Abstract
Aims: This study aimed to investigate the nuclear expression status of cyclin dependent kinase like 2 (CDKL2) in glioma and its correlation with the characteristics of clinical pathology, including patient survival. Methods and results: In the present study, the expression of CDKL2 mRNA was detected by real-time QPCR in freshly collected glioma and para-carcinoma tissues. Moreover, immunohistochemistry was used to identified nuclear expression of CDKL2, and the characteristics of clinical pathology from glioma cases (n = 144) and non-cancerous brain tissues (n = 32) were counted. Low mRNA and nuclear protein expression of CDKL2 was observed in glioma tissues compared to non-cancerous tissues. Glioma patients with negative nuclear expression of CDKL2 were correlated with histologic type, clinical World Health Organization (WHO) grade, tumor location, and KI-67 expression status. Negative nuclear expression of CDKL2 in glioma patients predicted an observably shorter overall survival time than did positive expression. However, as demonstrated by multivariate analysis, nuclear expression of CDKL2 was not an independent prognostic biomarker for the survival of patients with glioma. Conclusions: Our study indicated that negative nuclear expression of CDKL2 may represent a potential unfavorable marker for progression and poor prognostic in glioma.
Keywords: Glioma, CDKL2 nuclear, prognosis
Introduction
Glioma is the most common solid tumor of the central nervous system, comprising around one quarter of all newly diagnosed primary brain tumors annually. It is characterized by a diffusely infiltrating clinical phenotype [1]. According to the latest WHO classification of CNS tumors, gliomas are divided into noninvasive, lowly proliferative astrocytomas (grade I-II), anaplastic astrocytomas (grade III), and necrosis-prone, highly infiltrative, and mitotically active glioblastoma (grade IV) [2]. Despite intensive multimodal management involving surgery resection, adjuvant radiotherapy, and chemotherapy with temozolomide, prognosis is very poor [3]. For example, glioblastoma (WHO grade IV) is the most aggressive malignant glioma, with a median survival time of merely 12 to 15 months [4,5]. Further work is required to identify the precise molecular events underlying glioma initiation, progression, and prognosis. Glioma pathogenesis is clearly complex and may involve ionizing radiation, environmental factors, genetic susceptibility and alerted molecular expression. The latter may result in inhibition or deregulation of a series of genes such as EGFR, RTK and mTOR [1,6-10].
Cyclin-dependent kinase-like 2 (CDKL2), also known as protein kinase P56 KKIAMRE, has been molecularly cloned from human fetal brain. CDKL2 contains a conserved cdc2-related serine/threonine kinase domain in the N-terminus [11,12]. It belongs to the cyclin-dependent kinase-like (CDKL) family, which are considered to be more distant members of the cyclin-dependent kinase (CDK) family and are similar to the mitogen-activated protein kinase (MAPK) family [11-13]. Corresponding to the dual phosphorylation motif (Thr-Xaa-Tyr) of the MAPK family, CDKL2 has a similar amino acid sequence (Thr-Asp-Tyr). Previous studies suggested that the Thr and Tyr residues of CDKL2 are activated by MEK1. However, mutational removal of Thr or Tyr had no effect on epidermal growth factor stimulated kinase activity in this motif [11]. Thus, the role of CDKL2 activated regulation in signal transduction could be distinct from those of MAPK or cdc2 family members [14].
In recent years, CDKL2 has been demonstrated to be highly expressed in kidneys, testis, and lungs. It is also highly expressed in various regions of the brain from the forebrain to the spinal cord and including entorhinal cortex, cerebral cortex, amygdala, dorsal thalamus, hippocampus, and the deep cerebellar nuclei [14,15]. CDKL2 functions principally in mature neurons and may play an important function in maintenance of the normal adult brain [14]. A role for CDKL2 had been observed in a number of tumor types including breast and prostate cancer, and hepatocellular carcinoma [16-19]. However, the part played by CDKL2 nuclear expression in human gliomas has not been elucidated.
