Abstract
Background: Circular RNA is a novel type of RNAs and may regulate gene expression in cells. It is also involved in various biological processes. The high glucose stress is one of the major risk factors for cardiovascular diseases. It can induce vascular endothelial cell apoptosis. However, the role and biological function of circRNA is still unclear under high glucose. The purpose of this study is to investigate the role of circRNAs in human umbilical vein endothelial cells (HUVECs) induced by high glucose. Method and results: We investigated the expression pattern of circRNA-001175 and the cell proliferation, tubule formation and apoptosis of human umbilical vein endothelial cells (HUVECs) induced by high glucose. The real-time PCR results showed that the glucose treatments gradually decreased the expressions of circRNA-001175 in a concentration dependent manner. The CCK-8 assay showed that high glucose treatment significantly decreased the cell viability, while the decrease was reversed by the up-regulation of circRNA-001175. Also, the circRNA-001175 transfection showed protective effect on the proliferation decrease induced by high glucose treatment. The Hoechst staining and flow cytometry analysis showed that the Up-regulation of circRNA-001175 inhibits the HUVECs apoptosis induced by high glucose treatment. Furthermore, up-regulation of circRNA-001175 was observed to increases the tubule formation ability of HUVECs under high glucose. Conclusions: CircRNA-001175 may play a key role of protection on HUVECs from high glucose stress. CircRNA-001175 has great potential to become diagnostic or predictive biomarkers for high glucose disease and provide new insights into the treatment of diseases.
Keywords: Circular RNA, high glucose, proliferation, apoptosis
Introduction
High glucose has been considered as the risk factor for cardiovascular disease [1]. Also, it accounts for a lot of global mortality for diabetic patients [2]. Vascular endothelium plays an important role in high glucose-associated diabetes. Some researchers have demonstrated that high glucose can affect cell apoptosis level, which may increase the level of reactive oxygen species in endothelial cells [3,4], thus causing cellular dysfunction and even cell death [5,6]. However, the underlying mechanism of the effects of high glucose tress on human endothelial cells is still not fully clear.
Recently, it has been demonstrated that the circular RNAs (circRNAs) belong to a particular class of ubiquitous non-coding RNAs [7,8]. Because of the circular structure and lacking of a 5’cap, the circRNAs can not be translated to proteins. In addition, the remarkable feature of circRNAs is head-to-tail or backsplice junction, and the exons of the RNA arise in reversed order compared with chromosomal localization [9]. The circRNAs performed cellular functions by suppressing and binding to microRNAs (miRNAs) as miRNA sponges [10]. The circRNAs can also play a key role in the responses of human umbilical vein endothelial cells (HUVEC) to the glucose stress by circRNA-miRNA-mRNA pathway [11]. Some evidences have indicated that circRNAs mediate gene expression regulation [10]. It showed that circRNAs take part in diabetes mellitus diseases [7,12,13]. Therefore, we proposed that the expression of circRNAs may be involved in genes expression for the progress of diabetes mellitus, and the role of circRNA-001175 in the high glucose-induced HUVECs was investigated.
Materials and methods
Cell culture
The HUVEC cells were acquired from American Type Culture Collection and maintained in Dulbecco’s modified Eagle’s medium supplemented with 20% fetal bovine serum (Sijiqing, Hangzhou, China). The cultures were kept in a humidified air containing 5% carbon dioxide for 37°C. The cultures medium was refreshed every two days. HUVECs were treated with different concentrations of glucose (5 mM, 10 mM, 20 mM, 30 mM) and 25 mM mannitol (osmotic control) for 4 h, 12 h, 24 h, 48 h, and 72 h.
CircRNA-001175 mimic transfection
The circRNA-001175 mimics (GenePharma, Shanghai, China) transfection was carried out according to the manufacturer’s instructions. In brief, 5×105 cells were cultured onto culture plates for overnight at 37°C. CircRNA-001175 mimics or the negative control (NC) in 200 µl of culture medium were mixed with 5 µl of Lipofectamine 2000 transfection reagent (Thermo Fisher Scientific, USA) and incubated at room temperature for 20 min. After transfection, 2 mL of fresh culture medium containing 5 mM, 30 mM glucose, and 25 mM mannitol was added to each well. The cells were further incubated for cell proliferation, cell apoptosis, and the tubule formation assay analysis.
