Abstract
Background
Hepatocellular carcinoma (HCC) is the most common form of liver cancer. Radioresistance is a significant obstacle in HCC therapy. Long non-coding RNA 473 (LINC00473) has been found to impair the effect of radiotherapy. This study aimed to explore the function and molecular basis of LINC00473 in the radiosensitivity of HCC cells.
Methods
The levels of LINC00473, miR-345-5p and Forkhead Box P1 (FOXP1) were determined by quantitative real-time polymerase chain reaction. Cell viability was assessed by 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide (MTT) assay. Survival fraction was calculated by colony survival assay after exposure to different doses of radiation. Cell apoptosis was evaluated by flow cytometry. The interaction among LINC00473, miR-345-5p and FOXP1 was confirmed by dual-luciferase reporter assay. The protein level of FOXP1 was detected by Western blot assay.
Results
LINC00473 and FOXP1 were up-regulated, while miR-345-5p was down-regulated in HCC tissues and cells. Radiation elevated LINC00473 expression in a dose- and time-dependent manner. Depletion of LINC00473 inhibited proliferation and heightened radiosensitivity and apoptosis in HCC cells. In addition, LINC00473 was a sponge of miR-345-5p. Also, miR-345-5p overexpression sensitized HCC cells to radiation. Moreover, miR-345-5p directly targeted FOXP1. MiR-345-5p inhibition or FOXP1 up-regulation reversed the enhanced radiosensitivity caused by LINC00473 knockdown.
Conclusion
LINC00473 contributed to radioresistance in HCC via modulating the miR-345-5p/FOXP1 axis, which might provide a promising diagnostic marker for HCC radiotherapy.
Keywords: LINC00473, miR-345-5p, FOXP1, hepatocellular carcinoma, radioresistance
Introduction
Liver cancer is the fourth leading cause of cancer death worldwide.1 Hepatocellular carcinoma (HCC) accounts for 75–85% of liver cancer and is the most prevalent primary liver cancer.2 HCC is principally caused by hepatitis B virus (HBV) or hepatitis C virus (HCV) infection.3 Currently, radiotherapy has become one of the conventional methods of treating HCC.4,5 Nevertheless, the emergence of radioresistance has become an obstacle stone for HCC radiotherapy.6 Therefore, one of the ways to improve the effect of radiotherapy is to increase the radiosensitivity of HCC cells.
Long non-coding RNAs (lncRNAs) are transcripts over 200 nucleotides in length and have no protein-coding capacity.7 Growing evidence has elucidated that the dysregulation of lncRNAs is related to cancer progression.8 Besides, more and more investigations have manifested that lncRNAs exert a vital effect on the radiosensitivity of tumor cells by functioning as competing endogenous RNA (ceRNA).9 For example, PTPRG-AS1 facilitates radioresistance in nasopharyngeal carcinoma by sponging microRNA-194-3p and increasing PRC1 expression.10 Besides, LINC00483 silencing potentiates radiosensitivity in lung adenocarcinoma by regulating the microRNA-144/HOXA10 axis.11 Gao et al revealed that lncRNA GAS5 functioned as a microRNA-106b sponge to enhance radiosensitivity in cervical cancer (CC) by upregulating IER3.12 A previous study confirmed that knockdown of LINC00473 increased radiosensitivity in esophageal squamous cell carcinoma (ESCC) via modulating the microRNA-374a-5p/SPIN1 axis.13 However, the effect of LINC00473 on the radiosensitivity of HCC cells remains unknown.
MicroRNAs (miRNAs) are a group of highly conserved non-coding RNAs consisting of 19–25 nucleotides. A growing number of studies demonstrated that miRNAs silenced mRNA by base pairing with complementary sequences within the target mRNA, leading to the mediation of cellular mechanisms.14 Many miRNAs have been reported to strongly link to cancer development, including HCC.15 Moreover, some studies illustrated that miRNAs are predictors of radiotherapy in various types of cancers.16 For instance, miR-9-5p targets SOCS5 to expedite angiogenesis and radiosensitivity in CC.17 Wang et al presented that miR-27a heightened the radiosensitivity of ESCC cells via targeting Hsp90.18 In addition, miR-345 is a suppressing factor in many malignancies, including HCC.19 Nevertheless, the relationship between LINC00473 and miR-345-5p has not been investigated.
Forkhead Box P1 (FOXP1), a member of the FOX transcription factor family, is a promising therapeutic target for cancer.20 FOXP1 is an oncogene, and overexpression of FOXP1 leads to poor prognosis of lymphoma.21 On the other hand, FOXP1 functions as a tumor suppressor and loss of FOXP1 relates to poor prognosis in breast cancer.22 Meanwhile, a previous study indicated that FOXP1 is a prognostic biomarker of HCC by acting as an oncogene.23
In this study, we measured the levels of LINC00473, miR-345-5p and FOXP1 in HCC tissues and cells. More importantly, we investigated the function and molecular basis of LINC00473 in the radiosensitivity of HCC cells. These findings might provide a potential biomarker for HCC radiotherapy.
Materials and Methods
Specimen Collection
Thirty-six HCC tissues and paired adjacent normal tissues were collected from Shanxi Provincial People’s Hospital. No patients received any adjuvant treatment before surgery. All participants signed written informed consents. The study was permitted by the Ethics Committee of Shanxi Provincial People’s Hospital.
Cell Culture
Two HCC cell lines (SK-HEP-1 and Hep3B) and human hepatocyte cells THLE-2 were obtained from American Type Culture Collection (ATCC, Manassas, VA, USA). Two HCC cell lines (Huh-7 and Huh-1) were commercially acquired from Yu Bo Biotech (Shanghai, China). Huh-1 cells were cultured in RPMI-1640 medium (Gibco, Los Angeles, CA, USA) supplemented with 10% fetal bovine serum (FBS; Gibco), and other cell lines were cultured in Dulbecco’s Modified Eagle Medium (DMEM; Gibco) containing 10% FBS. All cells were incubated at 37°C with 5% CO2.
