Abstract
Glioma is one of the most malignant tumor types worldwide. Despite great efforts made in surgery, imaging, chemotherapy, and radiation, the overall prognosis for patients with glioma remains poor. The pathogenesis of glioma is an urgent problem that must be solved. Tripartite motif-containing protein 37 (TRIM37) is an E3 ubiquitin ligase that may be involved in the tumorigenesis of several types of cancer; however, its expression pattern and biological functions in glioma remain unknown. Our previous studies indicated that the expression levels of TRIM37 were upregulated in glioma samples and were associated with a higher glioma grade. The present study demonstrates that suppression of TRIM37 by RNA-mediated interference inhibits the growth of glioma cells in vitro, which is associated with induction of apoptosis and cell cycle arrest, and with inhibition of migration and invasion. These results provide insights into understanding the role of TRIM37 in regulating the biological behavior of glioma cells and may indicate TRIM37 as a candidate target for the treatment of glioma.
Keywords: TRIM37, glioma, siRNA, proliferation, migration, invasion, apoptosis
Introduction
Gliomas are the most common primary brain tumors diagnosed in adults, and are responsible for ~70% of adult malignant primary brain tumors [1]. Cancer research has made great strides in the treatment of most cancer types; however, despite the advances made in detection, radiation therapy, chemotherapy, and surgical techniques, the median survival for patients with glioma is still only ~14 months [2,3]. Since the overall prognosis of current therapies is poor, an understanding of gliomas at the molecular level is urgently required for the future development of more effective treatments for this malignant disease.
Tripartite motif-containing protein 37 (TRIM37) is a member of the TRIM subfamily (comprising RING, B-Box and coiled-coil domains) of zinc-finger proteins that is located on chromosome 17q22-23 [4]. Numerous previous studies have investigated the function of TRIM37 in various physiological processes and diseases. One study indicated that TRIM37 had ubiquitin E3 ligase activity [5]. Another study used small interfering RNA (siRNA)-based functional genomics in HeLa cells to demonstrated that TRIM37 prevented centriole reduplication during centriole biogenesis [6]. TRIM37 was also identified as an asthma-related gene in gene-expression profiling of peripheral blood mononuclear cells [7]. In addition, mutations in TRIM37 resulted in mulibrey nanism, a rare autosomal recessive disorder associated with severe primordial growth retardation and multi-organ involvement, which leads to a failure of sexual maturation and infertility [8,9]. Furthermore, human TRIM37 was demonstrated to have potent anti-human immunodeficiency virus type 1 activity by interfering with viral DNA synthesis [10]. Increasing evidence indicated that TRIM37 was involved in the tumorigenesis of several types of cancer. For example, TRIM37 was reported to be involved in the transformation of follicular lymphoma to a more aggressive disease, which is associated with rapid progression and mortality. A previous study also revealed that TRIM37 is an oncogenic H2A ubiquitin ligase, which is overexpressed in breast cancer and promotes transformation via silencing of tumor suppressors and other genes [11]. Another study indicated that overexpression of TRIM37 could promote cell migration and metastasis in hepatocellular carcinoma by activating Wnt/β-catenin signaling [12]. A recent study demonstrated that TRIM37 promoted the growth and migration of pancreatic cancer cells [13]. These studies indicated that TRIM37 is involved in cancer initiation and progression. However, the biological role and the molecular mechanisms of TRIM37 in glioma pathogenesis remain unclear.
The aim of the present study was to examine the expression of TRIM37 in human glioma tissues and to investigate the functions of TRIM37 in the progression of glioma. TRIM37 was upregulated in glioma tissues. The suppression of TRIM37 inhibited glioma cell proliferation, migration, and invasion and induced apoptosis. The results of the present study provide further evidence for the potential in targeting glioma growth through the TRIM37 pathway.
Material and methods
Tissue samples
Glioma samples (n=124), including 32 low-grade glioma (32 grade II tumors) and 92 high-grade glioma (38 grade III tumors and 54 grade IV tumors) samples were obtained from patients with primary glioma who underwent surgical treatment at the Department of Neurosurgery (Qilu Hospital, Shandong University, Jinan, China). A total of 20 normal brain tissue samples were collected from patients undergoing internal decompression surgery following severe traumatic brain injury and were examined to verify the absence of tumor. For all patients, the histological types and grade of the gliomas were evaluated by two experienced pathologists, according to the 2007 World Health Organization (WHO) Classification of Tumors of the Central Nervous System [14]. The present study protocol was approved by the Ethics Committee of Qilu Hospital. Informed consent was obtained from all patients included in the study. All patients had a well-documented clinical history.
Cell lines and cell culture
The human glioma cell line U87 was obtained from the Chinese Academy of Sciences (Beijing, China) and cultured in Dulbecco’s modified Eagle’s medium (DMEM; Hyclone, Beijing, China) supplemented with 10% fetal bovine serum (FBS; Hyclone, Beijing, China), 100 U/ml penicillin and 100 μg/ml streptomycin in a humidified chamber containing 5% CO2 at 37°C. U87 glioma cells were randomly divided into three groups: control (non-transfected U87 glioma cells), siRNA-control (transfected with scrambled negative control) and siRNA-TRIM37 (transfected with siRNA targeting TRIM37).
