Abstract
Gastric cancer is a commonly found malignant tumor, yet research on biomarkers of gastric cancer still face tremendous challenges. This study is the first to use gas chromatography-mass spectrometry (GC-MS) to measure and compare the metabolic profiles of gastric cancer cell lines with varying degrees of differentiation (MKN-28, SGC-7901, and AGS) with that of a normal gastric epithelial cell line (GES-1). OPLS-DA models were established to distinguish gastric cancer cell lines from a normal gastric epithelial cell line. In this study, we identified 278 metabolites, of which 111 show similarity scores greater than 700. Most notably, 6 metabolites (alanine, α-ketoisocaproic acid, proline, glyceric acid, pantothenic acid, and adenosine) showed varying expression levels between gastric cancer cell lines and a normal gastric epithelial cell line. These metabolites are potential biomarkers of gastric cancer and may be of great significance for the diagnosis, treatment and prognosis of gastric cancer patients.
Keywords: Gastric cancer cells, metabolomics, biomarker, GC-MS
Introduction
Gastric cancer is 4th most common and 2nd most deadly malignant tumor worldwide. Gastric cancer is asymptomatic in its early stages and there is no effective diagnostic method. Because of this, gastric cancer is usually only discovered at a more advanced stage and patients with advanced gastric cancer have poor prognoses. The five-year survival rate in advanced gastric cancer is lower than 10%, whereas gastric cancer detected at an earlier stage has a post-surgical survival rate of up to 90%.
Currently, endoscopy is one of the common screening methods for early stage gastric cancer. However, this method is invasive and the results are not definitive. There is a need for noninvasive methods and more effective tumor biomarkers in screening for early stage gastric cancer. Currently used biomarkers are pepsinogen, CEA, and CA199, all of which are not sufficiently accurate for early gastric cancer diagnosis.
Metabolomics is an extension of the fields of genomics, transcriptomics, and proteomics, and is considered as the final destination of “omics” research. It is a hot area of research in the post-genomics era. The growth and development of tumors are intimately linked to abnormal metabolic processes, as metabolic products are the final products of cellular gene expression. Small changes in gene expression can become amplified in the resulting metabolites and the study of metabolomics can further expand the scope of research in life sciences. Metabolomic techniques have been used in the study of ovarian, colon, and pancreatic tumors [1-3]. In recent years, the use of mass spectrometry (MS) and nuclear magnetic resonance (NMR) has increased in research on human gastric cancer tissue, blood, and urinary metabolites [4-6]. However, these techniques have thus far not been used in the study of gastric cancer cell lines at varying degrees of differentiation and their metabolic profiles in vitro.
This article reports for the first time that the use of GC-MS to compare the metabolic profiles of gastric cancer cell lines at varying degrees of differentiation and a normal gastric epithelial cell line. The OPLS-DA models were established to distinguish between gastric cancer cell lines with varying degrees of differentiation and a normal gastric epithelial cell line. The metabolites identified in this study may be potential markers of clinical gastric cancer.
Materials and methods
Cell culture and collection
The cell lines used in this study were gastric cancer cell lines MKN-28 (well-differentiated), SGC-7901 (moderately differentiated), and AGS (poorly differentiated), as well as the normal gastric epithelial cell line GES-1. The cell lines were obtained from Beijing Institute for Cancer Research (Beijing, China). The culture medium for MKN-28, SGC-7901, and GES-1 cells was RPMI 1640/10% Newborn Calf Serum/1% antibiotic (Gibco). The culture medium for AGS cells was F-12K/10% Newborn Calf Serum/1% antibiotic (Gibco). The incubation environment was 5% CO2, 37°C. The inoculation concentration was 1 × 106 cells/mL, and the cells were grown to approximately 90% confluence. Ten EP tubes were collected for each cell line (1 × 107 cells/tube). The tubes were frozen with liquid nitrogen and stored at -80°C.