Radiation Treatment
Huh-7 and SK-HEP-1 cells (5×103) were treated with gradient doses of irradiation (0 Gy, 2 Gy, 4 Gy, 6 Gy and 8 Gy) using a 6-MV X-ray linear accelerator (ELEKTA, Beijing, China) at 400 cGy/min dose rate. After 24 h, the cells were used for RNA extraction. In addition, cells were exposed to 6 Gy radiation and then were collected every 6 h for RNA extraction until 24 h after irradiation.
Cell Transfection
Small interfering RNA (siRNA) against LINC00473 (si-LINC00473#1, si-LINC00473#2 and si-LINC00473#3), siRNA against FOXP1 (si-FOXP1), the siRNA control (si-NC), miR-345-5p mimics (miR-345-5p), the mimics control (miR-NC), LINC00473 overexpression vector (LINC00473), FOXP1 overexpression vector (FOXP1), the empty overexpression vector (Vector), miR-345-5p inhibitor (anti-miR-345-5p) and the inhibitor control (anti-miR-NC) were synthesized from RiboBio (Guangzhou, China). The plasmids and oligonucleotides were transfected into HCC cells using Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA).
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
RNA was extracted with Trizol reagent (Invitrogen). Then, the first-stranded cDNA was synthesized by One-Step RT-PCR Kit (Takara, Shiga, Japan) or miScript II RT Kit (Qiagen, Frankfurt, Germany). AceQ qPCR SYBR Green Master Mix (Vazyme, Nanjing, China) was used for quantitative PCR. The internal control of LINC00473 and FOXP1 was glyceraldehyde-3-phosphate dehydrogenase (GAPDH), and the internal control of miR-345-5p was U6. Relative expression levels were calculated using 2−ΔΔCt method. The primers were as follows: LINC00473-F: 5ʹ-GATGGAAAGGAGGGAAGG-3ʹ, LINC00473-R: 5ʹ-CACAGTGGGTCCAGGGTT-3ʹ; miR-345-5p-F: 5ʹ-GCTGACTCCTAGTCCA-3ʹ, miR-345-5p-R: 5ʹ-TGGTGTCGTGGAGTCG-3ʹ; FOXP1-F: 5ʹ-ATGATGCAAGAATCTGGGACTG-3ʹ, FOXP1-R: 5ʹ-AGCTGGTTGTTTGTCATTCCTC-3ʹ; GAPDH-F: 5ʹ-GAAGGTGAAGGTCGGAGT −3ʹ, GAPDH-R: 5ʹ-GATGGCAACAATATCCACTT-3ʹ; U6-F: 5ʹ-CTCGCTTCGGCAGCACA-3ʹ, U6-R: 5ʹ-ACGCTTCACGAATTTGCGT-3ʹ.
Cell Viability Assay
Cells (2.0×103) were injected into 96-well plates. Next, 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide (MTT) solution (Solarbio, Beijing, China) was added to each well after incubation for 1 d, 2 d, 3 d and 4 d. Then, cells were treated with dimethyl sulfoxide (DMSO; Solarbio) for 15 min. Finally, cell viability was evaluated by monitoring the absorbance at 490 nm using Varioskan™ LUX Multimode Microplate Reader (Thermo Fisher Scientific, Waltham, MA, USA)
Colony Survival Assay
Transfected cells were seeded into six-well plates. After 48 h of incubation, cells were exposed to different doses of X-ray irradiation (0 Gy, 2 Gy, 4 Gy, 6 Gy and 8 Gy) at 400 cGy/min dose rate. Subsequently, the cells were seeded in 60 mm dishes and incubated at 37°C for 14–21 d until visible colonies appeared. Next, the colonies were stained with 1% crystal violet and counted under a microscope. Survival fraction was calculated.
Flow Cytometry
Cell apoptosis was assessed by AnnexinV-fluorescein isothiocyanate (AnnexinV-FITC)/propidium iodide (PI) Apoptosis Detection kit (Invitrogen). Briefly, transfected cells were treated with 6 Gy radiation. After radiation treatment for 24 h, the cells were stained with Annexin V-FITC and PI. Then, the apoptotic rate was monitored by Attune NxT Flow Cytometer (Thermo Fisher Scientific).
Dual-Luciferase Reporter Assay
The sequences of LINC00473 or FOXP1 3ʹUTR containing wild-type or mutant binding sites of miR-345-5p were cloned into the pmirGLO vector (Promega, Madison, WI, USA) to form LINC00473-WT, LINC00473-MUT, FOXP1-WT or FOXP1-MUT, respectively. Next, the corresponding luciferase reporter and miR-345-5p or miR-NC were co-transfected into HCC cells. The luciferase activity was examined by Dual-Lucy Assay Kit (Solarbio).
Western Blot Assay
After cells were lysed using RIPA buffer (Invitrogen), equal amounts of protein samples were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto polyvinylidene fluoride (PVDF) membranes (Millipore, Billerica, MA, USA). After blocking with 5% skim milk for 2 h, the membranes were incubated with primary antibodies against FOXP1 (ab16645, Abcam, Cambridge, UK) or GAPDH (ab9485, Abcam) at 4°C overnight and then interacted with secondary antibody (ab7090, Abcam) for 2 h at room temperature. The intensity was detected by enhanced chemiluminescence reagents (Millipore).
Statistical Analysis
Graphpad Prism 7.0 software (GraphPad, San Diego, CA, USA) was executed to analyze data. The data were represented as mean ± standard deviation. Student’s t-test or one-way analysis of variance was carried out for differences analysis. All experiments were repeated three times independently. P <0.05 was considered statistically significant.
