Abstract
Multiple chromosome aberrations are responsible for tumorigenesis of esophagus squamous cell carcinoma (ESCC). To characterize genetic alterations by comparative genomic hybridization (CGH) and their relation to ESCC, We enrolled 54 members with ESCC from Kazakh’s patients. We found that the deletions of 3p (P = 0.032), 17p (P = 0.004), 22q (P = 0.000) and gains of 5p (P = 0.000), 11q (P = 0.000) were significantly correlated with the location of tumors. Losses of 1p (P = 0.005), 3p (P = 0.006), 22q (P = 0.024) and gains of 3q (P = 0.043), 8q (P = 0.038), 18q (P = 0.046) were also found more frequently in patients with larger diameter disease. The loss of 19q (P = 0.005) and gains of l3q (P = 0.045), 18p (P = 0.018) were significantly correlated with pathologic grade. The gain of 7p (P = 0.009) and deletion of 19q (P = 0.018) were seen more frequently in patients with Grade III-IV tumors. Chromosome amplifications in ESCC at 1q (P = 0.008), 7p (P = 0.008), 8q (P = 0.018) and deletions at 3p (P = 0.021), 11q (P = 0.002), 17p (P = 0.012) were related to lymph node metastasis; the gains of 1q (P = 0.026) and 6q (P = 0.017) and the loss of 11q (P = 0.001) were significant in different isoforms of HPV infection. We identified some chromosomes in which the genes were related to the tumorgenesis of ESCC, which may be a theme for future investigation.
Keywords: Chromosomal aberration, esophagus squamous cell carcinoma, HPV infection, Xinjiang Kazakh, comparative genomic hybridization
Introduction
Esophagus squamous cell cancer (ESCC), the major histologic type of esophageal cancer in East Asian countries [1], remains one of the most common malignant diseases of digestive system. It ranks the sixth of the leading causes of cancer-related deaths worldwide [2], especially in Chinese Kazakh ethnic population who is residing in Xinjiang, Northwest China [3]. In recent years, advances in diagnosis and therapy for ESCC have improved the survival of cancer patients to some extent. However, the prognosis remains poor with an average 5-year survival rate ranging from 20% to 40% [4] due to its highly invasive nature [5]. Thus, it is of great clinical value to look for sensitive and specific biomarkers for the early detection and prognosis of this malignancy, as well as novel therapeutic targets. Genetic changes were often detected in tumor tissues and they could also be indicators of tumor progression, for examples, the loss of TUSC3 (8p22) gene may serve as a good indicator of malignancy of oral squamous cell carcinoma [7]; a significant association between FHIT (3p14.2) and invasive ductal carcinoma (IDC) histopathological type was observed in breast cancer [8], so on and so forth. Thus, we aim to understand the genetic changes that happened in these tumors, including those from major chromosomal variations as well as gene sequence changes.
Comparative genomic hybridization (CGH) was evolved from fluorescence in situ hybridization (FISH) by Kallioniemi et al. in 1992 [9], and CGH is more rapid for analyzing DNA copy number alters across the genome in a single sample than traditional methods. Genomic aberration was a common characteristic of human carcinomas and also was one of the basic mechanisms that result in overexpression of oncogenes or underexpression of tumor suppressor genes [10], thus genomic variation was one of the mechanisms that could lead to gene dysfunction and result in the carcinogens and tumor progression. Numerous studies had detected the chromosome aberrations in ESCC, such as gains at 3q26-qter, 5p15, 7p, 7q, 11q13.3, 8q24.3-qter and losses of 16p13.3 and 18q22-qter were most frequent aberrations in ESCC [11-13]; gains of 12p, 11q13.2, 8q24.21 and losses at 3p14.2 were related to poor prognosis [14]; Moreover, overexpression of IGHMBP2, which located in chromosome 11q13.2, may promote invasion and migration of ESCC cells through down-regulation of E-cadherin [15]; Cyclin D1 plays an important role in ESCC cases by amplification of the 11q13.3 locus, which, together with FBXO31, becomes a biomarker of prognosis of ESCC [16], and so on.
Although plenty of studies had demonstrated chromosome aberrations in ESCC were connected with tumorgenesis, progression and prognosis of ESCC, to the best of our knowledge, there was no research on the relationship between the location, size of ESCC, HPV infection and chromosome variations, especially in Xinjiang Kazakh nation. Herein, we for the first time report results of a CGH analysis performed in 54 cases of Xinjiang Kazakh ESCC in order to discover common regions of gains and losses in chromosome, which could provide evidence on the important role of chromosome changes in the tumorigenesis and progression of ESCC. Interestingly, not only did we find that losses of 19q and gains of l3q, 18p were significantly correlated with pathologic grade; the gain of 7p and deletion of 19q was seen more frequently in patients with Grade III-IV tumors; chromosome amplifications in ESCC at 1q, 7p, 8q and deletions at 3p, 11q, 17p were related to lymph node metastasis, but also discovered that deletions of 3p, 17p, 22q and gains at 5p, 11q were significantly correlated with the location of tumors; Losses of 1p, 3p, 22q and gains of 3q, 8q, 18q also were found more frequently in patients with larger diameter disease; the gains of 1q, 6q and the losses of 11q were in significantly different isoforms of HPV infection. Thus, we envision that the chromosome changes could be a potential biomarker of early diagnosis of ESCC.
Materials and methods
Samples and controls
A total of 54 paraffin-embedded tissues specimens with primary ESCC patients were all from the tissue bank of the Peoples’ Hospital reach center related to cancer in Xinjiang Uygur Autonomous Region. The clinical features and clinicopathological characteristics of the samples are shown in Table 1. The patients did not accept any preoperative chemotherapy or radiotherapy before their surgery. Diagnosis of every patient was confirmed to be esophagus squamous cell cancer by histological observation. And all of the samples were classified and graded by means of the criteria of the World Health Organization (WHO). Meanwhile, metaphase chromosome slices and reference DNA came from peripheral blood of an adult male volunteer.
Table 1.