Cell metabolite extraction
One mL of extraction solution, a mixture of methanol-chloroform (3:1, v/v) was added to each cell-containing EP tube. Twenty μL of L-2-chlorophenylalanine (1 mg/mL stock in dH2O) was added to each tube and the mixture was vortexed for 30 s. Porcelain beads were added and a 45 Hz mill was used to process the sample for 4 min. The sample was placed on an ice water bath and sonicated for 5 min and sonication was repeated five times. The sample was then centrifuged at 13,000 rpm for 15 min at 4°C and 0.89 mL of the supernatant was carefully pipetted into a 2 mL sample vial. The extract was dried in a vacuum concentrator and 30 μL methoxy amination hydrochloride (20 mg/mL in pyridine) was added to each tube of dried metabolites, gently mixed, then placed in an oven and incubated at 80°C for 30 min. 40 μL of BSTFA (containing 1% TMCS, v/v) was added to each sample and the mixture was incubated at 70°C for 1.5 h. The samples were then cooled to room temperature and 5 μL FAMEs (Standard mixture of fatty acid methyl esters, C8-C16: 1 mg/mL; C18-C24: 0.5 mg/mL in chloroform) was added to each vial. All samples were then analyzed by GC-MS.
GC-MS analysis
GC-MS analysis was performed using an Agilent 7890A gas chromatograph system coupled with a Pegasus HT time-of-flight mass spectrometer (LECO, St Joseph, MI, USA). The system was equipped with a DB-5MS capillary column coated with 5% diphenyl cross-linked with 95% dimethylpolysiloxane (30 m × 250 μm inner diameter, 0.25 μm film thickness; J&W Scientific, Folsom, CA, USA). Each 2-μL aliquot was injected in splitless mode with helium as the carrier gas at a flow rate of 1 mL/min. The temperature at 50°C was kept for 1 min in the beginning, and then it was gradually increased up to 310°C at a rate of 10°C/min, finally maintained at 310°C for 5 min. The temperatures of injection, transfer line, and ion source were 280°C, 270°C, and 220°C respectively. The electron energy was -70eV in impact mode. The MS data, which was with the m/z range of 50-500, and at a rate of 12/s spectra after a solvent delay of 370 s, were acquired in full-scan mode.
Data processing
Data were processed and analyzed using Chroma T0F4.3X software (LECO) and compared against the LECO-Fiehn Rtx5 database [7]. Comparison with standards in the database yielded a similarity score for identified metabolites. A perfect match between standard and sample spectra had a similarity score of 1000. If the similarity score for a metabolite was above 700, then the identification was considered credible. If the similarity score was below 200, then the identification was used as an “analyte”. If the similarity score was between 200 and 700, then the identification was inferred to be based on some evidence. The half of minimum value method was used to model any missing values in the raw data. The data were filtered data and noise removed by removing data containing over 80% null values. The filtered data was normalized using an internal standard and SIMCA-P + 14.0 software (Umetrics, Umea, Sweden) was used to further analyze the processed data. Principal component analysis (PCA) modeling was applied to visualize the data and to separate data from each sample, and an initial grouping was performed. In order to obtain a higher level of group separation and get a better understanding of variables responsible for classification, supervised orthogonal projections to latent structures-discriminate analysis (OPLS-DA) were applied. Then, we further validated the method using sevenfold cross-validation and a 200 permutation test, and the goodness of fit parameter (R2) and the goodness of prediction parameter (Q2) values were used to assess the quality of the models, respectively. Finally, the VIP value of the first principal component of variable importance in the projection was calculated and the P value was calculated using Student’s t-test. The standard selection threshold for differential metabolites was set at VIP > 1.0 and P < 0.05 [7,8].
Results
Development of a predictive model for gastric cancer cells with various differentiation grades
PCA showed the original distribution of two sets of data. The PCA model established in this experiment can fully separate the gastric cancer cells from the normal gastric epithelial cells (Figures 1 and 2A); indicating that when compared with normal gastric epithelial cells, gastric cancer cells show significant changes in intracellular metabolites.
Figure 1.
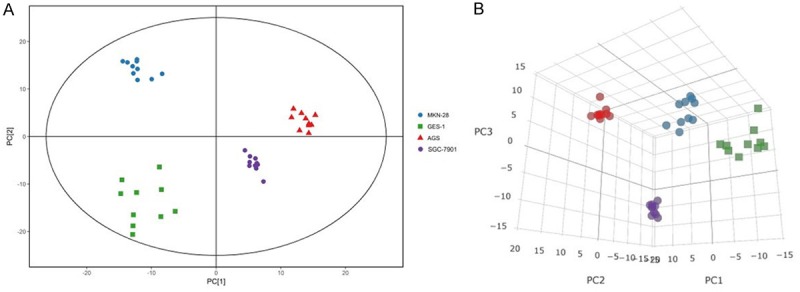
PCA analysis of GC-MS metabolite profiles. A. PCA score plot for gastric epithelial and cancer cells (n = 10 for each group). The X-axis, PC [1], and Y-axis, PC [2], indicate the first and second principal components, respectively. B. 3-D score plot of the PCA analysis.