In this work, we investigated the involvement of CDKL2 in gliomas by examining CDKL2 mRNA and nuclear protein expression in glioma specimens and in non-cancerous brain samples. We then analyzed the potential correlation between nuclear expression of CDKL2 and clinicopathologic characteristics including patient survival (n = 144). We discovered that CDKL2 mRNA and nuclear protein expression were lower in glioma specimens than in non-cancerous brain specimens. Furthermore, the comparably negative nuclear protein expression of CDKL2 was related to glioma progression and poor prognosis. Our findings support the concept that negative nuclear expression of CDKL2 is potentially an unfavorable prognostic factor for patients with glioma.
Materials and methods
Sample collection
Eleven fresh non-cancerous brain tissues and 48 fresh human primary glioma tissues were collected from the Southern Medical University/Nanfang Hospital (Guangzhou, China) between 2016 and 2017. The glioma samples were obtained at diagnosis and prior to treatment initiation. All fresh tissue samples were stored in liquid nitrogen immediately.
A further 144 glioma tissues and 32 non-cancerous brain tissues, collected between 2005 and 2009, were also obtained from Nanfang Hospital (Guangzhou, China). Clinical and prognostic details were available for the 85 male and 59 female glioma patients who ranged in age from 3 to 70 years (37 ± 17.39) (Table 2). All samples had confirmed pathological diagnosis of glioma. The hospital ethics committee approved the study, and prior consent was obtained from the patients for use of the clinical materials for research.
Table 2.
Correlation between the clinicopathologic characteristics and nuclear expression of CDKL2 protein in glioma
| Characteristics | n | CDKL2 (%) | P | |
|---|---|---|---|---|
|
| ||||
| Positive expression | Negative expression | |||
| Gender | 0.699 | |||
| Male | 85 | 39 (45.9) | 46 (54.1) | |
| Female | 59 | 29 (49.2) | 30 (50.8) | |
| Age | 0.230 | |||
| ≥ 50 | 34 | 13 (38.2) | 21 (61.8) | |
| < 50 | 110 | 55 (50.0) | 55 (50.0) | |
| Histologic Type | 0.029* | |||
| Astrocytic tumors | 98 | 39 (39.8) | 59 (60.2) | |
| Oligodendrogial tumors | 16 | 11 (68.8) | 5 (31.2) | |
| Other | 30 | 18 (60.0) | 12 (40.0) | |
| WHO Grade | 0.035* | |||
| I+II | 63 | 36 (57.1) | 27 (42.9) | |
| III+IV | 81 | 32 (39.5) | 49 (60.5) | |
| Tumor Location | 0.018* | |||
| Frontal | 44 | 19 (43.2) | 25 (56.8) | |
| Temporal | 26 | 8 (30.8) | 18 (69.2) | |
| Parietal | 18 | 12 (66.7) | 6 (33.3) | |
| Occipital | 9 | 1 (11.1) | 8 (88.9) | |
| Cerebellum | 9 | 6 (66.7) | 3 (33.3) | |
| Other | 38 | 22 (57.9) | 16 (42.1) | |
| Necrosis | 0.486 | |||
| Absent | 55 | 28 (50.9) | 27 (49.1) | |
| Present | 89 | 40 (44.9) | 49 (55.1) | |
| KI-67 expression | 0.038* | |||
| Positive | 87 | 35 (40.2) | 52 (59.8) | |
| Negative | 57 | 33 (57.9) | 24 (42.1) | |
P < 0.05 was considered significant.
Real-time fluorescence quantitative PCR
Total RNA was isolated from the tissue samples using Trizol (Takara, Shiga, Japan) and reverse transcription was undertaken using the reverse transcription reagents (Thermo Scientific, Waltham, MA, USA). According to the manufacturer’s instructions, real-time QPCR was conducted on a BIO-RAD CFX 96 detection system using a SYBR Premix Ex TaqTM II kit (TaKaRa Bio, Inc., Shiga, Japan). The specific sequence for the sense/antisense primer was (sense: 5’-TCTCCCAGTCTGGCGTTGT-3’, and antisense: 5’-ACCATCGGGTTGCCACATAAT-3’). ARF5, used as an inner control, had the primer sequence of (sense: 5’-ATCTGTTTCACAGTCTGGGAC-3’, and antisense: 5’-CCTGCTTGTTGGCAAATACC-3’) [20].
