Abstract
Lung cancer is one of the most preventable forms of cancer, which is the second leading cause of cancer-related death. Emerging evidence has suggested a possible link between the endogenous circadian clock and cell growth, and even cancer development. However, the role of important clock gene period 3 (PER3) in lung cancer remains fully understand, especially its regulatory mechanism. In this study, we investigated the biological role of PER3 in non-small cell lung cancer (NSCLC), and also demonstrated the possible mechanism that induces dysfunction of PER3 in lung cancer. QPCR was performed to measure the expression of PER3 in NSCLC tissues and cell lines. Bisulfite genomic sequencing PCR (BSP) and methylation specific PCR (MSP) were used to determine the methylation status of PER3. And a battery of cell biology assays was used to measure the effects of PER3 on cell proliferation, apoptosis, migration and invasion. We found that PER3 was downregulated in NSCLC tissues and cell lines compared with the adjacent or normal cells control. The promoter of PER3 was hypermethylated, which could be restored by demethylation drug 5-Aza. And overexpression of PER3 suppressed NSCLC cell proliferation, induced apoptosis and inhibited the ability of migration and invasion. These findings reveal that PER3 is a tumor suppressor in NSCLC and provide a promising target and a novel strategy to control cancer progression.
Keywords: Non-small cell lung cancer, circadian clock, period 3, methylation, tumor suppressor
Introduction
Lung cancer is one of the most preventable forms of cancer, which is the second leading cause of cancer-related death [1]. Non-small cell lung cancer (NSCLC) is the major type of lung cancer with about 90% of lung cancer cases. Identification of the biomarkers and potential mechanisms for NSCLC would help to overcome this malignant.
Lung cancer is closely related to the environment changes and individuals’ habits [2]. Biological circadian rhythms are intrinsic time-keeping systems that control a variety of biological processes and physiological functions in animals and human beings [3]. Emerging evidence has suggested a possible link between the endogenous circadian clock and cell growth, and even cancer development [4]. Dysregulation of circadian rhythm is associated with greater risk for cancer development and poor prognosis [5]. An important circadian pathway regulatory gene, Nuclear receptor subfamily 1, group D member 1 (Nr1d1), is downregulated by both whole smoke and filtered smoke that may play a complementary role in CS-induced lung tumorigenesis [6]. Recent study demonstrates that disruption of central circadian clock components, such as PER2 and Bmal1 by physiologic perturbation or genetic mutation significantly decreased survival and promoted lung tumor growth and progression, which was associated with increased c-Myc expression [7].
There is closed association between circadian clock genes and precocious lesions, cancer growth and cancer metastasis. For example, genetic ablation of the clock gene Bmal1 in bronchiolar cells disrupts rhythmic CXCL5 expression, resulting in exaggerated inflammatory responses to lipopolysaccharide [8]. Pulmonary fibrosis is an important step for lung cancer development. Pekovic-Vaughan V et al show that an E-box-mediated circadian rhythm of NRF2 protein is essential in regulating the rhythmic expression of antioxidant genes involved in glutathione redox homeostasis lung [9]. Logan RW et al suggest that chronic circadian disruption promotes tumor growth by altering the circadian rhythms of NK cell function [10]. Single nucleotide polymorphisms (SNP) within the circadian clock genes have been found to confer excess cancer risk or protection from cancer. Two central clock genes, PER1 and PER2, are tumor suppressors because decreasing their expression in cancer cells enhances cancer growth rate, while their overexpression decreases cancer growth rate and diminishes tumor numbers. In addition, clinical circadian disruption in patients with advanced lung cancer is associated with higher cancer incidence and shorter cancer patient survival [11].
Previous studies have reveal the key clock molecules such as PER1 and PER2 play critical role in lung cancer and other type cancers, however, the role of another important clock gene PER3 in lung cancer remains fully understand, especially its regulatory mechanism. The SNP (rs228729) of PER3 is significantly correlated with genotype and allele frequency in NSCLC, suggesting that dysfunction of PER3 represents a risk factor in the occurrence and development of NSCLC [12]. In this study, we investigated the biological role of PER3 in NSCLC, and also demonstrated the possible mechanism that induces dysfunction of PER3 in lung cancer.
Materials and methods
Tissue samples
Eight paired non-small cell lung cancer tissues and the matched adjacent normal tissues used in the study were collected during 2015 in Xiangya hospital. All the patients signed the informed consents. This work was approved by the Independent Ethical Committee of Central South University. All samples were stored at -80°C until tissue analysis.