RNA isolation and reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from frozen samples using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) according to the manufacturer’s protocol. cDNA was generated by reverse transcription of total RNA (1 μg) with primer oligonucleotides (RiboBio, Guangzhou, China). qPCR reactions were conducted in a 10 μl reaction volume with SYBR Green I (TaKaRa, Dalian, China), with a 1:25 dilution of the cDNA and 20 nmol primers as previously described [15]. The reactions were performed using a Lightcycler 2.0 instrument (Roche Life Science, Shanghai, China). β-actin was used as a reference gene. Primer sequences were as follows: TRIM37 forward, 5’-TCAGCTGTATTAGGCGCTGG-3’, reverse, 5’-ACTTCTTCTGCCCAACGACA-3’; and β-actin forward, 5’-CGTTGACATCCGTAAAGACC-3’ and reverse, 5’-TAGAGCCACCAATCCACACA-3’. TRIM37 mRNA levels were calculated based on the quantification cycle values and the relative quantification was determined by normalizing expression to β-actin [16]. All experiments were performed in triplicate.
Western blot analysis
U87 cell cultures and brain tissue samples were lysed in radioimmunoprecipitation assay buffer (Beyotime Institute of Biotechnology, Haimen, China) containing 50 mmol Tris (pH 7.4), 150 mmol NaCl, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS and protease inhibitor cocktail with 1 mmol phenylmethanesulfonyl fluoride (Beyotime Institute of Biotechnology). Lysates were centrifuged at 12,000× g for 15 min and the protein concentration of the supernatant was quantified using the bicinchoninic acid method (Beyotime Institute of Biotechnology). Sample proteins (20 μg) were separated by 10% SDS-PAGE and transferred to a polyvinylidene difluoride membrane by electroblot apparatus. Membranes were blocked with 5% nonfat milk in PBS-Tween-20 (0.5%) for 1 h at room temperature, followed by incubation with primary polyclonal antibodies overnight at 4°C. The membranes were washed three times with TBS containing 0.1% Tween-20 and incubated with horseradish peroxidase-conjugated secondary goat anti-rabbit antibody (A0545, 1:8000, Sigma-Aldrich, Shanghai, China) for 2 h at room temperature. Immunoreactive bands were visualized using the Immobilon Western Chemiluminescent HRP Substrate kit (EMD Millipore, Billerica, MA, USA), according to the manufacturer’s protocol. Densitometric analysis of protein expression was conducted with ImageJ 1.44 (National Institutes of Health, Bethesda, MD, USA) and standardized against β-actin. The following primary antibodies were used: TRIM37 (SAB2702009, 1:1000, Sigma-Aldrich, Shanghai, China), B-cell lymphoma 2 (Bcl-2, 4223S, 1:500, Cell Signaling Technology, Shanghai, China), Bcl-2-associated X protein (Bax, 5023S, 1:500, Cell Signaling Technology, Shanghai, China), matrix metalloproteinase 2 (MMP2, 13132S, 1:800, Cell Signaling Technology, Shanghai, China), matrix metalloproteinase 9 (MMP9, 13667S, 1:800, Cell Signaling Technology, Shanghai, China), cleaved-caspase 3 (9664S, 1:500, Cell Signaling Technology, Shanghai, China).
Immunohistochemistry
Tissue samples were fixed in 10% buffered formalin and embedded in paraffin. Serial sections of paraffinized tissues were cut in 4 μm increments. Slides were stained according to the manufacturer’s protocol (DAB detection kit, ZSGB-BIO, Beijing, China). Briefly, slides were deparaffinized using xylene and rehydrated through an ethanol series (100%, 95%, 90%, 80%, 70%, separately) to water. The dewaxed slides underwent antigen retrieval by heating in a microwave in citrate buffer for 30 min, followed by cooling to room temperature. Hydrogen peroxide (3%) was added to the slides to inhibit endogenous peroxidase activity. The slides were blocked with 10% normal goat serum (CWBIO, Beijing, China) for 1 h at 37°C and incubated with primary rabbit polyclonal anti-TRIM37 antibody (SAB2702009, 1:500, Sigma-Aldrich, Shanghai, China) overnight at 4°C, followed by incubation with anti-rabbit secondary antibody (PV-9000-D, ZSGB-BIO, Beijing, China) for 2 h at room temperature. Finally, 3,3’-diaminobenzdine was used as a chromogen, and the slides were counterstained with hematoxylin for 3 min at room temperature. Stained slides were dehydrated and mounted with neutral balsam according to the laboratory protocol. Staining with PBS instead of the primary antibody was used as the negative control.
Evaluation of immunohistochemistry
Stained slides were scored by two independent pathologists who were blinded to the clinical information. Ten low power fields (X100) were selected randomly and cells were counted at high power (X400) using a microscope (Olympus, Japan). TRIM37-positive cells were scored as follows: 0, no staining; 1, <25% cells stained; 2, 25-75% cells stained; and 3, >75% cells stained. Staining intensity was categorized as follows: 0, negative; 1, weak; 2, moderate; and 3, strong. Overall staining scores (ranging between 0 and 9) were obtained by multiplying the intensity score and the percentage score. The staining pattern of the slides was defined as follows: 0, -; 1-3, +; 4-6, ++; and 6-9, +++. Samples with scores >4 were regarded as positive.