Demographics of patients and tumor characteristics
| Characteristic | No. of patients (n = 54) |
|---|---|
| Age | |
| 40-50 | 19 |
| 51-61 | 23 |
| 62-72 | 12 |
| Sex | |
| Male | 35 |
| Female | 19 |
| Histological differentiation | |
| Well | 17 |
| Moderate | 29 |
| Poor | 8 |
| Location | |
| Middle | 33 |
| Low | 21 |
| Size | |
| <5 cm | 35 |
| ≥5 cm | 19 |
| TNM stage | |
| I | 0 |
| II | 24 |
| III | 27 |
| IV | 3 |
| LN | |
| N0 | 33 |
| N1 | 21 |
| HPV infection | |
| HPV16 | 14 |
| HPV18 | 8 |
| Both and others | 24 |
| None | 8 |
LN represents lymph node metastasis, N0-without, N1-with.
DNA extraction
Total DNA was extracted from the 54 samples with ESCC using a standard phenol/chloroform extraction method, and peripheral blood lymphocytes cells from a normal male volunteer was used as controls. DNA quality was checked on a 1.2% agarose gel, the length of the DNA were confirmed to be 300-3000 base pairs (bp) by electrophoresis and the amount of extracted DNA was measured spectrophotometrically at 260 nm (impurity and ratio of DNA to non-DNA were also cross-checked at 280 nm). Extractions were stored at -20°C prior to labeling by nick translation.
Comparative genomic hybridization
Comparative genomic hybridization used a commercially available kit on the basis of the manufacturer’s protocol (Vysis Inc., U.S). Briefly, tumor DNA was labeled with 0.2 mmol/L fluorescein isothiocyanate (FITC)-dUTP, while the normal reference DNA was labeled with cyanine 3 (Cy3)-dUTP. The hybridization mixture incorporated approximately 300 ng Spectrum Green-labeled test DNA and 300 ng Spectrum Red-labeled total genomic reference DNA coprecipitated with 10 μg human Cot-1 DNA (Invitrogen, Vysis, USA) and was dissolved in hybridization buffer before hybridization to metaphase chromosomes. Metaphase slides were denatured at 73°C for 5 min in 70% methanamide/2× SSC and dehydrated in an ethanol series (75%, 85%, and 100%). After washing, chromosomes were counterstained with 4,6-diamidino-2-phenylindole-2 HCl (DAPI II; Vysis) and embedded in an antifading agent to reduce photobleaching. To provide a control in case of artifacts, normal people DNA (labeled red) was used as a negative control. DNA extracted from the MPE600 breast cancer cell line (with known genetic aberrations that were easy to be detected by comparative genomic hybridization) was used as a positive control (labeled green).
Microscopy and digital image analysis
A fluorescence microscope (Olympus BX51) equipped with appropriate filters (DAPI, FITC, and Cy3) was used to bring out the signals. For each hybridization panel, raw images from at least 5 metaphases were captured through a computer-driven CCD camera and analyzed with ISIS image software (Carl Zeiss METASYSTEMS Inc., Germany). Chromosomes were identified by their DAPI banding patterns. Threshold levels of 1.25 and 0.8 were used to score gains and losses, respectively. High-level amplification was indicated by a ratio greater than 1.5.
Statistical analysis
To get categorical data, Pearson’s X2 test or Fisher’s exact test was used to interclass comparison. A value of P<0.05 was considered statistically significant. Data were analyzed by the software for SPSS13.0.
Results
CGH profiles of 54 ESCC samples in Chinese Kazakh and a case of representative image
The genetic alterations results detected by CGH in the 54 cases of ESCC showed that chromosome variations occurred in all cases. CGH displayed DNA sequence gains and losses in all 54 primary esophageal cancer cases, with 683 gains and 488 losses. The mean numbers of DNA gains and losses per tumor sample were 12.64 and 9.04, respectively, and the ratio was 1.4:1. Figure 1 showed the frequency of DNA copy number of gains or losses of every chromosome arm in 54 ESCC patients. Gains were most frequently detected on chromosomal 8q (65%), 1q (60%), 3q (56%), 18q (54%), 5p (52%), 11q (48%), 13q (44%), 6q (41%), 20q (39%), 5q (35%), 18p (35%) etc., in which 8q22-24 (60%), 1q13 (54%), 1q21-23 (51%), 3q21-qter (50%), 18q11-24 (42%) and 5p13.1-ter (30%) were the most frequent region, and the minimal overlapping region lay in 3q21-22, 8q22-23, 5p13-14, 1q21, 13q, 18q11-14, 20q11-12 and 11q13. Frequently occurring losses involved chromosome 22q (52%), 8p (44%), 13q (39%), 17p (39%), 5q (37%), 10q (33%), (the chromosomal aberrant frequency on chromosome 1p, 18q, 3q, 19q was similar, for all it was 30% followed by 11q (20%). The chromosome 22q11.2-14 (48%), 8p22-ter (35%), 13q13-21 (33%) and 17p11-13 (30%) were the most frequent losses region. The minimal regions of overlap were 22q11-13, 8p12-23, 13q12-14, 17p12-13, 5q31, 3p21, 1p36, 3p21, 18p13 and 18q21-23.
Figure 1.
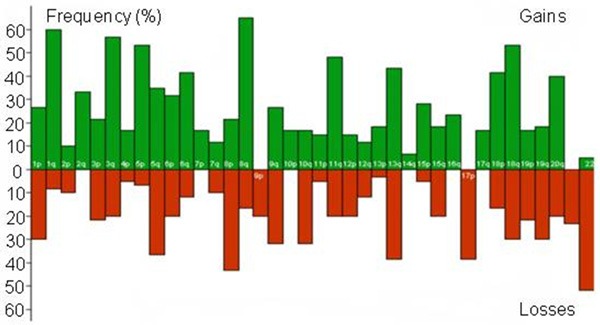
Frequency of DNA copy number gains and losses for each chromosome arm in 54 patients with primary ESCC of Xinjiang Kazakh. Green histograms are gains, red are deletions.