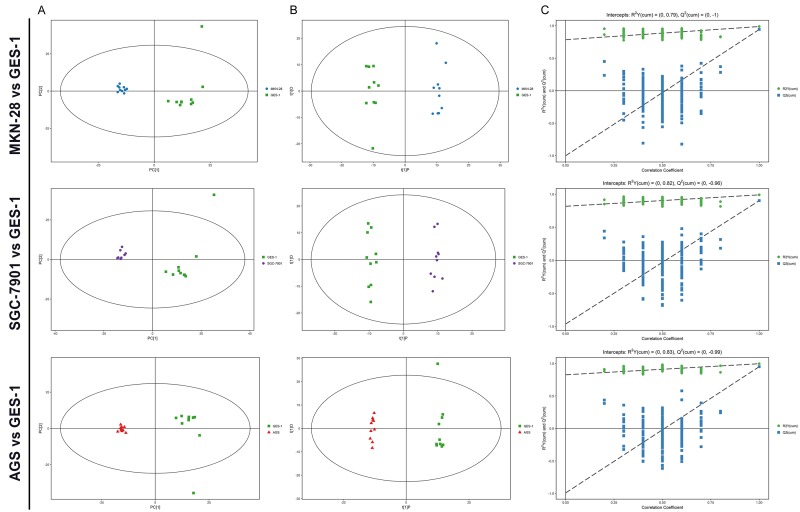
Figure 2.
Metabolic profiling between various differentiation grades of gastric cancer cells and normal controls. A. PCA score plots based on various differentiation grades of gastric cancer cells and normal controls. B. OPLS-DA scores plots based on various differentiation grades of gastric cancer cells and normal controls. C. Statistical validation of thecorresponding OPLS-DA models using permutation analysis (200 times).
Through multivariate statistical analysis of the metabolomic data from gastric cancer cells and that from normal gastric epithelial cells, many significant differences were found. Using these data, the OPLS-DA model could completely separate gastric cancer cells, regardless of degree of differentiation, from normal gastric epithelial cells (Figure 2B).
Permutation analysis of the corresponding OPLS-DA is shown in Figure 2C. The parameters for different gastric cancer cell lines were as follows: MKN-28: R2Y = 0.988, Q2 = 0.943; SGC-7901: R2Y = 0.991, Q2 = 0.903 and AGS: R2Y = 0.997, Q2 = 0.951, which indicated the excellence of the model.
Multivariate statistical analysis between gastric cancer cells with various differentiation grades and normal controls
A total of 138 distinct metabolites were identified by multivariate analysis (VIP > 1, P < 0.05). Of these, six metabolites showed significant differences between gastric cancer cells and normal gastric epithelial cells (Table 1). Alanine, α-ketoisocaproic acid and adenosine were elevated in all three gastric cancer cell lines. Glyceric acid was lower in all three gastric cancer cell lines. Proline was significantly increased in the moderately differentiated gastric cancer cell line, but was lower in highly differentiated and poorly differentiated gastric cancer cell lines. Pantothenic acid was lower in moderately differentiated gastric cancer cell lines, but elevated in highly differentiated and poorly differentiated gastric cancer cell lines.
Table 1.
Potential biomarkers of various differentiation grades of gastric cancer identified by GC-MS
| Name | MKN-28/GES-1 | SGC-7901/GES-1 | AGS/GES-1 | ||||||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| VIP* | Fold change | P-value | VIP* | Fold change | P-value | VIP* | Fold change | P-value | |
| Alanine | 1.75 | 1.91 | 0.000 | 2.00 | 1.66 | 0.000 | 2.02 | 2.25 | 0.000 |
| Proline | 1.49 | 0.66 | 0.002 | 1.44 | 1.28 | 0.006 | 1.53 | 0.68 | 0.001 |
| Adenosine | 2.17 | 9.66 | 0.000 | 2.33 | 3.15 | 0.000 | 2.22 | 10.66 | 0.000 |
| Pantothenic acid | 1.04 | 1.70 | 0.040 | 2.36 | 0.20 | 0.000 | 2.03 | 2.71 | 0.000 |
| Glyceric acid | 2.20 | 0.06 | 0.000 | 2.39 | 0.14 | 0.000 | 2.15 | 0.12 | 0.000 |
| α-ketoisocaproic acid | 1.36 | 2.35 | 0.003 | 1.37 | 2.19 | 0.007 | 1.30 | 2.12 | 0.010 |
VIP, variable importance for the projection.