Transfection
U87 cells were cultured in DMEM supplemented with 10% FBS in a humidified atmosphere of 5% CO2 at 37°C in 60 mm flasks. When cells reached 70% confluence, they were transfected with siRNA targeting TRIM37. siRNA sequences were as follows: TRIM37, 5’-GAGAGUCUGAAGGAUACUATT-3’ and 5’-UAGUAUCCUUCAGACUCUCTT-3’; scrambled negative control, 5’-UUCUCCGAACGUGUCACGUTT-3’ and 5’-ACGUGACACGUUCGGAGAATT-3’. RNA duplexes were synthesized by Shanghai Genepharma Co., Ltd. (Shanghai, China). Transfection of siRNAs was performed using Lipofectamine 2000 (Thermo Fisher Scientific, Inc.) according to the manufacturer’s protocol. Transfection efficiency was verified by Western blot analysis.
Cell proliferation assay
U87 cells were seeded in 96-well culture plates at a density of 3,000 cells/well, and cell proliferation was analyzed using a Cell Counting Kit-8 (CCK-8; Beyotime Institute of Biotechnology) at 24, 48, and 72 h after transfection. At the designated time, 10 μl CCK-8 solution was added to each well, and the cells were incubated for another 1.5 h in a humidified incubator at 37°C. Optical density was measured at 450 nm using a microplate reader (Bio-Rad Laboratories, Inc., Hercules, CA, USA). Each assay was performed in triplicate.
Cell apoptosis analysis
Cell apoptosis was evaluated with an Annexin V-fluorescein isothiocyanate (FITC)/propidium iodide (PI) kit (Thermo Fisher Scientific, Inc.), according to the manufacturer’s protocol. U87 cells were grown in DMEM supplemented with 10% FBS in a humidified atmosphere with 5% CO2 at 37°C. When cells reached 70% confluence, they were transfected with siRNA targeting TRIM37. After transfection, attached cells were collected and washed twice with PBS. Binding buffer (400 μl), Annexin V-FITC (5 μl) and PI (5 μl) were sequentially added to the cell suspension. Following 15 min incubation in the dark at room temperature, cells were subjected to a flow cytometer (BD Biosciences, San Jose, CA, USA). At least three independent experiments were carried out.
Cell cycle analysis
PI (Beyotime Institute of Biotechnology) staining was used for cell cycle analysis. Briefly, transfected cells were collected by the trypsin method, washed three times with cold PBS, and fixed overnight at 4°C in 70% ethanol. Cells were washed with PBS and 20 μl RNase A was added, followed by incubation at 37°C for 30 min to hydrolyze RNA. Cells were subsequently stained with PI in the dark for 30 min at 37°C, and the cell cycle was evaluated by flow cytometry. The experiments were repeated at least three times independently.
In vitro cell migration and invasion assay
Cell migration and invasion were determined using a modified Boyden chamber assay. Briefly, cells in each group (5×104 cells/well) in 0.1 ml serum-free DMEM were separately added to the wells of an 8 mm pore membrane Boyden chamber (Corning, Inc., Corning, NY, USA) that was either uncoated (for the migration assay) or coated with Matrigel (BD Biosciences; for the invasion assay). DMEM containing 10% FBS was added to the lower chamber and cells were allowed to invade for 12 hours at 37°C. Any cells that did not penetrate the filters were removed by scrubbing the filters with cotton swabs. Chambers were fixed for 30 min at room temperature with 4% formaldehyde in PBS, stained in 0.1% crystal violet for 15 min at room temperature, and rinsed in water. Cells that migrated to the bottom surface of the filter were counted under a light microscope. Assays were repeated three times using triplicate wells.
Statistical analysis
Data are expressed as the mean ± SD. Data were analyzed by two-tailed Student’s t-test and one-way analysis of variance, followed by Dunnett’s test for multiple comparisons of the means. χ2 test was used to analyze categorical data. P<0.05 was considered to indicate a statistically significant difference.
Results
TRIM37 expression is markedly increased in glioma tissues
qRT-PCR was performed to determine the difference in TRIM37 expression between glioma tissues and normal brain tissues. Glioma tissue samples had a significantly higher expression of TRIM37 compared to normal brain samples (Figure 1A). Western blot and immunohistochemistry analyses were conducted to determine whether an upregulation of TRIM37 mRNA expression resulted in a commensurate increase in TRIM37 protein expression. Western blot analysis revealed that the levels of TRIM37 expression in glioma samples were markedly higher than those in normal brain samples (Figure 1B). TRIM37 immunoreactivity was stronger in glioma samples than in normal brain samples. Immunohistochemical staining revealed TRIM37 to be localized in the cytoplasm of glial cells and glioma cells (Figure 2). Approximately 83.1% (103 of 124 samples) of glioma samples demonstrated increased levels of TRIM37 expression, whereas only 15% (3 of 20 samples) of normal brain samples demonstrated increased expression (Table 1). These results indicate that upregulated expression of TRIM37 is significantly associated with glioma.
Figure 1.