Cells culture
Lung cancer cell lines, A549 and H1650, and human bronchial epithelial cells (HBE) were obtained from American type culture collection (ATCC). All the cells are cultured in RPMI1640 (Gibco) basic medium supplemented with 10% FBS at the conditions: 95% air and 5% CO2 at 37°C.
Lentivirus infection
In order to investigate the role of PER3 in lung cancer cells, A549 and H1650 cells were infected with negative control lentivirus (NC) or PER3 lentivirus (PER3) for 48 h. QPCR and western blot were used to detect the expression of PER3.
Real time quantitative PCR (qPCR) analysis
Trizol reagent (Invitrogen, USA) was used to extract total RNA from the indicated cells according to the manufacturer’s instructions. FastLane Cell SYBR® Green Kit (Qiagen, Valencia, CA) was used for real time PCR to detect the expression of PER3. The primers of PER3 and β-actin are as follow: PER3, sense, GCAGGTCTATGCCAGTGTGA, antisense, TGCCTTGTGGTTCTGTTTGT; β-actin, sense, AGGGGCCGGACTCGTCATACT, antisense, GGCGGCACCACCATGTACCCT. PER3 expression was normalized by β-actin. All the qPCR data were processed using 2-ΔΔCT method.
Western blot
RIPA lysis buffer (Boster, Wuhan, China) was used to extract the total protein was extracted from the tissues or cells. BCA Protein Assay Kit (Thermo, Waltham, MA) was used to measure the protein concentration. Sixty μg of total protein was then separated with 10% SDS-PAGE and transferred to a nitrocellulose membrane. The membranes were blocked and incubated with indicated primary antibody (mouse monoclonal, anti-PER3 from Abcam, 1:1000; mouse monoclonal, anti-cleaved caspase 3 from Abcam, 1:500; mouse monoclonal, anti- cleaved caspase 8 from Santa Cruze, 1:2000; Rabbit polyclonal, anti-Bcl-2 from Abcam, 1:500; mouse monoclonal, anti-GAPDH from Santa Cruze, 1:3000) overnight at 4°C. The membrane was washed and incubated with secondary antibody for 90 min at 37°C. The signals on the membrane were detected by enhanced chemiluminescence reagent. Data was analyzed by densitometry using Image-Pro plus software 6.0 and normalized to GAPDH expression.
CCK-8 cell proliferation assay
Cell growth was measured by CCK-8 assay. 2000 indicated cells were seeded in each 96-well plate for 12 h, and further incubated for 0 h, 24 h, 48 h and 72 h respectively. 1 hour before the ending of incubation, 10 μl CCK-8 reagents (Boster, Wuhan, China) were added to each well. OD 570 nm value in each well was determined by an enzyme immunoassay analyzer.
Colonal formation
For the colony formation assay, following treatment, adherent cells were trypsinised and 1000 viable cells were subcultured in six-well plates (in triplicate). Cells were allowed to adhere and colonise for two weeks. To visualise colonies, media were removed and cells were fixed in 96% ethanol for 10 min, and then stained with crystal violet staining solution. The colonies were captured and counted. The colonal formation rate was calculated as the number of colonies/1000.
Scratch assay
Cells in each group were collected and resuspended in completed medium containing 10% FBS. Then, each well of a 6-well plate was seeded with 5×105 cells. When the cells were cultured into about 100% confluence, and were scratched with the head of a 200 μl tip, and washed these cells with serum-free medium. Then, these cells were further cultured for 24 h in serum-free medium. After that, serum-free medium were replaced with H-DMEM medium containing 10% FBS and continued to culture for 72 h, then these cells in each group were photographed for analysis.
Transwell assay
The indicated cells were starved in serum-free medium for 24 h. The cells were collected and then resuspended in serum-free medium. The cells were added to the upper chamber, while the lower chamber was filled with completed medium containing 10% FBS. After 24 hours incubation, cells attached to the bottom of upper chamber were fixed and stained with crystal violet for 30 min at 37°C, and dried in air. Acetic acid was used to dissolve the crystal violet. The optical density (OD) at 570 nm was detected by an enzyme immunoassay analyzer.
Flow cytometric analysis of apoptosis
Cells from each group were trypsinizated and washed with cold PBS, and then stained by Annexin V-FITC/PI Apoptosis Detection Kit (BD Biosciences) according to the manufacturer’s instructions. The cell apoptosis was analyzed by flow cytometry (Beckman Coulter, USA). The experiments were independently performed in triplicate.