Cell viability and proliferation assay
HUVECs were incubated with 5 mM (normal control), 10 mM, 20 mM, 30 mM glucose, and 25 mM mannitol (osmotic control) for 4 h, 12 h, 24 h, 48 h and 72 h. Cell viability was determined with the Cell Counting Kit-8 (Bogoo, Shanghai, China) according to the manufacturer’s instructions. Briefly, 10 μl of CCK-8 solution (5 mg/ml) was added to each well of plate and incubated with the cells for 2 h at 37°C. The optical density was detected at the wave length of 450 nm by using a microplate reader (Thermo Fisher Scientific, Kalamazoo, MI, USA). For detecting the cell proliferation of the transfected HUVECs, the optical density at 450 nm was calculated every 24 h culturing period.
EdU staining
For EdU staining, the cells were fixed with methanol, and washed twice with PBS, then incubated in 3% bovine serum albumin (BSA) in PBS. The cells were then incubated with freshly prepared Click-iT reaction cocktail (Thermo Fisher Scientific, USA) for 60 min at room temperature in the dark. The cells were further stained and then mounted in standard mounting media. The stained cells were examined with Nikon Eclipse E600 fluorescence microscope.
Apoptosis assay
The apoptosis level of treated HUVECs was performed by cell apoptosis detection kit (Boehringer Mannheim, USA). The cells were collected and fixed in methanol/acetone solution for 8 minutes and washed with PBS. A 100 mL sample of cells was incubated for 20 min in the dark. The cells were then observed and photographed by a Nikon fluorescence microscope.
Flow cytometry analysis
The cell apoptosis rate was identified using the FITC Annexin V Apoptosis Detection Kit I (BD, USA) according to the manufacturer’s instructions. The HUVECs were collected and washed with PBS. The cells were stained with 5 μl annexin v-FITC for 15 min and then 5 μl propidium iodide (PI) for 10 min in the dark. The cell apoptosis rates were determined using a FACSCanto II cytometer (BD Biosciences, Germany), and the percentage of apoptotic cells was calculated.
Tubule formation assay
After circRNA-001175 mimics transfection, the HUVECs were plated on Matrigel in a 24-well plate. The degree of the angiogenic response was evaluated after 20 h of incubation in complete medium at 37°C. The length of tubes in 10 randomly chosen low-power fields from each well was evaluated, and the number of branching points was counted. Each well was photographed, and the relative acquired images were quantified using Image-Pro Plus 6.0 software (Media Cybernetics, Rockville, MD, USA).
RNA extraction and real-time quantitative RT-PCR
We used RNAiso™ Plus kit (TaKaRa Biotechnology Inc., Japan) to extract the total RNA from cells according to the manufacturer’s protocol. The real time PCR was performed using SYBR Premix Ex Taq™ II (TaKaRa, Japan). The Premier 5.0 biological software (PREMIER Biosoft, Palo Alto, CA) was used to design the specific primers for circRNA-001175 and β-Actin, and the sequences were as follows: circRNA-001175: 5’-CCTGTAGGAAGCAACCAGTC-3’ (forward) and 5’-ACCTCCACAATGAACTACACC-3’ (reverse); β-Actin: 5’-TGTTCGTCATGGGTGTGAAC-3’ (forward) and 5’-ATGGCATGGACTGTGGTCAT-3’ (reverse). The PCR conditions were as follows: 94°C denaturation for 35 s, followed by 35 cycles of 15 s at 94°C, and 30 s at 59°C. The PCR products were electrophoresed on a 1% agarose gel containing ethidiumbromide. The expression levels of target genes were normalized to β-actin (endogenous control). The relative expression of circRNAs was calculated using the 2-ΔΔct method.
Statistical analysis
The Data were represented as mean ± SD. The statistical difference was evaluated by Student’s t tests between two groups. P<0.05 was considered statistically significant.