We showed one representative CGH analysis image of case No.17 in Figure 2. The gained chromosome of case 17 were 1p, 1q, 3q, 5q, 7q, 8q, 18q and lost chromosome were 6p, 6q, 8p, 18p, 22p, 22q. The amplifications of 1q were focused on 1q13 and 1q21-23, the gains of 3q occurred on 3q21-qter, and the minimal overlapping region was 3q21-22. While the amplifications of 5q, 18q, 8q mostly located on 5q13-14, 18q11-14, 8q23-q25, 8q27, respectively. The chromosomal profile of DNA copy number losses was characterized as follows: 22q11-13 of 22q, 8p12-23 on chromosome 8p. And the chromosome alterations on other chromosomal regions were scattered, showing not high frequency of altered DNA copy number. Thus they belong to small probability events.
Figure 2.
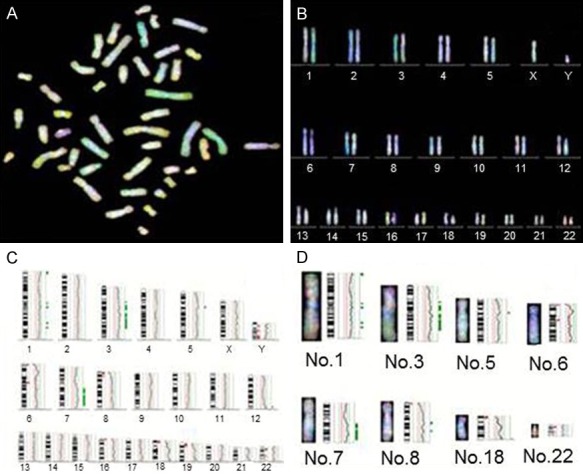
Representative image of chromosome variations by CGH analysis in case No.17. Red-reference DNA, green-DNA of tumor, blue-DAPI staining. A. CGH detected metaphase spreads. B. Chromosome arrangement by karyotype. C. Fluorescence ratio figure. D. Image of several chromosome aberration regions. Green to red fluorescence thresholds (represented by the green/red line) are 0.85 and 1.25, respectively. The curve shows DNA copy status. Curves to the left of the red line indicate losses, while curves to the right indicate gains.
The correlation between the chromosome aberration of Kazakh patients with ESCC and clinical pathology
The association was detected by small sample Fisher’s exact test to confirm the correlation between ESCC patients with chromosome variation, age, sex, tumorigenic position, tumor size and tumor classification. The chromosomal alteration of 54 ESCC had bearing on tumorigenic position and tumor site except age, sex, tumor classification (Table 2). For example, the gains of 5p (P = 0.000), 11q (P = 0.000) and the losses on chromosome 3p (P = 0.032), 17p (P = 0.004), 22q (P = 0.000) were found to be positive correlation with the location of tumor. Especially the gain of 5p, 11q and the deletion of 22q had significant positive relationship with the size of tumor (P = 0.000); while the gains of 3q (P = 0.043), 8q (P = 0.038), 18q (P = 0.046) and the losses of 1p (P = 0.005), 3p (P = 0.006), 22q (P = 0.024) were discovered to be connected with tumor size, in which the gains of 8q, 18q and the losses of 1p, 3p, 22q had positive correlation with tumor size. Moreover, tumor size of those (patients with ESCC) with the amplification of chromosome 3q was commonly less than 5 cm.
Table 2.
The relationship between chromosome aberration of 54 ESCC in Xinjiang Kazak and location, size
| Clinical character | No. | DNA Gains (%) | DNA Losses (%) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||||
| 3q+ | 5p+ | 8q+ | 11q+ | 18q+ | 1p- | 3p- | 17p- | 22q- | ||
| Location | ||||||||||
| Middle | 33 | 20 (60) | 5 (15.1) | 18 (54) | 9 (27.3) | 17 (51.5) | 10 (30.3) | 6 (18.2) | 8 (24.2) | 8 (24.2) |
| Low | 21 | 11 (52) | 14 (66) | 17 (80) | 17 (80.9) | 14 (66.7) | 6 (28.6) | 10 (47.6) | 14 (66.7) | 20 (95.2) |
| p-value | 0.584 | 0.000*** | 0.079 | 0.000*** | 0.398 | 1.000 | 0.032* | 0.004** | 0.000*** | |
| Size | ||||||||||
| <5 cm | 35 | 24 (68) | 14 (40) | 19 (54) | 19 (54.3) | 15 (42.9) | 6 (17.1) | 7 (20.0) | 13 (37.1) | 14 (40.0) |
| ≥5 cm | 19 | 7 (36.8) | 5 (26.3) | 16 (84) | 7 (36.8) | 14 (73.7) | 11 (57.9) | 9 (47.4) | 9 (47.4) | 14 (73.7) |
| p-value | 0.043* | 0.381 | 0.038* | 0.264 | 0.046* | 0.005** | 0.006** | 0.566 | 0.024* | |
The X2 test (Fisher’s exact test);
P<0.05;
P<0.01;
P<0.001.
In the present study we evaluated the association between the profile of the chromosomal variation and histological grade. The relationship between chromosomal aberrations and histological differentiation was evaluated for 54 Kazakh patients with ESCC (Table 3). Amplifications at 13q (P = 0.045), 18p (P = 0.018) and deletions at 19q (P = 0.005) were compared between the patients with high differentiation and those with middle-low differentiation, which was found to be significant differences. Whereas, with the decreased degree of tumor differentiation the chromosome 13q gained more, thus it may significantly associated with the malignancy of ESCC. The deletion of 19q was much frequent in squamous carcinoma, which might be a potential marker of well differentiation.
Table 3.
The relationship between chromosomal aberration and tissue differentiation gradation
| Chromosome change | High differentiation | Middle-low differentiation | p value |
|---|---|---|---|
| 1q+ | 14/18 (77.8) | 18/36 (50) | 0.078 |
| 13q+ | 8/18 (33.3) | 23/36 (63.9) | 0.045* |
| 17p- | 5/18 (27.8) | 17/36 (47.2) | 0.242 |
| 18p+ | 3/18 (16.7) | 19/36 (52.8) | 0.018* |
| 19q- | 10/18 (55.6) | 6/36 (16.7) | 0.005** |
The X2 test (Fisher’s exact test);
P<0.05;
P<0.01.
