Abstract
The aim of this study was to clarify the clinicopathological features and role of zinc finger protein 10 (ZNF10) in breast invasive ductal cancer (IDC). Our data first showed that ZNF10 expression was higher in 8 pairs of fresh breast IDC and breast cancer cell lines compared with their respective adjacent non-tumor breast tissues. ZNF10 expression was significantly higher in IDC compared with DCIS and fibroadenoma of the breast. ZNF10 expression was significantly associated with patients’ age, tumor stage, and breast cancer molecular subtype. ZNF10 knockdown inhibited breast cancer cell proliferation, colony formation, cell cycle progression, cell migration, and invasion but induced apoptosis. ZNF10 knockdown also suppressed the tumorigenicity of breast cancer in vivo. The underlying mechanism study showed that ZNF10 regulated the β-catenin signaling pathway in breast cancer. ZNF10 might bind to the region (nucleotides -300 to +100) of the β-catenin promoter. In conclusion, our results first suggest that ZNF10 promotes the carcinogenesis and progression of breast IDC via the β-catenin signaling pathway. Targeting ZNF10 might be a novel treatment strategy for breast cancer.
Keywords: ZNF10, β-catenin, breast cancer, carcinogenesis, clinicopathologic significance
Introduction
The catenins (α, β, γ) are composed of a group of cytoplasmic proteins with common structural and functional features [1]. The interaction between E-cadherin and the catenins is essential for the adhesive function of E-cadherin and depends largely on β-catenin expression and function. β-catenin associates directly with E-cadherin and through α-catenin with the cellular cytoskeleton producing stable cell-cell adhesion [2]. In addition, pro-oncogenic factors including ras, epidermal growth factor, and c-erbB-2 release β-catenin from the adherens complex and encourage translocation to the nucleus. Association of nuclear β-catenin with the T cell factor/lymphoid enhancer factor family of transcription factors promotes the expression of several compounds including c-myc, cyclin D1, and matrix metalloproteinase (MMP)-7 that have important roles in the development and progression of colorectal carcinoma [3]. The loss of β-catenin expression is associated with metastasis and poor prognosis in invasive breast cancer [4].
A conserved sequence is found at the N-terminus of many human zinc finger proteins of the Krüppel type (C2H2 zinc fingers), referred to as Krüppel-associated box (KRAB) domain. The KRAB domain consists of ~75 amino acids, that can be further subdivided into an A box and a B box. The A box, but not the B box, is present in every KRAB domain and essential for transcriptional repression [5,6]. KRAB-containing zinc finger (ZNF) genes constitute the single largest gene family of transcriptional repressors in the genomes of higher organisms. ZNF121 expression is upregulated in human breast cancer, and the up-regulation significantly associates with worse patient survival in the luminal A subtype of breast cancer [7]. ZNF224 induces cell growth and apoptosis-resistance by downregulating p21 and p53 via miR-663a in breast cancer [8]. MED28 increases the colony-forming ability of breast cancer cells by stabilizing the ZNF224 protein upon DNA damage [9]. However, the ectopic expression of ZNF23 induces cell apoptosis by the activation of caspase-3, p27, p53 expression and the downregulation of Bcl-2 through the mitochondria-dependent pathway [10]. ZNF10 inhibits HIV-1 LTR activity by interacting with NF-kappaB and Sp1 binding motifs [11]. Kretschmer et al. identified early molecular markers in breast cancer by gene expression profiling analysis [12]. However, the role of ZNF10 remains unclear in breast cancer. Our current study aims to clarify the clinicopathological significance of ZNF10 in breast IDC.
Materials and methods
Cell lines and small interfering RNA (siRNA) sequences
The human mammary cancer cell lines MCF7, BT549 and MDA-MB-231 were purchased from ATCC and maintained in Dulbecco’s modified Eagle’s medium (Invitrogen, Carlsbad, CA). All media were supplemented with 10% (v/v) fetal bovine serum (Invitrogen), 1× antibiotic/antimycotic (100 units/mL streptomycin, 100 units/mL penicillin, and 0.25 mg/mL amphotericin B). All cell lines were cultured in a humidified incubator at 37°C with 5% CO2.
ZNF10 siRNA sequences followed: 5’-GGA GAG CAG TTC TTA ACA T dTdT-3’. The siRNA duplexes were chemically synthesized and purified by Ribobio Co. Ltd (Guangzhou, China). The siRNA was transfected using the Lipofectamine RNAiMAX transfection reagent (Invitrogen). Scrambled siRNA (NC-siRNA) was used as a negative control group.
Patient information and tissue specimen
Eight pairs of fresh female breast cancer tissues and their respective adjacent non-tumor tissue samples were collected from our institute in 2015, and a total of 121 samples of breast IDC, 14 DCIS and 29 fibroadenoma of the breast paraffin-embedded archived tumor tissues were collected between 2000 and 2009 from our Institute. All tissues were fixed in 10% neutral buffered formalin within 30 minutes after tumor resection for 24-48 hours. For the research purposes of these clinical materials, prior patient consent and approval from the first affiliated Hospital, Sun Yat-sen University Research Ethics Committee were obtained. No patients had received chemotherapy or radiotherapy before their operations. Pertinent follow-up information was available for all patients. Detailed clinical information is summarized in Table 1.
Table 1.
