, stained with 0.1% crystal violet solution. The numbers of the migrated or invaded cells were then counted and imaged under an Olympus IX-81 inverted microscope (Olympus, Japan).
Western Blotting Analysis
RIPA lysis buffer was used to isolate the total proteins from CC cells. Equal amounts of proteins were separated on a 10% SDS-PAGE gel, transferred onto the PVDF membranes and incubated with the diluted primary antibodies overnight at 4°C, followed by incubation with secondary antibody for 1 hrs at room temperature. The membranes were washed three times with TBST and visualized using SuperSignal West Dura Extended Duration Substrate according to the manufacturer’s instructions.
Statistical Analysis
Statistical analyses were achieved by SPSS 20.0 (IBM, SPSS, Chicago, IL, USA) and GraphPad Prism (GraphPad, La Jolla, USA). Student’s t-test and one-way ANOVA followed by Tukey’s post hoc test were utilized to compare two or multiple groups, respectively. Pearson correlation analysis was used to evaluate the association between genes. p< 0.05 was statistical significance.
Results
Amplified circ-HIPK3 Was Found in CC Patients
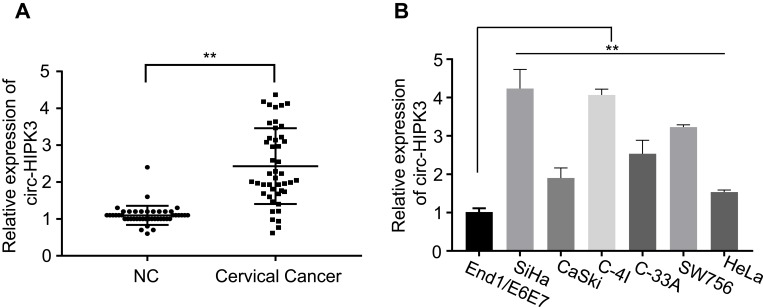
To explore the clinical implication of circ-HIPK3 expression in CC patients, we collected 45 pairs of CC and sef-matched adjacent normal tissues from the CC patients. As the expression of circ-HIPK3 in CC patients has not been reported, we first evaluated the expression of circ-HIPK3 in these 45 CC patients by qRT-PCR. Our results demonstrated that circ-HIPK3 expression was drastically amplified in the CC tissues than that in the adjacent normal tissues (Figure 1A, p < 0.01).
Figure 1.
circ-HIPK3was up-regulated in CC. (A), circ-HIPK3 expression was statistically significantly higher in the CC tissues than in the paired normal adjacent tissues from 45 CC patients; (B), circ-HIPK3 expression was statistically significantly higher in the CC cells (HeLa, CaSki, SiHa, C-33A, C-4I, SW756) than in the normal human cervical epithelial End1/E6E7 cells. **p< 0.01.
By comparing the circ-HIPK3 expression levels between the CC cells and the normal human cervical epithelial cells, we further proved the circ-HIPK3 expression status in CC. As shown in Figure 1B, circ-HIPK3 expression levels were significantly higher in all tested CC cells (HeLa, CaSki, SiHa, C-33A, C-4I, SW756) than that in the normal human cervical epithelial End1/E6E7 cells (Figure 1B, p < 0.01).