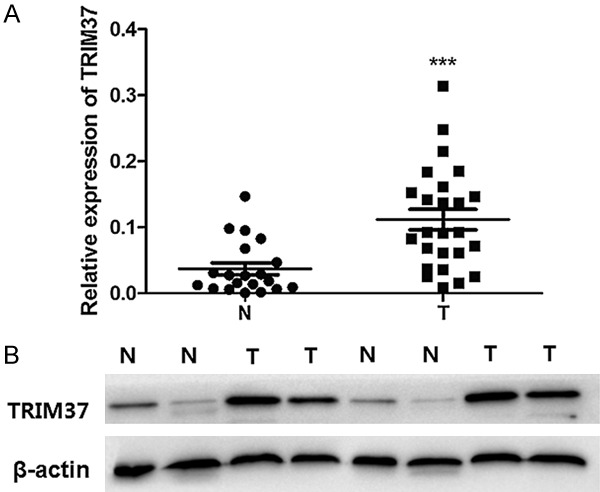
Expression of TRIM37 in glioma and normal brain tissues. A. Quantitative reverse transcription-polymerase chain reaction analysis of TRIM37 mRNA expression levels. B. Western blot analyses of glioma samples and normal brain samples; β-actin served as an internal control. Data are presented as the mean ± standard deviation of three independent experiments (***P<0.001 vs. N). N, normal brain tissue; T, glioma tissue; TRIM37, tripartite motif-containing protein 37.
Figure 2.
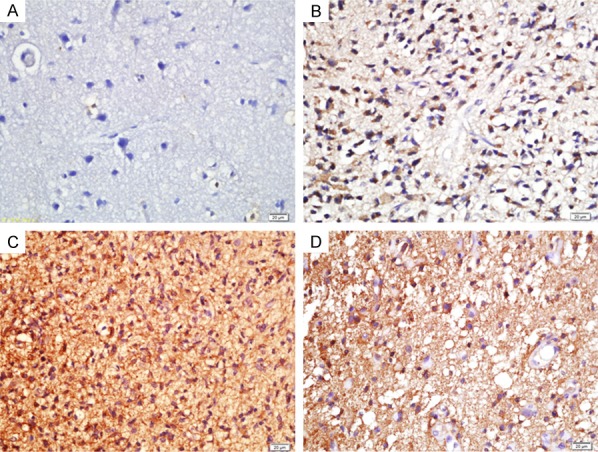
TRIM37 expression in different grades of glioma tissues. Immunohistochemical analysis indicated that an upregulation of TRIM37 in gliomas was closely associated with glioma grades. A. Normal brain tissue; B. Grade II glioma; C. Grade III glioma; D. Grade IV glioma. Scale bar, 20 μm. TRIM37, tripartite motif-containing protein 37.
Table 1.
TRIM37 protein expression in glioma tissues and normal brain tissues was assessed by immunohistochemistry
| Tissue | Total | -/+ (%) | ++/+++ (%) | P-value |
|---|---|---|---|---|
| Normal | 20 | 17 (85.0) | 3 (15.0) | |
| Glioma | 124 | 21 (16.9) | 103 (83.1) | P<0.05 |
TRIM37, tripartite motif-containing protein 37.
An association between the expression of TRIM37 and clinical pathological features was also investigated (Table 2). TRIM37 expression was high in 62.5% of low-grade glioma cases (grade II tumors) and in an average of 82.45% of high-grade gliomas (grade III and grade IV tumors). These data indicate that high expression levels of TRIM37 are significantly associated with high-grade glioma. However, there was no association between the level of TRIM37 expression and either age or gender (Table 2).
Table 2.
Association between TRIM37 expression and clinicopathological features of patients with glioma
| Factor | Total | -/+ (%) | ++/+++ (%) | P-value |
|---|---|---|---|---|
| Age (years) | ||||
| <50 | 67 | 32 (47.8) | 35 (52.2) | |
| ≥50 | 57 | 26 (45.6) | 31 (54.4) | P>0.05 |
| Gender | ||||
| Male | 69 | 31 (44.9) | 38 (55.1) | |
| Female | 55 | 27 (49.1) | 28 (50.9) | P>0.05 |
| WHO grade | ||||
| II | 32 | 13 (37.5) | 19 (62.5) | |
| III | 38 | 7 (18.4) | 31 (81.6) | P<0.05a |
| IV | 54 | 9 (16.7) | 45 (83.3) | P<0.05a |
vs. WHO grade II.
TRIM37, tripartite motif-containing protein 37; WHO, World Health Organization.
TRIM37 knockdown inhibits glioma cell growth
siRNA-TRIM37 was transiently transfected into U87 glioma cells and the transfection efficiency was determined by Western blot analysis. We confirmed downregulation of the TRIM37 protein level with TRIM37 siRNA transfection as compared with control (Figure 3). The CCK-8 assay indicated that cell proliferation was suppressed by siRNA-TRIM37 at each time point, and the proliferation ratio was decreased to 55.3% at 72 h post-transfection, compared with the control group (Figure 4).
Figure 3.
