Abstract
The aim of this study is to explain the effects and mechanism of CtBP2 in the development of esophageal squamous cell carcinoma. In this study, we first evaluated CtBP2 protein expression of ESCC tumor and adjacent normal tissues by immunohistochemistry (IHC) and Western blot (WB) assay. Meanwhile, the number of vessels of ESCC and adjacent normal tissues were measured by immunofluorescence. In cell experiments, the effects of CtBP2 were evaluated by wound healing assay, flow cytometry detection, and EPC tube formation. The mechanisms of CtBP2 were investigated by immunofluorescence, qRT-PCR, WB, and EdU incorporation assay. In conclusion, CtBP2 inhibits ESCC in vitro and CtBP2 has a key role in the development of ESCC.
Keywords: CtBP2, ESCC, biological activity
Introduction
Esophageal carcinoma is the eighth most common cancer around the world contributing to cancer-related mortality. Esophageal squamous cell carcinoma (ESCC) is the key form of esophageal carcinoma [1]. Epidemiological evidence shows that environmental risk factors, linking heavy alcohol drinking, micronutrient deficiency, and so on, may participate in the development of ESCC. While, only a few individuals develop ESCC, showing that host genetic change may also promote ESCC etiology [2]. Recent research found a transcription gene as a novel lineage survival oncogene during the development of ECSS [3]. However, the precise molecular mechanism underlying ESCC still remains unknown. Therefore, it is important to identify functional metastasis genes and their intrinsic molecular mechanisms in the ESCC field.
Angiogenesis plays an essential role in metastasis and growth of ESCC, and endothelial growth factors have been reported to be important molecules during the development of tumor angiogenesis [4]. Two principal signaling pathways participate in angiogenesis: vascular endothelial growth factor (VEGF) signaling and Notch-Delta signaling [5,6]. Previous studies found that VEGF, a vital angiogenetic factor, leads to tumor angiogenesis and tumor progression in ESCC. VEGF may be a promising factor for anti-angiogenic co-treatment in ESCC by therapeutic interference with VEGF [7]. In 53.8% of esophageal adenocarcinoma patients, VEGFR expression was observed and was correlated with poor survival [8]. Moreover, VEGF promotes Notch signaling through inducing dll4 expression, which ameliorates VEGF signaling [9-11]. Furthermore, previous research found that the translocation leukemia associated gene, Tel, has a vital role in the analogous process of angiogenesis [12]. Lacking Tel inhibits development of angiogenesis [13].
C-terminal binding protein (CtBP) interacts with the C-terminus of the adenovirus E1A oncoprotein, which has been identified as a potential target for treating cancer [14]. CtBP2 modulates proliferation and cell cycle in an ESCC cell line through interaction with p16INK4A, finally promoting progression of ESCC [15]. Moreover, CtBP2 induces migration of ESCC cells and the transition of epithelial-mesenchymal (EMT) in ESCC in a CCNG/CDK7-dependent manner [16]. However, participation of CtBP2 in angiogenesis under ESCC is still unknown. Thus, this study aims to detect the underlying mechanism of CtBP2 during progression of angiogenesis with ESCC insults, and may provide a novel therapeutic target for the treatment of ESCC.
Materials and methods
Cell lines and cell cultures
ECA109, a human esophageal cancer cell line, was purchased from the American Type Culture Collection (ATCC, USA). Cells were cultured in RPMI 1640 (GibCo BRL, Grand Island, NY), containing 10% heat-inactivated fetal calf serum, 2 mM L-glutamine, and 100 U/ml penicillin-streptomycin mixture (GibCo BRL), at 37°C in a humidified atmosphere of 5% CO2.
Clinical specimens
Fifty clinical specimens with ESCC were collected from the Department of Pathology, Affiliated Hospital of Nantong University. None of the patients was treated with preoperative therapies, such as radiation and immunotherapy. After surgical removal, all tumorous and surrounding non-tumorous tissues were fixed in 10% buffered formalin and then embedded in paraffin for sectioning. Furthermore, ten paired tumorous and adjacent non-tumorous tissue specimens were immediately stored at -80°C for further analysis.
Plasmid construction
pcDNA3.1 CtBP2 and CtBP2 siRNA vector were kindly donated by Dr. J. P. Blaydes (School of Medicine, University of Southampton). ECA109 cells were seeded the day before transfection using 1,640 with 10% FBS but without antibiotics. Transient transfection of siRNA vectors and the non-silencing vectors was carried out employing lipofectamine 2000 and plus reagent in Opti-MEM as suggested by the manufacturer. The cells were incubated with the pSilencer vectors and lipofectamine plus reagent complexes for 4 h at 37°C. Transfected cells were used for subsequent experiments for 48 h after transfection. The experiments were repeated at least three times.
Histopathological examination
Sections of tumor tissue were removed and immediately fixed in 10% phosphate buffered formalin solution and embedded in paraffin. Sections (4-μm) from each sample were cut and stained with hematoxylin and eosin (H&E) and Masson’s trichrome. The prepared kidney sections were observed under an Olympus BX51 light microscope (Olympus, Japan) at a magnification of ×400 with an OlympusDP70 digital imaging system (Olympus, Japan). The results of H&E stain were graded according to epithelial proliferation, alveolitis, edema, inflammatory cell infiltration, and interstitial fibrosis, and the results of Masson’s trichrome stain were graded according to the levels of ECM. Criteria for grading were as follows: grade 0, normal; grade 0.5, slight; grade 1, mild; grade 2, moderate; and grade 3, severe.