Results
LINC00473 Expression Was Increased in HCC Tissues and Cells and Related to Radiation
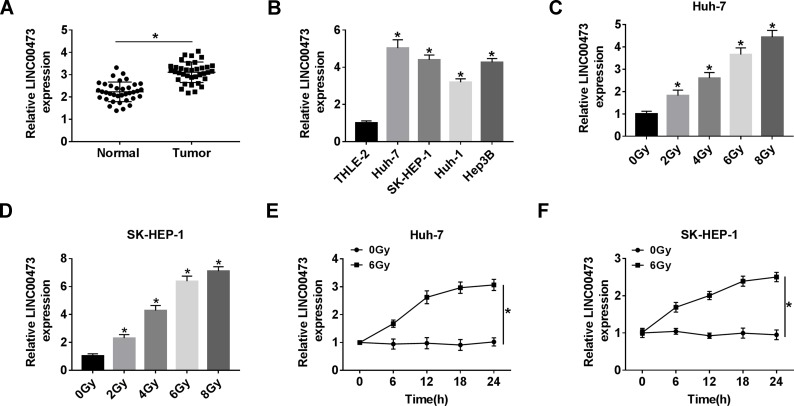
First, compared with adjacent tissues, LINC00473 expression was distinctly increased in CRC tissues (Figure 1A). Also, the expression level of LINC00473 was remarkably higher in HCC cells (Huh-7, SK-HEP-1, Huh-1 and Hep3B) than that in THLE-2 cells (Figure 1B). Huh-7 and SK-HEP-1 cells were exposed to gradient doses of radiation, and the results showed that LINC00473 expression was strikingly elevated in a dose-dependent manner after radiation treatment (Figure 1C and D). We chose 6 Gy radiation that caused a significant upregulation of LINC00473 expression for subsequent experiments, because its promoting effect was close to 8 Gy that maximized the promoting effect. In addition, the expression of LINC00473 was significantly increased in a time-dependent manner after HCC cells were exposed to 6 Gy radiation (Figure 1E and F). These data indicated that radiation increased the expression of LINC00473 in HCC cells.
Figure 1.
LINC00473 expression was increased in HCC tissues and cells and related to radiation. (A) LINC00473 expression in 36 pairs of HCC tissues and adjacent normal tissues was measured by qRT-PCR. (B) LINC00473 expression in THLE-2 cells and HCC cells (Huh-7, SK-HEP-1, Huh-1 and Hep3B) was detected by qRT-PCR. (C, D) The level of LINC00473 was examined in Huh-7 and SK-HEP-1 cells under various doses (0 Gy, 2 Gy, 4 Gy, 6 Gy and 8 Gy) of radiation treatment for 24 h. (E, F) The level of LINC00473 was measured at the indicated time points after radiation treatment. *P < 0.05.
Knockdown of LINC00473 Repressed Proliferation and Increased Radiosensitivity and Apoptosis of HCC Cells
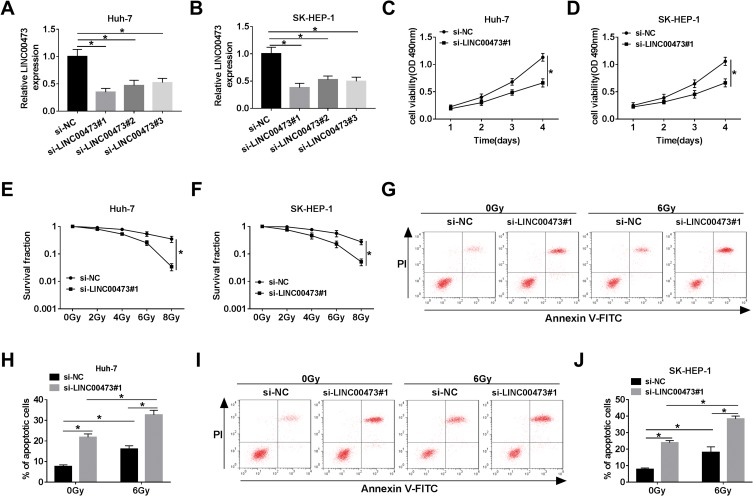
To investigate whether LINC00473 could modulate the radiosensitivity of HCC cells, si-LINC00473 was introduced into Huh-7 and SK-HEP-1 cells to inhibit LINC00473 expression. First of all, the results revealed that si-LINC00473#1 had the highest knockdown efficiency (Figure 2A and B). MTT assay suggested that depletion of LINC00473 prominently suppressed the viability of HCC cells compared to the control group (Figure 2C and D). Besides, colony survival assay exhibited that transfection with si-LINC00473#1 resulted in a sharp reduction in survival fraction compared to the si-NC group, indicating that LINC00473 silenced HCC cells were more sensitive to radiation (Figure 2E and F). Moreover, Huh-7 and SK-HEP-1 cells introduced with si-NC or si-LINC00473#1 were exposed to 6 Gy radiation. Flow cytometry showed that LINC00473 silencing or 6 Gy radiation treatment induced HCC cell apoptosis, and LINC00473 knockdown combined with radiation stimulation dramatically increased the apoptosis rate induced by LINC00473 depletion or 6 Gy radiation therapy (Figure 2G–J). These data implied that knockdown of LINC00473 impeded proliferation and promoted radiosensitivity and apoptosis of HCC cells.
Figure 2.
Knockdown of LINC00473 repressed proliferation and increased radiosensitivity and apoptosis of HCC cells. (A, B) The level of LINC00473 in Huh-7 and SK-HEP-1 cells transduced with si-NC, si-LINC00473#1, si-LINC00473#2 and si-LINC00473#3 was detected using qRT-PCR. (C, D) MTT assay was used to assess the viability of Huh-7 and SK-HEP-1 cells introduced with si-NC or si-LINC00473#1. (E, F) Colony survival assay was performed in Huh-7 and SK-HEP-1 cells transduced with si-NC or si-LINC00473#1 with various doses of X-ray radiation. (G–J) The apoptosis rate of HCC cells transfected with si-NC or si-LINC00473#1 was measured after exposure to 6 Gy radiation for 24 h. *P < 0.05.