Results
High glucose treatment decreases the circRNA-001175 expression and HUVEC viability
To study the effect of high glucose on HUVEC viability, we treated HUVECs with high glucose in different concentrations for 4 h, 12 h, 24 h, 48 h, and 72 h. The cell viability was determined by the CCK-8 assay. The results showed that, with 25 mM mannitol treatment, the cell viability was not obviously influenced, while the cell viability was decreased when cells were treated with high concentrations of glucose (Figure 1A). Specifically, the cell viability was significantly decreased when the cells were 48 h post-treated with 10 mM and 20 mM glucose, and 24 h post-treated with 30 mM glucose (P<0.05 versus the osmotic control group). Furthermore, the real-time PCR results showed that mannitol as an osmotic control showed no effect on circRNA-001175 expression in HUVECs, while the glucose treatments gradually decreased the expressions of circRNA-001175 in a concentration dependent manner (Figure 1B). 72 h post-treated with 10 mM, 20 mM, and 30 mM glucose, the expressions of circRNA-001175 were significantly decreased compared with the osmotic control group (P<0.05). The 30 mM glucose treatment was applied for the following studies.
Figure 1.
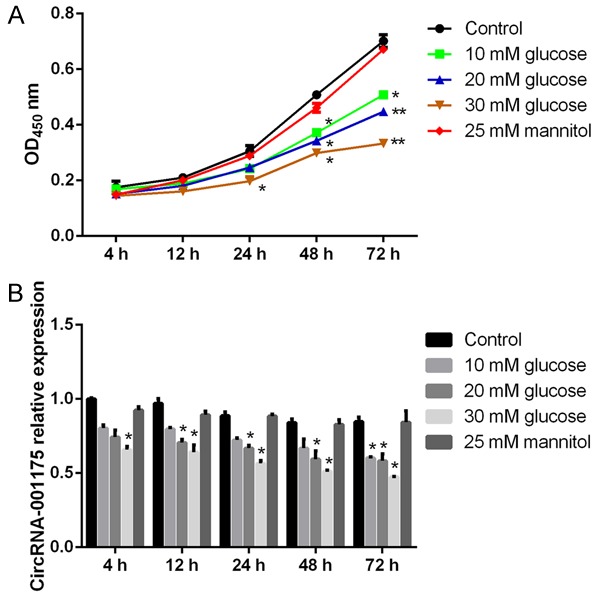
Cell viability of HUVECs and the expression levels of circRNA-001175 in HUVECs. A: Cell viabilities after glucose (10 mM, 20 mM, and 30 mM) and mannitol (25 mM) treatments for 4 h, 12 h, 24 h, 48 h, and 72 h were measured by the CCK-8 method and normalized to the control. B: The expression levels of circRNA-001175 after glucose (10 mM, 20 mM, and 30 mM) and mannitol (25 mM) treatments for 4 h, 12 h, 24 h, 48 h, and 72 h detected by RT-PCR. *P<0.05; **P<0.01; versus the osmotic control group.
Up-regulation of circRNA-001175 reverses the HUVECs viability and proliferation decreases induced by high glucose treatment
The HUVECs were cultured in different concentration glucose medium, and then transfected with circRNA-001175 mimics or NC mimics. We found that, after the transfection with circRNA-001175 mimics, the expressions of circRNA-001175 mimics were significantly up-regulated in all groups (P<0.01 versus the NC group; Figure 2B). Moreover, the CCK-8 assay showed that 30 mM glucose treatment significantly decreased the cell viability, while the decrease was reversed by the up-regulation of circRNA-001175 (P<0.05 after 48 h and 72 h treatment compared with the NC group; Figure 2A). It also indicates that the up-regulation of circRNA-001175 induced significant enhance on cell growth after transfection. Furthermore, the proliferation ability of HUVECs was analyzed by examined the Edu incorporation (Figure 2C). As the results indicated, the cell proliferation for the 30 mM glucose and NC treated group was decreased compared with the control and mannitol treated groups. However, the circRNA-001175 transfection showed protective effect on the proliferation decrease induced by high glucose treatment (Figure 2C).
Figure 2.
