Abstract
Aims: The chief aim of this study was to correlate expression of the nasopharyngeal epithelium protein (CCDC19) with clinicopathological characteristics and survival prognosis in lung squamous cancer patients. Methods and results: Using real-time PCR, we detected the mRNA expression of CCDC19 in lung squamous cell carcinoma tissues and lung tissue. CCDC19 mRNA expression was confirmed to be downregulated compared to normal lung tissues. Furthermore, we analyzed CCDC19 protein expression using immunohistochemical analysis and observed CCDC19 protein in 136 paraffin-embedded squamous cell carcinoma tissues and 47normal paraffin-embedded lung tissues. CCDC19 protein was downregulated in lung squamous carcinoma, but overexpressed in normal lung tissues. Furthermore, correlation between the level of CCDC19 expression and clinical features, including survival prognosis was analyzed. Decreased expression of CCDC19 protein was significantly associated with N stage (P = 0.024) and gender (P = 0.022). Furthermore, decreased CCDC19 expression was associated with poorer overall survival rates than high expression of CCDC19 (P = 0.01). Multivariate analysis showed expression of CCDC19 to be an independent prognostic indicator of survival. Conclusions: Our findings demonstrate that decreased CCDC19 expression facilitates disease progression and poor outcome in lung squamous cell carcinoma patients.
Keywords: CCDC19, lung squamous cell carcinoma, prognosis, tumor suppressor
Introduction
Lung cancer is one of the most common malignancies in the world [1]. In China, lung cancer-associated mortality for both male and female patients has yet to reach its peak [2,3], and the five-year survival rate of these patients is only 15% [4]. Non-small cell lung cancer (NSCLC) is the most common type of lung cancer, and is mostly comprised of squamous cell carcinoma and adenocarcinoma. Several factors influence the occurrence and development of lung cancer including smoking, genetic susceptibility, and environmental factors that can lead to abnormal gene expression [5,6].
CCDC19/NESG1 was cloned and revised in 1999 and 2005, respectively [7,8]. The revised CCDC19 gene encodes 551 amino acids and has a predicted molecular weight of 65729.7 Da. The expression, function, and molecular mechanism of the revised CCDC19 gene have also been investigated [8-10]. CCDC19 is mainly expressed in the nasopharyngeal mucosa, the cytoplasm of columnar epithelial squamous cells, and in the nuclei of individual cells. In comparison with non-cancerous nasopharyngeal tissues,CCDC19 is downregulated in nasopharyngeal carcinoma (NPC) tissues and cells, and its decreased expression is an unfavorable factor, promoting NPC progression and resulting in a poor prognosis. CCDC19 acts as a tumor suppressor gene in NPC by inhibiting expression of CCNA1 and upregulating p21, which are involved in the regulation of cell cycle progression [8]. In addition, CCDC19 is involved in multiple signaling pathways that affect tight junctions and mitogen-activated protein kinase (MAPK) expression [11-14]. In earlier work, we also observed that decreased expression of CCDC19 correlated with the prognostic status of NSCLC and lung adenocarcinoma, but not squamous cell carcinoma [15]. This could have been due to the limited number of samples available for that work. Therefore, in the current study, 136 squamous cell tissue samples were collected and analyzed, to determine whether reduced expression of CCDC19 is associated with the clinical features and prognosis of squamous cell carcinoma. Our results indicate that decreased CCDC19 expression correlates with the stage of N classification of squamous cell carcinoma. Furthermore, downregulated CCDC19 expression is a prognostic factor for squamous cell carcinomacarcinoma progression and patient survival. These data were inconsistent with a previous investigation in NSCLC [15].
Materials and methods
Sample collection
Overall, one hundred and thirty-six paraffin-embedded undifferentiated squamous cell carcinoma specimens were obtained at the time of diagnosis from the First Affiliated Hospital of Guangdong Medical University, Zhenjiang, China, and the Tumor Hospital of Yunnan Province. None of the cases had received therapy prior to the diagnosis of NSCLC. Written informed consent was obtained from all the tissue donors, and the study was approved by the Ethics Committee of these two hospitals.