Measurement of PER3 promoter CpG island methylation status by bisulfite genomic sequencing PCR (BSP) and methylation specific PCR (MSP)
TaKaRa Genomic DNA Extraction Kit (TaKaRa Co., China) was used to extract Genomic DNA. Total 1 μg of Genomic DNA was modified with bisulfite using the Epitect Bisulfite Kit Protocol (Qiagen) according to the manufacturer’s instructions. The primers of PER3 used to amplify were as following: forward, TTTTTATTAGGGTTGTTTTATGGTT, and reverse, ATCCTAAAATTTATAAATAACTCCC. The PCR products were gel extracted (Qiagen) to confirm that a single band had been obtained and were then sequenced by Beijing Genomics Institute (Wuhan, China).
Methylation-specific PCR (MS-PCR) was performed on bisulfate-treated DNA. The primers used were Un-methylated PER3 forward, AAAATTGTGGGAATTTAGAAAGATG, and reverse, AAAAAAAACAAAAACTAAAAACAAA; and methylated PER3 forward: AAAATCGTGGGAATTTAGAAAGAC, and reverse, AAAAAAAACGAAAACTAAAAACGAA. The annealing temperature was 58°C for Methylated-PCR, and Un-methylated-PCR, with 35 cycles used for each.
Statistical analysis
Student t-tests or one-way ANOVA were used to analyze statistical data by Graphpad prism5 software, depending on the experimental conditions. Statistical significance was considered when P values of <0.05.
Results
Hypermethylation induces downregulation of PER3 in NSCLC
To investigate the role of PER3 in lung cancer, we examined the expression of PER3 in the paired NSCLC tissues and adjacent tissues by real time q-PCR. And we found that PER3 expression was significantly lower in NSCLC tissues than in the adjacent tissues (Figure 1A). Similar results also appear in cell lines. The expression of PER3 was significantly decreased in the NSCLC cell lines (A549 and H1650) compared with HEB cells (Figure 1B). We further investigated the underlying mechanisms of PER3 silence. We performed MSP to examine the promoter methylation status of PER3 in 8 NSCLC and paired adjacent tissues. The results showed that PER3 exhibited unmethylation status in adjacent tissues, while the methylation of PER3 promoter was observed in 62.5% cancer tissues (5/8) (Figure 2A). In addition, a significantly higher level of methylation was also observed in A549 (86.2%) and H1650 cells (80%) compared with in the HBE cells (20%) evaluated by BSP assay (Figure 2B). Furthermore, The A549 and H1650 cells were treated with demethylation agents Aza. We found that PER3 expression was significantly increased in a time-dependent manner in A549 and H1650 cells after Aza treatment (Figure 2C, 2D). These results indicate that downregulation of PER3 in NSCLC tissues and cells is induced by hypermethylation of its promoter.
Figure 1.
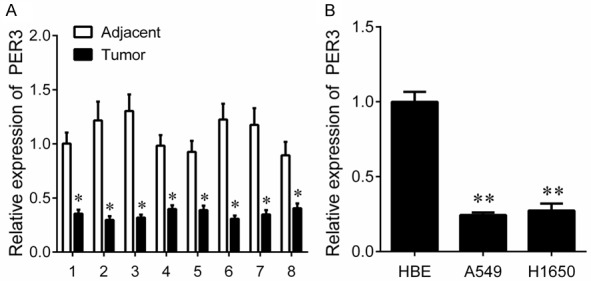
The expression of PER3 in NSCLC tissues and cells. A: QPCR was used to measure the expression of PER3 in NSCLC tissues and their matched adjacent tissues. B: QPCR was used to measure the expression of PER3 in normal human bronchial epithelial cells (HBE), and cancer cells (A549 and H1650). *P<0.05, **P<0.01.
Figure 2.