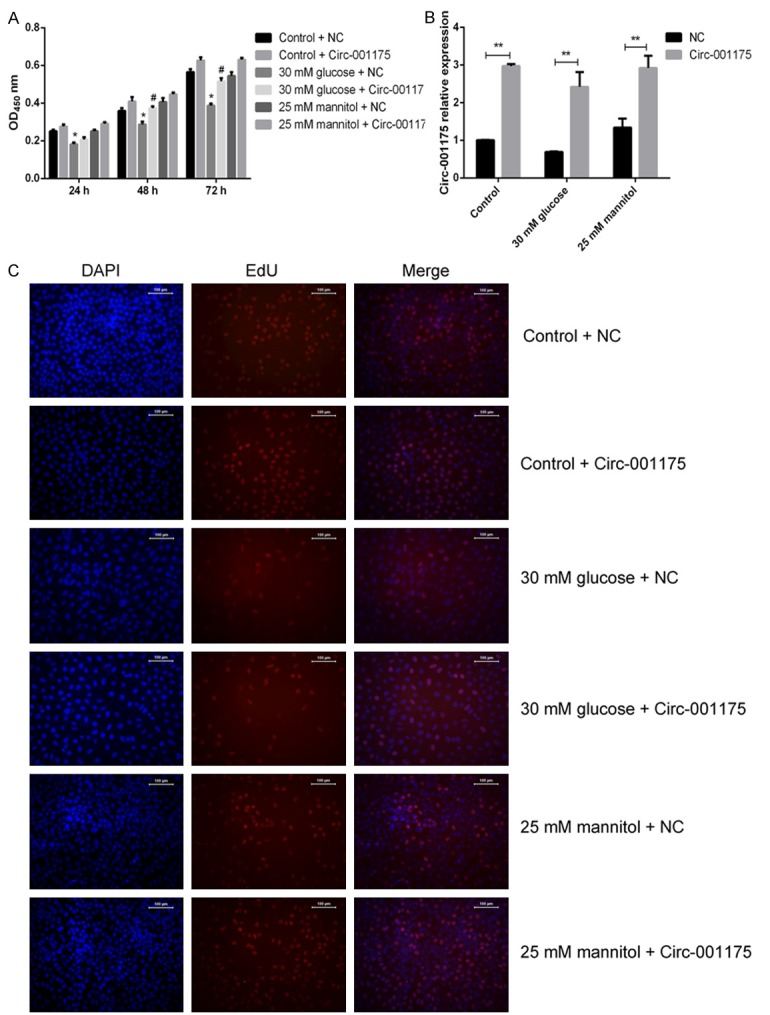
Up-regulation of circRNA-001175 reverses the HUVECs viability and proliferation decreases induced by high glucose treatment. A: Effect of up-regulation of circRNA-001175 on HUVECs viability and proliferation. B: CircRNA-001175 expression in HUVECs after transfected with circRNA-001175 mimics. C: Detection of EdU incorporated into DNA of HUVECs by fluorescence microscopy. *P<0.05, versus the osmotic control group. **P<0.01, versus the NC groups as indicated in the figure; #P<0.05, versus the 30 mM glucose + NC group.
Up-regulation of circRNA-001175 inhibits the HUVECs apoptosis induced by high glucose treatment
As shown in Figure 3A and 3B, 30 mM glucose treatment significantly increased the Hoechst staining ratio of HUVECs (P<0.01 versus the osmotic control group), while the ratio was dramatically decreased by the up-regulation of circRNA-001175 (P<0.01 versus the NC control group). Moreover, the cell apoptosis rate was further identified using the FITC Annexin V Apoptosis assay by flow cytometric analysis (Figure 3C and 3D). 30 mM glucose treatment significantly increased the apoptosis rate of HUVECs (P<0.01 versus the osmotic control group), while the rate was significantly decreased by the up-regulation of circRNA-001175 (P<0.05 versus the NC control group). Interestingly, the up-regulation of circRNA-001175 also significantly decreased the apoptosis rate of HUVECs in the control and mannitol treated groups (P<0.05 versus the NC group). All these indicate that up-regulation of circRNA-001175 can inhibit the cell apoptosis induced by high glucose treatment.
Figure 3.