Immunohistochemistry
Paraffin sections (3 μm thick) from the 144 stored glioma and 32 stored non-cancerous brain tissues were deparaffinized in 100% xylene and rehydrated with a descending ethanol series (100%, 90%, 80% and 70%). The sections were then washed in water according to a standard protocol, and citrate 10 mM buffer was used for heat-induced antigen retrieval (2 min at 100°C). Endogenous peroxidase activity was inactivated with peroxidase blocking reagent (hydrogen peroxide 3%) and non-specific antigen was blocked with serum. The sections were then incubated with Primary antibodies (anti-human CDKL2 antibody, 1/100, Novus, Littleton, USA; anti-human KI-67 antibody, 1/200, Abcam, Cambridge, UK) at 4°C overnight. After washing three times with PBS to remove unbound antibody, the sections were incubated with biotin-labeled goat anti-mouse antibody at room temperature for 30 min. After further washing, streptavidin-conjugated horseradish peroxidase (HRP) (Maixin Inc, China) was added to the sections and incubated. The peroxidase reaction was proved using 3,3-diaminobenzidine (DAB) chromogen solution in DAB buffer substrate. Sections were visualized with DAB developer and counterstained with hematoxylin. Finally, the sections were mounted in neutral gum and analyzed under a bright field microscope.
Evaluation of staining
The immunohistochemically stained sections of CDKL2 or KI-67 in nuclei were reviewed and scored by two pathologists blinded to the clinical parameters. Sections with 10% or more nuclear stained cells were considered positive for nuclear expression, whereas less than 10% was regarded as negative [20,21].
Statistical analyses
All statistical analyses were undertaken with SPSS 20.0 software. Data were expressed as mean ± SD. The relationship between CDKL2 nuclear expression levels and clinicopathologic characteristics were analyzed using the χ2 test. The Kaplan-Meier method was used to plot survival curves, which were compared using the log-rank test. The statistical significances of univariate and multivariate variables for survival were conducted using the Cox proportional hazards model. A p-value of less than 0.05 was considered statistically significant.
Results
CDKL2 mRNA expression is low in human glioma
To clarify the role of CDKL2 in human glioma, CDKL2 mRNA expression was assessed using real-time QPCR in freshly collected glioma and non-cancerous brain tissues. CDKL2 mRNA expression was significantly lower in low-grade glioma (WHO grade I+II) compared to non-tumor samples (P = 0.0009). Furthermore, CDKL2 mRNA expression was significantly lower in high-grade glioma (WHO grade III+IV) versus low-grade glioma samples (P < 0.0001) (Figure 1).
Figure 1.
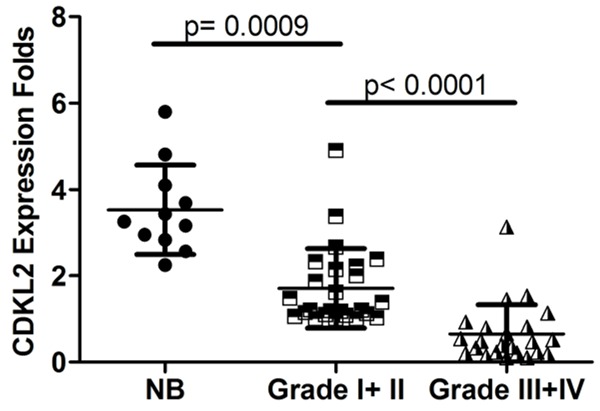
Scatter plots showed that expression of CDKL2 mRNA was decreased in glioma tissues compared with non-cancerous brain (NB) tissues and decreased CDKL2 mRNA levels significantly correlated with clinical WHO grades of human gliomas.
Immunohistochemical analysis of CDKL2 protein nuclear expression in glioma and non-cancerous brain tissues
To determine whether the subcellular location and nuclear expression of CDKL2 was different in glioma, we measured nuclear expression of CDKL2 in 144 archived paraffin-embedded glioma and 32 non-cancerous tissue samples. CDKL2 was mainly expressed in the nucleus and partially in the cytoplasm of both non-cancerous brain cells (Figure 2A) and in malignant glioma cells (Figure 2B, 2C), but was significantly lower in the glioma cells (P < 0.001) (Table 1).
Figure 2.