CCDC19 mRNA measurement
Total RNA from squamous cell carcinoma samples and their partly matched lung tissue samples was used to examine the differential expression of CCDC19, using real-time RT-PCR. The RT-PCR results were normalized to the invariant housekeeping gene, ACTG1. The primer sequences of CCDC19 were as follows: forward 5’-CGCCTGTGAGTGAGTGC-3’ and reverse 5’-CTTATCCATCCTTTCGGTCTT-3’. The SYBR Green Mix reagent (Takara, Japan) was used to perform the PCR reaction. The analysis was repeated three times.
Immunohistochemistry
Immunohistochemical detection of CCDC19 expression in lung cancer and normal tissue samples was performed, as previously described [7,8]. Two pathologists blinded to the clinical parameters, independently assessed and scored the stained tissue sections. The staining was scored based on a previously described protocol [7,8]. CCDC19 expression was compared between non-cancerous and cancerous lung tissues. Scores between 0 and 4 were indicative of low expression, whereas scores between 5 and 6 were considered high expression.
Statistical analysis
All statistical analyses were performed using the SPSS software version 13.0 (SPSS, Inc, USA). For comparison of two independent groups, the two-tailed Student’s t-test was used. Spearman’s correlation coefficient was calculated to evaluate the correlation between CCDC19 expression levels and normal or cancerous tissues. The relationship between CCDC19 expression and the clinicopathological characteristics of squamous cell carcinoma was analyzed with the χ2 test. Survival curves were plotted using the Kaplan-Meier method and compared using the log-rank test. The multivariate Cox proportional hazards model was applied to evaluate the impact of various variables on survival. P values of less than 0.05 were considered statistically significant.
Results
Differential expression of CCDC19 in squamous cell lung carcinoma and normal tissues
Real-time PCR was used to quantitatively measure CCDC19 mRNA expression in normal lung tissue samples and squamous cell carcinoma tissue samples. The results indicated that CCDC19 mRNA expression was significantly decreased in squamous cell carcinoma tissues compared to normal lung tissues (P = 0.004; Figure 1). Furthermore, CCDC19 protein expression was also detected in 136 archived paraffin-embedded squamous cell carcinoma samples and 47 normal lung samples using immunohistochemical staining (Figure 2). Expression of CCDC19 was significantly decreased in squamous cell lung carcinoma tissues (66/136 [48.5%]) compared to non-cancerous tissues (34/47 [72.3%]; Table 1).
Figure 1.
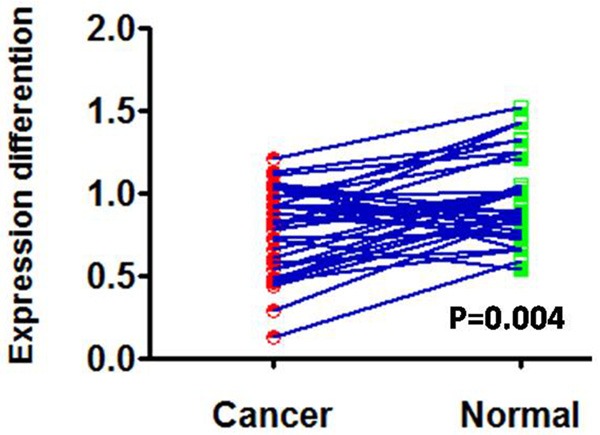
Differentiation of CCDC19 mRNA expression in lung squamous carcinoma tissues and lung tissues.
Figure 2.
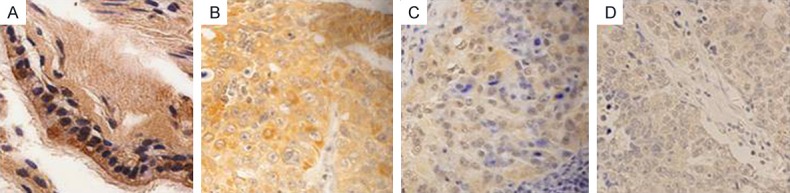
Immunohistochemical staining of CCDC19 protein in lung squamous cell carcinoma and lung tissues. A. High expression of CCDC19 protein in lung tissues; B. High expression of CCDC19 protein in lung squamous cell carcinoma tissues with a high differentiation state. C and D. Low expression of CCDC19 protein in lung squamous cell carcinoma tissues with medium and low differentiation degrees, respectively.
Table 1.
CCDC19 protein expression in lung adenocarcinoma and lung tissues
| Group | CCDC19 Protein Expression | P value | |
|---|---|---|---|
|
| |||
| High expression | Low expression | ||
| Cancer tissues | 66 | 70 | 0.006* |
| Lung tissues | 34 | 13 | |
Statistically significant.