LINC00473 Targeted miR-345-5p
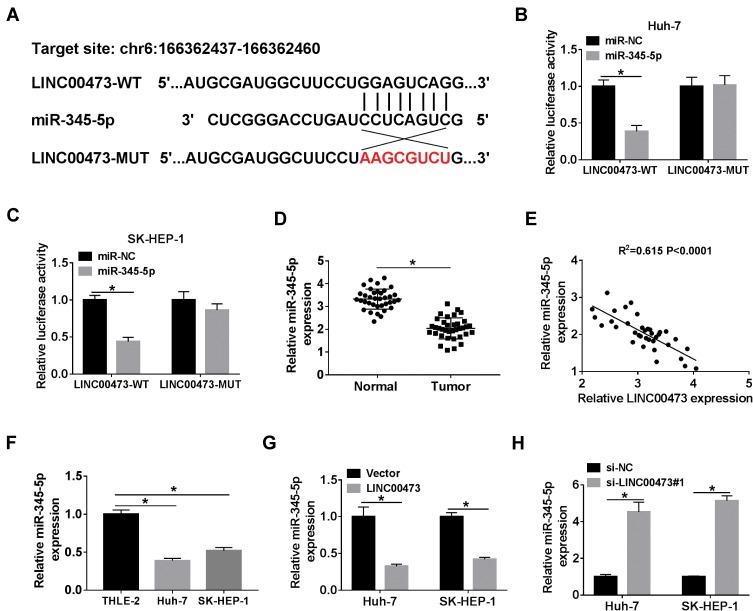
The starBase online database predicted putative miRNAs that might bind to LINC00473. As shown in Figure 3A, miR-345-5p has complementary binding sites with LINC00473. Next, dual-luciferase reporter assay displayed that co-transfection with miR-345-5p and LINC00473-WT strikingly reduced the luciferase activity in Huh-7 and SK-HEP-1 cells (Figure 3B and C). Besides, miR-345-5p expression in HCC tissues was conspicuously decreased in comparison with the normal tissues (Figure 3D). The level of LINC00473 was negatively correlated with miR-345-5p level in HCC tissues (Figure 3E). Consistently, miR-345-5p expression was markedly lower in Huh-7 and SK-HEP-1 cells than that in THLE-2 cells (Figure 3F). Also, to clarify LINC00473 modulated miR-345-5p expression, Huh-7 and SK-HEP-1 cells were transduced with Vector, LINC00473, si-NC or si-LINC00473#1. The results showed that up-regulation of LINC00473 overtly decreased miR-345-5p level, and knockdown of LINC00473 remarkably enhanced miR-345-5p level (Figure 3G and H). Altogether, these results identified that miR-345-5p was a target of LINC00473 and was negatively regulated by LINC00473 in HCC cells.
Figure 3.
LINC00473 targeted miR-345-5p. (A) The binding sites of LINC00473 and miR-345-5p were shown. (B, C) Luciferase activity was detected in Huh-7 and SK-HEP-1 cells co-transfected with LINC00473-WT or LINC00473-MUT and miR-345-5p or miR-NC by dual-luciferase reporter assay. (D) The level of miR-345-5p was determined in HCC tissues and adjacent normal tissues. (E) Spearman correlation analysis was used to analyze the correlation between LINC00473 and miR-345-5p levels in HCC tissues. (F) The expression of miR-345-5p was examined in THLE-2 cells and HCC cell lines (Huh-7 and SK-HEP-1). (G, H) The level of miR-345-5p was detected in Huh-7 and SK-HEP-1 cells transfected with Vector, LINC00473, si-NC or si-LINC00473#1. *P < 0.05.
miR-345-5p Inhibited Proliferation and Enhanced Radiosensitivity and Apoptosis of HCC Cells
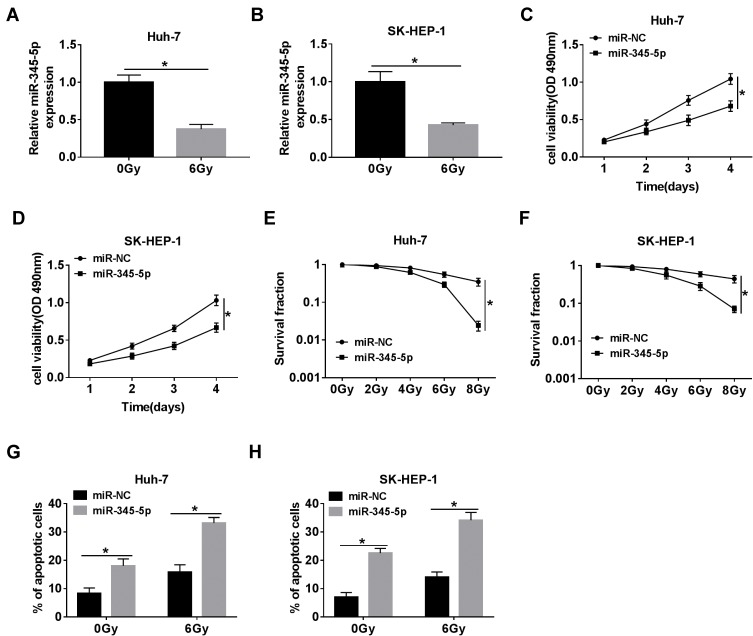
To explore the role of miR-345-5p in the radiosensitivity of HCC cells, Huh-7 and SK-HEP-1 cells were transduced with miR-NC or miR-345-5p. After treatment with 6 Gy radiation, the expression of miR-345-5p was drastically decreased in Huh-7 and SK-HEP-1 cells compared with the control group (Figure 4A and B). The viability of Huh-7 and SK-HEP-1 cells transfected with miR-345-5p was distinctly reduced in comparison with the miR-NC group (Figure 4C and D). Also, Huh-7 and SK-HEP-1 cells introduced with miR-NC or miR-345-5p were exposed to different doses of radiation and the results of colony survival assay uncovered that HCC cells transfected with miR-345-5p were more sensitive to radiation (Figure 4E and F). Additionally, up-regulation of miR-345-5p significantly increased the apoptotic rate of Huh-7 and SK-HEP-1 cells (Figure 4G and H). These data concluded that overexpression of miR-345-5p increased the radiosensitivity of HCC cells.
Figure 4.
MiR-345-5p inhibited proliferation and enhanced radiosensitivity and apoptosis of HCC cells. (A, B) The expression of miR-345-5p was detected in Huh-7 and SK-HEP-1 cells treated with 6 Gy radiation for 24 h. (C, D) Cell viability was monitored in Huh-7 and SK-HEP-1 cells transfected with miR-NC or miR-345-5p by MTT assay. (E, F) The survival fraction of Huh-7 and SK-HEP-1 cells transduced with miR-NC or miR-345-5p at different doses of radiation was evaluated by colony survival assay. (G, H) Cell apoptosis was assessed in Huh-7 and SK-HEP-1 cells transfected with miR-NC or miR-345-5p after exposure to 6 Gy radiation for 24 h. *P < 0.05.