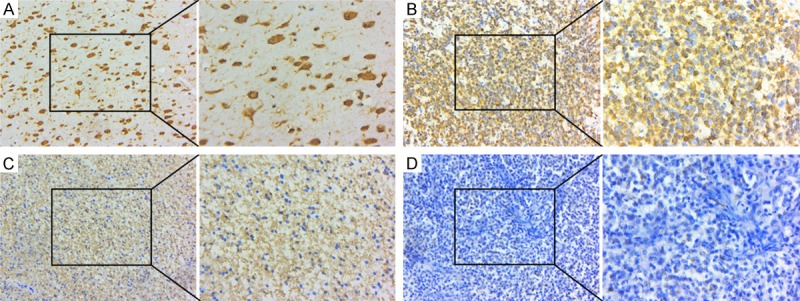
Decreased CDKL2 protein nuclear expression was examined in glioma tissues compared to non-cancerous brain samples (original magnifications are:×200). A: Positive nuclear expression of CDKL2 in non-cancerous brain tissues. B: CDKL2 positive nuclear expression in the glioma tissues. C: High CDKL2 expression in cytoplasm of glioma tissues, but low expression in nuclei. D: Negative expression of CDKL2 in glioma tissues. (insets displaying cdkl2 expression under high magnification ×400).
Table 1.
Nuclear expression of CDKL2 between glioma and non-cancerous brain tissues
| Group | Cases | Nuclear expression | P value | |
|---|---|---|---|---|
|
| ||||
| Positive expression | Negative expression | |||
| Glioma | 144 | 68 (47.2) | 76 (52.8) | |
| Non-cancerous | 32 | 27 (84.4) | 5 (15.6) | < 0.001* |
P < 0.05 was considered significant.
Analysis of the relationship between clinicopathological characteristics and CDKL2 nuclear expression in glioma patients
Table 2 outlines the associations between CDKL2 nuclear expression and clinicopathological characteristics in patients with glioma. We found that nuclear expression of CDKL2 was inversely correlated with histologic type (P = 0.029), clinical WHO stage (I-II vs. III-IV) (P = 0.035), tumor location (P = 0.018), and KI-67 expression status (P = 0.038), but not with patient age or gender.
Association of CDKL2 nuclear expression with overall survival of glioma patients
To examine the prognostic value of CDKL2 nuclear expression for glioma, Kaplan-Meier analysis with the log-rank test was performed to examine the relationship between the nuclear expression of CDKL2 and patient survival (n = 144). We observed a significant correlation with overall survival, with positive nuclear expression of CDKL2 correlating with better survival than negative expression (P = 0.0312) (Figure 3), and patients in the CDKL2 positive nuclear expression group having longer overall survival in the necrosis present group (P = 0.0068) and KI-67 positive expression group (P = 0.0026) (Figure 4D, 4F). Furthermore, we found that survival was also significantly correlated with clinical WHO grade, tumor location, KI-67 expression status, necrosis status, and nuclear expression status of CDKL2 in univariate analyses (P < 0.001, P = 0.039, P < 0.001, P = 0.001, and P = 0.032 respectively) (Table 3).
Figure 3.
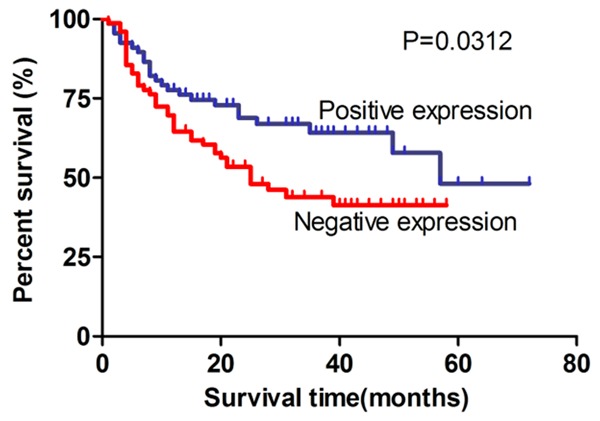
Kaplan-Meier survival analysis of overall survival duration in 144 glioma patients according to CDKL2 nuclear expression level. Patients with CDKL2 positive nuclear expression had better survival than those with negative nuclear expression of CDKL2 (P = 0.0312).
Figure 4.