Correlation between CCDC19 expression and the clinicopathological parameters of squamous cell carcinoma patients
Univariate analysis indicated that the overall survival of patients with squamous cell carcinoma was significantly associated with certain clinicopathological variables. Specifically, CCDC19 expression was associated with gender and N classification (P = 0.022 and P = 0.024, respectively; Table 2) in patients with squamous cell carcinoma. However, no significant relationship was observed between CCDC19 expression and patient age, T classification, degree of differentiation, clinical stage, or distant metastasis. These results suggest that change in CCDC19 expression significantly affects tumor cell lymph metastasis for squamous cell lung carcinoma.
Table 2.
Correlation between the clinicopathologic characteristics and expression of CCDC19 protein in lung squamous cancer
| Characteristics | n | CCDC19 (%) | P value | |
|---|---|---|---|---|
|
| ||||
| High expression | Low expression | |||
| Gender | 0.022* | |||
| Male | 99 | 42 | 57 | |
| Female | 37 | 24 | 13 | |
| Age (y) | 0.607 | |||
| ≥60 | 70 | 34 | 32 | |
| <60 | 66 | 32 | 38 | |
| Differentiated degree | 0.067 | |||
| High | 22 | 15 | 7 | |
| Middle | 36 | 19 | 17 | |
| Low or undifferentiated | 78 | 32 | 46 | |
| T classification | 0.479 | |||
| T1+T2 | 115 | 54 | 61 | |
| T3+T4 | 21 | 12 | 9 | |
| N classification | 0.024* | |||
| N0+N1 | 79 | 48 | 31 | |
| N2+N3 | 57 | 23 | 34 | |
| Distant metastasis | 0.496 | |||
| Negative | 134 | 66 | 68 | |
| Positive | 2 | 0 | 2 | |
| Clinical stage | 0.124 | |||
| I~II | 75 | 41 | 34 | |
| III~IV | 61 | 25 | 36 | |
Statistically significant.
Decreased expression of CCDC19 is associated with poor overall survival
Kaplan-Meier analysis with the log-rank test was used to evaluate the association between CCDC19 expression and patient survival. The results obtained indicated that the level of CCDC19 protein expression was positively associated with overall survival time of patients with squamous cell carcinoma. Patients with low CCDC19 expression had a worse prognosis than patients with high expression of CCDC19 (P = 0.01; Figure 3).
Figure 3.
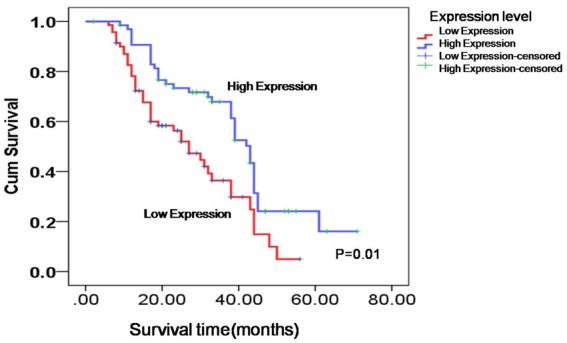
Low expression of CCDC19 protein was unfavorable for lung squamous carcinoma patients’ overall survival time.
Low expression of CCDC19 is an independent prognostic factor for patients with squamous cell lung carcinoma
To investigate whether expression of CCDC19 is an independent prognostic factor, we used univariate and multivariate Cox proportional hazards models to analyze the significance of various variables for the survival of lung squamous cell carcinoma patients. The results suggested that CCDC19 expression and N classification were significantly associated with survival of squamous cell lung cancer patients (P = 0.001 and P = 0.037, respectively). Low expression of CCDC19 was shown as an independent prognostic marker for squamous cell lung cancer patients (Table 3).
Table 3.