Discussion
To our knowledge, this is the first study to undertake a metabolomic study of gastric cancer cell lines at varying degrees of differentiation (highly, moderately, and poorly differentiated) and to compare their metabolic profile to that of a normal gastric epithelial cell line.
Amino acid levels in gastric cancer cell lines showed serious metabolic disruptions. Alanine and α-ketoisocaproic acid levels were elevated in all three gastric cancer cell lines when compared with the normal gastric epithelial cell line. α-ketoisocaproic acid may be produced through the action of reversible aminotransferase (AT) on leucine. Proline was significantly elevated in moderately differentiated gastric cancer cell line, but was reduced in highly differentiated and poorly differentiated gastric cancer cell lines. Previous studies have reported that alanine, leucine and proline levels in tissue from gastric cancer patients are significantly higher than those found in healthy individuals [9]. However, patients with different types of tumors can also have different blood amino acid levels [10]. In our experiments, the elevated levels of three amino acids observed may be due to a lack of amino acids in the tumor microenvironment, resulting in high rates of protein breakdown. The reduction of proline levels in the highly and poorly differentiated gastric cancer cell lines may be due to excessive energy consumption by tumor cells, shunting proline into the tricarboxylic acid cycle. Therefore, we speculate that in gastric cancer patients, the amino acid changes are not only associated with tumor type, but also with the degree of tumor differentiation.
The level of glyceric acid decreased in all three gastric cancer cell lines. Glyceric acid is an intermediate of serine degradation and is phosphorylated to produce glycerate 3-phosphate, which is involved in glycolysis, an importantsource of energy for tumor cells. Researchers have found that glyceric acid is present at lower levels in highly metastatic breast cancer cells when compared to low metastatic breast cancer cells [11]. Glyceric acid levels are also decreased in the blood of patients with advanced pancreatic cancer [12]. Therefore, we can ascertain that in tumor development, different stages of tumor differentiation will involve changes in glyceric acid content.
Adenosine levels were elevated in all three differentiated gastric cancer cell lines. Previous studies suggest that adenosine levels were also increased in blood from gastric cancer patients, in tissues and urine from colon cancer patients, and in the urine of lung cancer patients [13,14]. Adenosine is one of the four major mononucleotides, and the metabolite via which energy is transferred in the body. Tumor cells consume a large quantity of adenosine triphosphate (ATP) to maintain their growth, resulting in the accumulation of excess adenosine, and the production of adenosine also provides raw materials for tumor nucleic acid synthesis.
Pantothenic acid levels were increased in highly differentiated and poorly differentiated gastric cancer cell lines, but decreased in the moderately differentiated gastric cancer cell line. Increased pantothenic acid in tumor cells can increase the production of CoA, promote the production of phospholipids, and protect against lipid peroxidation. At the same time, pantothenic acid also increases the content of GSH in cells. Both metabolic routes have a protective effect for tumor cells [15,16]. The reduction of pantothenic acid in the moderately differentiated gastric cancer cell line may be due to the over-proliferation of tumor cells, resulting in depletion of pantothenic acid.
Tumor prediction models and biomarkers have been reported in a number of studies. Recent studies have shown that circulating tumor cells are linked to different types of tumor metastasis and spread [17,18]. Certain studies claim that circulating tumor DNA can be used as a diagnostic marker for early stage gastric cancer [19]. In metabolomics research, Chen and colleagues [20] used GC-MS in combination with surface-enhanced Raman scattering to test the composition of exhaled gas from gastric cancer patients (early and advanced stage) and from healthy individuals, and found differences in 14 volatile organic compounds which may be used to diagnose early stage gastric cancer. Wang [4] and colleagues used 1HNMR to study the metabolic profiles of gastric cancer tissues from different TNM stages. Forty-eight different metabolites were found, of which 13 expressed changes with the progression of gastric cancer. The numerical model established in that study showed that the AUC value of gastric cancer diagnosis was 0.945.