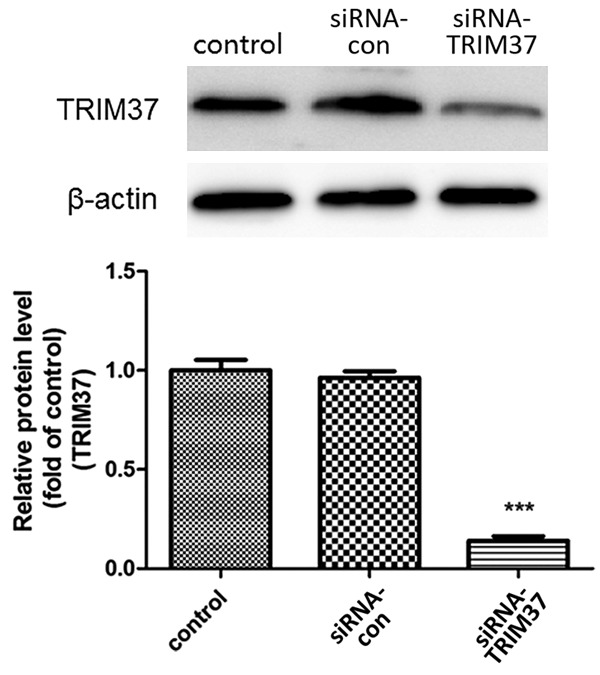
Transfection efficiency of siRNA-TRIM37. TRIM37 protein expression was determined by Western blot analysis. Data are presented as the mean ± standard deviation of three independent experiments (***P<0.001 vs. control). siRNA, small interfering RNA; siRNA-con, control siRNA; TRIM37, tripartite motif-containing protein 37.
Figure 4.
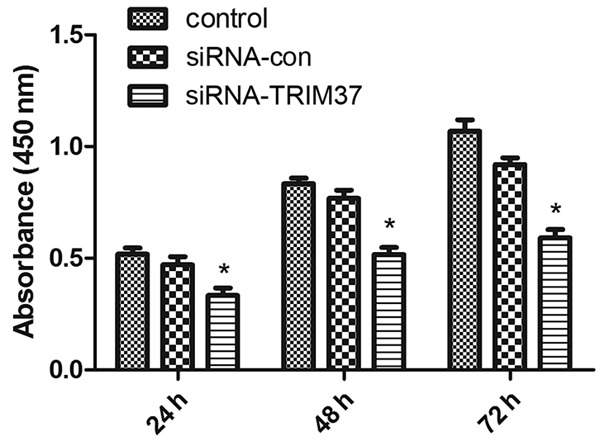
Anti-proliferative effects of siRNA-TRIM37. Inhibition of cell growth by siRNA knockdown of TRIM37 was in a time-dependent manner and the proliferation rate was decreased to 55.3% of the control at 72 h post-transfection. (*P<0.05 vs. control). siRNA, small interfering RNA; siRNA-con, control siRNA; TRIM37, tripartite motif-containing protein 37.
TRIM37 knockdown induces apoptosis in glioma cells
The effects of TRIM37 knockdown on apoptosis in glioma cells were evaluated by flow cytometry using Annexin V-FITC/PI double staining. The total apoptotic rate reached 32.7% in the siRNA-TRIM37 group compared with 1.7 and 1.9% for the control and siRNA-con groups, respectively (Figure 5A). Several known regulators of apoptosis were also examined (Figure 5B), which revealed that TRIM37 knockdown increased the ratio of Bax/Bcl-2 (Figure 5C). In addition, cleaved-caspase 3 activity was significantly upregulated in the siRNA-TRIM37 group compared with the siRNA-con group (Figure 5D). These results indicated an anti-apoptotic role of TRIM37 in glioma cells.
Figure 5.
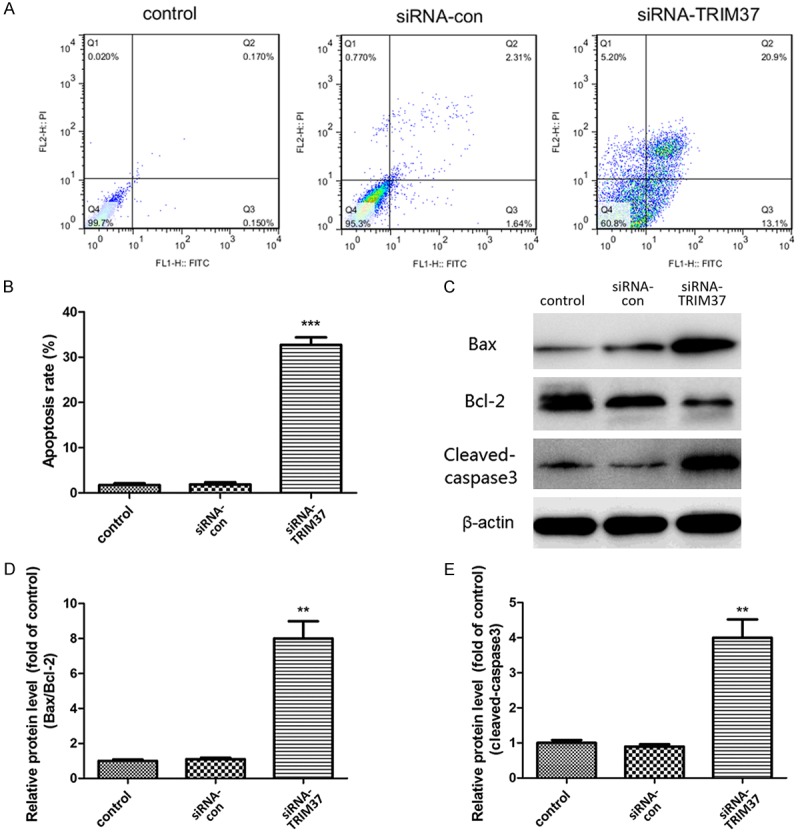
Evaluation of apoptotic rate of U87 cells. (A, B) The proportion of apoptotic cells was analyzed by flow cytometry. (C) Western blot analysis of the effect of siRNA-TRIM37 transfection on (D) the ratio of Bax/Bcl-2, and (E) the protein expression of cleaved-caspase 3 in U87 cells. β-actin served as an internal control. Data are presented as the mean ± standard deviation of three independent experiments (**P<0.01 vs. control; ***P<0.001 vs. control). siRNA, small interfering RNA; siRNA-con, control siRNA; TRIM37, tripartite motif-containing protein 37.