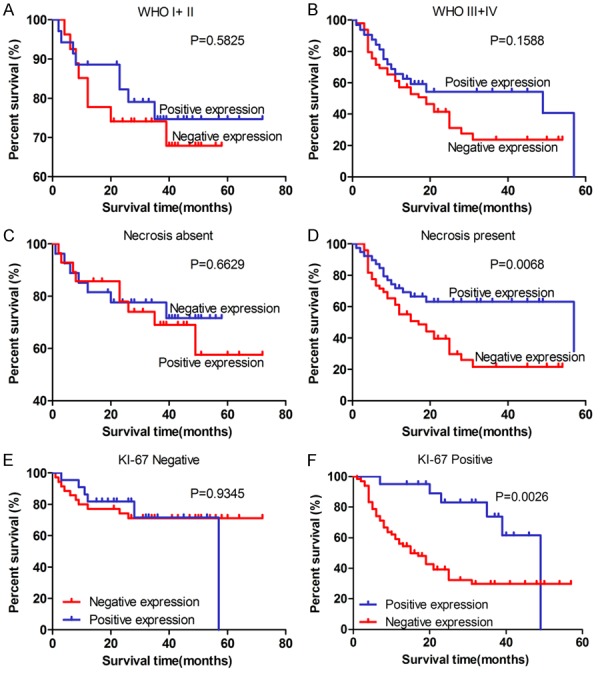
Kaplan-Meier survival analysis of overall survival duration in 144 glioma patients according to WHO grade classification, necrosis status, and KI-67 expression status in glioma. A, B: Correlation of CDKL2 protein nuclear expression with glioma patients’ survival time in strata analysis in WHO grade (I+II and III+IV). C, D: Correlation of CDKL2 protein nuclear expression with glioma patients’ survival time in strata analysis in necrosis status. Patients in the CDKL2 positive nuclear expression group had longer overall survival in the necrosis present group (P = 0.0068). E, F: The correlation of CDKL2 protein nuclear expression with glioma patients’ survival time in strata analysis in KI-67 expression status, and patients with CDKL2 positive nuclear expression had longer survival time (P = 0.0026). The log-rank test was used to calculate P values.
Table 3.
Summary of univariate and multivariate Cox regression analysis of overall survival duration
| Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|
|
|
||||||
| P | RR | 95% CI | P | RR | 95% CI | |
| Age | ||||||
| < 50 vs. ≥ 50 years | 0.216 | 1.412 | 0.818-2.438 | |||
| Gender | ||||||
| Male vs. female | 0.466 | 0.833 | 0.510-1.360 | |||
| Histologic Type | ||||||
| AT+OT vs. other* | 0.067 | 1.937 | 0.956-3.924 | |||
| Tumor Location | 0.039 | 2.173 | 1.039-4.545 | 0.126 | 1.818 | 0.846-3.908 |
| Necrosis | ||||||
| Absent vs. present | 0.001 | 0.371 | 0.209-0.657 | 0.729 | 1.182 | 0.459-3.042 |
| KI-67 expression | ||||||
| Positive vs. negative | 0.000 | 0.295 | 0.154-0.568 | 0.022 | 0.431 | 0.211-0.883 |
| CDKL2 nuclear expression | ||||||
| Negative vs. positive | 0.032 | 1.744 | 1.050-2.896 | 0.302 | 1.322 | 0.778-2.247 |
| WHO Grade | ||||||
| I vs. II vs. III vs. IV | 0.000 | 0.275 | 0.155-0.488 | 0.017 | 0.280 | 0.098-0.800 |
Abbreviation: AT: Astrocytic tumors, OT: Oligodendrogial tumors.
P < 0.05 was considered significant.
Subsequently, to determine whether CDKL2 nuclear expression is an independent prognostic factor for glioma, multivariate analysis was performed to analyze expression status adjusted for clinical WHO grade, tumor location, KI-67 expression status, necrosis status, and nuclear expression status of CDKL2. The results showed that CDKL2 nuclear expression was not an independent prognostic factor for human glioma (P = 0.302) (Table 3).
Discussion
CDKL2 has been observed in three types of tumors including breast, prostate cancer, and hepatocellular carcinoma. However, the role of CDKL2 in human glioma development and progression has not been elucidated. In this study, we present evidence that expression of CDKL2 mRNA is lower in glioma samples than in non-cancerous brain tissue. Consistent with this, CDKL2 nuclear protein expression was significantly decreased in glioma tissue compared to non-cancerous brain tissue. Our data support the concept that CDKL2 is downregulated in a subset of patients with glioma and this suggests that CDKL2 may be an important regulator of glioma.