ZNF10 expression and its association with clinicopathological features and β-catenin expression of breast IDC
| Characteristics | ZNF10 expression | p-value | ||
|---|---|---|---|---|
|
| ||||
| High No. (%) | Low No. (%) | |||
| Age (years) | ≤50 | 46 (69.7) | 20 (30.3) | 0.002 |
| >50 | 23 (41.8) | 32 (58.2) | ||
| TNM | Stage I | 17 (70.8) | 7 (29.2) | 0.127 |
| Stage II-III | 52 (53.6) | 45 (46.4) | ||
| T classification | T1-2 | 65 (63.0) | 43 (37) | 0.043 |
| T3-4 | 4 (30.8) | 9 (69.2) | ||
| Lymph node metastasis | Yes | 33 (55.9) | 26 (44.1) | 0.813 |
| No | 36 (58.1) | 26 (41.9) | ||
| ER | Positive | 44 (53.7) | 38 (46.3) | 0.278 |
| Negative | 25 (64.1) | 14 (35.9) | ||
| PR | Positive | 50 (57.5) | 37 (42.5) | 0.874 |
| Negative | 19 (55.9) | 15 (44.1) | ||
| HER2 | Positive | 24 (63.2) | 14 (36.8) | 0.357 |
| Negative | 45 (54.2) | 38 (45.8) | ||
| Histological grade | I-II | 48 (57.8) | 35 (42.2) | 0.791 |
| III | 21 (55.3) | 17 (44.7) | ||
| Molecular subtype | Luminal A | 23 (74.2) | 8 (25.8) | 0.023 |
| Luminal B1 | 20 (40.0) | 30 (60.0) | ||
| Luminal B2 | 11 (61.1) | 7 (38.9) | ||
| HER2 positive | 9 (75.0) | 3 (25.0) | ||
| Triple negative | 6 (60.0) | 4 (40.0) | ||
| β-catenin expression | Normal | 40 (72.7) | 15 (27.3) | 0.001 |
| Abnormal | 29 (43.9) | 37 (56.1) | ||
Immunohistochemical staining
As we previously described [14], the primary antibodies including ZNF10 (Abcam, Cambridge, MA, USA), β-catenin (Santa Cruz Biotechnology, Santa Cruz, CA), estrogen receptor (ER), progesterone receptor (PR), human epidermal growth factor receptor type 2 (HER2) from the Roche Company, and Ki-67 (Dako) were used. The positive signal of ZNF10 was localized in the cell nucleus. The positive signal of β-catenin was located in the cell membrane and/or cytoplasm. The degree of ZNF10 immunostaining was based on both the proportion of positively stained tumor cells and the intensity of staining. The proportion of positively stained tumor cells was scored as follows: 0 (0-5% positive tumor cells), 1 (>5-25% positive tumor cells), 2 (>25-50% positive tumor cells), 3 (>50-75% positive tumor cells), and 4 (>75% positive tumor cells). Staining intensity was classified according to the following criteria: 0 (no staining); 1 (weak staining = light yellow), 2 (moderate staining = yellow-brown), and 3 (strong staining = brown). The staining index was calculated as the staining intensity score × the proportion score. We evaluated ZNF10 expression in the breast tumor specimens by determining the staining index, which scores as 0, 1, 2, 3, 4, 6, 8, 9, and 12. The staining index score of 2 (the cutoff point) was used to distinguish between low and high expressions of ZNF10.
The expression of β-catenin was evaluated according to the method established by Dillon et al. [15]. There are three expression patterns: 1) normal, cases not reproducibly distinguishable from normal in either pattern or intensity; 2) altered, a broken or discontinuous staining pattern, or a patchy staining pattern with or without a decrease in intensity; and 3) loss, a complete loss of staining as the predominant pattern in the section examined. Altered and loss of β-catenin staining was considered as abnormal β-catenin expression. The HER2 status was assessed by immunohistochemistry and the tumors were considered HER2-positive when IHC staining was 3+ (uniform, intense membrane staining of >10% of invasive tumor cells) and 2+ (uniform, weak membrane staining of >10% of invasive tumor cells), in which the HER2 gene amplification was confirmed by fluorescence in situ hybridization (FISH).
Breast carcinoma molecular subtyping was performed according to the method established by Goldhirsh et al. (2011) [16] and classified as 5 subtypes using the expression or amplification of ER, PR, HER2 and Ki-67: luminal A (ER and/or PR-positive/HER2-negative/low Ki-67), luminal B (ER- and/or PR-positive/HER2-negative/high Ki-67), HER2-positive luminal B (ER- and/or PR-positive/HER2 overexpression/any Ki-67), non-luminal HER2-positive (ER and PR absent/HER2 overexpression), and triple negative (ER and PR absent/HER2-negative). The cutoff value for Ki-67 expression is 14% in breast cancer.
Cell proliferation assay
MCF7 and BT549 cells (1×103) were plated onto 96-well plates with a medium containing 10% FBS and incubated overnight. After transfection with 100 nM ZNF10 siRNA, cell proliferation was determined at 0, 24, 48, 72, and 96 hours using the Cell Counting Kit-8 (CCK8) (Keygene, China). The absorbance (OD) was measured at a wavelength of 450 nm using a Microplate Autoreader (Bio-Tek Instruments, VT).
Transwell migration and invasion assays
Migration and invasion assays were carried out in transwell chambers containing polycarbonate filters (8 μm pore size; Corning Incorporated, Life Sciences, NY). After being transfected with 100 nM ZNF10 siRNA for 48 hours, 2×104 (migration assay) or 2×105 (invasion assay) MCF7 and BT549 cells in a 500 μl serum-free medium, the samples were placed in the upper chambers and incubated at 37°C with 5% CO2 for 24 hours, respectively. At the same time, a 200 μl medium containing 15% FBS was added to the lower chamber as a chemoattractant. The cells were allowed to invade through the matrigel (BD Biosciences) or migrate for 24 hours at 37°C with 5% CO2. Following invasion or migration, cells were fixed with 4% formaldehyde and stained with 1% crystal violet. Cells on the upper surface of the filters were removed by wiping with a cotton swab. Cells counts were the mean number of cells per fields of view. Three independent experiments were performed and the data were presented as mean ± standard deviation (SD).