circ-HIPK3 Promoted CC Growth
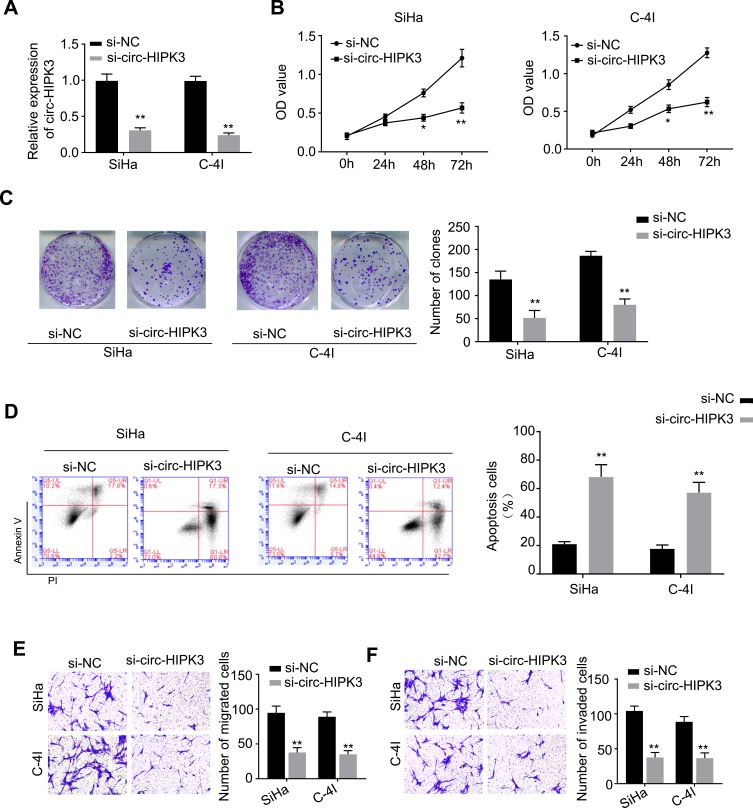
To explore the biological functions of circ-HIPK3 in CC, the siRNA targeting circ-HIPK3 (assigned as si-circ-HIPK3) was transfected into SiHa and C-4I cells, the two CC cell lines with the highest circ-HIPK3 expression level, to silence the circ-HIPK3 expression. Our results showed that transfection of si-circ-HIPK3 into SiHa and C-4I cells resulted in a statistically significant reduction in circ-HIPK3 expression level (Figure 2A, p < 0.01); compared with the negative control (si-NC), si-circ-HIPK3 silenced over 50% of circ-HIPK3 expression in both SiHa and C-4I cells detected by qRT-PCR assay (Figure 2A). Furthermore, the CCK-8 assay was utilized to identify the cell viability of SiHa and C-4I cells at 0, 24, 48 and 72hrs after transfection of si-NC or si-circ-HIPK3, we found that circ-HIPK3 silence statistically significantly reduced the viability (OD value) of both SiHa (Figure 2B, left panel) and C-4I (Figure 2B, right panel) cells detected at 450 nm (p< 0.05 at 48hrs, p< 0.01 at 72hrs); clone formation assay showed that circ-HIPK3 silence statistically significantly restrained the clone formation ability (p < 0.01), the images (Figure 2C, the left and middle panels) and numbers (Figure 2C, the right panel) of the clones were acquired and counted under the microscope; meanwhile, flow cytometric assay demonstrated that the apoptosis proportion was statistically significantly increased in the SiHa and C-4I cells (both p< 0.01) after circ-HIPK3 silence (Figure 2D).
Figure 2.
circ-HIPK3 silence inhibited proliferation, migration and invasion, while induced apoptosis of CC cells. (A) qRT-PCR analysis confirmed that circ-HIPK3 was successfully silenced with si-circ-HIPK3 in the SiHa and C-4I cells; the following phenotypes were then evaluated in the SiHa and C-4I cells transfected with the si-NC or si-circ-HIPK3: (B) viability, (C) clone formation ability, (D) apoptosis, (E) migration, (F) cell invasion. *p < 0.05; **p< 0.01.
circ-HIPK3 Advanced EMT of CC Cells
As we all have known, EMT is an important procedure to stimulate the aggressiveness in cancer cells. Therefore, the transwell migration and invasion assays were utilized to estimate whether circ-HIPK3 could enhance the EMT characteristics in CC cells. Our results showed that circ-HIPK3 silence statistically significantly restrained the migration (Figure 2E) and invasive (Figure 2F) abilities of the CC cells (p< 0.01), the images (Figure 2E and F, left panels) and numbers of the migrated (Figure 2E, right panel) and invaded (Figure 2F, right panel) cells were acquired and counted under the microscope.