The association between chromosomal aberrations and TNM classification was detected in 54 patients with ESCC in Kazakh. The average DNA copy numbers of amplifications and losses in patients with early-stage tumor (TNM I and II) were 12.4 and 8.5, respectively, Whereas the numbers in advanced-stage tumor (TNM III and IV) were 15.7 and 10.3, respectively. Furthermore, the frequency of chromosome gains and losses in advanced-stage tumor were found to be higher than those in early-stage tumor, and the ratio was 1.27:1 and 1.21:1, respectively. In our current study, the gain of 7p (7p13-14) (P = 0.009) and the loss of 19q (19q13) (P = 0.018) were observed to be significantly higher in advanced-stage than early-stage (Table 4).
Table 4.
The relationship between chromosomal aberration and clinical stage in 54 ESCC of Xinjiang Kazak
| Chromosome aberrations | II (IIa+IIb) | III-IV | p value |
|---|---|---|---|
| 7p+ | 3/24 (12.5) | 14/30 (46.7) | 0.009** |
| 13q+ | 14/24 (58.3) | 15/30 (50) | 0.592 |
| 19q- | 3/24 (12.5) | 13/30 (43.3) | 0.018* |
The X2 test (Fisher’s exact test);
P<0.05;
P<0.01.
Using Fisher’s exact test, the regions with a variation copy number were compared between patients with lymph node metastasis and those without (Table 5). As a result, amplifications at 1q (P = 0.008), 7p (P = 0.008), 8q (P = 0.018) and deletions at 3p (P = 0.021), 11q (P = 0.002), 17p (P = 0.012) were related to lymph node metastasis, and the gains of 1q had negative relationship with lymph node metastasis, which could be a good mark of prognosis of ESCC in Kazakh. On the contrary, the gain of 8q and the losses on 11q and 17p may be a symbol of poor prognosis.
Table 5.
The relationship between chromosome aberration of 54 ESCC in Kazak and lymphatic metastasis
| Chromosomal aberration | Non lymphatic metastasis (%) | Lymphatic metastasis (%) | p value |
|---|---|---|---|
| 1q+ | 27/33 (81.8) | 10/21 (47.6) | 0.008** |
| 3p- | 6/33 (18.2) | 10/21 (47.6) | 0.021* |
| 7p+ | 6/33 (18.2) | 11/21 (52.4) | 0.008** |
| 8q+ | 17/33 (51.5) | 18/21 (85.7) | 0.018* |
| 11q- | 2/33 (15.2) | 9/21 (42.9) | 0.002** |
| 17p- | 9/33 (27.3) | 13/21 (61.9) | 0.012* |
The X2 test (Fisher’s exact test);
P<0.05;
P<0.01.
Chromosome aberration in relation to the infection of HPV
In addition, to analyze the correlation between the infection of HPV and chromosome alternation, we previously examined 54 ESCC patients with or without HPV infection (Table 6). The statistical results showed that the gains of 1q (P = 0.026), 6q (P = 0.017) and deletions at 11q (P = 0.001) in different type of HPV infection were in significant difference. Notably the gains of 1q were the highest aberration in the ESCC patients with infection of HPV16, and amplifications at 6q and deletions at 11q were with high frequency in the patients infected with HPV18. The ratio was 87.5% and 62.5%, respectively, and significant differences were observed compared with other infected types. Simply put, the results indicated that amplifications at 1q would be a representative characteristic of infection with HPV16, nevertheless the gain of 6p and the loss of 11q might have much to do with infection of HPV18.
Table 6.
The relationship between chromosome aberration of 54 ESCC in Xinjiang Kazak and HPV infection
| Chromosome aberration | HPV16 | HPV18 | Multiple infection | No infection | p value |
|---|---|---|---|---|---|
| 1q+ | 13/14 (92.9) | 1/8 (12.5) | 12/24 (50) | 3/8 (37.5) | 0.026* |
| 6q+ | 6/14 (42.8) | 7/8 (87.5) | 9/24 (37.5) | 1/8 (12.5) | 0.017* |
| 8q+ | 10/14 (71.4) | 8/8 (100) | 14/24 (58.3) | 3/8 (37.5) | 0.053 |
| 11q- | 3/14 (21.4) | 6/8 (62.5) | 3/24 (4.2) | 1/8 (12.5) | 0.001** |
The X2 test (Fisher’s exact test);
P<0.05;
P<0.01.
Discussion
ESCC is one of the most aggressive malignancies with a very poor survival in China, particularly in the Chinese Kazakh ethnic population residing in Xinjiang, Northwest China. Compared with other ethnic groups and parts of China, the incidence rate and mortality rate of patients with ESCC in the Chinese Kazakh were detected higher in malignant tumors of Kazak [17]. Thus the early diagnosis of ESCC in Kazakh is very urgent for better prognosis. As far as we know, genomic variations are one of the mechanisms that could result in gene dysfunction and lead to carcinogenesis and tumor progression. Differentially expressed genes correlated with altered DNA copy number may be candidate targets of amplifications or deletions. Thus a CGH analysis on 54 Kazak patients with ESCC resided in Xinjiang had been performed for their chromosome variations. We found that many chromosome changes significantly correlated with tumorgenesis, progression, and prognosis. For instance, deletions of 3p, 17p, 22q and gains of 5p, 11q were significantly correlated with the location of tumors; losses of 19q and gain of l3q, 18p were significantly correlated with pathologic grade; the gains of 1q, 6q and the loss of 11q were in significant different isoforms of HPV infection with ESCC and so on. Therefore we made an assumption that the changes of chromosome could be a biomarker of early diagnosis in ESCC.
Since invented in 1990s, CGH was widely used to detect the changes of chromosome in all kinds of tumors [9,18]. Numerous previous studies had been performed in patients with ESCC, and the results of these researches also differ in different degrees. Anna Hirasaki et al. [12] applied Array-CGH to study the chromosome changes of patients with EC and they found that the chromosomal variations with high incidence were different, whereas the chromosomal aberrations of different isoforms in EC (squamous cancers and adenocarcinomas) either had similarity or imparity. Simultaneously, the use of Array-CGH may also find some genes that could code some proteins. For examples, genes that code cell adhesion molecule (CAM), cytoskeleton proteins, transcription factor and cyclin-related proteins might affect the occurrence and development of ESCC and discovery of some new cancer-related genes not reported before. The relationship between the chromosome changes of ESCC and age, gender, cancer types, histological grade and clinical stage were not the same in different studies [19,20]. Nonetheless, the relationship between ESCC chromosome aberrations and HPV infection, tumor size and position has not been reported.