In this study, the OPLS-DA model that we established using GC-MS was able to distinguish the metabolic profiles of gastric cancer cells at different degrees of differentiation from normal gastric epithelial cells. All metabolite data were used to establish the model. Multivariate analysis established that our model has better predictive value and accuracy than traditional univariate analysis.
Our study also found that gastric cancer cell lines show significant changes in their metabolic processes when compared to a normal gastric epithelial cell line, including metabolism of carbohydrates, amino acids, purines, fatty acids, and other small molecules. With the establishment of our model, six metabolites were screened as potential tumor biomarkers for gastric cancer, which is of great significance for the early diagnosis and treatment of gastric cancer. However, in the end, whether these putative metabolic markers of gastric cancer are clinically relevant will require further validation in large multicenter clinical studies.
Metabolomic analysis can be used to discover potential tumor biomarkers and complement genomic and proteomic research. The technique used in this study was GC-MS. In the future, metabolic profiling of gastric cancer cells can also be performed using techniques such as LC-MS and NMR. Different detection technologies have different sensitivity and accuracy for the detection of metabolites. The range of detection of metabolites will not be the same. “Precision Medicine” is gaining more and more attention in cancer treatment. By combining the areas of metabolomics, proteomics, and genomics, individualized treatment programs can tailored to guide clinical practice. Therefore, accelerating metabolomic research in cancer is of great significance for the screening, treatment, and prevention of tumors.
Acknowledgements
This work was supported by National Natural Science Foundation of China (no. 81672410).
Disclosure of conflict of interest
None.
References
- 1.Chan EC, Kho PK, Mal M, Cheah PY, Eu KW, Backshall A, Cavill R, Nicholson JK, Keun HC. Metabolic profiling of human colorectal cancer using high-resolution magic angle spinning nuclear magnetic resonance (HR-MAS NMR) spectroscopy and gas chromatography mass spectrometry (GC/MS) J Proteome Res. 2008;8:352–361. doi: 10.1021/pr8006232. [DOI] [PubMed] [Google Scholar]
- 2.Denkert C, Budczies J, Kind T, Weichert W, Tablack P, Sehouli J, Niesporek S, Konsgen D, Dietel M, Fiehn O. Mass spectrometry-based metabolic profiling reveals different metabolite patterns in invasive ovarian carcinomas and ovarian borderline tumors. Cancer Res. 2006;66:10795–10804. doi: 10.1158/0008-5472.CAN-06-0755. [DOI] [PubMed] [Google Scholar]
- 3.Urayama S, Zou W, Brooks K, Tolstikov V. Comprehensive mass spectrometry based metabolic profiling of blood plasma reveals potent discriminatory classifiers of pancreatic cancer. Rapid Commun Mass Spectrom. 2010;24:613–620. doi: 10.1002/rcm.4420. [DOI] [PubMed] [Google Scholar]
- 4.Wang H, Zhang H, Deng P, Liu C, Li D, Jie H, Zhang H, Zhou Z, Zhao YL. Tissue metabolic profiling of human gastric cancer assessed by 1H NMR. BMC Cancer. 2016;16:371–383. doi: 10.1186/s12885-016-2356-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuligowski J, Sanjuan-Herráez D, Vázquez-Sánchez MA, Brunet-Vega A, Pericay C, Ramírez-Lázaro MJ, Lario S, Gombau L, Junquera F, Calvet X, Quintas G. Metabolomic analysis of gastric cancer progression within the Correa’s cascade using ultra performance liquid chromatography-mass spectrometry. J Proteome Res. 2016;15:2729–2738. doi: 10.1021/acs.jproteome.6b00281. [DOI] [PubMed] [Google Scholar]
- 6.Liang Q, Wang C, Li B. Metabolomic analysis using liquid chromatography/mass spectrometry for gastric cancer. Appl Biochem Biotechnol. 2015;176:2170–2184. doi: 10.1007/s12010-015-1706-z. [DOI] [PubMed] [Google Scholar]