Summary of univariate and multivariate Cox regression analysis of overall survival duration
| Parameter | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| P | HR | 95% CI | P | HR | 95% CI | |
| Age | ||||||
| ≥60 vs. <60 years | 0.148 | 1.377 | 0.892-2.127 | 0.222 | 1.333 | 0.840-2.114 |
| Gender | ||||||
| Male vs. female | 0.201 | 1.376 | 0.844-2.244 | 0.624 | 1.145 | 0.666-1.968 |
| Differentiation degree | ||||||
| High vs. Middle vs. Low | 0.830 | 1.031 | 0.779-1.365 | 0.073 | 1.338 | 0.973-1.841 |
| TNM Classification | ||||||
| TNM I-II vs. TNM III-IV | 0.415 | 1.198 | 0.776-1.848 | 0.41 | 3.661 | 1.051-12.743 |
| T classification | ||||||
| T1-T2 vs. T3-T4 | 0.786 | 1.085 | 0.602-1.953 | 0.280 | 1.445 | 0.741-2.819 |
| N classification | ||||||
| N0-N1 vs. N2-N3 | 0.713 | 1.086 | 0.700-1.683 | 0.037* | 0.261 | 0.074-0.923 |
| M classification | ||||||
| M0 vs. M1 | 0.409 | 1.811 | 0.442-7.414 | 0.449 | 1.781 | 0.399-7.947 |
| CCDC19 expression | ||||||
| High vs. Low | 0.002* | 0.490 | 0.314-0.765 | 0.001* | 0.439 | 0.265-0.728 |
Statistically significant.
Discussion
In a study by Liu et al. CCDC19 was shown to act as a tumor suppressor for the occurrence and development of NPC. The researchers concluded that decreased expression of CCDC19 in patients promoted NPC pathogenesis [8,9]. To further investigate whether CCDC19 participates in the pathogenesis of NSCLC, Liu et al. explored expression of CCDC19 in NSCLC and observed that CCDC19 expression was down-regulated in NSCLC compared to normal tissues [15]. In this study, we used real-time PCR and immunohistochemistry to compare CCDC19 mRNA and protein expression between normal lung tissue and squamous cell carcinoma tissue. Our results indicatethat CCDC19 is significantly downregulated in squamous cell carcinoma tissues, which is consistent with results obtained in NPC and NSCLC [8,9,15]. Therefore, abnormal expression of CCDC19 is a significant factor in the pathogenesis of squamous cell lung cancer.
Previously, Liu et al. only explored the correlation of CCDC19 expression with clinical features of NSCLC, without particular focus on the clinical features of squamous cell lung cancer alone. Therefore, this was a major focus of our investigation in the present report. We found that decreased CCDC19 expression negatively associated with N classification of patients with squamous cell lung cancer. These results were consistent with our previous observations in NPC [8], where CCDC19 protein levels were higher at N0-N1 classification than those at N2-N3 classification. This result suggests that reduced CCDC19 expression promotes metastasis of squamous cell lung cancer. Interestingly, we also observed that CCDC19 expression correlated with gender in patients with squamous cell lung cancer, with reduced CCDC19 protein levels in male patients. However, detailed molecular mechanisms underlying this finding are currently unknown.
Reduced CCDC19 expression was previously shown to be positively correlated with overall survival of patients with NPC and NSCLC or lung adenocarcinoma, but not with squamous cell lung cancer [15]. In this study, we demonstrate that low expression of CCDC19 protein in squamous cell lung cancer tissues is inversely correlated with overall survival time. We suggest that low expression of CCDC19 is a significant clinical biomarker in the prognosis of squamous cell lung cancer. However, this data is not in accordance with previous work in squamous cell carcinoma tissues [15], which further supported the significance of CCDC19 in squamous cell lung cancer.
Finally, we verified that CCDC19 expression was an independent prognostic factor for squamous cell lung cancer based on univariate and multivariate Cox analyses. Overall, our results indicate that decreased expression of CCDC19 promotes development of squamous cell lung cancer, therefore, affecting its prognosis.
In conclusion, we demonstrate that decreased expression of CCDC19 plays a role in the clinical progression and is associated with poor prognosis in patients with squamous cell lung cancer. Moreover, reduced CCDC19 expression is an independent prognostic factor for squamous cell lung cancer.
Acknowledgements
This study was supported by the National Natural Science Foundation of China (No. 81572247).
Disclosure of conflict of interest
None.