The overall chromosome aberrations of 54 primary ESCC patients from Xinjiang Kazakh in our study were very complex. The overall chromosome gains and losses were 683 and 488, respectively; the average gains and losses in every specimen was 12.68 and 9.04, respectively, and the ratio was 1.4:1. Our result of chromosome amplifications and deletions were similar to previous investigations. The gains of 3q, 5p, 8q, 1q, 11q and the losses of 11q, 18q, 5q were detected in many researches related to esophagus cancer [10], carcinoma in other organs and we had known that some genes related to tumorigenesis and lymph node metastasis were located in those places, such as PI3K and ALG3 in 3q, FHIT at 3p, BCL1 and CCND1 in 11q13 [8,10,21-23]. Except for the similarity, our results also revealed that 13q gained and 22q deleted with high frequency, and the common sites were involved in 13q13-14, 13q13-21, 13qq33, 13q32, 13q33-ter, 13q14 and 22q13, in which the minimal overlapping region was 13q13-14. RB1 and BRAC2 located in 13q14 were related to tumorigenesis of numerous tumors, and it was also deserved further researches to identify whether it related to ESCC or not. The deletion of tumor suppressor gene (such as BAM22, Hsnf5/INI, NF2, LGALLS1, and BZRP) that lay in the chromosome 22q13 also took part in many kinds of cancer’s tumorigenesis [24]. These different results might be connected with race and region of patients, and it could be the characteristic chromosome changes of patients with ESCC in Chinese Kazakh.
The relationship between chromosome aberrations of ESCC and clinicopathologic features had been reported in many researches, but different researches showed various outcomes. The previous studies have shown that chromosome aberrations of ESCC had no significant relationship with age, gander, clinical tumor type [11,19], which was coincident with our study. Whereas we also discovered some new special genome changes in Kazakh patients with ESCC. In our study, the gains of 5p, 11q and the losses of 3p, 17p, 22q had significant correlation with the location of the tumor, and the change of genome was more frequent in the ESCC patients whose tumor was located in middle lower of esophagus; in addition, the amplifications in chromosome 3q, 8q, 18q and the deletions at 1p, 3p, 22q had remarkable relationship with tumor size, and when the tumor size ≥5 cm, the patients would reveal more complex variation in chromosome, which had not been reported ever. In our study, the changes at chromosome 3q were most located in 3q21, 3q24-ter, and 3q27-29. Chromosome 3q was aberrant in high frequency in plenty of cancers, such as cervical cancer [25], lung squamous carcinoma [26], ovarian carcinoma [27], and so on. MBD4 (3q21-q22), ECT2 (3q26.2-q26), p63 (3q27-q29), PIK3CA (3q24-qter) gene had been detected in chromosome 3q at present [25,26], and all this gene belong to oncogene of db1 family. They could code growth factors with guanine nucleotide exchange factor activity thus to active Rho-family GTPases, and then took part in cell growth, differentiation, and cycle regulating. In our study, the chromosomal 3q had high frequent amplifications and had significant correlation with invasive growth of ESCC, which might play a great role in the development of ESCC. The mechanism was via chromosome aberration by activating oncogene or suppressing anti-oncogene. The change of chromosome 8q lay in 8q13, 8q23-ter, which was also a research hotspot of many types of tumor in their cancer genetics. And the oncogene of MYC located in chromosome 8q might participate in the progression of ESCC [14].
Previous studies of CGH analysis in ESCC indicated that the alternations of DNA copy numbers had significant relationship with prognosis of ESCC: the amplifications of 3q, 8q, 12p, 16p, 17q and the deletions of 18q correlated with pathologic stage in ESCC [10]; the gains of 2q12-14, 3q24-26, 7q21-31 [28], 3q27.1 (ALG3), 11q13.3 (CCND1) [10] related to lymph node metastasis; and the amplifications in chromosome 7p13-21 was connected with distance metastasis [28]. They presumed that the gains of 8q, 2p and the losses of 4pq, 11q14-qter was an event of the advanced stage. And the deletion of chromosome 8q indicated poor prognosis of patients with ESCC [19]. Other investigates revealed that the gains of 3q, 5p, 7p14.1, 11q13.2, 13q12, 14q11-32, 18q11.2, 19p and the losses at 2q37.3, 3p, 11q23.1, 13q12.2, 16q23.1, 17p had relationship with pathologic type of ESCC [29]; the chromosome 8q24-qter amplified frequently in low-differentiation, in addition in high-differentiation of ESCC the losses of chromosome 8p22-pter, 9p was more common [11]. Our results of CGH analysis indicated that the changes of DNA copy numbers had crucial relationship with histological grade, clinical stage and lymph node metastasis. The amplifications of 13q, 17p and the deletion of 19q were related to histological grade, and the gains of chromosome 13q was obvious higher in middle-low differentiation than in high differentiation. The regions of chromosome 13q changed were often located in 13q13-21 and 13q33-ter, in which RB1 and BRCA2 genes took part in tumorigenesis in numerous carcinomas (for instance, chronic lymphocytic leukemia, brain malignant glioma, and so on [30,31]. The deletion level of chromosome 19q was higher in high differentiation, thus we supposed that it might be a mark of cancer differentiation.