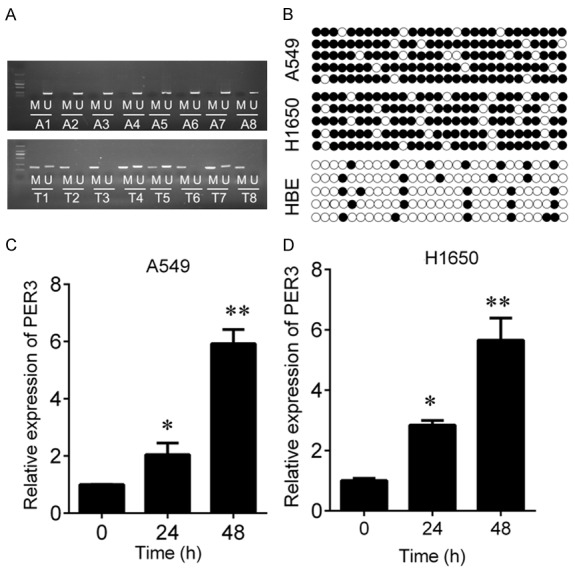
PER3 is hypermethylated in NSCLC tissues and cells. A: MSP was used to measure the hypermethylation status of PER3 in NSCLC tissues and their matched adjacent tissues. M, methylation; U, unmethylation. B: BSP was used to measure the hypermethylation status of PER3 in HBE, A549 and H1650 cells. C, D: QPCR was used to measure the expression of PER3 in A549 and H1650 cells after 5-Aza treatment for 24 or 48 h. *P<0.05, **P<0.01.
Overexpression of PER3 represses cell proliferation and induces apoptosis in A549 and H1650 cells
In order to investigate the biological function of PER3 in lung cancer cells, we infected the A549 and H1650 cells with lentivirus to overexpress PER3 (Figure 3A). CCK-8 was used to detect the A549 and H1650 cells viability. We found that upregulation of PER3 significantly reduced cell viability (Figure 3B and 3C). And colonal formation assay further confirmed that upregulation of PER3 significantly suppressed the ability of colonal formation in A549 and H1650 cells (Figure 3D). In addition, flow cytometric analysis was used to analyze cell apoptosis alterations. The results revealed that upregulation of PER3 increased the apoptosis rate compared with negative control (Figure 4A). And the increased apoptosis was accompanied with upregulated cleaved caspase 3/8 and decreased Bcl-2 expression (Figure 4B).
Figure 3.
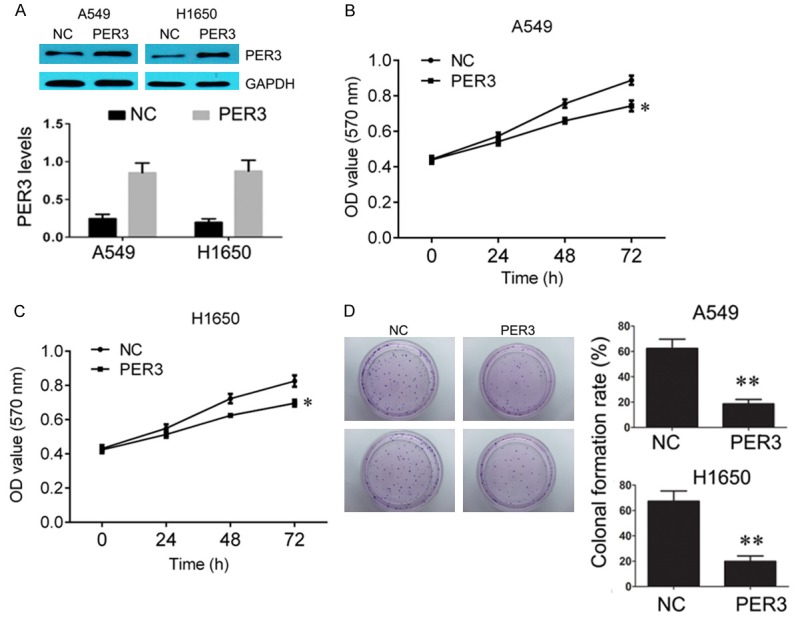
Overexpression of PER3 inhibits A549 and H1650 cells growth. A: Western blot was used to measure the expression of PER3 in A549 and H1650 cells after transfection with PER3 lentivirus, and quantification of the bands. B, C: CCK-8 was used to measure the cell viability of HBE, A549 and H1650 cells after overexpression of PER3. D: Colonal formation was used to measure the ability of colonal formation in HBE, A549 and H1650 cells after overexpression of PER3. *P<0.05, **P<0.01 vs. negative control (NC).
Figure 4.