circ-HIPK3 Was Proved to Sponge miR-338-3p in CC Cells
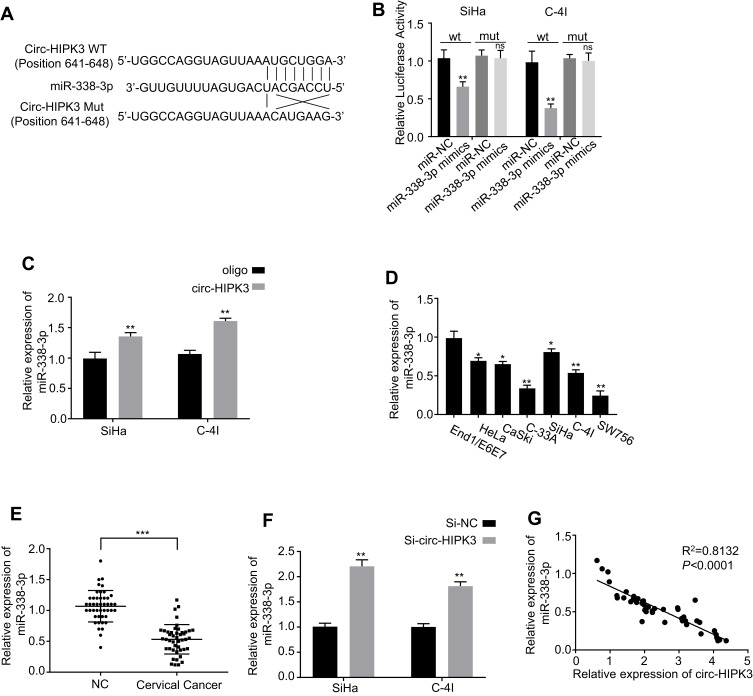
As previously reported, circRNAs predominantly functioned as miRNA sponges to control gene expressions. Consequently, the potential miRNAs related to circ-HIPK3 were investigated. The circular RNA Interactome (https://circinteractome.nia.nih.gov/) was utilized to forecast the prospective target miRNAs to bind with the circ-HIPK3 sequence, followed by the luciferase reporter gene assay, and the RNA in vivo precipitation (RIP) in SiHa and C-4I cells, respectively. Our results of the following experiments demonstrated that miR-338-3p was the potential target of circ-HIPK3 (Figure 3A). Over-expression of miR-338-3pwith the mimics restrained luciferase activity of the wild-type circ-HIPK3 reporter gene in SiHa and C-4I cells (Figure 3B, p< 0.01), while this restrained luciferase activity of circ-HIPK3 reporter gene was rescued when the forecasted binding site of circ-HIPK3 with miR-338-3p was mutated (Figure 3B); to further verify this result, the interact between circ-HIPK3 and miR-338-3p was identified by RNA in vivo precipitation (RIP), it was found that the circ-HIPK3 probe could pull down more miR-338-3p versus the control oligo probe in SiHa and C-4I cells, the difference was statistically significant (Figure 3C, p < 0.01). Furthermore, the practical relationship between the circ-HIPK3 and the miR-338-3p was investigated by detecting miR-338-3p expression in the CC cells and tissues, our results proved that miR-338-3p expression was statistically significantly lower in the CC cells (HeLa, CaSki, SiHa, C-33A, C-4I, SW756) than in the normal human cervical epithelial End1/E6E7 cells (p < 0.05 or p< 0.01, Figure 3D); in the CC tissues than in the paired adjacent normal tissues from 45 CC patients (same samples as in Figure 1A) detected by qRT-PCR (p< 0.001, Figure 3E); in the meantime, the miR-338-3p expression in circ-HIPK3-silenced SiHa and C-4I cells was statistically significantly up-regulated (p< 0.01, Figure 3F). Pearson correlation coefficient was further used to evaluate the relationship between circ-HIPK3 and miR-338-3p expression, which illustrated that circ-HIPK3 expression was considerably negatively associated with miR-338-3p expression in the CC tissues from 45 CC patients (same samples as in Figure 1A) (Figure 3G, p< 0.0001). These results proposed that circ-HIPK3 directly interacted with miR-338-3p, leading to CC development.
Figure 3.