Chromosomal variations in advanced-stages of ESCC were significantly higher than it in early stage, which revealed that with the progression of ESCC the more aberration regions would occur and most of them were amplifications. The gains of 7p and the deletions of 19q had important correlation with clinical stages in ESCC, and it was more common in patients with ESCC of stage II and stage III, which were strongly linked with progression of ESCC. Similarly, in squamous cell carcinomas of the oral cavity, the gain of 7p focused on 7p12 related to clinical stage, which may because it is the band harboring the epidermal growth factor receptor (EGFR) [32]. But the amplification regions of chromosome 7p were located in 7p13-14, which was rarely reported in ESCC. Therefore further researches of the gains of 7p needed to be performed in ESCC. The position of chromosome 19q deleted was 19q13 where PDCD5, KISS1R, BRG1 genes were located. These genes played a role in ovarian cancer, for instance, PDCD5 might lead to the tumor via disturbing the apoptosis progression of cancer cell [33]. Further studies should be performed to identify if they induce tumor via the same approach in ESCC.
Previous studies indicated that the deletions of 3p, 11q, 17p would be a remark of prognosis of ESCC, which was coincident with our research [10,34]. In patients with ESCC, the amplification of chromosomal 1q of those without lymph node metastasis was more frequent than those with lymph node metastasis, and the region of which was 1q21-22 (S100), indicating a well prognosis of ESCC. In addition, the gains in chromosome 7p, 8q and the losses in chromosome 3p, 11q, 17p in ESCC of those with lymph node metastasis were greater than those without lymph node metastasis, and the minimum region was involved,, such as 7p13-p11 (CCM2), 7p15.2 (LOC100129.36), 8q22 (CDH17), 8q23 (MAPK6PS5), 8q23-q24.1 (COLEC10, myc). And it had significant differences among two groups. By comparing the three above-mentioned indexes and their homologous chromosomal variations, we could come to a conclusion that the gains of 7p and the loss of 19q had important correlation with the progression of ESCC, in which the amplification of 7p could reveal poor prognosis and the deletion of 19q was related to the late-progression of ESCC. However, it was too early to summarize the characteristic aberrations at present. We need more patients with ESCC to analyze the relationship between chromosome changes and clinical index. Even so we could also conclude that with the development of ESCC cancer-related chromosome variations occurred more frequently and common chromosomal changes could be detected in different stage of ESCC, which indicated that common gene changes were existed in ESCC.
Human papilloma viruses (HPV) were addicted to squamous epithelium and separated into high-risk HPV (HPV16, 18, 31, 45) and low-risk HPV (HPV11, 6, 41) depending on the relationship between viruses and clinic. High-risk HPV was significantly related to carcinoma, and the mechanism was its participation in cell cycle control, thus preventing cell self-healing or stopping cells to go into programmed cell death, which caused cell immortalization. Compared with HPV16 infection, HPV18 infection enhanced imbalance significantly in chromosome [35]. However further researches of the relationship between regions of chromosome alternations and various types of HPV infection had not been carried out. The investigation of the correlation between chromosome variations of ESCC and HPV infection was rarely investigated for now. It was found that the level of amplification in chromosome 6q was higher in ESCC, whereas it had nothing to do with isolation of HPV. Further it indicated that chromosomal aberrations and HPV infection were likely to be mutual independence factors in the process of esophageal canceration course [36], and HPV-related simultaneously companied with the loss of heterozygosity at 11q22 in studies of oral squamous cell carcinoma and its derivative cell line [37].
Studies on HPV-induced chromosomal aberrations in Chinese Xinjiang Kazakh, had not been reported at home and abroad. HPV infection in Kazakh Xinjiang had vital correlations with chromosome variations, in which the gains of 1q (1q13, 1q21-23), 6q (6q24) and the losses at 11q (11q22) were related to HPV infection, and it had significant difference in the four isoforms of HPV. We came to a conclusion that the amplifications of 1q and 6q would be a characteristic feature of chromosome changes of HPV16 and HPV18 infection, respectively. Particularly worth mentioning was that the frequency of the deletions at 11q was 20%, which showed important difference with different types of HPV infection. The H-ras gene located in 11q existed variations in previous studies on oral squamous cell carcinoma and head-neck cancer, and the mutation of H-ras played a crucial role in HPV-induced cancer [37]. Nevertheless the relationship between gains at 1q, 6q in ESCC and HPV infection had not been investigated in previous studies, thus the mechanism of 1q and 6q amplified promoting tumorigenesis of ESCC might be similar to the changes of 11q or not.
Conclusion
In summary, we did preliminary analysis with our results and referred to numerous literatures related chromosome aberrations of ESCC. Thus we thought that chromosomal variations were common in ESCC patients residing in Xinjiang Kazakh. And some special chromosome changes may focus on patients with ESCC in Xinjiang Kazakh. For instance, the gains of 1q, 6q and the losses of 11q were significant in different isoforms of HPV infection, which not only provided the tumorgenesis, progression, diagnosis and prognosis of ESCC, but also offered a foundation for our further research related to gene identification and analysis of chromosome aberrations by array-CGH.
Acknowledgements
This work was supported by Grants from the National Natural Science Foundation of China (No. 81560399, 81360358, 81460362), the Major Science and Technology Projects of Shihezi University (No. gxjs2014 zdgg06), the Applied Basic Research Projects of Xinjiang Production and Construction Corps (No. 2016AG020), the High-Level Talent Project of Shihezi University (No. RCZX201533), and the Foundation for Distinguished Young Scholars of Shihezi University (No. 2015ZRKXJQ02).
Disclosure of conflict of interest
None.