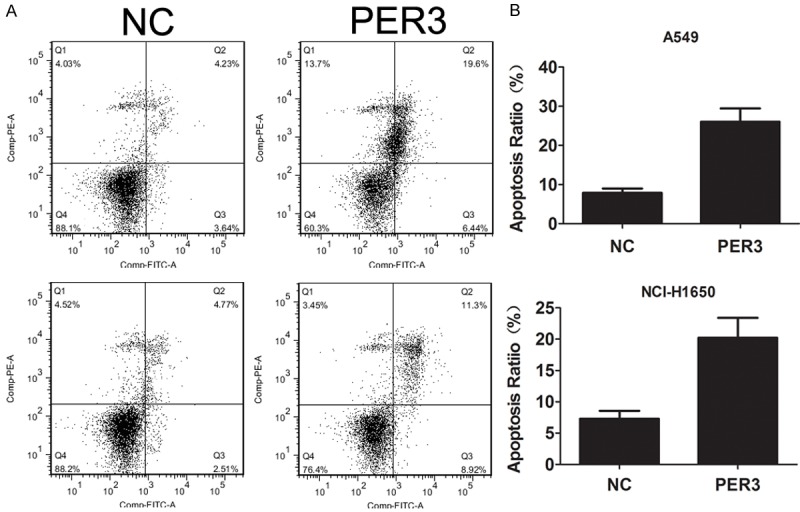
Overexpression of PER3 induces apoptosis in A549 and H1650 cells. A: Flow cytometry was used to measure the cell apoptosis of A549 and H1650 cells after overexpression of PER3. B: Western blot was used to measure the expression of cleaved caspase 3, cleaved caspase 8 and Bcl2 in A549 and H1650 cells after transfection with PER3 lentivirus. GAPDH was used a loading control. **P<0.01 vs. negative control (NC).
Overexpression of PER3 inhibits cell migration and invasion in A549 and H1650 cells
Scratch assay was used to measure the migration ability of A549 and H1650 cells after PER3 overexpression. The results indicated that overexpression of PER3 dramatically decreased the migration ability compared with negative control (Figure 5). The results of transwell assay showed that the invasive ability was significantly reduced by PER3 compared with negative control (Figure 6).
Figure 5.

Overexpression of PER3 inhibits cell migration in A549 and H1650 cells. Scratch assay was used to measure the cell migration of A549 and H1650 cells after overexpression of PER3. *P<0.05 vs. negative control (NC).
Figure 6.
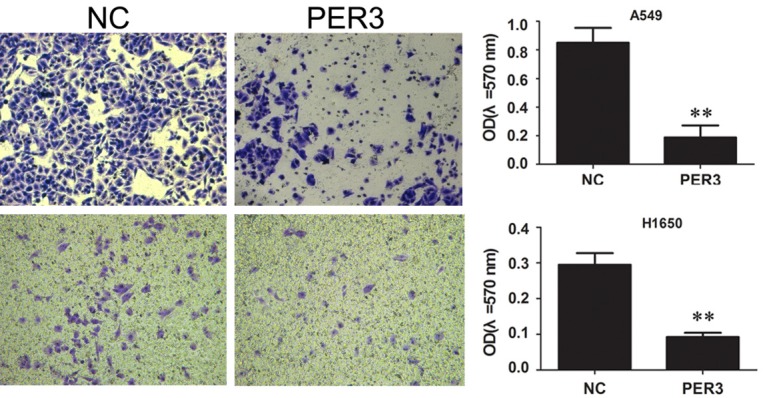
Overexpression of PER3 inhibits cell invasion in A549 and H1650 cells. Scratch assay was used to measure the ability of cell invasion of A549 and H1650 cells after overexpression of PER3. **P<0.01 vs. negative control (NC).
Discussion
In this study, we find that PER3 is downregulated in NSCLC tissues and cell lines, which is induced by hypermethylation. And overexpression of PER3 suppresses NSCLC cell proliferation, induces apoptosis and inhibits the ability of migration and invasion. These findings reveal that PER3 is a tumor suppressor in NSCLC.
Disruption of the circadian rhythm has been demonstrated to be one of the endogenous factors contributing to tumorigenesis of various human malignancies, including breast cancer, colorectal cancer [13,14]. It was shown that circadian clock genes (PER1 and PER2) could control the expression of multidrug resistance-associated protein 2 (Mrp2) in mouse liver, which might be associated with drug resistance in cancer [15]. Kiessling S et al found that clock genes were suppressed in melanoma cells and tumors. A stimulating circadian rhythmicity by dexamethasone, forskolin and heat shock could trigger rhythmic clock and cell cycle gene expression, resulting inhibition of cell proliferation in vitro and tumor growth in vivo [16].