- 7.Kind T, Wohlgemuth G, Lee DY, Lu Y, Palazoglu M, Shahbaz S, Fiehn O. FiehnLib: mass spectral and retention index libraries for metabolomics based on quadrupole and time-of-flight gas chromatography/mass spectrometry. Anal Chem. 2009;81:10038–10048. doi: 10.1021/ac9019522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hirayama A, Kami K, Sugimoto M, Sugawara M, Toki N, Onozuka H, Kinoshita T, Saito N, Ochiai A, Tomita M, Esumi H, Soga T. Quantitative metabolome profiling of colon and stomach cancer microenvironment by capillary electrophoresis time-of-flight mass spectrometry. Cancer Res. 2009;69:4918–4925. doi: 10.1158/0008-5472.CAN-08-4806. [DOI] [PubMed] [Google Scholar]
- 10.Miyagi Y, Higashiyama M, Gochi A, Akaike M, Ishikawa T, Miura T, Saruki N, Bando E, Kimura H, Imamura F, Moriyama M, Ikeda I, Chiba A, Oshita F, Imaizumi A, Yamamoto H, Miyano H, Horimoto K, Tochikubo O, Mitsushima T, Yamakado M, Okamoto N. Plasma free amino acid profiling of five types of cancer patients and its application for early detection. PLoS One. 2011;6:e24143. doi: 10.1371/journal.pone.0024143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim HY, Lee KM, Kim SH, Kwon YJ, Chun YJ, Choi HK. Comparative metabolic and lipidomic profiling of human breast cancer cells with different metastatic potentials. Oncotarget. 2016;7:67111–67128. doi: 10.18632/oncotarget.11560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nishiumi S, Shinohara M, Ikeda A, Yoshie T, Hatano N, Kakuyama S, Mizuno S, Sanuki T, Kutsumi H, Fukusaki E, Azuma T, Takenawa T, Yoshida M. Serum metabolomics as a novel diagnostic approach for pancreatic cancer. Metabolomics. 2010;6:518–528. [Google Scholar]
- 13.Ong ES, Zou L, Li S, Cheah PY, Eu KW, Ong CN. Metabolic profiling in colorectal cancer reveals signature metabolic shifts during tumorigenesis. Mol Cell Proteomics. 2010 doi: 10.1074/mcp.M900551-MCP200. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 14.Hsu WY, Chen CJ, Huang YC, Tsai FJ, Jeng LB, Lai CC. Urinary nucleosides as biomarkers of breast, colon, lung, and gastric cancer in taiwanese. PLoS One. 2013;8:e81701. doi: 10.1371/journal.pone.0081701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Slyshenkov VS, Omelyanchik SN, Moiseenok AG, Trebukhina RV, Wojtczak L. Pantothenol protects rats against some deleterious effects of gamma radiation. Free Radic Biol Med. 1998;24:894–899. doi: 10.1016/s0891-5849(97)00378-x. [DOI] [PubMed] [Google Scholar]
- 16.Slyshenkov VS, Moiseenok AG, Wojtczak L. Noxious effects of oxygen reactive species on energy-coupling processes in Ehrlich ascites tumor mitochondria and the protection by pantothenic acid. Free Radic Biol Med. 1996;20:793–800. doi: 10.1016/0891-5849(95)02210-4. [DOI] [PubMed] [Google Scholar]
- 17.Allard WJ, Matera J, Miller MC, Repollet M, Connelly MC, Rao C, Tibbe AG, Uhr JW, Terstappen LW. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin Cancer Res. 2004;10:6897–6904. doi: 10.1158/1078-0432.CCR-04-0378. [DOI] [PubMed] [Google Scholar]
- 18.Paterlini-Brechot P, Benali NL. Circulating tumor cells (CTC) detection: clinical impact and future directions. Cancer Lett. 2007;253:180–204. doi: 10.1016/j.canlet.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 19.Elshimali YI, Khaddour H, Sarkissyan M, Wu Y, Vadgama JV. The clinical utilization of circulating cell free DNA (CCFDNA) in blood of cancer patients. Int J Mol Sci. 2013;14:18925–18958. doi: 10.3390/ijms140918925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen Y, Zhang Y, Pan F, Liu J, Wang K, Zhang C, Cheng S, Lu Lg, Zhang Z, Zhi X, Zhang Q, zhang W, Chen D, Alfranca G, De La Fuente MJ, Cui D. Breath analysis based on surface enhanced rama scattering sensors distinguishes early and advanc gastric cancer patients from healthy persons. ACS Nano. 2016;10:8169–8179. doi: 10.1021/acsnano.6b01441. [DOI] [PubMed] [Google Scholar]