TRIM37 knockdown induces glioma cell cycle arrest
To examine if the loss of TRIM37 expression induced cell cycle arrest, PI staining was used to analyze cell cycle progression by flow cytometry. TRIM37 knockdown increased the population of cells in the S phase and decreased the number of cells in the G2/M phase (Figure 6). The proportion of cells in the S phase increased to 41.5% in the siRNA-TRIM37 group compared with 18.3 and 19.3% in the control and siRNA-control groups, respectively (Figure 6A). The proportion of cells in the G2/M phase decreased from 14.1% in the siRNA-con group to 5.7% in the siRNA-TRIM37 group. These results suggest that knockdown of TRIM37 expression in glioma cells suppressed cell cycle transition, thus inhibiting cell proliferation.
Figure 6.
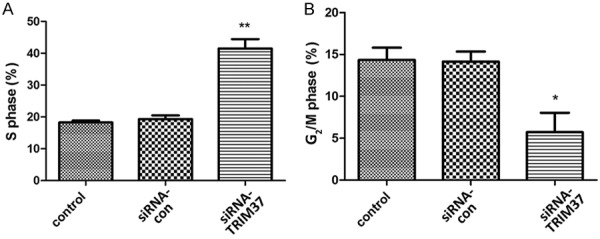
Cell cycle analysis of U87 cells. U87 cells were stained with propidium iodide and subjected to flow cytometry for cell cycle analysis. A. TRIM37 knockdown resulted in an increase in the population of cells in S phase compared with the control. B. TRIM37 knockdown resulted in a decrease in the number of cells in G2/M phase. Data are presented as the mean ± standard deviation of three independent experiments (*P<0.05 vs. control; **P<0.01 vs. control). siRNA, small interfering RNA; siRNA-con, control siRNA; TRIM37, tripartite motif-containing protein 37.
TRIM37 knockdown inhibits glioma cell migration and invasion
Transwell migration and invasion assays were performed in Boyden chambers to evaluate the effect of TRIM37 on U87 cell mobility (Figure 7). Decreased expression of TRIM37 caused a marked reduction in the migration of U87 cells (Figure 7A and 7B). The in vitro invasion assay indicated that the invasive ability of TRIM37-knockdown cells was significantly inhibited (Figure 7A and 7C). Furthermore, the expression of MMP2 and MMP9 was significantly decreased in TRIM37-knockdown U87 cells (Figure 7D and 7E). These data suggest that knockdown of TRIM37 inhibited glioma cell migration and invasion.
Figure 7.
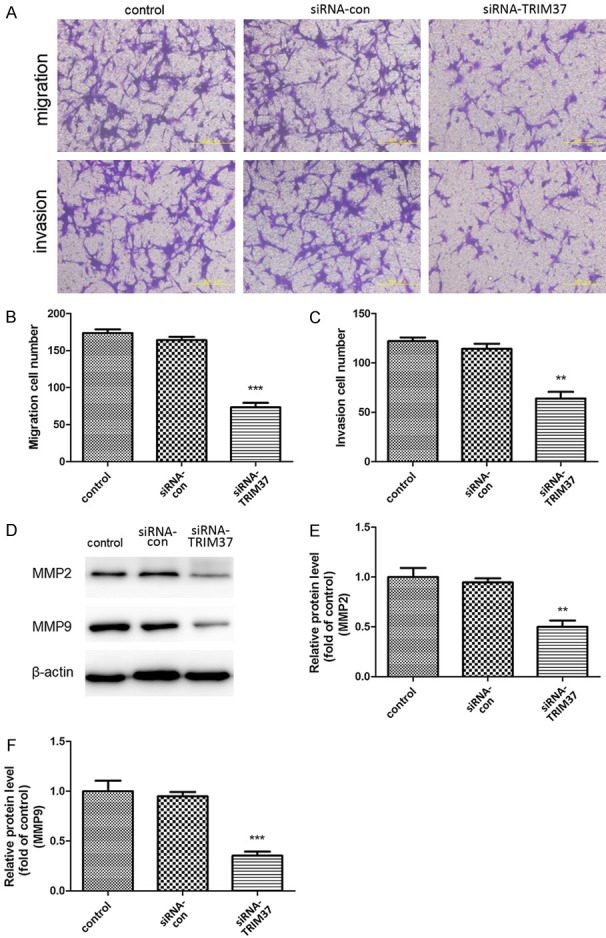
TRIM37 knockdown affects cell motility. (A-C) TRIM37 knockdown inhibited U87 cell migration and invasion. Scale bar, 200 μm. (D) Western blot and densitometric analysis of the expression levels of (E) MMP2 and (F) MMP9 proteins. Data are presented as the mean ± standard deviation of three independent experiments (**P<0.01 vs. control; ***P<0.001 vs. control). MMP, matrix metalloproteinase; siRNA, small interfering RNA; siRNA-con, control siRNA; TRIM37, tripartite motif-containing protein 37.