CDKL2 has been shown to have high expression in various adult brain regions including the entorhinal cortex, cerebral cortex, amygdala, hippocampus, and the deep cerebellar nuclei [14,15], CDKL2 functions principally in mature neurons, and the regulatory mechanisms of CDKL2 expression in mature neurons differs from those of immediate-early genes [14]. Recent studies have discovered that CDKL2 has a positive role in cognition, emotion, and learning, with mutations in the CDKL family kinases potentially causing neurodevelopmental and neuropsychiatric disorders [22,23]. Collectively, these studies suggest that CDKL2 may play an important function in normal adult brain maintenance.
CDKL2 has been reported to play dual roles and to be differentially expressed in several types of tumors. In hepatocellular carcinoma, Yongchang Zheng et al. collected three genome-wide DNA methylation datasets with 800 clinical samples and the corresponding gene expression datasets by analyzing the DNA methylation patterns in cancer across multiple datasets. They observed that CDKL2 was a candidate molecular marker of hepatocellular carcinoma [16]. Interestingly, paradoxical findings were observed in a recent prostate cancer study. Rohina Rubicz et al. determined that the CDKL2 gene with higher promotor region methylation and a corresponding decrease in mRNA level occurred in patients with more aggressive prostate cancer [17]. In breast cancer, Linna Li et al. reported high expression of CDKL2 in mesenchymal subtypes of breast cancer cells, human mesenchymal stem cells, and fibroblasts compared to epithelial subtypes of breast cancer cells. Their results suggested that CDKL2 might be an unfavorable prognostic factor and a potential therapeutic target for human malignant breast cancers [18]. However, Lindqvist et al. used methylation-sensitive RFLP analysis and observed that CDKL2 had higher promotor region methylation in breast cancer samples and a corresponding decrease in mRNA levels in the patients (P = 0.019) [19].
Our study of glioma tissues had consistent findings, in terms of CDKL2 mRNA expression, to studies on other tumors. In addition, we demonstrated that CDKL2 nuclear expression was significantly decreased in glioma tissue compared to non-cancerous brain tissue. This result further supports the role of CDKL2 as a candidate suppressor in glioma. Moreover, we analyzed the relationship between the nuclear protein expression of CDKL2 and clinical features of patients with glioma. Although nuclear expression of CDKL2 was not related to patient age or gender, it was correlated with histologic type, clinical WHO stage (I-II vs. III-IV), tumor location, and KI-67 expression status of glioma. Ki-67 positive expression is associated with cellular growth and proliferation in glioma [24]. Evidence was also provided that nuclear expression of CDKL2 negatively correlates with overall survival time. Overall, our study indicates that CDKL2 protein nuclear expression is a tumor suppressor that represses the pathogenesis and progression of glioma.
Finally, to further determine whether CDKL2 nuclear expression is an independent prognostic factor for glioma, univariate analyses were undertaken. We found that survival was positively correlated with CDKL2 nuclear expression, but correlated negatively with KI-67 expression status, necrosis status, and WHO grade classification in glioma. Multivariate analyses showed that nuclear expression of CDKL2 was not an independent predictor of prognosis for glioma patients regardless of KI-67 expression status and WHO grade classification.
In summary, our research shows that negative CDKL2 nuclear expression may be involved in progression and poor prognosis, but was not an independent prognostic factor for patients with glioma.
Acknowledgements
This study was supported by National Nature Science Foundation of China (NO. 81372692, 81502178, 81502177) (http://www.nsfc.gov.cn), Natural Science Fund of Guangdong Province, China (NO. 2014A030313303, 2014A030313282, 2016A030313549) (http://www.gdstc.gov.cn), Science and Technology Program of Guangzhou, China (NO. 201607010350) (http://wsbs.gzsi.gov.cn/login.html), The funders had no role in study design, data collection, data analysis, decision to publish, or preparation of the manuscript.
Disclosure of conflict of interest
None.