Xenograft tumor model
Female BALB/c-nude mice (4-5 weeks old and weighing 15-18 g) were housed under pathogen-free conditions. MCF7 cells were trypsinized, washed twice with a serum-free medium and reconstituted in the serum-free medium DMEM, mixed 1:1 with Matrigel (Becton Dickinson) and then inoculated subcutaneously into the right flank of each nude mouse. A local ZNF10 siRNA treatment was initiated when the tumor was palpable at a volume of approximately 30 mm3. The mice were randomly assigned into treatment and negative control groups (n=6 mice/group) and given intratumor injections with 2 nM ZNF10 siRNA or NC-siRNA dissolved in 30 μl PBS every 3 days. We modified siRNA with 2-O-methyl and conjugated cholesterol to the ends of the siRNA, which can retain the full potency of the siRNA, confers substantial nuclease resistance, improves bio-distribution, and facilitates entry into cells. The treatment time was 12 days. Tumor size was measured every 3 days using a digital caliper, and the tumor volume was calculated according to the formula: tumor volume (mm3) = length × width2 × 0.5. At the end of the experiment, all the mice were sacrificed and their total weights, the tumor weights, and the tumor volumes were recorded. All the experiments were performed following the Guide for the Care and Use of Laboratory Animals (National Institutes of Health publication).
Statistical analyses
The groups from the cell culture and in vivo experiments were compared using an unpaired, two-tailed Student’s t test, and the results were presented as mean ± SD. For the CCK8 assay, the comparison was done by univariate variance analysis (two-way ANOVA). Statistical analyses were performed using SPSS version 16.0 statistical software. P<0.05 was considered to be statistically significant.
Results
ZNF10 expression and its relationship with clinicopathological features and β-catenin expression of breast IDC
To identify the potential markers in breast cancer, we first analyzed the mRNA expression profiles of patients with IDC, DCIS, and healthy tissues of the human breast in the Gene Expression Omnibus (GEO) database [12]. We found ZNF10 mRNA expression was significantly higher in breast IDC compared with DCIS and healthy breast tissues (Figure 1A). Interestingly, there was a significantly positive correlation between ZNF10 mRNA expression and β-catenin protein expression by TCGA database analysis (P<0.05) (Figure 1B) [13]. To further determine the ZNF10 and β-catenin expression levels in breast cancer, our data showed that ZNF10 and β-catenin expressions were higher in 8 samples of fresh breast IDC compared with their respective adjacent, non-tumor breast tissues by Western blot analysis (Figure 1C). Meanwhile, ZNF10 and β-catenin protein expression was also higher in breast cancer cell lines than in human normal breast tissue (Figure 1D). Further study showed that high ZNF10 expression was found in 42.1% (69/121) IDC, 3% (5/14) in DCIS and 3.7% (6/29) in fibroadenoma of the breast. ZNF10 expression was significantly higher in IDC compared with DCIS and fibroadenoma of the breast (P=0.0012). Abnormal β-catenin expression was found in 54.5% (66/121) IDC. Normal β-catenin expression was found in 45.5% (55/121) IDC (Figure 1E). ZNF10 expression was significantly associated with patients’ age (P=0.002), tumor stage (P=0.043), breast cancer molecular subtype (P=0.023), and β-catenin expression (P=0.001). There was no significant association of ZNF10 expression with TNM, lymph node status, ER, PR, HER2, and histological grade of IDC (Table 1). In addition, there was a negative correlation between ZNF10 expression and normal β-catenin expression in breast IDC (P=0.001, r=-0.2782). Patients with high ZNF10 expression had a lower survival rate than patients with low ZNF10 expression in breast IDC, although there was no significant difference (P>0.05, Figure 1F).
Figure 1.
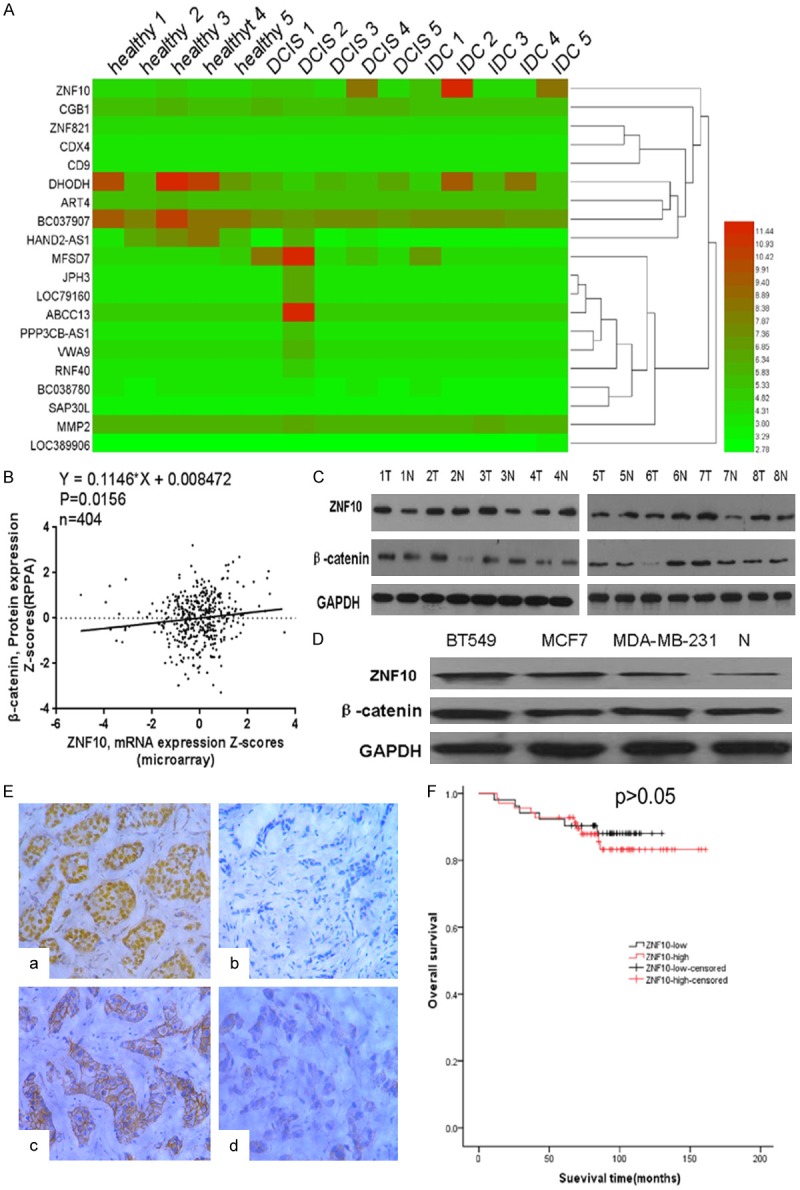
ZNF10 is overexpressed in breast IDC cells and tissues. A. ZNF10 mRNA expression was significantly higher in breast IDC compared with DCIS and healthy breast tissues from the GEO database. B. Positive correlation between ZNF10 mRNA expression and β-catenin protein expression from TCGA database analysis. C. ZNF10 and β-catenin expression level in 8 paired of fresh breast IDC samples (T) and adjacent non-tumor breast tissues (N) by Western blot analysis. D. ZNF10 and β-catenin expression in breast IDC cell lines and non-tumor breast tissue (N) by Western blot analysis. E. ZNF10 (a: high expression; b: low expression) and β-catenin (c: normal expression; d: abnormal expression) expression in breast IDC by immunohistochemical staining ×200. F. Overall survival of breast IDC patients with different ZNF10 expression level.