References
- 1.Baba Y, Watanabe M, Murata A, Shigaki H, Miyake K, Ishimoto T, Iwatsuki M, Iwagami S, Yoshida N, Oki E, Sakamaki K, Nakao M, Baba H. LINE-1 hypomethylation, DNA copy number alterations, and CDK6 amplification in esophageal squamous cell carcinoma. Clin Cancer Res. 2014;20:1114–1124. doi: 10.1158/1078-0432.CCR-13-1645. [DOI] [PubMed] [Google Scholar]
- 2.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 3.Cui X, Li S, Li T, Pang X, Zhang S, Jin J, Hu J, Liu C, Yang L, Peng H, Jiang J, Liang W, Suo J, Li F, Chen Y. Significance of elevated ERK expression and its positive correlation with EGFR in Kazakh patients with esophageal squamous cell carcinoma. Int J Clin Exp Pathol. 2014;7:2382–2391. [PMC free article] [PubMed] [Google Scholar]
- 4.Ma Q, Liu W, Jia R, Long H, Zhang L, Lin P, Zhao H, Ma G. Alcohol and survival in ESCC: prediagnosis alcohol consumption and postoperative survival in lymph node-negative esophageal carcinoma patients. Oncotarget. 2016;7:38857–38863. doi: 10.18632/oncotarget.8754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen YZ, Cui XB, Hu JM, Zhang WJ, Li SG, Yang L, Shen XH, Liu CX, Pan QF, Yu SY, Yuan XL, Yang L, Gu WY, Chen JZ, Wang LD, Li F. Overexpression of PLCE1 in Kazakh esophageal squamous cell carcinoma: implications in cancer metastasis and aggressiveness. APMIS. 2013;121:908–918. doi: 10.1111/apm.12095. [DOI] [PubMed] [Google Scholar]
- 6.Jamieson GG, Mathew G, Ludemann R, Wayman J, Myers JC, Devitt PG. Postoperative mortality following oesophagectomy and problems in reporting its rate. Br J Surg. 2004;91:943–947. doi: 10.1002/bjs.4596. [DOI] [PubMed] [Google Scholar]
- 7.Ribeiro IP, Marques F, Caramelo F, Pereira J, Patricio M, Prazeres H, Ferrao J, Juliao MJ, Castelo-Branco M, de Melo JB, Baptista IP, Carreira IM. Genetic gains and losses in oral squamous cell carcinoma: impact on clinical management. Cell Oncol (Dordr) 2014;37:29–39. doi: 10.1007/s13402-013-0161-5. [DOI] [PubMed] [Google Scholar]
- 8.Ismail HM, Medhat AM, Karim AM, Zakhary NI. FHIT gene and flanking region on chromosome 3p are subjected to extensive allelic loss in Egyptian breast cancer patients. Mol Carcinog. 2011;50:625–634. doi: 10.1002/mc.20797. [DOI] [PubMed] [Google Scholar]
- 9.Kallioniemi A, Kallioniemi OP, Sudar D, Rutovitz D, Gray JW, Waldman F, Pinkel D. Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors. Science. 1992;258:818–821. doi: 10.1126/science.1359641. [DOI] [PubMed] [Google Scholar]
- 10.Shi ZZ, Jiang YY, Hao JJ, Zhang Y, Zhang TT, Shang L, Liu SG, Shi F, Wang MR. Identification of putative target genes for amplification within 11q13.2 and 3q27.1 in esophageal squamous cell carcinoma. Clin Transl Oncol. 2014;16:606–615. doi: 10.1007/s12094-013-1124-z. [DOI] [PubMed] [Google Scholar]
- 11.Yen CC, Chen YJ, Chen JT, Hsia JY, Chen PM, Liu JH, Fan FS, Chiou TJ, Wang WS, Lin CH. Comparative genomic hybridization of esophageal squamous cell carcinoma: correlations between chromosomal aberrations and disease progression/prognosis. Cancer. 2001;92:2769–2777. doi: 10.1002/1097-0142(20011201)92:11<2769::aid-cncr10118>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 12.Hirasaki S, Noguchi T, Mimori K, Onuki J, Morita K, Inoue H, Sugihara K, Mori M, Hirano T. BAC clones related to prognosis in patients with esophageal squamous carcinoma: an array comparative genomic hybridization study. Oncologist. 2007;12:406–417. doi: 10.1634/theoncologist.12-4-406. [DOI] [PubMed] [Google Scholar]
- 13.Shi ZZ, Liang JW, Zhan T, Wang BS, Lin DC, Liu SG, Hao JJ, Yang H, Zhang Y, Zhan QM, Zhang KT, Wang MR. Genomic alterations with impact on survival in esophageal squamous cell carcinoma identified by array comparative genomic hybridization. Genes Chromosomes Cancer. 2011;50:518–526. doi: 10.1002/gcc.20875. [DOI] [PubMed] [Google Scholar]
- 14.Miyawaki Y, Kawachi H, Ooi A, Eishi Y, Kawano T, Inazawa J, Imoto I. Genomic copy-number alterations of MYC and FHIT genes are associated with survival in esophageal squamous-cell carcinoma. Cancer Sci. 2012;103:1558–1566. doi: 10.1111/j.1349-7006.2012.02329.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chunli W, Jiajie H, Lifei W, Beiqing P, Xin X, Yan C, Mingrong W, Xuemei J. [IGHMBP2 overexpression promotes cell migration and invasion in esophageal squamous carcinoma] . Yi Chuan. 2015;37:360–366. doi: 10.16288/j.yczz.14-371. [DOI] [PubMed] [Google Scholar]
- 16.Kogo R, Mimori K, Tanaka F, Komune S, Mori M. FBXO31 determines poor prognosis in esophageal squamous cell carcinoma. Int J Oncol. 2011;39:155–159. doi: 10.3892/ijo.2011.1018. [DOI] [PubMed] [Google Scholar]
- 17.Shi ZZ, Shang L, Jiang YY, Hao JJ, Zhang Y, Zhang TT, Lin DC, Liu SG, Wang BS, Gong T, Zhan QM, Wang MR. Consistent and differential genetic aberrations between esophageal dysplasia and squamous cell carcinoma detected by array comparative genomic hybridization. Clin Cancer Res. 2013;19:5867–5878. doi: 10.1158/1078-0432.CCR-12-3753. [DOI] [PubMed] [Google Scholar]
- 18.Thallinger CM, Raderer M, Hejna M. Esophageal cancer: a critical evaluation of systemic second-line therapy. J. Clin. Oncol. 2011;29:4709–4714. doi: 10.1200/JCO.2011.36.7599. [DOI] [PubMed] [Google Scholar]
- 19.Qin YR, Wang LD, Kwong D, Gao SS, Guan XY, Zhuang ZH, Fan ZM, Deng W, Hu L. [Comparative genomic hybridization: the profile of chromosomal imbalances in esophageal squamous cell carcinoma] . Zhonghua Bing Li Xue Za Zhi. 2005;34:80–83. [PubMed] [Google Scholar]