PER3 is an important member of clock genes. It functions as tumor suppressor in various cancers. PER3 was decreased in primary colorectal cancer (CRC), colorectal liver metastases (CRLM) with a significant correlation with the number of CRLM [17,18]. Up-regulation of PER3 is correlated with a better clinical outcome in patients with acute leukemia [19]. PER3 was also found to be associated with survival outcome independent of established clinicopathological parameters in breast cancer [20]. Downregulated PER3 correlated with larger tumor size and lower expression of PER3 correlated with deeper tumor invasion [21]. Some observations supporting the tumor suppressor role of PER3 in NSCLC were found by Liu B et al. They found that the positive rate of Per3 was reduced in human lung cancer samples as measured by immunohistochemical staining. Loss of PER3 was correlated with poor differentiation, tumor status, high p-TNM stage and lymph node metastasis. In addition, patients with lower expression of PER3 had a shorter survival time than those with higher expression, indicating that PER3 may serve as a novel prognostic biomarker of NSCLC [22]. Zhang F et al found that overexpression of PER3 inhibits self-renewal capability and chemoresistance of colorectal cancer stem-like cells via inhibition of notch and β-catenin signaling [23]. PER3 knockdown had an inhibitory effect on the apoptosis of human gingival cancer cells induced by cisplatin treatment, suggesting that PER3 has pro-apoptotic effects in human gingival cancer [24]. The disruption of PER3 in various cancers is involved in genetic polymorphisms. Qu F et al found that SNP (rs228729) from PER3 was significantly associated with overall survival of gastric cancer in the training set, and verified in the validation set and pooled analysis [25]. And PER3 rs10462020 variant showed significant difference in overall survival between patients containing mutated genotypes and those with non-mutated genotypes [26].
PER3 is also regulated by transcript factor, non-coding RNA or epigenetic modifications. Hong Z et al found that PER3 was negatively regulated by miR-103 [27]. Epigenetic regulation on PER3 also frequently observed. Twenty-one loci located in the circadian genes such as PER3 were found to be significantly hypomethylated among nightshift workers, most of which were located in a CpG island and near the transcription start site of the gene. Methylation changes in PER3 may lead to altered expression of genes that were associated with cancer [28]. By genome-wide methylation profiling and vertical integration with array-based comparative genomic hybridization, as well as expression data from a cohort of well-characterized human hepatocellular carcinomas (HCCs), Neumann O et al revealed 537 CpG sites were hypermethylated in the tumor DNA, including PER3. And they confirmed that PER3 was down-regulated in human HCCs, while 5-Aza treatment restored PER3 expression in HCC cell lines, indicating that promoter hypermethylation was indeed responsible for gene silencing [29]. These results strongly supports our findings that PER3 is hypermethylated in NSCLC, which is reversed by 5-Aza. Similar results were also found in chronic myeloid leukemia (CML) that downregulated PER3 expression in CML was correlated with the inactivation of PER3 by methylation [30].
Summary, recent study demonstrates that PER3 functions as a tumor suppressor in lung cancer, and the decrease of PER3 induced by promoter hypermethylation can be restored by demethylation drug 5-Aza. These findings provide a promising target and a novel strategy to control cancer progression.
Acknowledgements
This study was supported by Guangdong Provincial Natural Science Foundation of China (2015A030313773).
Disclosure of conflict of interest
None.