Identification of the potential miRNA sponged by circ-HIPK3. (A) Schematic representation of the potential binding sites of miR-338-3p with circ-HIPK3 (https://circinteractome.nia.nih.gov/) and the mutation sites for specific assay; (B), double luciferase reporter assay in the SiHa and C-4I cells co-tranfected with circ-HIPK3WT or mutated reporter with or without miR-338-3p mimics; (C), miR-338-3p in the CC cell lysates was pulled down and enriched with biotin-labeled miR-338-3p specific probe; (D) the miR-338-3p expression level in the human CC cells (HeLa, CaSki, SiHa, C-33A, C-4I, SW756) and the normal human cervical epithelial End1/E6E7 cells; (E), the miR-338-3p expression levels in paired CC and adjacent normal tissues from 45 CC patients (same samples as in Figure 1A) detected by qRT-PCR; (F) the miR-338-3p expression levels in circ-HIPK3-silenced SiHa and C-4I cells detected by qRT-PCR; (G) Pearson correlation analysis of the association between miR-338-3p with circ-HIPK3 in the CC tissues from the 45 CC patients (same samples as in Figure 1A). *p < 0.05; **p< 0.01, ***p< 0.001.
circ-HIPK3 Sponged miR-338-3p to Up-Regulatehypoxia Inducible Factor-1α (HIF-1α) in CC Cells
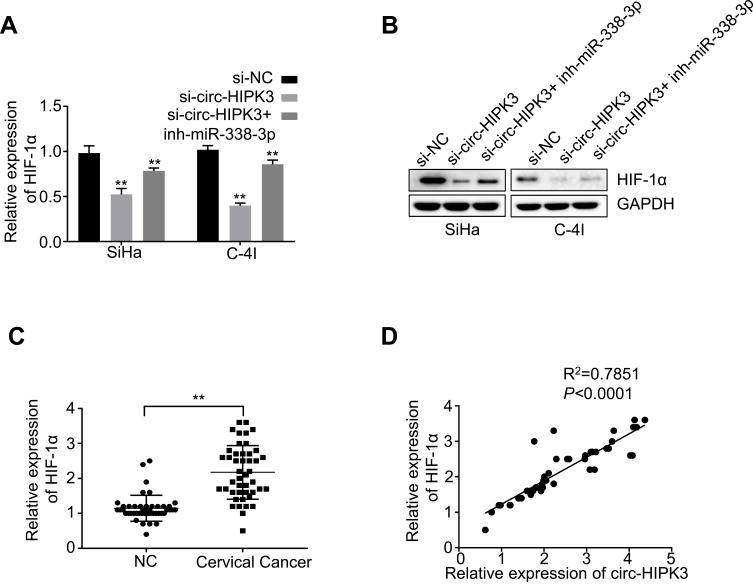
It has been reported that HIF-1α was the downstream target of miR-338-3p.28–30 To find out whether circ-HIPK3 sponges miR-338-3p, thus to regulate HIF-1α expression, we first detected the expression levels of HIF-1α in the CC cells by qRT-PCR. Our results revealed that circ-HIPK3 silence significantly down-regulated HIF-1α expression at both the mRNA (p< 0.01, Figure 4A) and protein (Figure 4B) levels in CC cells, and these effects were partially rescued by co-transfection of the miR-338-3p inhibitor (inh-miR-338-3p) (Figure 4A and B). HIF-1α expression levels were further quantified using qRT-PCR in the CC tissues from 45 CC patients (same samples as in Figure 1A), which proved that HIF-1α expression was considerably increased in the CC tissues than that in the adjacent normal tissues (Figure 4C, p< 0.01). Meanwhile, the Pearson correlation analysis of the association between circ-HIPK3 and HIF-1α expression in the CC tissues from 45 CC patients (same samples as in Figure 1A) showed that circ-HIPK3 expression was statistically significantly and positively associated with HIF-1α expression (Figure 4D, p< 0.0001).
Figure 4.
circ-HIPK3 increased the expression of oncogenic HIF-1α by sponging miR-338-3p in CC. (A) The mRNA expression levels of HIF-1α in the SiHa and C-4I cells after circ-HIPK3 knockdown (si-circ-HIPK3) with or without miR-338-3p inhibitor (inh-miR-338-3p) detected by qRT-PCR; (B) the protein expression levels of HIF-1α in the SiHa and C-4I cells after circ-HIPK3 knockdown with or without inh-miR-338-3p detected by Western-Blot; (C) the relative expression levels of HIF-1α in paired CC and adjacent normal tissues from 45 patients (same samples as in Figure 1A) detected by qRT-PCR; (D) Pearson correlation analysis of the association between HIF-1α with circ-HIPK3 in the CC tissues from 45 CC patients (same samples as in Figure 1A). **p< 0.01.