ZNF10 knock-down inhibited cell proliferation, colony formation, and cell cycle progression but induced apoptosis of breast cancer
To determine the biological significance of ZNF10 in breast cancer, ZNF10 expression was significantly decreased in MCF7 and BT549 cells transfected with ZNF10 siRNA at 48 hours compared with the control group, respectively (Figure 2A). CCK8 assay showed that ZNF10 knockdown significantly suppressed MCF7 and BT549 cell proliferation compared with the NC-siRNA control group and in a time-dependent manner (P<0.01 and P<0.01), respectively (Figure 2B). Furthermore, a cell colony formation assay demonstrated that the mean number (n=223) of colony formation in MCF7 cells transfected with ZNF10-siRNA was significantly less than the mean number (n=436) of colony formation in the NC-siRNA transfection group (P<0.01). Likewise, the mean number (n=263) of colony formation in BT549 transfected with ZNF10-siRNA was significantly less than mean number (n=443) of colony formation in NC-siRNA transfection group (P<0.01) (Figure 2C).
Figure 2.
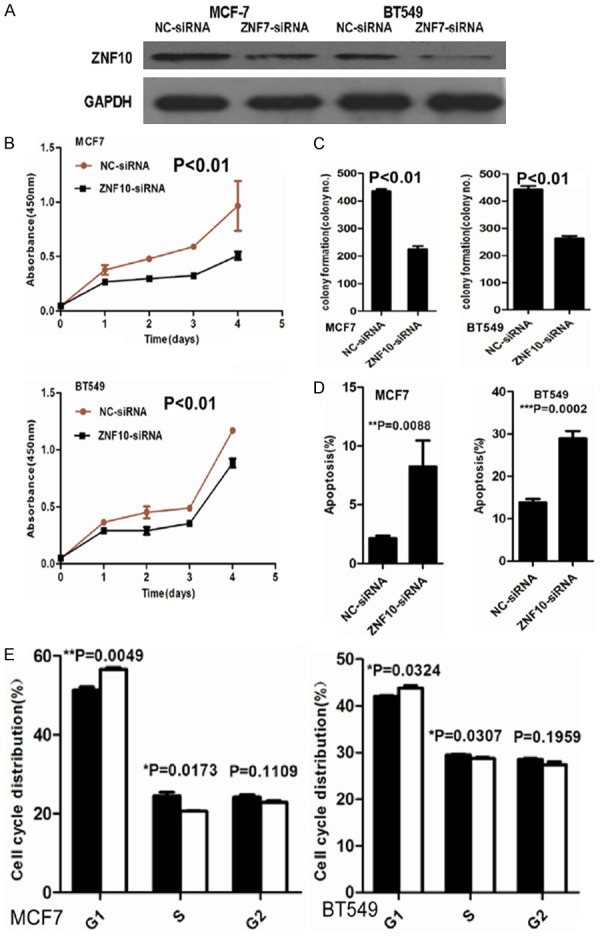
ZNF10 promotes proliferation and inhibits apoptosis of breast IDC cells. (A) ZNF10 expression decreased in MCF7 and BT549 cell lines transfected with ZNF10 siRNA compared with the control group by Western blot analysis, respectively. (B, C) ZNF10 knockdown significantly suppressed cell proliferation (B) and colony formation (C) in MCF7 and BT549 cells. (D, E) ZNF10 knockdown significantly induced cell apoptosis (D) and cell cycle G-S phase arrest (E) in MCF7 and BT549.
After 48 hours transfection, the proportion of apoptotic cells (5.7%) in MCF7 cells transfected with ZNF10-siRNA was significantly more than that of apoptotic cells (0.9%) in the control group (P<0.01). The proportion of apoptotic cells (19.5%) in BT549 cells transfected with ZNF10 si-RNA was significantly higher compared with 9.6% in the control group (P<0.01). Further cell cycle analysis showed that ZNF10 knock-down significantly induced G1-S phase arrest in MCF7 and BT549 compared with the control group (P=0.005 and P=0.032), respectively (Figure 2D, 2E).