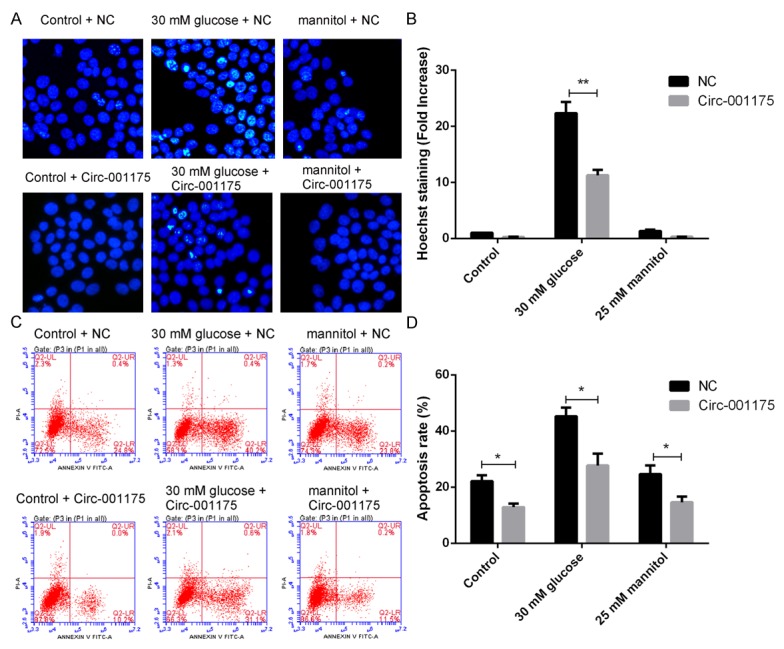
Up-regulation of circRNA-001175 inhibits the HUVECs apoptosis induced by high glucose treatment. A: The apoptosis of HUVECs was showed by Hoechst staining. B: Quantitative analysis of the Hoechst staining ratio of HUVECs. C: The apoptosis cycle analysis of HUVEC cells by flow cytometry. D: Quantitative analysis of the apoptosis rate of HUVECs. *P<0.05, **P<0.01, versus the NC groups as indicated in the figure.
Up-regulation of circRNA-001175 increases the tubule formation ability of HUVECs
The Matrigel tubule formation experiment results (Figure 4) showed that up-regulation of circRNA-001175 significantly increased the average tubule length compared with those of the NC groups. The tubule formation ability of high glucose treated HUVECs was significantly increased when the circRNA-001175 was up-regulated (P<0.01 versus the NC group).
Figure 4.
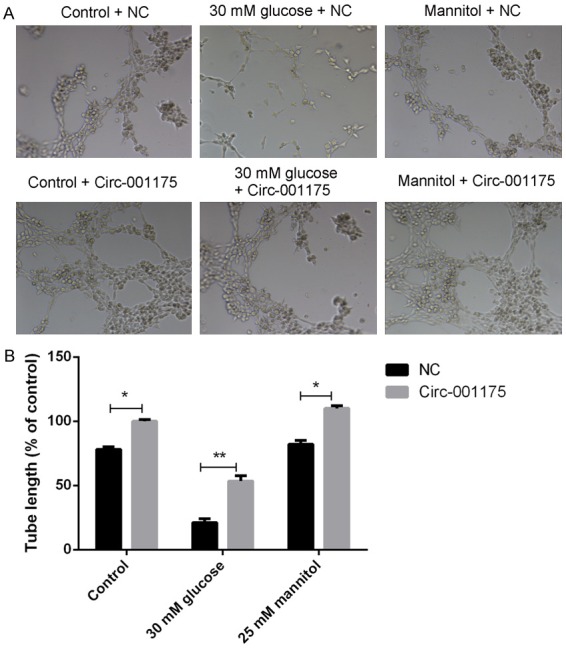
Up-regulation of circRNA-001175 increases the tubule formation ability of HUVECs. A: Representative images of the tube structures of HUVECs under different conditions. B: Quantitative analysis of the tube length of HUVECs. *P<0.05, **P<0.01, versus the NC groups as indicated in the figure.
Discussion
High glucose has long been considered as a risk factor for several forms of cardiovascular disease (CVD). Some researchers have attempted to build the novel targeted therapies and explain the molecular mechanism of cells under high glucose conditions [14-17]. Although considerable progress has been made, the molecular mechanisms for high glucose stress are still obscure.