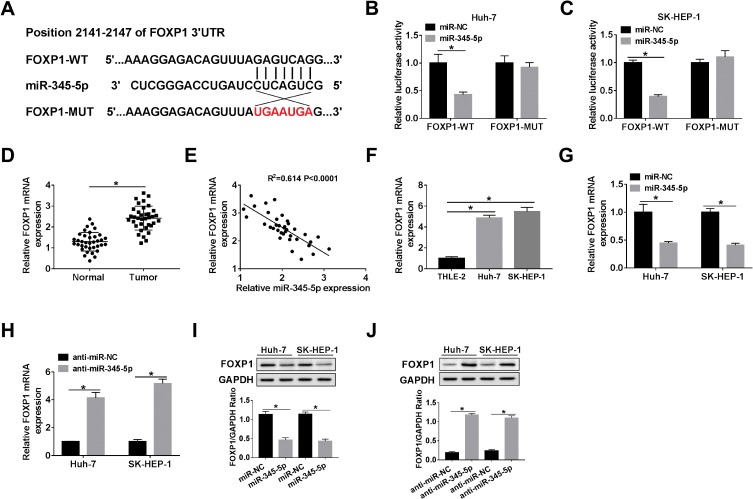
FOXP1 Was a Target of miR-345-5p
We further explored the potential regulatory mechanism by which LINC00473 induced radioresistance in Huh-7 and SK-HEP-1 cells. TargetScan online database predicted that miR-345-5p and FOXP1 3ʹUTR had putative binding sites (Figure 5A). More importantly, dual-luciferase reporter assay revealed that miR-345-5p mimics dramatically repressed the luciferase activity of FOXP1-WT reporter (Figure 5B and C). Besides, the mRNA level of FOXP1 was markedly increased in HCC tissues and negatively correlated with miR-345-5p expression (Figure 5D and E). Also, FOXP1expression was predominantly higher in Huh-7 and SK-HEP-1 cells than that in THLE-2 cells (Figure 5F). Further, we detected the mRNA and protein levels of FOXP1 in Huh-7 and SK-HEP-1 cells transduced with miR-345-5p or anti-miR-345-5p. The results indicated that up-regulation of miR-345-5p dramatically reduced the mRNA and protein expression of FOXP1, and inhibition of miR-345-5p overtly increased the mRNA and protein expression of FOXP1 (Figure 5G–J). These data disclosed that FOXP1 was a target of miR-345-5p in HCC cells.
Figure 5.
FOXP1 was a target of miR-345-5p. (A) The binding sites of miR-345-5p and FOXP1 3ʹUTR were displayed. (B, C) Dual-luciferase reporter assay was performed to confirm the relationship between miR-345-5p and FOXP1. (D) FOXP1 expression was tested in HCC tissues and adjacent normal tissues. (E) Spearman correlation curve depicted a negative correlation between miR-345-5p and FOXP1 levels in HCC tissues. (F) The level of FOXP1 was measured in THLE-2 cells and HCC cell lines (Huh-7 and SK-HEP-1). (G–J) The mRNA and protein levels of FOXP1 were detected in Huh-7 and SK-HEP-1 cells introduced with miR-NC, miR-345-5p, anti-miR-NC or anti-miR-345-5p by qRT-PCR and Western blot assay. *P < 0.05.
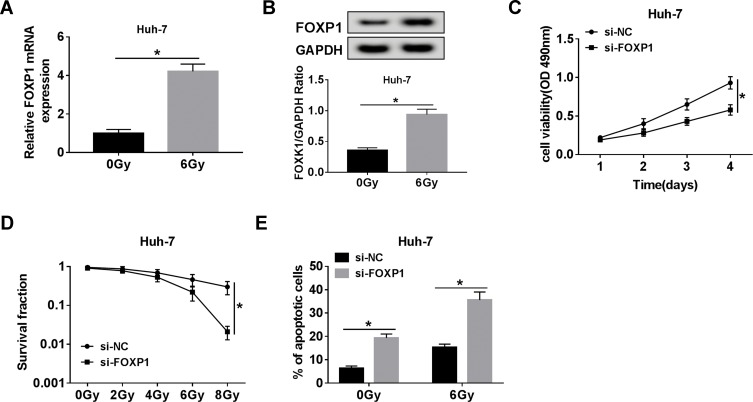
FOXP1 Silencing Increased the Radiosensitivity of HCC Cells
First, 6 Gy radiation caused a marked increase in FOXP1 mRNA and protein levels compared to the control group (Figure 6A and B). To elucidate the effect of FOXP1 on the radiosensitivity of HCC cells, Huh-7 cells were introduced with si-NC or si-FOXP1. MTT assay exhibited that transfection of si-FOXP1 drastically inhibited the viability of Huh-7 cells compared with si-NC group (Figure 6C). Moreover, colony survival assay revealed that Huh-7 cells transfected with si-FOXP1 were more sensitive to radiation (Figure 6D). In addition, knockdown of FOXP1 expedited the apoptosis of Huh-7 cells (Figure 6E). These data evidenced that FOXP1 silencing increased the radiosensitivity of HCC cells.
Figure 6.
FOXP1 silencing increased the radiosensitivity of HCC cells. (A, B) The mRNA and protein levels of FOXP1 were examined in Huh-7 cells stimulated with 6 Gy radiation for 24 h. (C) Cell proliferation was assessed by MTT assay in Huh-7 cells transfected with si-NC or si-FOXP1. (D) The survival fraction of Huh-7 cells introduced with si-NC or si-FOXP1 was detected by colony survival assay at different doses of radiation. (E) Cell apoptosis was monitored in Huh-7 cells transfected with si-NC or si-FOXP1 after exposure to 6 Gy radiation for 24 h. *P < 0.05.