ZNF10 knock-down inhibited breast cancer cell migration and invasion
A scratch wound assay showed that cell migration was dramatically inhibited in MCF7 and BT549 cells transfected with ZNF10-siRNA compared with NC-siRNA control groups at 12, 24, and 48 hours, respectively (Figure 3A). Furthermore, a transwell migration assay showed that the mean number (n=143) of migrated cells per field of view was significantly decreased in MCF7 transfected with ZNF10-siRNA than that (mean number =186) in the NC-siRNA control group (P=0.003). The mean number (n=45) of migrated cells per field of view was significantly decreased in BT549 transfected with ZNF10-siRNA than that (mean number =65) in the NC-siRNA control group (P<0.01) (Figure 3B). A transwell invasion assay showed that the mean number (n=57) of invasive cells in the MCF7 cells transfected with ZNF10-siRNA was significantly less than that (mean number =132) in the NC-siRNA control group (P<0.01). The mean number (n=31) of invasive cells in the BT549 cells transfected with ZNF10-siRNA was significantly less than that (mean number =42) in the NC-siRNA transfection group (P=0.02) (Figure 3C).
Figure 3.
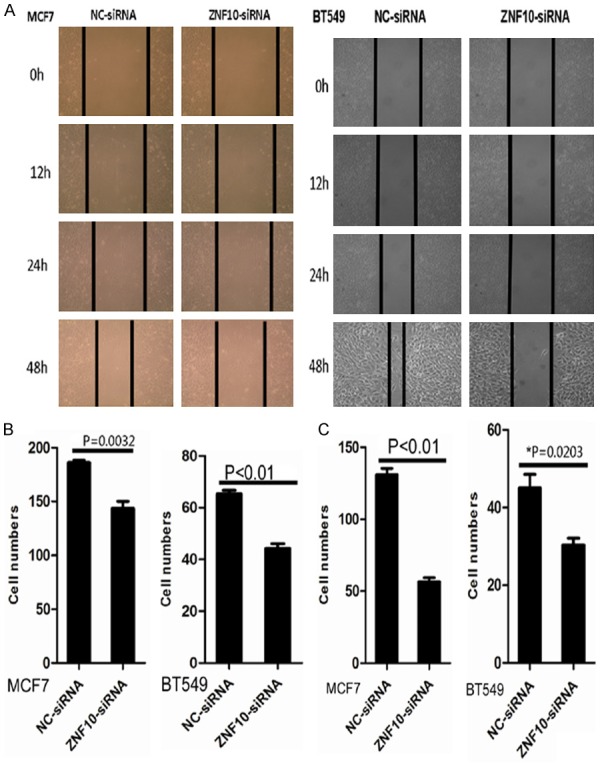
ZNF10 promotes migration and invasion of breast IDC cells. (A, B) ZNF10 knockdown suppressed MCF7 and BT549 cell migration by scratch wound healing assay (A) and cell migration assay (B), respectively. (C) ZNF10 knockdown significantly suppressed MCF7 and BT549 cell invasion by cell invasion assay.
ZNF10 knock-down inhibited the tumorigenicity of breast cancer in vivo
To determine whether ZNF10 affects the tumorigenicity of breast cancer in vivo, we performed a tumor growth experiment in nude mice using MCF7 cells. Tumor growth was significantly inhibited in the ZNF10-siRNA treatment group compared with the control group (P<0.01). The control group exhibited a rapid increase in tumor volume over 12 days. However, the ZNF10 siRNA treatment group showed a low increase in tumor volume. Concomitantly, the average tumor weight in the ZNF10 siRNA treatment group was significantly less than that in the control group upon termination of the experiment (P=0.029). Histological features of the tumor xenograft from the NC-siRNA group and the ZNF10-siRNA group were similar to human breast cancer tissues by haematoxylin and eosin staining. However, ZNF10 expression in enucleated tumors with ZNF10 siRNA treatment was dramatically lower than that in enucleated tumors with the NC-siRNA control group by immunohistochemical staining (Figure 4).
Figure 4.
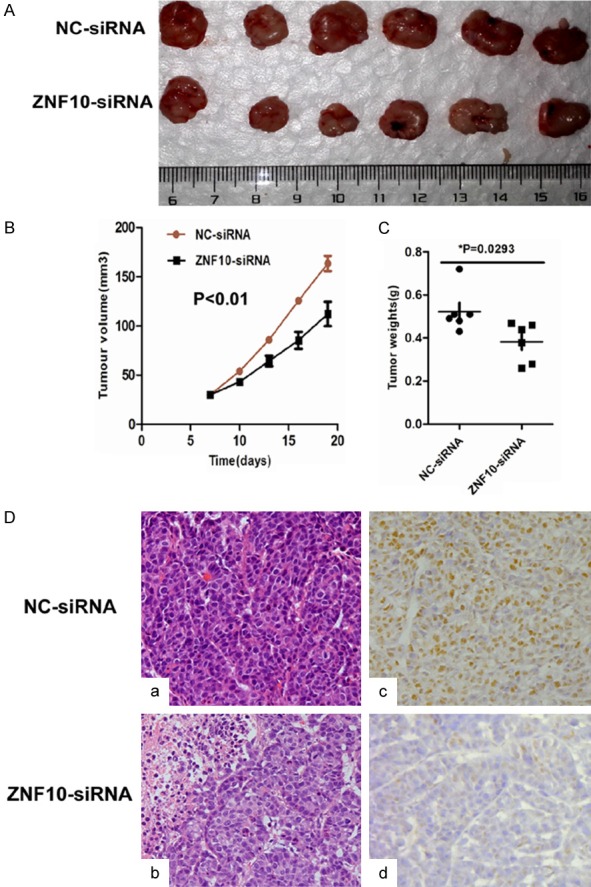
ZNF10 enhances the tumorigenicity of breast IDC in vivo. (A-C) ZNF10 knockdown significantly suppressed breast cancer growth (A), tumor weight (B), and the tumor volume (C) of MCF7 cells implanted subcutaneously in BALB/c-nu mice compared with the control group, respectively. (D) Histological morphology and ZNF10 expression in tumor xenografts generated by MCF7 cells transfected with ZNF10 siRNA compared with NC-siRNA group by hematoxylin and eosin staining (a, b) and immunohistochemistry staining (c, d) ×200.