References
- 1.Zakaria N, Satar NA, Abu HN, Ngalim SH, Yusoff NM, Lin J, Yahaya BH. Targeting lung cancer stem cells: research and clinical impacts. Front Oncol. 2017;7:80. doi: 10.3389/fonc.2017.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersen ME, Black MB, Campbell JL, Pendse SN, Clewell IH, Pottenger LH, Bus JS, Dodd DE, Kemp DC, McMullen PD. Combining transcriptomics and PBPK modeling indicates a primary role of hypoxia and altered circadian signaling in dichloromethane carcinogenicity in mouse lung and liver. Toxicol Appl Pharmacol. 2017;332:149–158. doi: 10.1016/j.taap.2017.04.002. [DOI] [PubMed] [Google Scholar]
- 3.Wang RH, Zhao T, Cui K, Hu G, Chen Q, Chen W, Wang XW, Soto-Gutierrez A, Zhao K, Deng CX. Negative reciprocal regulation between Sirt1 and Per2 modulates the circadian clock and aging. Sci Rep. 2016;6:28633. doi: 10.1038/srep28633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benna C, Helfrich-Forster C, Rajendran S, Monticelli H, Pilati P, Nitti D, Mocellin S. Genetic variation of clock genes and cancer risk: a field synopsis and meta-analysis. Oncotarget. 2017;8:23978–23995. doi: 10.18632/oncotarget.15074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Truong KK, Lam MT, Grandner MA, Sassoon CS, Malhotra A. Timing matters: circadian rhythm in sepsis, obstructive lung disease, obstructive sleep apnea, and cancer. Ann Am Thorac Soc. 2016;13:1144–1154. doi: 10.1513/AnnalsATS.201602-125FR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vasu VT, Cross CE, Gohil K. Nr1d1, an important circadian pathway regulatory gene, is suppressed by cigarette smoke in murine lungs. Integr Cancer Ther. 2009;8:321–328. doi: 10.1177/1534735409352027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Papagiannakopoulos T, Bauer MR, Davidson SM, Heimann M, Subbaraj L, Bhutkar A, Bartlebaugh J, Vander HM, Jacks T. Circadian rhythm disruption promotes lung tumorigenesis. Cell Metab. 2016;24:324–331. doi: 10.1016/j.cmet.2016.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gibbs J, Ince L, Matthews L, Mei J, Bell T, Yang N, Saer B, Begley N, Poolman T, Pariollaud M, Farrow S, DeMayo F, Hussell T, Worthen GS, Ray D, Loudon A. An epithelial circadian clock controls pulmonary inflammation and glucocorticoid action. Nat Med. 2014;20:919–926. doi: 10.1038/nm.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pekovic-Vaughan V, Gibbs J, Yoshitane H, Yang N, Pathiranage D, Guo B, Sagami A, Taguchi K, Bechtold D, Loudon A, Yamamoto M, Chan J, van der Horst GT, Fukada Y, Meng QJ. The circadian clock regulates rhythmic activation of the NRF2/glutathione-mediated antioxidant defense pathway to modulate pulmonary fibrosis. Genes Dev. 2014;28:548–560. doi: 10.1101/gad.237081.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Logan RW, Zhang C, Murugan S, O’Connell S, Levitt D, Rosenwasser AM, Sarkar DK. Chronic shift-lag alters the circadian clock of NK cells and promotes lung cancer growth in rats. J Immunol. 2012;188:2583–2591. doi: 10.4049/jimmunol.1102715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hrushesky WJ, Grutsch J, Wood P, Yang X, Oh EY, Ansell C, Kidder S, Ferrans C, Quiton DF, Reynolds J, Du-Quiton J, Levin R, Lis C, Braun D. Circadian clock manipulation for cancer prevention and control and the relief of cancer symptoms. Integr Cancer Ther. 2009;8:387–397. doi: 10.1177/1534735409352086. [DOI] [PubMed] [Google Scholar]
- 12.Couto P, Miranda D, Vieira R, Vilhena A, De Marco L, Bastos-Rodrigues L. Association between CLOCK, PER3 and CCRN4L with nonsmall cell lung cancer in Brazilian patients. Mol Med Rep. 2014;10:435–440. doi: 10.3892/mmr.2014.2224. [DOI] [PubMed] [Google Scholar]
- 13.Shostak A. Circadian clock, cell division, and cancer: from molecules to organism. Int J Mol Sci. 2017;18 doi: 10.3390/ijms18040873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Y, Sun N, Lu C, Bei Y, Qian R, Hua L. Upregulation of circadian gene ‘hClock’ contribution to metastasis of colorectal cancer. Int J Oncol. 2017;50:2191–2199. doi: 10.3892/ijo.2017.3987. [DOI] [PubMed] [Google Scholar]
- 15.Oh JH, Lee JH, Han DH, Cho S, Lee YJ. Circadian clock is involved in regulation of hepatobiliary transport mediated by multidrug resistance-associated protein 2. J Pharm Sci. 2017;106:2491–2498. doi: 10.1016/j.xphs.2017.04.071. [DOI] [PubMed] [Google Scholar]