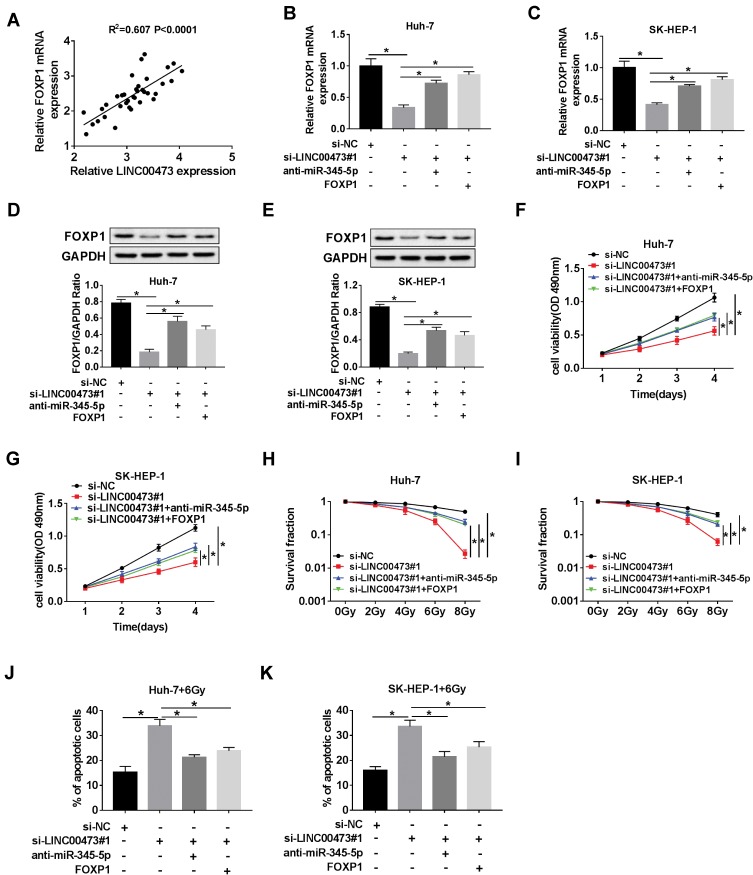
LINC00473 Regulated Radioresistance of HCC Cells via miR-345-5p/FOXP1 Axis
First, Spearman correlation analysis discovered that LINC00473 was positively correlated with FOXP1 in HCC cells (Figure 7A). To investigate the potential mechanism of LINC00473 on radiosensitivity of HCC cells, rescue experiments were performed by transfecting Huh-7 and SK-HEP-1 cells with si-LINC00473#1+anti-miR-345-5p or si-LINC00473#1+FOXP1. The results illustrated that inhibition of miR-345-5p or up-regulation of FOXP1 in HCC cells reversed the decrease in FOXP1 mRNA expression triggered by knockdown of LINC00473 (Figure 7B and C). Similarly, suppression of miR-345-5p or overexpression of FOXP1 reversed the reduction in FOXP1 protein level induced by LINC00473 silencing (Figure 7D and E). Additionally, down-regulation of LINC00473 inhibited the viability of Huh-7 and SK-HEP-1 cells, while the effect was rescued after transfection with anti-miR-345-5p or FOXP1 (Figure 7F and G). Colony survival assay demonstrated that inhibition of miR-345-5p or overexpression of FOXP1 recuperated the inhibitory effect of LINC00473 depletion on the survival fraction of HCC cells (Figure 7H and I). Moreover, silencing of LINC00473 led to a significant increase in apoptotic rate under radiation treatment, while down-regulation of miR-345-5p or up-regulation of FOXP1 reversed this effect (Figure 7J and K). These data concluded that LINC00473 knockdown hindered cell proliferation and radioresistance in HCC via regulating the miR-345-5p/FOXP1 axis.
Figure 7.
LINC00473 regulated radiosensitivity of HCC cells via miR-345-5p/FOXP1 axis. (A) The correlation between LINC00473 and FOXP1 expression in HCC tissues was confirmed by Spearman correlation analysis. (B–K) Huh-7 and SK-HEP-1 cells were introduced with si-NC, si-LINC00473#1, si-LINC00473#1+anti-miR-345-5p or si-LINC00473#1+FOXP1, respectively. (B, C) The mRNA level of FOXP1 was examined by qRT-PCR. (D, E) The protein level of FOXP1 was detected using Western blot analysis. (F, G) MTT assay was conducted to detect the viability of Huh-7 and SK-HEP-1 cells. (H, I) Colony survival assay was utilized to assess the radiosensitivity of Huh-7 and SK-HEP-1 cells after transfection. (J, K) Flow cytometry was used to evaluate cell viability after treatment with 6 Gy radiation for 24 h. *P < 0.05.
Discussion
Radioresistance remains a major obstacle to radiation therapy. Numerous studies have confirmed that lncRNAs regulate the radiosensitivity of cancer cells, including HCC cells.9 For instance, NEAT1_2 depletion enhanced HCC cell radiosensitivity by modulating miR-101-3p and repressing WEE1.24 Additionally, lncRNA H19 triggered the radioresistance of HCC cells by acting as a ceRNA for miR-193a-3p and upregulating PSEN1.25 Furthermore, knockdown of lncRNA ROR sensitized HCC cells to radiation via regulating the microRNA-145/RAD18 axis.26 Similar to previous studies, LINC00473 expression was prominently up-regulated in HCC tissues and cells. Also, silenced LINC00473 attenuated HCC cell viability and radioresistance. Further, we explored the mechanism of LINC00473-driving radioresistance in HCC.
Growing evidence has manifested that lncRNA acts as a “miRNA sponge” to regulate gene expression and participates in the progression of various tumors.27 Besides, a large number of lncRNAs are abnormally expressed in HCC and involved in HCC progression.28 For example, Wu et al suggested that lncSNHG3 accelerated the propagation and metastasis in HCC via sponging miR-139-5p and increasing BMI1 expression.29 Fen et al unveiled that silencing of RHPN1-AS1 impeded the malignant phenotypes of HCC cells via regulating the miR-596/IGF2BP2 axis.30 Dai et al discovered that lncRNA-SNHG15 expedited HCC development via inhibiting miR-490-3p and enhancing HDAC2.31 In addition, this study testified that LINC00473 was a sponge of miR-345-5p, suggesting the role of miR-345-5p in the radioresistance of HCC cells driven by LINC00473.