So far, the circRNAs as a new special class of endogenous noncoding RNA have been recently discovered and studied [8,18-20]. Moreover, the circRNAs can function as miRNA sponges to regulate the expression of genes and further affect disease initiation [21-23]. Recently, more and more studies uncovered that circRNA is a transcriptional product in various tissues and cell types of archaea, human, and mouse [24,25]. In this study, we studied the expression of circRNA-001175 in HUVECs treated with high glucose. Our results showed that high glucose treatment decreases the circRNA-001175 expression and HUVEC viability. This indicates that the circRNA-001175 may be involved in the cell responses to the high glucose. The circRNAs may be related to their involvement in the transcription level regulation on the high glucose resistance of HUVEC cells.
To further understand the role of circRNA-001175 in HUVECs, additional studies on the biological function were conducted. As we expected, overexpression of circRNA-001175 affected the cell proliferation of in HUVECs. The results demonstrated that circRNA-001175 play a stimulative role on cellular proliferation. We further investigated the effect of circRNA on cell apoptosis induced by high glucose treatment. Previous studies have reported that circRNAs can serve as molecular markers of cell apoptosis. Such as circRNA Foxo3 could inhibits progression of the cell cycle and apoptosis [26,27]. In the present study, we found that circRNA-001175 was involved in the regulation of the cell cycle in HUVECs, and it could effectively depressed apoptosis rate elicited by high glucose. Our findings also suggested that the up-regulation of circRNA-001175 promotes tubule formation of HUVECs. It was indicated that the circRNA-001175 has positive effect on tubule formation and angiogenesis.
In conclusion, all these studies indicate that circRNA-001175 may play a key role of protection on HUVECs from high glucose stress. CircRNA-001175 have great potential to become diagnostic or predictive biomarkers for high glucose disease and provide new insights into the treatment of diseases.
Acknowledgements
This work was supported by the National Science Foundation of Education Department of Anhui province (NO.KJ2015BO55by).
Disclosure of conflict of interest
None.
References
- 1.Haring R, Wallaschofski H, Nauck M, Felix SB, Schmidt CO, Dörr M, Sauer S, Wilmking G, Völzke H. Total and cardiovascular disease mortality predicted by metabolic syndrome is inferior relative to its components. Exp Clin Endocrinol Diabetes. 2010;118:685–691. doi: 10.1055/s-0030-1261876. [DOI] [PubMed] [Google Scholar]
- 2.Ziegler D. Type 2 diabetes as an inflammatory cardiovascular disorder. Curr Mol Med. 2005;5:309–322. doi: 10.2174/1566524053766095. [DOI] [PubMed] [Google Scholar]
- 3.Boyle PJ. Diabetes Mellitus and macrovascular disease: mechanisms and mediators. Am J Med. 2007;120(Suppl 2):S12–S17. doi: 10.1016/j.amjmed.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 4.Versari D, Daghini E, Virdis A, Ghiadoni L, Taddei S. Endothelial dysfunction as a target for prevention of cardiovascular disease. Diabetes Care. 2009;32(Suppl 2):S314–S321. doi: 10.2337/dc09-S330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ho FM, Liu SH, Liau CS, Huang PJ, Lin-Shiau SY. High glucose-induced apoptosis in human endothelial cells is mediated by sequential activations of c-Jun NH2-terminal kinase and caspase-3. Circulation. 2000;101:2618–2624. doi: 10.1161/01.cir.101.22.2618. [DOI] [PubMed] [Google Scholar]
- 6.Baumgartner-Parzer SM, Wagner L, Pettermann M, Grillari J, Gessl A, Waldhäusl W. High glucose triggered apoptosis in cultured endothelial-cells. Diabetes. 1995;44:A221–A221. doi: 10.2337/diab.44.11.1323. [DOI] [PubMed] [Google Scholar]
- 7.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 8.Chen LL. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol. 2016;17:205–211. doi: 10.1038/nrm.2015.32. [DOI] [PubMed] [Google Scholar]