Discussion
Gliomas are the most common form of malignant brain tumors in adults. They are tumors of the neoplastic glial cells and are categorized by the WHO as astrocytoma, oligodendroglioma, mixed oligoastrocytoma and ependymoma [17]. Although the cause of glioma is unknown, complex genetic abnormalities in combination with unknown environmental factors may predispose individuals to glioma. The current standard of care for newly diagnosed patients with glioma includes surgery to excise as much of the tumor as safely possible, and a combination of radiotherapy and chemotherapy [3]; however, therapeutic efficacy is not satisfactory [2]. The present study hypothesized that the characterization of a novel molecular biomarker that is significantly upregulated in glioma tissues (compared with normal brain tissues) may aid in the identification of a novel therapeutic or diagnostic marker. Results from the present study describe a potential oncogenic role for TRIM37.
qRT-PCR analyses demonstrated that the mRNA expression levels of TRIM37 were significantly increased in the glioma samples examined compared with the normal brain samples. This is consistent with previous studies [12,13,18] that demonstrated that TRIM37 is highly expressed in tumor cells. Immunohistochemical analyses also indicated an increase in the protein expression levels of TRIM37 in tumor samples compared with normal brain tissues. Notably, high expression levels of TRIM37 had a significant correlation with high-grade gliomas. Significant upregulation of TRIM37 in glioma tissues revealed that TRIM37 may function as a tumor promoter in glioma.
A series of in vitro experiments with siRNA-TRIM37 transfection were conducted to assess the effects of TRIM37 knockdown on U87 glioma cells. Knockdown of TRIM37 significantly inhibited cell growth in U87 glioma cells and induced cell apoptosis. TRIM37 knockdown also markedly induced cell apoptosis in vitro via upregulating the ratio of Bax/Bcl-2 and activating caspase 3 in U87 cells. In addition, the present study demonstrated that the inhibition of TRIM37 significantly increased the cell population in the S phase and decreased that in the G2/M phase, resulting in cell cycle arrest.
Results from the present study also revealed that TRIM37 has a specific role in U87 cell migration and invasion. Following TRIM37 knockdown, cell migration, and invasion rates were significantly decreased compared with the control groups, suggesting that loss of TRIM37 expression is detrimental to U87 cell migration and invasion. Notably, inhibition of TRIM37 led to decreased expression of MMP2 and MMP9, two markers for cell migration and invasion. These data support our hypothesis that TRIM37 may be a crucial molecular marker in glioma tumorigenesis, particularly in the process of malignant transformation.
Recent developments concerning the role of TRIM37 may provide novel directions towards elucidation of the mechanisms by which TRIM37 may induce tumorigenesis and other diseases. TRIM37 has been revealed as an oncogenic H2A ubiquitin ligase that is overexpressed in breast cancers [11,18]. The same study demonstrated that TRIM37 is associated with Polycomb repressive complex 2 and promotes tumor formation by facilitating silencing of tumor suppressors and other genes. They also found a statistically significant correlation between high levels of TRIM37 and decreased survival in patients with estrogen-receptor-positive breast cancer. Another study demonstrated that the expression level of TRIM37 was significantly higher in pancreatic cancerous tissues compared with adjacent normal tissues [13]. Notably, knocking down the expression of TRIM37 inhibited the growth and migration of pancreatic cancer cells. The study suggested that TRIM37 interacted with β-catenin and activated the transcriptional activity of β-catenin/T cell factor (TCF) complex as well as expression of its downstream target genes to promote the progression of pancreatic cancer. These data suggested that TRIM37 may act as a therapeutic target to inhibit β-catenin/TCF signaling. Another study observed higher expression of TRIM37 in hepatocellular carcinoma, which was associated with advanced stage and tumor volume, which indicated poor outcome [12]. TRIM37 was also revealed to induce aberrant activation of Wnt/b-catenin signaling, which was responsible for promoting cell migration and invasion. However, the specific mechanism remained unknown. In addition, mutations in TRIM37 have been demonstrated to result in mulibrey nanism, further supporting the theory that TRIM37 serves a significant role in the pathogenesis of disease.
In conclusion, the present study demonstrates that TRIM37 is significantly upregulated in human glioma. The expression level of TRIM37 was closely associated with glioma grade. In addition, TRIM37 knockdown inhibited proliferation and migration of glioma cells. Although further research using TRIM37-knockout mice or xenografts may provide novel insight into its function in glioma progression, the present study suggests an oncogenic role for TRIM37 in glioma.
Acknowledgements
The present study was supported by the National Natural Science Foundation of China (grant no. 81401867), Research and Development Plan of Shandong Province (grant no. 2016GSF201230).