- 16.Kiessling S, Beaulieu-Laroche L, Blum ID, Landgraf D, Welsh DK, Storch KF, Labrecque N, Cermakian N. Enhancing circadian clock function in cancer cells inhibits tumor growth. BMC Biol. 2017;15:13. doi: 10.1186/s12915-017-0349-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huisman SA, Ahmadi AR, IJzermans JN, Verhoef C, van der Horst GT, de Bruin RW. Disruption of clock gene expression in human colorectal liver metastases. Tumour Biol. 2016;37:13973–13981. doi: 10.1007/s13277-016-5231-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang X, Yan D, Teng M, Fan J, Zhou C, Li D, Qiu G, Sun X, Li T, Xing T, Tang H, Peng X, Peng Z. Reduced expression of PER3 is associated with incidence and development of colon cancer. Ann Surg Oncol. 2012;19:3081–3088. doi: 10.1245/s10434-012-2279-5. [DOI] [PubMed] [Google Scholar]
- 19.Yang MY, Lin PM, Hsiao HH, Hsu JF, Lin HY, Hsu CM, Chen IY, Su SW, Liu YC, Lin SF. Up-regulation of PER3 expression is correlated with better clinical outcome in acute leukemia. Anticancer Res. 2015;35:6615–6622. [PubMed] [Google Scholar]
- 20.Cadenas C, van de Sandt L, Edlund K, Lohr M, Hellwig B, Marchan R, Schmidt M, Rahnenfuhrer J, Oster H, Hengstler JG. Loss of circadian clock gene expression is associated with tumor progression in breast cancer. Cell Cycle. 2014;13:3282–3291. doi: 10.4161/15384101.2014.954454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hsu CM, Lin SF, Lu CT, Lin PM, Yang MY. Altered expression of circadian clock genes in head and neck squamous cell carcinoma. Tumour Biol. 2012;33:149–155. doi: 10.1007/s13277-011-0258-2. [DOI] [PubMed] [Google Scholar]
- 22.Liu B, Xu K, Jiang Y, Li X. Aberrant expression of Per1, Per2 and Per3 and their prognostic relevance in non-small cell lung cancer. Int J Clin Exp Pathol. 2014;7:7863–7871. [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang F, Sun H, Zhang S, Yang X, Zhang G, Su T. Overexpression of PER3 inhibits self-renewal capability and chemoresistance of colorectal cancer stem-like cells via inhibition of notch and beta-catenin signaling. Oncol Res. 2017;25:709–719. doi: 10.3727/096504016X14772331883976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sato F, Wu Y, Bhawal UK, Liu Y, Imaizumi T, Morohashi S, Kato Y, Kijima H. PERIOD1 (PER1) has anti-apoptotic effects, and PER3 has proapoptotic effects during cisplatin (CDDP) treatment in human gingival cancer CA9-22 cells. Eur J Cancer. 2011;47:1747–1758. doi: 10.1016/j.ejca.2011.02.025. [DOI] [PubMed] [Google Scholar]
- 25.Qu F, Qiao Q, Wang N, Ji G, Zhao H, He L, Wang H, Bao G. Genetic polymorphisms in circadian negative feedback regulation genes predict overall survival and response to chemotherapy in gastric cancer patients. Sci Rep. 2016;6:22424. doi: 10.1038/srep22424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gutierrez-Monreal MA, Villela L, Baltazar S, Perfecto-Avalos Y, Cardineau GA, Scott SP. A PER3 polymorphism is associated with better overall survival in diffuse large B-cell lymphoma in Mexican population. Cancer Biomark. 2015;15:699–705. doi: 10.3233/CBM-150511. [DOI] [PubMed] [Google Scholar]
- 27.Hong Z, Feng Z, Sai Z, Tao S. PER3, a novel target of miR-103, plays a suppressive role in colorectal cancer in vitro. BMB Rep. 2014;47:500–505. doi: 10.5483/BMBRep.2014.47.9.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bhatti P, Zhang Y, Song X, Makar KW, Sather CL, Kelsey KT, Houseman EA, Wang P. Nightshift work and genome-wide DNA methylation. Chronobiol Int. 2015;32:103–112. doi: 10.3109/07420528.2014.956362. [DOI] [PubMed] [Google Scholar]
- 29.Neumann O, Kesselmeier M, Geffers R, Pellegrino R, Radlwimmer B, Hoffmann K, Ehemann V, Schemmer P, Schirmacher P, Lorenzo BJ, Longerich T. Methylome analysis and integrative profiling of human HCCs identify novel protumorigenic factors. Hepatology. 2012;56:1817–1827. doi: 10.1002/hep.25870. [DOI] [PubMed] [Google Scholar]
- 30.Yang MY, Chang JG, Lin PM, Tang KP, Chen YH, Lin HY, Liu TC, Hsiao HH, Liu YC, Lin SF. Downregulation of circadian clock genes in chronic myeloid leukemia: alternative methylation pattern of hPER3. Cancer Sci. 2006;97:1298–1307. doi: 10.1111/j.1349-7006.2006.00331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]