References
- 1.Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD, Kleihues P, Ellison DW. The 2016 world health organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 2.Liu N, Wang Z, Cheng Y, Zhang P, Wang X, Yang H, Liu H, Zhang Y, Tu Y. Acylglycerol kinase functions as an oncogene and an unfavorable prognostic marker of human gliomas. Hum Pathol. 2016;58:105–112. doi: 10.1016/j.humpath.2016.07.034. [DOI] [PubMed] [Google Scholar]
- 3.Mostafa H, Pala A, Hogel J, Hlavac M, Dietrich E, Westhoff MA, Nonnenmacher L, Burster T, Georgieff M, Wirtz CR, Schneider EM. Immune phenotypes predict survival in patients with glioblastoma multiforme. J Hematol Oncol. 2016;9:77. doi: 10.1186/s13045-016-0272-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li B, Huang MZ, Wang XQ, Tao BB, Zhong J, Wang XH, Zhang WC, Li ST. TMEM140 is associated with the prognosis of glioma by promoting cell viability and invasion. J Hematol Oncol. 2015;8:89. doi: 10.1186/s13045-015-0187-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu X, Chong Y, Tu Y, Liu N, Yue C, Qi Z, Liu H, Yao Y, Liu H, Gao S, Niu M, Yu R. CRM1/XPO1 is associated with clinical outcome in glioma and represents a therapeutic target by perturbing multiple core pathways. J Hematol Oncol. 2016;9:108. doi: 10.1186/s13045-016-0338-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu Z, Xie G, Zhou G, Cheng Y, Zhang G, Yao G, Chen Y, Li Y, Zhao G. NVP-BEZ235, a novel dual PI3K-mTOR inhibitor displays anti-glioma activity and reduces chemoresistance to temozolomide in human glioma cells. Cancer Lett. 2015;367:58–68. doi: 10.1016/j.canlet.2015.07.007. [DOI] [PubMed] [Google Scholar]
- 7.Wen PY, Yung WK, Lamborn KR, Dahia PL, Wang Y, Peng B, Abrey LE, Raizer J, Cloughesy TF, Fink K, Gilbert M, Chang S, Junck L, Schiff D, Lieberman F, Fine HA, Mehta M, Robins HI, DeAngelis LM, Groves MD, Puduvalli VK, Levin V, Conrad C, Maher EA, Aldape K, Hayes M, Letvak L, Egorin MJ, Capdeville R, Kaplan R, Murgo AJ, Stiles C, Prados MD. Phase I/II study of imatinib mesylate for recurrent malignant gliomas: North American brain tumor consortium study 99-08. Clin Cancer Res. 2006;12:4899–4907. doi: 10.1158/1078-0432.CCR-06-0773. [DOI] [PubMed] [Google Scholar]
- 8.Wen PY, Lee EQ, Reardon DA, Ligon KL, Alfred YW. Current clinical development of PI3K pathway inhibitors in glioblastoma. Neuro Oncol. 2012;14:819–829. doi: 10.1093/neuonc/nos117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Galanis E, Buckner JC, Maurer MJ, Kreisberg JI, Ballman K, Boni J, Peralba JM, Jenkins RB, Dakhil SR, Morton RF, Jaeckle KA, Scheithauer BW, Dancey J, Hidalgo M, Walsh DJ. Phase II trial of temsirolimus (CCI-779) in recurrent glioblastoma multiforme: a north central cancer treatment group study. J. Clin. Oncol. 2005;23:5294–5304. doi: 10.1200/JCO.2005.23.622. [DOI] [PubMed] [Google Scholar]
- 10.Batchelor TT, Sorensen AG, di Tomaso E, Zhang WT, Duda DG, Cohen KS, Kozak KR, Cahill DP, Chen PJ, Zhu M, Ancukiewicz M, Mrugala MM, Plotkin S, Drappatz J, Louis DN, Ivy P, Scadden DT, Benner T, Loeffler JS, Wen PY, Jain RK. AZD2171, a pan-VEGF receptor tyrosine kinase inhibitor, normalizes tumor vasculature and alleviates edema in glioblastoma patients. Cancer Cell. 2007;11:83–95. doi: 10.1016/j.ccr.2006.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taglienti CA, Wysk M, Davis RJ. Molecular cloning of the epidermal growth factor-stimulated protein kinase p56 KKIAMRE. Oncogene. 1996;13:2563–2574. [PubMed] [Google Scholar]