ZNF10 knockdown inhibited β-catenin signaling in breast cancer
To determine the effect of ZNF10 on β-catenin signaling pathway in breast cancer, our data showed that ZNF10 knockdown dramatically inhibited β-catenin and its down-stream genes including c-Myc, cyclin D1 and MMP9 and up-stream gene p-GSK3β (Ser9) expression in MCF7 and BT549 cells compared with the NC-siRNA control group by Western blot analysis, respectively. Further cellular fractionation and Western blot analysis showed that decreased nuclear and cytoplasmic β-catenin expression was found in MCF7 and BT549 cells transfected with ZNF10 siRNA compared with the control group. Moreover, suppression of β-catenin expression was much more in cytoplasmic fraction compared with nuclear fraction of MCF7 and BT549 with ZNF10 knockdown, respectively. Further dual-luciferase reporter assay showed that transcriptional activity of β-catenin significantly increased in MCF7 and BT549 cells transfected with the ZNF10 expression plasmid compared with the empty vector control group at 48 hours (P<0.01 and P<0.01), respectively (Figure 5A-C).
Figure 5.
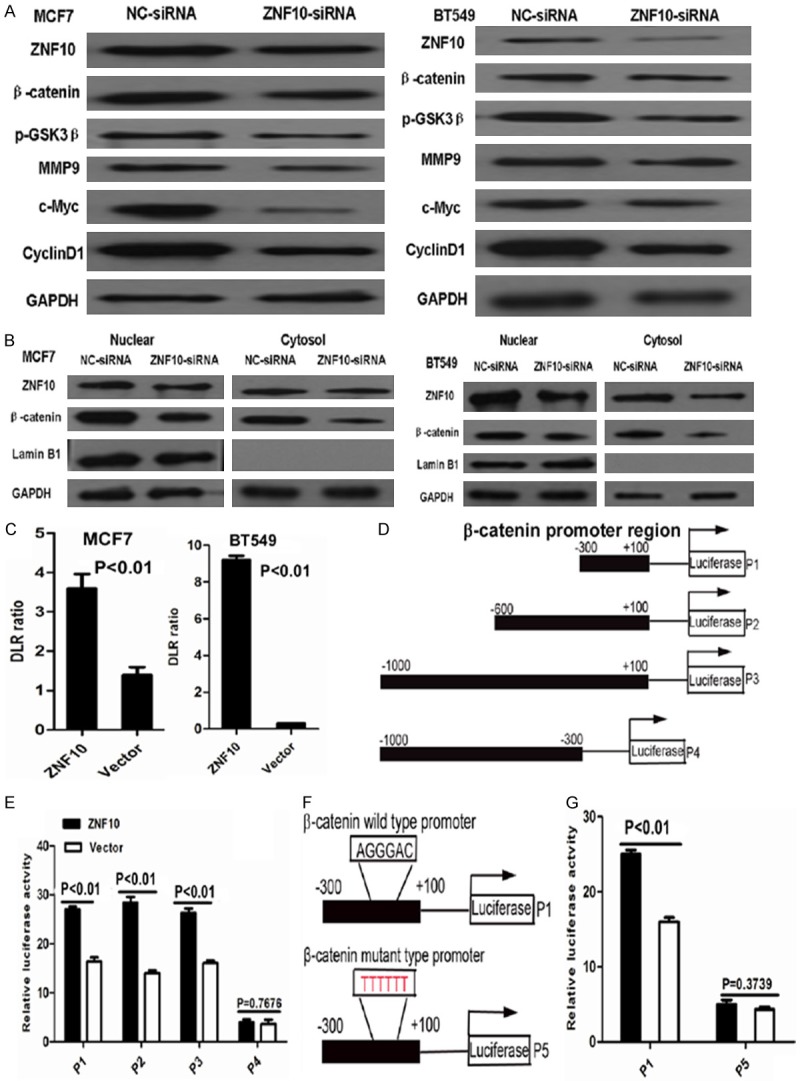
ZNF10 activates Wnt/β-catenin signaling in breast IDC cells. A. ZNF10 knockdown dramatically inhibited β-catenin protein expression and its downstream genes including c-Myc, cyclin D1 and MMP9 and upstream gene p-GSK3β (Ser9) expression in MCF7 and BT549 cells compared with the control group by Western blot analysis, respectively. B. Decreased nuclear and cytoplasmic β-catenin expression was found in MCF7 and BT549 cells transfected with ZNF10 siRNA compared with the control group by Western blot analysis. C. A dual-luciferase reporter assay showed that transcriptional activity of β-catenin significantly increased in MCF7 and BT549 cells transfected with ZNF10 expression plasmid compared with the empty vector group. D. The schematic diagram depicts different β-catenin promoter regions as indicated by P1, P2, P3, and P4. E. The luciferase activity in 293T cells transfected with ZNF10 expression plasmid and different β-catenin promoter fragments. F. The schematic diagram depicts β-catenin wild (P1) and mutant type (P5) promoter sequence. G. The binding efficiency of ZNF10 with β-catenin promoter dramatically decreased in 293T cells transfected with mutant β-catenin promoter sequence (P5) by luciferase reporter analysis.