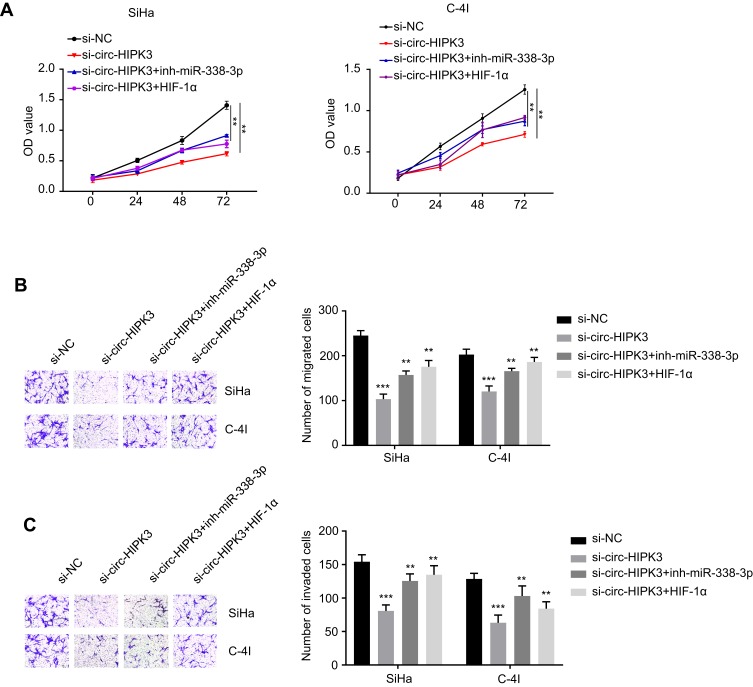
Cell viability in SiHa and C-4I cells (transfected with si-NC, si-circ-HIPK3, si-circ-HIPK3+inh-miR-338-3p or si-circ-HIPK3+HIF-1α) were identified with CCK-8 assay, our results demonstrated that circ-HIPK3 knockdown time dependently (from 0 to 72hrs) reduced the OD value at 450 nm, and this effect was partially rescued by miR-338-3p inhibition or HIF-1α over-expression (Figure 5A, p< 0.01 at 72hrs).
Figure 5.
MiR-338-3p silence or HIF-1α over-expression rescued the circ-HIPK3 knockdown caused malignant phenotype inhibition in CC. (A) cell viability in SiHa and C-4I cells after circ-HIPK3 knockdown with or without inh-miR-338-3p detected by CCK-8 assay; (B), cell migration ability in SiHa and C-4I cells after circ-HIPK3 knockdown with or without inh-miR-338-3p detected by the transwell assay without matrigel; (C) cell invasion ability in SiHa and C-4I cells after circ-HIPK3 knockdown with or without inh-miR-338-3p detected by the transwell assay with matrigel. **p< 0.01, ***p< 0.001.
Cell migration and invasion abilities in SiHa and C-4I cells (transfected with si-NC, si-circ-HIPK3, si-circ-HIPK3+inh-miR-338-3p or si-circ-HIPK3+HIF-1α) were verified by the Transwell assay without or with the matrigel, our results confirmed that circ-HIPK3 silence reduced both the migration (Figure 5B, p< 0.001) and invasion (Figure 5C, p< 0.001) abilities, and this effect was partially rescued by inhibition of miR-338-3p or over-expression of HIF-1α.
Discussion
circ-HIPK3 is a newly identified circRNA, which can promote the proliferation and invasion of glioma cells via the miR-124-3p/STAT3 axis.21 However, its function and clinical significance in CC remain unknown. Since the wider existence of the cell origin and histology in different cancer types, it’s necessary to further explore the function, molecular mechanisms and clinical implications of the circ-HIPK3 in CC.
In our current study, we examined the CC and paired adjacent normal tissues from 45 CC patients. Here, we first reported a significantly higher circ-HIPK3 expression in the CC tissues than the matched adjacent normal tissues, implying its potential oncogenic function in CC. Second, from a biological view, we showed that the up-regulation of circ-HIPK3 destroyed the normal function of miR-338-3p, as a consequence, the growth and EMT of CC cells were promoted. Third, we revealed a novel mechanism that circ-HIPK3 functioned as a competing endogenous RNA (ceRNA) of miR-338-3p to raise the HIF-1α expression. Considering the vital function of HIF-1α in CC development, our results for the first time found out the therapeutic meaning of circ-HIPK3 in CC patients.
miRNA, an essential part of the ncRNA family with a length of less than 22 nucleotides, plays intricate roles in managing the cellular functions through degradation of the target genes, thus to promote the cell growth, signaling transduction and oxidative response.