- 12.Malumbres M, Harlow E, Hunt T, Hunter T, Lahti JM, Manning G, Morgan DO, Tsai LH, Wolgemuth DJ. Cyclin-dependent kinases: a family portrait. Nat Cell Biol. 2009;11:1275–1276. doi: 10.1038/ncb1109-1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tang D, Lee KY, Qi Z, Matsuura I, Wang JH. Neuronal Cdc2-like kinase: from cell cycle to neuronal function. Biochem Cell Biol. 1996;74:419–429. doi: 10.1139/o96-046. [DOI] [PubMed] [Google Scholar]
- 14.Sassa T, Gomi H, Itohara S. Postnatal expression of Cdkl2 in mouse brain revealed by LacZ inserted into the Cdkl2 locus. Cell Tissue Res. 2004;315:147–156. doi: 10.1007/s00441-003-0828-8. [DOI] [PubMed] [Google Scholar]
- 15.Gomi H, Sassa T, Thompson RF, Itohara S. Involvement of cyclin-dependent kinase-like 2 in cognitive function required for contextual and spatial learning in mice. Front Behav Neurosci. 2010;4:17. doi: 10.3389/fnbeh.2010.00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng Y, Huang Q, Ding Z, Liu T, Xue C, Sang X, Gu J. Genome-wide DNA methylation analysis identifies candidate epigenetic markers and drivers of hepatocellular carcinoma. Brief Bioinform. 2018;19:101–108. doi: 10.1093/bib/bbw094. [DOI] [PubMed] [Google Scholar]
- 17.Rubicz R, Zhao S, Geybels M, Wright JL, Kolb S, Klotzle B, Bibikova M, Troyer D, Lance R, Ostrander EA, Feng Z, Fan JB, Stanford JL. DNA methylation profiles in African American prostate cancer patients in relation to disease progression. Genomics. 2016 doi: 10.1016/j.ygeno.2016.02.004. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li L, Liu C, Amato RJ, Chang JT, Du G, Li W. CDKL2 promotes epithelial-mesenchymal transition and breast cancer progression. Oncotarget. 2014;5:10840–10853. doi: 10.18632/oncotarget.2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lindqvist BM, Wingren S, Motlagh PB, Nilsson TK. Whole genome DNA methylation signature of HER2-positive breast cancer. Epigenetics. 2014;9:1149–1162. doi: 10.4161/epi.29632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang Q, Mai C, Yang H, Wu Q, Hua S, Yan C, Long Y, Zhang Y, Long X, Fang W, Liu Z. Nuclear expression of CDK4 correlates with disease progression and poor prognosis in human nasopharyngeal carcinoma. Histopathology. 2014;64:722–730. doi: 10.1111/his.12319. [DOI] [PubMed] [Google Scholar]
- 21.Nagy B, Toth L, Molnar P, Mehes G, Thurzo L, Poka R, Hernadi Z. Nuclear beta-catenin positivity as a predictive marker of long-term survival in advanced epithelial ovarian cancer. Pathol Res Pract. 2017;213:915–921. doi: 10.1016/j.prp.2017.05.011. [DOI] [PubMed] [Google Scholar]
- 22.Dubos A, Pannetier S, Hanauer A. Inactivation of the CDKL3 gene at 5q31.1 by a balanced t(X;5) translocation associated with nonspecific mild mental retardation. Am J Med Genet A. 2008;146A:1267–1279. doi: 10.1002/ajmg.a.32274. [DOI] [PubMed] [Google Scholar]
- 23.Weaving LS, Christodoulou J, Williamson SL, Friend KL, McKenzie OL, Archer H, Evans J, Clarke A, Pelka GJ, Tam PP, Watson C, Lahooti H, Ellaway CJ, Bennetts B, Leonard H, Gecz J. Mutations of CDKL5 cause a severe neurodevelopmental disorder with infantile spasms and mental retardation. Am J Hum Genet. 2004;75:1079–1093. doi: 10.1086/426462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao C, Ban N, Dai S, Zhang X, Zhang L, Xu P, Chen W, Sun J, Bao Z, Chang H, Wang D, Ren J. The role of Alix in the proliferation of human glioma cells. Hum Pathol. 2016;52:110–118. doi: 10.1016/j.humpath.2015.09.046. [DOI] [PubMed] [Google Scholar]