- 9.Wilusz JE, Sharp PA. Molecular biology. A circuitous route to noncoding RNA. Science. 2013;340:440–441. doi: 10.1126/science.1238522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang YH, Yu XH, Luo SS, Han H. Comprehensive circular RNA profiling reveals that circular RNA100783 is involved in chronic CD28-associated CD8(+)T cell ageing. Immun Ageing. 2015;12:17. doi: 10.1186/s12979-015-0042-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lukiw WJ, Rogaev EI, Zhao YH. Circular RNA (circRNA) ciRS-7 targets miRNA-7 trafficking and ubiquitin-conjugase E2A (UBE2A)-mediated protein degradation in Alzheimer’s disease (AD) and age-related macular degeneration (AMD) Invest Ophth Vis Sci. 2016;57 [Google Scholar]
- 12.Wang K, Long B, Liu F, Wang JX, Liu CY, Zhao B, Zhou LY, Sun T, Wang M, Yu T, Gong Y, Liu J, Dong YH, Li N, Li PF. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur Heart J. 2016;37:2602–2611. doi: 10.1093/eurheartj/ehv713. [DOI] [PubMed] [Google Scholar]
- 13.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, Loewer A, Ziebold U, Landthaler M, Kocks C, le Noble F, Rajewsky N. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 14.Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32:453–461. doi: 10.1038/nbt.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lasda E, Parker R. Circular RNAs: diversity of form and function. RNA. 2014;20:1829–1842. doi: 10.1261/rna.047126.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang PL, Bao Y, Yee MC, Barrett SP, Hogan GJ, Olsen MN, Dinneny JR, Brown PO, Salzman J. Circular RNA is expressed across the eukaryotic tree of life. PLoS One. 2014;9:e90859. doi: 10.1371/journal.pone.0090859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liang DM, Wilusz JE. Short intronic repeat sequences facilitate circular RNA production. Gene Dev. 2014;28:2233–2247. doi: 10.1101/gad.251926.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9:e1003777. doi: 10.1371/journal.pgen.1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Starke S, Jost I, Rossbach O, Schneider T, Schreiner S, Hung LH, Bindereif A. Exon circularization requires canonical splice signals. Cell Rep. 2015;10:103–111. doi: 10.1016/j.celrep.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 21.Lu TT, Cui LL, Zhou Y, Zhu CR, Fan DL, Gong H, Zhao Q, Zhou CC, Zhao Y, Lu DF, Luo J, Wang YC, Tian QL, Feng Q, Huang T, Han B. Transcriptome-wide investigation of circular RNAs in rice. RNA. 2015;21:2076–2087. doi: 10.1261/rna.052282.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qin ML, Liu G, Huo XS, Tao XM, Sun XM, Ge ZH, Yang J, Fan J, Liu L, Qin WX. Hsa_ circ_0001649: a circular RNA and potential novel biomarker for hepatocellular carcinoma. Cancer Biomark. 2016;16:161–169. doi: 10.3233/CBM-150552. [DOI] [PubMed] [Google Scholar]
- 23.Shang XC, Li GZ, Liu H, Li T, Liu J, Zhao Q, Wang CX. Comprehensive circular RNA profiling reveals that hsa_circ_0005075, a new circular RNA biomarker, is involved in hepatocellular Crcinoma development. Medicine. 2016;95:e3811. doi: 10.1097/MD.0000000000003811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zheng C, Niu H, Li M, Zhang H, Yang Z, Tian L, Wu Z, Li D, Chen X. Cyclic RNA hsacirc000595 regulates apoptosis of aortic smooth muscle cells. Mol Med Rep. 2015;12:6656–6662. doi: 10.3892/mmr.2015.4264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang C, Wu H, Wang Y, Zhao Y, Fang X, Chen C, Chen H. Expression patterns of circular RNAs from primary kinase transcripts in the mammary glands of lactating rats. J Breast Cancer. 2015;18:235–241. doi: 10.4048/jbc.2015.18.3.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu WY. Roles of the circular RNA circ-Foxo3 in breast cancer progression. Cell Cycle. 2017;16:589–590. doi: 10.1080/15384101.2017.1278935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhong Z, Lv M, Chen J. Screening differential circular RNA expression profiles reveals the regulatory role of circTCF25-miR-103a-3p/miR-107-CDK6 pathway in bladder carcinoma. Sci Rep. 2016;6:30919. doi: 10.1038/srep30919. [DOI] [PMC free article] [PubMed] [Google Scholar]