References
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 2.Chen WQ, Zhang SW, Zou XN, Zhao P. Cancer incidence and mortality in China, 2006. Chin J Cancer Res. 2011;23:3–9. doi: 10.1007/s11670-011-0003-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen WQ, Zhang SW, Zou XN, Zhao P. [An analysis of lung cancer mortality in China, 2004-2005] . Zhonghua Yu Fang Yi Xue Za Zhi. 2010;44:378–382. [PubMed] [Google Scholar]
- 4.Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ. Cancer statistics, 2007. CA Cancer J Clin. 2007;57:43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- 5.Sun M, Nie FQ, Zang C, Wang Y, Hou J, Wei C, Li W, He X, Lu KH. The pseudogene DUXAP8 promotes non-small-cell lung cancer cell proliferation and invasion by epigenetically silencing EGR1 and RHOB. Mol Ther. 2017;25:739–751. doi: 10.1016/j.ymthe.2016.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 6.Zabarovsky ER, Lerman MI, Minna JD. Tumor suppressor genes on chromosome 3p involved in the pathogenesis of lung and other cancers. Oncogene. 2002;21:6915–6935. doi: 10.1038/sj.onc.1205835. [DOI] [PubMed] [Google Scholar]
- 7.Li Z, Yao K, Cao Y. Molecular cloning of a novel tissue-specific gene from human nasopharyngeal epithelium. Gene. 1999;237:235–240. doi: 10.1016/s0378-1119(99)00234-6. [DOI] [PubMed] [Google Scholar]
- 8.Liu Z, Li X, He X, Jiang Q, Xie S, Yu X, Zhen Y, Xiao G, Yao K, Fang W. Decreased expression of updated NESG1 in nasopharyngeal carcinoma: its potential role and preliminarily functional mechanism. Int J Cancer. 2011;128:2562–2571. doi: 10.1002/ijc.25595. [DOI] [PubMed] [Google Scholar]
- 9.Liu Z, Luo W, Zhou Y, Zhen Y, Yang H, Yu X, Ye Y, Li X, Wang H, Jiang Q, Zhang Y, Yao K, Fang W. Potential tumor suppressor NESG1 as an unfavorable prognosis factor in nasopharyngeal carcinoma. PLoS One. 2011;6:e27887. doi: 10.1371/journal.pone.0027887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Z, Chen C, Yang H, Zhang Y, Long J, Long X, Fang W. Proteomic features of potential tumor suppressor NESG1 in nasopharyngeal carcinoma. Proteomics. 2012;12:3416–3425. doi: 10.1002/pmic.201200146. [DOI] [PubMed] [Google Scholar]
- 11.Ohtani S, Terashima M, Satoh J, Soeta N, Saze Z, Kashimura S, Ohsuka F, Hoshino Y, Kogure M, Gotoh M. Expression of tight-junction-associated proteins in human gastric cancer: downregulation of claudin-4 correlates with tumor aggressiveness and survival. Gastric Cancer. 2009;12:43–51. doi: 10.1007/s10120-008-0497-0. [DOI] [PubMed] [Google Scholar]
- 12.Martinho O, Gouveia A, Viana-Pereira M, Silva P, Pimenta A, Reis RM, Lopes JM. Low frequency of MAP kinase pathway alterations in KIT and PDGFRA wild-type GISTs. Histopathology. 2009;55:53–62. doi: 10.1111/j.1365-2559.2009.03323.x. [DOI] [PubMed] [Google Scholar]
- 13.VanBrocklin MW, Verhaegen M, Soengas MS, Holmen SL. Mitogen-activated protein kinase inhibition induces translocation of Bmf to promote apoptosis in melanoma. Cancer Res. 2009;69:1985–1994. doi: 10.1158/0008-5472.CAN-08-3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gulmann C, Sheehan KM, Conroy RM, Wulfkuhle JD, Espina V, Mullarkey MJ, Kay EW, Liotta LA, Petricoin EF 3rd. Quantitative cell signalling analysis reveals down-regulation of MAPK pathway activation in colorectal cancer. J Pathol. 2009;218:514–519. doi: 10.1002/path.2561. [DOI] [PubMed] [Google Scholar]
- 15.Liu Z, Mai C, Yang H, Zhen Y, Yu X, Hua S, Wu Q, Jiang Q, Zhang Y, Song X, Fang W. Candidate tumour suppressor CCDC19 regulates miR-184 direct targeting of C-Myc thereby suppressing cell growth in non-small cell lung cancers. J Cell Mol Med. 2014;18:1667–1679. doi: 10.1111/jcmm.12317. [DOI] [PMC free article] [PubMed] [Google Scholar]