Previous researches indicated that miR-345 targeted IRF1 or YAP1 to block HCC progression.19,32 In the present study, the gain-of-function experiments disclosed that up-regulation of miR-345-5p suppressed proliferation and strengthened radiosensitivity and apoptosis in HCC cells. Furthermore, miR-345-5p was an inhibitor of many types of cancers. For example, miR-345-5p up-regulation restrained tumor progression in pancreatic cancer by repressing CCL8 expression.33 In cholangiocarcinomas, miR-345-5p was sponged by LMCD1-AS1 to play an anticancer role.34
Fox proteins are a set of transcriptional factors involved in various cellular functions, such as apoptosis and proliferation.35 Recently, Fox proteins participated in the progression of various cancers, including acute myeloid leukemia,36 colorectal cancer37 and osteosarcoma.38 FOXP1 is a transcription factor with broad functions.22,39 Wang et al suggested that inhibition of FOXP1 suppressed HCC cell proliferation through regulation of cell cycle.40 Nevertheless, the role of FOXP1 in HCC radiotherapy remains unknown. In this research, we revealed that LINC00473 modulated the radiosensitivity of HCC cells by regulating miR-345-5p and FOXP1.
In summary, we discovered that radiation could up-regulate LINC00473 expression and down-regulate miR-345-5p expression in HCC cells. Further, LINC00473 potentiated the radioresistance of HCC cells by functioning as a ceRNA of miR-345-5p and elevating FOXP1. These findings contributed to our understanding of HCC and might provide a promising biomarker for HCC radiotherapy.
Disclosure
The authors report no conflicts of interest in this work
References
- 1.Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 2.Harris PS, Hansen RM, Gray ME, et al. Hepatocellular carcinoma surveillance: an evidence-based approach. World J Gastroenterol. 2019;25(13):1550–1559. doi: 10.3748/wjg.v25.i13.1550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ben Ari Z, Weitzman E, Safran M. Oncogenic viruses and hepatocellular carcinoma. Clin Liver Dis. 2015;19(2):341–360. doi: 10.1016/j.cld.2015.01.006 [DOI] [PubMed] [Google Scholar]
- 4.Donadon M, Solbiati L, Dawson L, et al. Hepatocellular carcinoma: the role of interventional oncology. Liver Cancer. 2016;6(1):34–43. doi: 10.1159/000449346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abdel-Rahman O, Elsayed Z. External beam radiotherapy for unresectable hepatocellular carcinoma. Cochrane Database Syst Rev. 2017;3:CD011314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kalogeridi MA, Zygogianni A, Kyrgias G, et al. Role of radiotherapy in the management of hepatocellular carcinoma: a systematic review. World J Hepatol. 2015;7(1):101–112. doi: 10.4254/wjh.v7.i1.101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fan C, Tang Y, Wang J, et al. Role of long non-coding RNAs in glucose metabolism in cancer. Mol Cancer. 2017;16(1):130. doi: 10.1186/s12943-017-0699-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arun G, Diermeier SD, Spector DL. Therapeutic targeting of long non-coding RNAs in cancer. Trends Mol Med. 2018;24(3):257–277. doi: 10.1016/j.molmed.2018.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu J, Chen S, Yang B, et al. Molecular mechanisms of lncRNAs in regulating cancer cell radiosensitivity. Biosci Rep. 2019;39(8). doi: 10.1042/BSR20190590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yi L, Ouyang L, Wang S, et al. Long noncoding RNA PTPRG-AS1 acts as a microRNA-194-3p sponge to regulate radiosensitivity and metastasis of nasopharyngeal carcinoma cells via PRC1. J Cell Physiol. 2019;234(10):19088–19102. doi: 10.1002/jcp.28547 [DOI] [PubMed] [Google Scholar]
- 11.Yang QS, Li B, Xu G, et al. Long noncoding RNA LINC00483/microRNA-144 regulates radiosensitivity and epithelial-mesenchymal transition in lung adenocarcinoma by interacting with HOXA10. J Cell Physiol. 2019;234(7):11805–11821. doi: 10.1002/jcp.27886 [DOI] [PubMed] [Google Scholar]
- 12.Gao J, Liu L, Li G, et al. LncRNA GAS5 confers the radio sensitivity of cervical cancer cells via regulating miR-106b/IER3 axis. Int J Biol Macromol. 2019;126:994–1001. doi: 10.1016/j.ijbiomac.2018.12.176 [DOI] [PubMed] [Google Scholar]
- 13.Chen W, Zhang Y, Wang H, et al. LINC00473/miR-374a-5p regulates esophageal squamous cell carcinoma via targeting SPIN1 to weaken the effect of radiotherapy. J Cell Biochem. 2019;120(9):14562–14572. doi: 10.1002/jcb.28717 [DOI] [PubMed] [Google Scholar]
- 14.Liu B, Li J, Cairns MJ. Identifying miRNAs, targets and functions. Brief Bioinform. 2014;15(1):1–19. doi: 10.1093/bib/bbs075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anwar SL, Lehmann U. MicroRNAs: emerging novel clinical biomarkers for hepatocellular carcinomas. J Clin Med. 2015;4(8):1631–1650. doi: 10.3390/jcm4081631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hummel R, Hussey DJ, Haier J. MicroRNAs: predictors and modifiers of chemo- and radiotherapy in different tumour types. Eur J Cancer. 2010;46(2):298–311. doi: 10.1016/j.ejca.2009.10.027 [DOI] [PubMed] [Google Scholar]
- 17.Wei YQ, Jiao XL, Zhang SY, et al. MiR-9-5p could promote angiogenesis and radiosensitivity in cervical cancer by targeting SOCS5. Eur Rev Med Pharmacol Sci. 2019;23(17):7314–7326. doi: 10.26355/eurrev_201909_18837 [DOI] [PubMed] [Google Scholar]