Disclosure of conflict of interest
None.
References
- 1.Ricard D, Idbaih A, Ducray F, Lahutte M, Hoang-Xuan K, Delattre JY. Primary brain tumours in adults. Lancet. 2012;379:1984–1996. doi: 10.1016/S0140-6736(11)61346-9. [DOI] [PubMed] [Google Scholar]
- 2.Johnson DR, O’Neill BP. Glioblastoma survival in the United States before and during the temozolomide era. J Neurooncol. 2012;107:359–364. doi: 10.1007/s11060-011-0749-4. [DOI] [PubMed] [Google Scholar]
- 3.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn U, Curschmann J, Janzer RC, Ludwin SK, Gorlia T, Allgeier A, Lacombe D, Cairncross JG, Eisenhauer E, Mirimanoff RO European Organisation for Research and Treatment of Cancer Brain Tumor and Radiotherapy Groups; National Cancer Institute of Canada Clinical Trials Group. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 4.Kallijarvi J, Lahtinen U, Hamalainen R, Lipsanen-Nyman M, Palvimo JJ, Lehesjoki AE. TRIM37 defective in mulibrey nanism is a novel RING finger ubiquitin E3 ligase. Exp Cell Res. 2005;308:146–155. doi: 10.1016/j.yexcr.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 5.Avela K, Lipsanen-Nyman M, Idanheimo N, Seemanova E, Rosengren S, Makela TP, Perheentupa J, de la Chapelle A, Lehesjoki AE. Gene encoding a new RING-B-box-Coiled-coil protein is mutated in mulibrey nanism. Nat Genet. 2000;25:298–301. doi: 10.1038/77053. [DOI] [PubMed] [Google Scholar]
- 6.Balestra FR, Strnad P, Fluckiger I, Gonczy P. Discovering regulators of centriole biogenesis through siRNA-based functional genomics in human cells (vol 25, pg 555, 2013) Dev Cell. 2013;26:220–220. doi: 10.1016/j.devcel.2013.05.016. [DOI] [PubMed] [Google Scholar]
- 7.Shin SW, Oh TJ, Park SM, Park JS, Jang AS, Park SW, Uh ST, An S, Park CS. Asthma-predictive genetic markers in gene expression profiling of peripheral blood mononuclear cells. Allergy Asthma Immun. 2011;3:265–272. doi: 10.4168/aair.2011.3.4.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kumpf M, Hamalainen RH, Hofbeck M, Baden W. Refractory congestive heart failure following delayed pericardectomy in a 12-year-old child with mulibrey nanism due to a novel mutation in TRIM37. Eur J Pediatr. 2013;172:1415–1418. doi: 10.1007/s00431-013-1962-2. [DOI] [PubMed] [Google Scholar]
- 9.Karlberg S, Toppari J, Karlberg N, Nurmio M, Karikoski R, Jalanko H, Lipsanen-Nyman M. Testicular failure and male infertility in the monogenic mulibrey nanism disorder. J Clin Endocr Metab. 2011;96:3399–3407. doi: 10.1210/jc.2011-1493. [DOI] [PubMed] [Google Scholar]
- 10.Tabah AA, Tardif K, Mansky LM. Anti-HIV-1 activity of Trim 37. J Gen Virol. 2014;95:960–967. doi: 10.1099/vir.0.057653-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bhatnagar S, Gazin C, Chamberlain L, Ou JH, Zhu XC, Tushir JS, Virbasius CM, Lin L, Zhu LJ, Wajapeyee N, Green MR. TRIM37 is a new histone H2A ubiquitin ligase and breast cancer oncoprotein. Nature. 2014;516:116–20. doi: 10.1038/nature13955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang JX, Yu C, Chen MY, Tian S, Sun CY. Over-expression of TRIM37 promotes cell migration and metastasis in hepatocellular carcinoma by activating Wnt/beta-catenin signaling. Biochem Bioph Res Co. 2015;464:1120–1127. doi: 10.1016/j.bbrc.2015.07.089. [DOI] [PubMed] [Google Scholar]
- 13.Jiang J, Tian S, Yu C, Chen M, Sun C. TRIM37 promoted the growth and migration of the pancreatic cancer cells. Tumour Biol. 2016;37:2629–34. doi: 10.1007/s13277-015-4078-7. [DOI] [PubMed] [Google Scholar]
- 14.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Scheithauer BW, Kleihues P. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114:97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu Z, Gong J, Wang C, Wang Y, Song Y, Xu W, Liu Z, Liu Y. The diagnostic value and functional roles of phosphoglycerate mutase 1 in glioma. Oncol Rep. 2016;36:2236–2244. doi: 10.3892/or.2016.5046. [DOI] [PubMed] [Google Scholar]
- 16.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 17.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Scheithauer BW, Kleihues P. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114:97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bhatnagar S, Gazin C, Chamberlain L, Ou J, Zhu X, Tushir JS, Virbasius CM, Lin L, Zhu LJ, Wajapeyee N, Green MR. TRIM37 is a new histone H2A ubiquitin ligase and breast cancer oncoprotein. Nature. 2014;516:116–120. doi: 10.1038/nature13955. [DOI] [PMC free article] [PubMed] [Google Scholar]