By circular RNA interactome, one of the online tools, we predicted the potential miRNAs binding to circ-HIPK3, and miR-338-3p was found to be the best possible candidate. In our current study, we further found that miR-338-3p was down-regulated in both the CC cell lines and tissues, and acted as an oncosuppressor in CC patients. MiR-338-3p expression was statistically considerably up-regulated when circ-HIPK3 was silenced in SiHa and C-4I cells; overexpression of miR-338-3p decreased the luciferase activities of the wild-type circ-HIPK3 reporter gene in CC cells, while this inhibited luciferase activity of circ-HIPK3 reporter gene was reversed when the predicted binding site of circ-HIPK3 with miR-338-3p was mutated; Pearson correlation coefficient analysis demonstrated a negative correlation between the circ-HIPK3 and the miR-338-3p expression, these results advised that circ-HIPK3 sponged miR-338-3p in CC cells, and circ-HIPK3 directly interacted with miR-338-3p to allow the metastasis and invasion of the CC cells.
Hypoxia, a common characteristic of solid tumors in the middle-late stages, plays a noteworthy role in oncogenesis through promoting the formation of a neoplastic environment.31 Cancer cells adapt to hypoxia partially via a transcriptional program coordinated by the HIF family of transcription factors.32 Among the three isoforms of HIFα (HIF-1α, −2α and −3α), HIF-1α is universally expressed in diverse cells and considered as the most important regulator of oxygen homeostasis.33 HIF-1α is an essential molecule involved in mediating the cellular processes. HIF-1α change and hypoxically regulated miRNAs are related to the outcome of patients with a variety of cancers, indicating their essential roles on cancer progress, and the miRNAs/HIF-1α circle plays complicated role in carcinogenesis.34
It has been reported that, as a tumor suppressor, miR-338-3p reduced the migration and proliferation through targeting HIF-1α in nasopharyngeal carcinoma.29 Our further research showed that the mRNA expression level of HIF-1α was significantly increased; Pearson correlation analysis illustrated that circ-HIPK3 expression was statistically significantly and positively related with HIF-1α expression in the CC tissues; circ-HIPK3 silence significantly down-regulated HIF-1α expression at both the mRNA and protein levels, decreased the cell viability, clone formation, cell migration and invasion abilities, while statistically significantly increased the percentage of apoptosis in the CC cells. These effects were partially rescued by inhibition of miR-338-3p or over-expression of HIF-1α, suggesting HIF-1α directly interacted with miR-338-3p through the sponge activity of circ-HIPK3 to promote the progress of CC. Therefore, circ-HIPK3 seemed to serve as a competing endogenous RNA of miR-338-3p that decreased the degradation of HIF-1α, thus promoted CC cell growth and metastasis.
In summary, our results disclosed that circ-HIPK3 expression was considerably amplified in CC patients and cells. Functionally, circ-HIPK3 promoted CC cell growth, clone formation, migration and invasion, while inhibited apoptosis by sponging miR-338-3p to up-regulate the HIF-1α expression, and contributed to the CC cell EMT. Therefore, our findings highlighted the prospective role of circ-HIPK3 as a new oncogenic non-coding RNA that promoted the tumorigenesis and metastasis of CC, dual targeting circ-HIPK3 and miR-338-3p may provide a novel therapeutic strategy to overcome this oncogenic pathway for CC patients. However, the effects of circ-HIPK3/miR-338-3p/HIF-1α axis among other type of cancers need to be further investigated.
Acknowledgments
We wish to appreciate Xuejin Song and Na An, who have contributed to the revision process of our manuscript.