- 20.Hu N, Wang C, Ng D, Clifford R, Yang HH, Tang ZZ, Wang QH, Han XY, Giffen C, Goldstein AM, Taylor PR, Lee MP. Genomic characterization of esophageal squamous cell carcinoma from a high-risk population in China. Cancer Res. 2009;69:5908–5917. doi: 10.1158/0008-5472.CAN-08-4622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu M, Zhang F, Liu S, Zhao W, Zhu J, Zhang X. Loss of heterozygosity analysis of microsatellites on multiple chromosome regions in dysplasia and squamous cell carcinoma of the esophagus. Exp Ther Med. 2011;2:997–1001. doi: 10.3892/etm.2011.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y, Yang D, Cogdell D, Hu L, Xue F, Broaddus R, Zhang W. Genomic characterization of gene copy-number aberrations in endometrial carcinoma cell lines derived from endometrioid-type endometrial adenocarcinoma. Technol Cancer Res Treat. 2010;9:179–189. doi: 10.1177/153303461000900207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ni IB, Ching NC, Meng CK, Zakaria Z. Translocation t(11;14) (q13;q32) and genomic imbalances in multi-ethnic multiple myeloma patients: a Malaysian study. Hematol Rep. 2012;4:e19. doi: 10.4081/hr.2012.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rickert CH, Korshunov A, Paulus W. Chromosomal imbalances in clear cell ependymomas. Mod Pathol. 2006;19:958–962. doi: 10.1038/modpathol.3800614. [DOI] [PubMed] [Google Scholar]
- 25.Wright TC, Compagno J, Romano P, Grazioli V, Verma Y, Kershnar E, Tafas T, Kilpatrick MW. Amplification of the 3q chromosomal region as a specific marker in cervical cancer. Am J Obstet Gynecol. 2015;213:51, e51–58. doi: 10.1016/j.ajog.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 26.Qian J, Zou Y, Wang J, Zhang B, Massion PP. Global gene expression profiling reveals a suppressed immune response pathway associated with 3q amplification in squamous carcinoma of the lung. Genom Data. 2015;5:272–274. doi: 10.1016/j.gdata.2015.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Micci F, Haugom L, Abeler VM, Davidson B, Trope CG, Heim S. Genomic profile of ovarian carcinomas. BMC Cancer. 2014;14:315. doi: 10.1186/1471-2407-14-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sakai N, Kajiyama Y, Iwanuma Y, Tomita N, Amano T, Isayama F, Ouchi K, Tsurumaru M. Study of abnormal chromosome regions in esophageal squamous cell carcinoma by comparative genomic hybridization: relationship of lymph node metastasis and distant metastasis to selected abnormal regions. Dis Esophagus. 2010;23:415–421. doi: 10.1111/j.1442-2050.2009.01026.x. [DOI] [PubMed] [Google Scholar]
- 29.Bandla S, Pennathur A, Luketich JD, Beer DG, Lin L, Bass AJ, Godfrey TE, Litle VR. Comparative genomics of esophageal adenocarcinoma and squamous cell carcinoma. Ann Thorac Surg. 2012;93:1101–1106. doi: 10.1016/j.athoracsur.2012.01.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rouault A, Banneau G, Macgrogan G, Jones N, Elarouci N, Barouk-Simonet E, Venat L, Coupier I, Letouze E, de Reynies A, Bonnet F, Iggo R, Sevenet N, Longy M. Deletion of chromosomes 13q and 14q is a common feature of tumors with BRCA2 mutations. PLoS One. 2012;7:e52079. doi: 10.1371/journal.pone.0052079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang SJ, Gillan TL, Gerrie AS, Hrynchak M, Karsan A, Ramadan K, Smith AC, Toze CL, Bruyere H. Influence of clone and deletion size on outcome in chronic lymphocytic leukemia patients with an isolated deletion 13q in a population-based analysis in British Columbia, Canada. Genes Chromosomes Cancer. 2016;55:16–24. doi: 10.1002/gcc.22294. [DOI] [PubMed] [Google Scholar]
- 32.Gebhart E, Ries J, Wiltfang J, Liehr T, Efferth T. Genomic gain of the epidermal growth factor receptor harboring band 7p12 is part of a complex pattern of genomic imbalances in oral squamous cell carcinomas. Arch Med Res. 2004;35:385–394. doi: 10.1016/j.arcmed.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 33.Gao L, Ye X, Ma RQ, Cheng HY, Han HJ, Cui H, Wei LH, Chang XH. Low programmed cell death 5 expression is a prognostic factor in ovarian cancer. Chin Med J (Engl) 2015;128:1084–1090. doi: 10.4103/0366-6999.155100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qin YR, Fu L, Sham PC, Kwong DL, Zhu CL, Chu KK, Li Y, Guan XY. Single-nucleotide polymorphism-mass array reveals commonly deleted regions at 3p22 and 3p14.2 associate with poor clinical outcome in esophageal squamous cell carcinoma. Int J Cancer. 2008;123:826–830. doi: 10.1002/ijc.23577. [DOI] [PubMed] [Google Scholar]
- 35.Rao PH, Arias-Pulido H, Lu XY, Harris CP, Vargas H, Zhang FF, Narayan G, Schneider A, Terry MB, Murty VV. Chromosomal amplifications, 3q gain and deletions of 2q33-q37 are the frequent genetic changes in cervical carcinoma. BMC Cancer. 2004;4:5. doi: 10.1186/1471-2407-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Braakhuis BJ, Snijders PJ, Keune WJ, Meijer CJ, Ruijter-Schippers HJ, Leemans CR, Brakenhoff RH. Genetic patterns in head and neck cancers that contain or lack transcriptionally active human papillomavirus. J Natl Cancer Inst. 2004;96:998–1006. doi: 10.1093/jnci/djh183. [DOI] [PubMed] [Google Scholar]
- 37.Steenbergen RD, Hermsen MA, Walboomers JM, Joenje H, Arwert F, Meijer CJ, Snijders PJ. Integrated human papillomavirus type 16 and loss of heterozygosity at 11q22 and 18q21 in an oral carcinoma and its derivative cell line. Cancer Res. 1995;55:5465–5471. [PubMed] [Google Scholar]