ZNF10 regulates β-catenin promoter activity in breast cancer cells
To determine whether ZNF10 regulates β-catenin promoter activity, serial fragments from the β-catenin promoter region including nucleotides -300 to +100 (P1), -600 to +100 (P2), -1000 to +100 (P3), -1000 to -300 (P4) were cloned and co-transfected with ZNF10 expression plasmid into 293T cells, and the luciferase activity was significantly increased by ectopic overexpression of ZNF10 in 293T cells transfected with β-catenin promoter fragments including nucleotides -300 to +100 (P1), -600 to +100 (P2), and -1000 to +100 (P3) compared with the vector control group (P<0.01, P<0.01, and P<0.01), respectively. However, the luciferase activity displayed no significant difference in 293T cells transfected with nucleotides -1000 to -300 (P4) in β-catenin promoter fragment and ZNF10 expression plasmid compared with the vector control group (P=0.76). Further study showed that the binding efficiency of ZNF10 with the β-catenin promoter dramatically decreased in 293T cells transfected with mutant β-catenin promoter sequence TTTTTT compared with the wild β-catenin promoter sequence AGGGAC within nucleotides -300 to +100 by luciferase reporter analysis (Figure 5D-G) .
Discussion
In this study, our data first showed that ZNF10 expression was higher in breast IDC and breast cancer cell lines compared with their respective adjacent non-tumor breast tissues, DCIS and fibroadenoma of the breast. The result was concordant with ZNF10 mRNA expression in breast cancer from the GEO database [12]. Further study showed that ZNF10 expression was significantly associated with patients’ age, tumor stage, and breast cancer molecular subtype. Patients with a high ZNF10 expression had less survival than patients with a low ZNF10 expression in breast IDC although there was no significant difference. This needs further study in large breast cancer samples.
As for the role of ZNF10 in breast cancer, our data showed that ZNF10 knock-down inhibited cell proliferation, colony formation, cell cycle progression, cell migration and invasion but induced the apoptosis of breast cancer. ZNF10 knockdown suppressed the tumorigenicity of breast cancer in vivo. Our results suggest that ZNF10 plays an important role in the carcinogenesis and progression of breast IDC.
Several studies have reported alteration of β-catenin expression in breast cancer [15,17,18]. lncRNA PVT1 promotes KLF5/beta-catenin signaling to drive TNBC tumorigenesis [19]. The Wnt-β-catenin signaling regulated MRTF-A transcription to activate migration-related genes in human breast cancer cells [20]. miR-27a may activate the Wnt/β-catenin signaling pathway by negatively regulating SFRP1 to promote the proliferation, migration and invasion of breast cancer cells [21]. Our data showed that abnormal β-catenin expression including altered and loss staining was found in 54.5% of IDC by immunohistochemical staining. To study whether ZNF10 regulates the β-catenin signaling pathway in breast cancer, our data showed that there was a significantly negative correlation between ZNF10 and β-catenin normal expression in breast IDC. Moreover, ZNF10 knockdown dramatically inhibited β-catenin and its down-stream genes including c-Myc, cyclin D1 and MMP9, and the up-stream gene p-GSK3β (Ser9) expression. Further study showed that ZNF10 regulates β-catenin promoter activity in breast cancer cells. The association of ZNF10 with gene promoter elements might be cooperating with other transcription factors such as NF-κB through binding to AGGGAC sequence [11]. Our results showed that the binding efficiency of ZNF10 with the β-catenin promoter dramatically decreased in 293T cells transfected with mutant β-catenin promoter sequence TTTTTT compared with the wild β-catenin promoter sequence AGGGAC within nucleotides -300 to +100 by luciferase reporter analysis. These results indicate that ZNF10 regulates the β-catenin promoter activity in breast cancer and ZNF10 might bind to the region (nucleotides -300 to +100) of β-catenin promoter. Nuclear β-catenin is an inducer of epithelial-mesenchymal transition (EMT) and supports the cancer stem cell phenotype, which promotes tumor progression and metastasis. Diverse targets of β-catenin during the EMT define cancer stem cells and predict disease relapse [22]. Autophagy is an important factor in the pathobiology of cancer and may offer new possibilities for the development of novel anti-cancer strategies. Autophagy is a signature of a signaling network that maintains hematopoietic stem cells [23]. Fu et al. have reported that resveratrol inhibits breast cancer stem-like cells and induces autophagy via suppressing Wnt/β-catenin signaling pathway [24]. Elevated CRB3 expression suppresses breast cancer stemness by inhibiting β-catenin signaling to restore tamoxifen sensitivity [25]. Whether targeting ZNF10 inhibits breast cancer stem-like cells and induces autophagy via β-catenin signaling pathway needs further study.
Acknowledgements
This study was supported by the National Natural Science Foundation of China (grant nos. 81472251, 81272636 and 81502021).
Disclosure of conflict of interest
None.