Abbreviations
CC, Cervical cancer; miR-338-3p, miRNA-338-3p; HPV, Human papilloma virus; EMT, Epithelial-mesenchymal transition; ncRNAs, Non-coding RNAs; miRNA, MicroRNA; lncRNA, Long non-coding RNA; circRNA, Circular RNA; MREs, miRNA response elements; RBPs, RNA binding proteins; FBS, Fetal Bovine Serum; si-circ-HIPK3, siRNA targeting circ-HIPK3; si-NC, Control siRNA; inh-miR-338-3p, miR-338-3p inhibitor; qRT-PCR, Quantitative real-time PCR; RIP, RNA in vivo precipitation; HIF-1α, Hypoxia inducible factor-1α; ceRNA, Competing endogenous RNA.
Disclosure
The authors stated no conflict of interests for current study.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. [DOI] [PubMed] [Google Scholar]
- 2.Haie-Meder C, Morice P, Castiglione M. Cervical cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2010;21(Suppl 5):v37–v40. doi: 10.1093/annonc/mdq162 [DOI] [PubMed] [Google Scholar]
- 3.Eiben GL, da Silva DM, Fausch SC, Le Poole IC, Nishimura MI, Kast WM. Cervical cancer vaccines: recent advances in HPV research. Viral Immunol. 2003;16:111–121. doi: 10.1089/088282403322017866 [DOI] [PubMed] [Google Scholar]
- 4.Tewari KS, Sill MW, Long HJ 3rd, et al. Improved survival with bevacizumab in advanced cervical cancer. N Engl J Med. 2014;370:734–743. doi: 10.1056/NEJMoa1309748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Petignat P, Roy M. Diagnosis and management of cervical cancer. BMJ. 2007;335:765–768. doi: 10.1136/bmj.39337.615197.80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Allemani C, Weir HK, Carreira H, et al. Global surveillance of cancer survival 1995-2009: analysis of individual data for 25,676,887 patients from 279 population-based registries in 67 countries (CONCORD-2). Lancet. 2015;385:977–1010. doi: 10.1016/S0140-6736(14)62038-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Natsuizaka M, Whelan KA, Kagawa S, et al. Interplay between Notch1 and Notch3 promotes EMT and tumor initiation in squamous cell carcinoma. Nat Commun. 2017;8:1758. doi: 10.1038/s41467-017-01500-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Singh M, Yelle N, Venugopal C, Singh SK. EMT: mechanisms and therapeutic implications. Pharmacol Ther. 2018;182:80–94. doi: 10.1016/j.pharmthera.2017.08.009 [DOI] [PubMed] [Google Scholar]
- 9.Wang BG, Xu Q, Lv Z, et al. Association of twelve polymorphisms in three onco-lncRNA genes with hepatocellular cancer risk and prognosis: A case-control study. World J Gastroenterol. 2018;24:2482–2490. doi: 10.3748/wjg.v24.i23.2482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao M, Wang S, Li Q, Ji Q, Guo P, Liu X. MALAT1: a long non-coding RNA highly associated with human cancers. Oncol Letters. 2018;16:19–26. doi: 10.3892/ol.2018.8613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9:e1003777. doi: 10.1371/journal.pgen.1003777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157. doi: 10.1261/rna.035667.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928 [DOI] [PubMed] [Google Scholar]
- 14.Cortes-Lopez M, Miura P. Emerging functions of circular RNAs. Yale J Biol Med. 2016;89:527–537. [PMC free article] [PubMed] [Google Scholar]
- 15.Barrett SP, Salzman J. Circular RNAs: analysis, expression and potential functions. Development. 2016;143:1838–1847. doi: 10.1242/dev.128074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Z, Huang C, Bao C, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22:256–264. doi: 10.1038/nsmb.2959 [DOI] [PubMed] [Google Scholar]