- 18.Wang X, An D, Liu X, et al. MicroRNA-27a downregulates the expression of Hsp90 and enhances the radiosensitivity in esophageal squamous cell carcinoma. Onco Targets Ther. 2019;12:5967–5977. doi: 10.2147/OTT.S197456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang H, Liu H, Bi H. MicroRNA-345 inhibits hepatocellular carcinoma metastasis by inhibiting YAP1. Oncol Rep. 2017;38(2):843–849. doi: 10.3892/or.2017.5772 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20.Koon HB, Ippolito GC, Banham AH, et al. FOXP1: a potential therapeutic target in cancer. Expert Opin Ther Targets. 2007;11(7):955–965. doi: 10.1517/14728222.11.7.955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Musilova K, Devan J, Cerna K, et al. miR-150 downregulation contributes to the high-grade transformation of follicular lymphoma by upregulating FOXP1 levels. Blood. 2018;132(22):2389–2400. doi: 10.1182/blood-2018-06-855502 [DOI] [PubMed] [Google Scholar]
- 22.Ijichi N, Ikeda K, Horie-Inoue K, et al. FOXP1 and estrogen signaling in breast cancer. Vitam Horm. 2013;93:203–212. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Y, Zhang S, Wang X, et al. Prognostic significance of FOXP1 as an oncogene in hepatocellular carcinoma. J Clin Pathol. 2012;65(6):528–533. doi: 10.1136/jclinpath-2011-200547 [DOI] [PubMed] [Google Scholar]
- 24.Chen X, Zhang N. Downregulation of lncRNA NEAT1_2 radiosensitizes hepatocellular carcinoma cells through regulation of miR-101-3p/WEE1 axis. Cell Biol Int. 2019;43(1):44–55. doi: 10.1002/cbin.v43.1 [DOI] [PubMed] [Google Scholar]
- 25.Ma H, Yuan L, Li W, et al. The LncRNA H19/miR-193a-3p axis modifies the radio-resistance and chemotherapeutic tolerance of hepatocellular carcinoma cells by targeting PSEN1. J Cell Biochem. 2018;119(10):8325–8335. doi: 10.1002/jcb.26883 [DOI] [PubMed] [Google Scholar]
- 26.Chen Y, Shen Z, Zhi Y, et al. Long non-coding RNA ROR promotes radioresistance in hepatocelluar carcinoma cells by acting as a ceRNA for microRNA-145 to regulate RAD18 expression. Arch Biochem Biophys. 2018;645:117–125. doi: 10.1016/j.abb.2018.03.018 [DOI] [PubMed] [Google Scholar]
- 27.Tam C, Wong JH, Tsui SKW, et al. LncRNAs with miRNAs in regulation of gastric, liver, and colorectal cancers: updates in recent years. Appl Microbiol Biotechnol. 2019;103(12):4649–4677. [DOI] [PubMed] [Google Scholar]
- 28.Lim LJ, Wong SYS, Huang F, et al. Roles and regulation of long noncoding RNAs in hepatocellular carcinoma. Cancer Res. 2019;79(20):5131–5139. doi: 10.1158/0008-5472.CAN-19-0255 [DOI] [PubMed] [Google Scholar]
- 29.Wu J, Liu L, Jin H, et al. LncSNHG3/miR-139-5p/BMI1 axis regulates proliferation, migration, and invasion in hepatocellular carcinoma. Onco Targets Ther. 2019;12:6623–6638. doi: 10.2147/OTT.S196630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fen H, Hongmin Z, Wei W, et al. RHPN1-AS1 drives the progression of hepatocellular carcinoma via regulating miR-596/IGF2BP2 axis. Curr Pharm Des. 2019;25. doi: 10.2174/1381612825666191105104549 [DOI] [PubMed] [Google Scholar]
- 31.Dai W, Dai JL, Tang MH, et al. lncRNA-SNHG15 accelerates the development of hepatocellular carcinoma by targeting miR-490-3p/histone deacetylase 2 axis. World J Gastroenterol. 2019;25(38):5789–5799. doi: 10.3748/wjg.v25.i38.5789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu M, Xue H, Wang Y, et al. miR-345 inhibits tumor metastasis and EMT by targeting IRF1-mediated mTOR/STAT3/AKT pathway in hepatocellular carcinoma. Int J Oncol. 2017;50(3):975–983. doi: 10.3892/ijo.2017.3852 [DOI] [PubMed] [Google Scholar]
- 33.Mou T, Xie F, Zhong P, et al. MiR-345-5p functions as a tumor suppressor in pancreatic cancer by directly targeting CCL8. Biomed Pharmacother. 2019;111:891–900. doi: 10.1016/j.biopha.2018.12.121 [DOI] [PubMed] [Google Scholar]
- 34.Yu J, Zhang B, Zhang H, et al. E2F1-induced upregulation of long non-coding RNA LMCD1-AS1 facilitates cholangiocarcinoma cell progression by regulating miR-345-5p/COL6A3 pathway. Biochem Biophys Res Commun. 2019;512(2):150–155. doi: 10.1016/j.bbrc.2019.03.054 [DOI] [PubMed] [Google Scholar]
- 35.Bach DH, Long NP, Luu TT, et al. The dominant role of forkhead box proteins in cancer. Int J Mol Sci. 2018;19(10):3279. doi: 10.3390/ijms19103279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gurnari C, Falconi G, De Bellis E, et al. The role of forkhead box proteins in acute myeloid leukemia. Cancers (Basel). 2019;11(6):865. doi: 10.3390/cancers11060865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laissue P. The forkhead-box family of transcription factors: key molecular players in colorectal cancer pathogenesis. Mol Cancer. 2019;18(1):5. doi: 10.1186/s12943-019-0938-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang W, Duan N, Song T, et al. The emerging roles of Forkhead Box (FOX) proteins in osteosarcoma. J Cancer. 2017;8(9):1619–1628. doi: 10.7150/jca.18778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gascoyne DM, Banham AH. The significance of FOXP1 in diffuse large B-cell lymphoma. Leuk Lymphoma. 2017;58(5):1037–1051. doi: 10.1080/10428194.2016.1228932 [DOI] [PubMed] [Google Scholar]
- 40.Wang X, Sun J, Cui M, et al. Downregulation of FOXP1 inhibits cell proliferation in hepatocellular carcinoma by inducing g1/S phase cell cycle arrest. Int J Mol Sci. 2016;17:9. [DOI] [PMC free article] [PubMed] [Google Scholar]