References
- 1.McCrea PD, Turck CW, Gumbiner B. A homolog of the armadillo protein in Drosophila (plakoglobin) associated with E-cadherin. Science. 1991;254:1359–61. doi: 10.1126/science.1962194. [DOI] [PubMed] [Google Scholar]
- 2.Hinck L, Nathke IS, Papkoff J, Nelson WJ. Dynamics of cadherin/catenin complex formation: novel protein interactions and pathways of complex assembly. J Cell Biol. 1994;125:1327–40. doi: 10.1083/jcb.125.6.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wong NA, Pignatelli M. Beta-catenin--a linchpin in colorectal carcinogenesis? Am J Pathol. 2002;160:389–401. doi: 10.1016/s0002-9440(10)64856-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yoshida R, Kimura N, Harada Y, Ohuchi N. The loss of E-cadherin, alpha- and beta-catenin expression is associated with metastasis and poor prognosis in invasive breast cancer. Int J Oncol. 2001;18:513–20. [PubMed] [Google Scholar]
- 5.Thiesen HJ. Multiple genes encoding zinc finger domains are expressed in human T cells. New Biol. 1990;2:363–74. [PubMed] [Google Scholar]
- 6.Vissing H, Meyer WK, Aagaard L, Tommerup N, Thiesen HJ. Repression of transcriptional activity by heterologous KRAB domains present in zinc finger proteins. FEBS Lett. 1995;369:153–7. doi: 10.1016/0014-5793(95)00728-r. [DOI] [PubMed] [Google Scholar]
- 7.Luo A, Zhang X, Fu L, Zhu Z, Dong JT. Zinc finger factor ZNF121 is a MYC-interacting protein functionally affecting MYC and cell proliferation in epithelial cells. J Genet Genomics. 2016;43:677–685. doi: 10.1016/j.jgg.2016.05.006. [DOI] [PubMed] [Google Scholar]
- 8.Cho JG, Park S, Lim CH, Kim HS, Song SY, Roh TY, Sung JH, Suh W, Ham SJ, Lim KH, Park SG. ZNF224, Kruppel like zinc finger protein, induces cell growth and apoptosis-resistance by down-regulation of p21 and p53 via miR-663a. Oncotarget. 2016;7:31177–90. doi: 10.18632/oncotarget.8870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cho JG, Lim KH, Park SG. MED28 increases the colony-forming ability of breast cancer cells by stabilizing the ZNF224 protein upon DNA damage. Oncol Lett. 2018;15:3147–3154. doi: 10.3892/ol.2017.7718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang X, Ding C, Tian H, Dong X, Meng X, Zhu W, Liu B, Wang L, Huang M, Li C. ZNF23 suppresses cutaneous melanoma cell malignancy via mitochondria-dependent pathway. Cell Physiol Biochem. 2017;43:147–157. doi: 10.1159/000480333. [DOI] [PubMed] [Google Scholar]
- 11.Nishitsuji H, Sawada L, Sugiyama R, Takaku H. ZNF10 inhibits HIV-1 LTR activity through interaction with NF-kappaB and Sp1 binding motifs. FEBS Lett. 2015;589:2019–25. doi: 10.1016/j.febslet.2015.06.013. [DOI] [PubMed] [Google Scholar]
- 12.Kretschmer C, Sterner-Kock A, Siedentopf F, Schoenegg W, Schlag PM, Kemmner W. Identification of early molecular markers for breast cancer. Mol Cancer. 2011;10:15. doi: 10.1186/1476-4598-10-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lian J, Tang J, Shi H, Li H, Zhen T, Xie W, Zhang F, Yang Y, Han A. Positive feedback loop of hepatoma-derived growth factor and beta-catenin promotes carcinogenesis of colorectal cancer. Oncotarget. 2015;6:29357–74. doi: 10.18632/oncotarget.4982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dillon DA, D’Aquila T, Reynolds AB, Fearon ER, Rimm DL. The expression of p120ctn protein in breast cancer is independent of alpha- and beta-catenin and E-cadherin. Am J Pathol. 1998;152:75–82. [PMC free article] [PubMed] [Google Scholar]
- 16.Goldhirsch A, Wood WC, Coates AS, Gelber RD, Thurlimann B, Senn HJ. Strategies for subtypes--dealing with the diversity of breast cancer: highlights of the St. Gallen international expert consensus on the primary therapy of early breast cancer 2011. Ann Oncol. 2011;22:1736–47. doi: 10.1093/annonc/mdr304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karayiannakis AJ, Nakopoulou L, Gakiopoulou H, Keramopoulos A, Davaris PS, Pignatelli M. Expression patterns of beta-catenin in in situ and invasive breast cancer. Eur J Surg Oncol. 2001;27:31–6. doi: 10.1053/ejso.1999.1017. [DOI] [PubMed] [Google Scholar]
- 18.Hashizume R, Koizumi H, Ihara A, Ohta T, Uchikoshi T. Expression of beta-catenin in normal breast tissue and breast carcinoma: a comparative study with epithelial cadherin and alpha-catenin. Histopathology. 1996;29:139–46. doi: 10.1046/j.1365-2559.1996.d01-499.x. [DOI] [PubMed] [Google Scholar]
- 19.Tang J, Li Y, Sang Y, Yu B, Lv D, Zhang W, Feng H. LncRNA PVT1 regulates triple-negative breast cancer through KLF5/beta-catenin signaling. Oncogene. 2018 doi: 10.1038/s41388-018-0310-4. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 20.He H, Du F, He Y, Wei Z, Meng C, Xu Y, Zhou H, Wang N, Luo XG, Ma W, Zhang TC. The Wnt-beta-catenin signaling regulated MRTF-A transcription to activate migration-related genes in human breast cancer cells. Oncotarget. 2018;9:15239–15251. doi: 10.18632/oncotarget.23961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kong LY, Xue M, Zhang QC, Su CF. In vivo and in vitro effects of microRNA-27a on proliferation, migration and invasion of breast cancer cells through targeting of SFRP1 gene via Wnt/beta-catenin signaling pathway. Oncotarget. 2017;8:15507–15519. doi: 10.18632/oncotarget.14662. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22.Chang YW, Su YJ, Hsiao M, Wei KC, Lin WH, Liang CL, Chen SC, Lee JL. Diverse targets of beta-catenin during the epithelial-mesenchymal transition define cancer stem cells and predict disease relapse. Cancer Res. 2015;75:3398–410. doi: 10.1158/0008-5472.CAN-14-3265. [DOI] [PubMed] [Google Scholar]
- 23.Nguyen-McCarty M, Klein PS. Autophagy is a signature of a signaling network that maintains hematopoietic stem cells. PLoS One. 2017;12:e0177054. doi: 10.1371/journal.pone.0177054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fu Y, Chang H, Peng X, Bai Q, Yi L, Zhou Y, Zhu J, Mi M. Resveratrol inhibits breast cancer stem-like cells and induces autophagy via suppressing Wnt/beta-catenin signaling pathway. PLoS One. 2014;9:e102535. doi: 10.1371/journal.pone.0102535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li P, Feng C, Chen H, Jiang Y, Cao F, Liu J, Liu P. Elevated CRB3 expression suppresses breast cancer stemness by inhibiting beta-catenin signalling to restore tamoxifen sensitivity. J Cell Mol Med. 2018;22:3423–3433. doi: 10.1111/jcmm.13619. [DOI] [PMC free article] [PubMed] [Google Scholar]