- 17.Gao YL, Zhang MY, Xu B, et al. Circular RNA expression profiles reveal that hsa_circ_0018289 is up-regulated in cervical cancer and promotes the tumorigenesis. Oncotarget. 2017;8:86625–86633. doi: 10.18632/oncotarget.21257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang J, Zhao X, Zheng X, Li F. Circular RNA hsa_circ_0023404 exerts an oncogenic role in cervical cancer through regulating miR-136/TFCP2/YAP pathway. Biochem Biophys Res Commun. 2018;501:428–433. doi: 10.1016/j.bbrc.2018.05.006 [DOI] [PubMed] [Google Scholar]
- 19.Cai H, Zhang P, Xu M, Yan L, Liu N, Wu X. Circular RNA hsa_circ_0000263 participates in cervical cancer development by regulating target gene of miR-150-5p. J Cell Physiol. 2019;234:11391–11400. doi: 10.1002/jcp.v234.7 [DOI] [PubMed] [Google Scholar]
- 20.Song T, Xu A, Zhang Z, et al. CircRNA hsa_circRNA_101996 increases cervical cancer proliferation and invasion through activating TPX2 expression by restraining miR-8075. J Cell Physiol. 2019;234:14296–14305. doi: 10.1002/jcp.v234.8 [DOI] [PubMed] [Google Scholar]
- 21.Zhuang C, Ma Q, Ye J, Zhang F, Gui Y. LncRNA GClnc1 promotes proliferation and invasion of bladder cancer through activation of MYC. FASEB J. 2019;33:11045–11059. [DOI] [PubMed] [Google Scholar]
- 22.Zhang L, Ding F. Hsa_circ_0008945 promoted breast cancer progression by targeting miR-338-3p. Onco Targets Ther. 2019;12:6577–6589. doi: 10.2147/OTT.S213994 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23.Zhang R, Shi H, Ren F, et al. Down-regulation of miR-338-3p and up-regulation of MACC1 indicated poor prognosis of epithelial ovarian cancer patients. J Cancer. 2019;10:1385–1392. doi: 10.7150/jca.29502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Y, Zhang D, Lv J, Wang S, Zhang Q. LncRNA SNHG15 acts as an oncogene in prostate cancer by regulating miR-338-3p/FKBP1A axis. Gene. 2019;705:44–50. doi: 10.1016/j.gene.2019.04.033 [DOI] [PubMed] [Google Scholar]
- 25.Lu M, Huang H, Yang J, et al. miR-338-3p regulates the proliferation, apoptosis and migration of SW480 cells by targeting MACC1. Exp Ther Med. 2019;17:2807–2814. doi: 10.3892/etm.2019.7260 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26.Ding Z, Zhu J, Zeng Y, et al. The regulation of Neuropilin 1 expression by miR-338-3p promotes non-small cell lung cancer via changes in EGFR signaling. Mol Carcinog. 2019;58:1019–1032. doi: 10.1002/mc.v58.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Y, Qin H. miR-338-3p targets RAB23 and suppresses tumorigenicity of prostate cancer cells. Am J Cancer Res. 2018;8:2564–2574. [PMC free article] [PubMed] [Google Scholar]
- 28.Yang X, Zi XH. LncRNA SNHG1 alleviates OGD induced injury in BMEC via miR-338/HIF-1alpha axis. Brain Res. 2019;1714:174–181. doi: 10.1016/j.brainres.2018.11.003 [DOI] [PubMed] [Google Scholar]
- 29.Shan Y, Li X, You B, Shi S, Zhang Q, You Y. MicroRNA-338 inhibits migration and proliferation by targeting hypoxia-induced factor 1alpha in nasopharyngeal carcinoma. Oncol Rep. 2015;34:1943–1952. doi: 10.3892/or.2015.4195 [DOI] [PubMed] [Google Scholar]
- 30.Xu H, Zhao L, Fang Q, et al. MiR-338-3p inhibits hepatocarcinoma cells and sensitizes these cells to sorafenib by targeting hypoxia-induced factor 1alpha. PLoS One. 2014;9:e115565. doi: 10.1371/journal.pone.0115565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Semenza GL. Targeting HIF-1 for cancer therapy. Nat Rev Cancer. 2003;3:721–732. doi: 10.1038/nrc1187 [DOI] [PubMed] [Google Scholar]
- 32.Semenza GL. Hypoxia-inducible factor 1 (HIF-1) pathway. Sci STKE. 2007;2007:cm8. [DOI] [PubMed] [Google Scholar]
- 33.Semenza GL. Hydroxylation of HIF-1: oxygen sensing at the molecular level. Physiology (Bethesda). 2004;19:176–182. doi: 10.1152/physiol.00001.2004 [DOI] [PubMed] [Google Scholar]
- 34.Yang W, Ma J, Zhou W, et al. Reciprocal regulations between miRNAs and HIF-1alpha in human cancers. Cell Mol Life Sci. 2019;76:453–471. doi: 10.1007/s00018-018-2941-6 [DOI] [PMC free article] [PubMed] [Google Scholar]