ELISA
Tumor tissue sections of rat were prepared for ELISA. 50 mg renal tissues were homogenized in 400 μl PBS and centrifuged at 800 g for 20 min. After centrifugation, the supernatants were then collected and stored at -80°C. The contents of VEGF, bFGF, IL-8, and PD-ECGF were detected by using the enzyme-linked immunosorbent assay (ELISA) kits according to the manufacturer’s instructions. The absorbance was measured at 450 nm.
Immunohistochemical staining
Tumor tissue sections of rat were fixed in 10% formalin, embedded in paraffin, sectioned, and then prepared for immunohistochemical staining. Sections were mounted on slides, dewaxed, and hydrated. Slides were boiled in 10 mM sodium citrate buffer (pH 6) for 20 min and cooled at room temperature about 30 min. After treatment with 0.1% triton-100 for 10 min, sections were incubated in 3% hydrogen peroxide for 10 min. After blocking with normal goat serum for 20 min at 37°C, sections were incubated with, anti-CtBP2 antibodies at 4°C overnight. They were washed and incubated with peroxidase-conjugated secondary antibodies at 37°C for 1 h. After incubation with 3, 3’-diaminobenzidine tetrahydrochloride and hydrogen peroxide, the sections were counterstained with hematoxylin.
Immunofluorescence
Tissues or cells were fixed in 10% formalin for 30 min at room temperature, and then treated with 0.1% triton-100 for 10 min and incubated in 3% hydrogen peroxide for 10 min. After blocking with normal goat serum for 20 min at 37°C, sections were incubated with Iso-B4 at 4°C overnight. They were washed and incubated with FITC-conjugated secondary antibodies at 37°C for 1 h. Tissues or cells were counterstained with Hoechst 33258.
Quantitative real-time polymerase chain reaction (qRT-PCR)
Cells were seeded into 6-well plates and incubated with serum-free medium for 2 h. Subsequently, cells were transfected with Lipo3000 and plasmids for 72 h. Total RNA was extracted using TRIzol (1 mL) according to the manufacturer’s instructions. The concentrations were determined using optical density measurements at 260 nm on a spectrophotometer. cDNAs were synthesized according to the manuscript. Then, 20-μL reaction mixtures containing SYBR green, nuclease-free water, forward primer, reverse primer and cDNA were transferred to different PCR tubes. Mefor every run to ensure that only the correct product was amplified. The Ct values were normalized to the corresponding values of GAPDH. The primer sequence as following: bFGF: F: 5’-CCTCACATCAAGCTACAACTTC-3’; R: 5’-GCCAGTAATCTTCCATCTTCCT-3’. IL-8: F: 5’-TTAAATCTGGCAACCCTAGTCTG-3’; R: 5’-TGTGAGGTAAGATGGTGGCTAA-3’. VEGF: F: 5’CGCTTACTCTCACCTGCTTCT-3’; R: 5’-GCTTCTTCCAACAATGTGTCTCT-3’. PD-ECGF: F: 5’-GCGGACGGAATCCTATATGC-3’; R: 5’-TACTGAGAATGGAGGCTGTGAT-3’. GAPDH: F: 5’-AGATCATCAGCAATGCCTCCT-3’; R: 5’-TGAGTCCTTCCACGATACCAA-3’.
Western blot analysis
Cells were seeded into 6-well plates and incubated with serum-free medium for 2 h. Subsequently, cells were transfected with Lipo3000 and plasmids for 72 h. Total protein was extracted with RIPA lysis buffer containing PMSF for 30 min on ice. The extracts were harvested using centrifugation at 12,000 rounds per minute for 5 min at 4°C. The contents of proteins were measured by BCA. Twenty μg of proteins were loaded and separated by 10% SDS-PAGE. Then, proteins were transferred to PVGF members (0.22 μm). Membranes were blocked by 3% BSA in TBS-Tween 20 (0.1%) and then incubated with the corresponding antibodies. Finally, the membranes were incubated in ECL reagent for 2-10 min and then exposed to X-ray film.
Wound healing assay
Cells were seeded into 6-well plates and incubated with serum-free medium for 2 h. Subsequently, cells were transfected with Lipo3000 and plasmids for 72 h. Cells (1×106 cells/ml) were seeded into a 6-well plate and incubated for 24 h. The center of the cell monolayer was then scraped with a sterile micropipette tip to create a straight gap of constant width. The wells were washed with PBS. Wound closure was imaged at 0 and 24 h using an inverted light microscope (magnification, ×200).
Flow cytometry detection
Cells were seeded into 6-well plates and incubated with serum-free medium for 2 h. Subsequently, cells were transfected with Lipo3000 and plasmids for 72 h. Cells were harvest by 0.25% trypsin without EDTA. Cells were washed by PBS for three times and then incubated with 500 μL binding buffer, 5 μL Annexin V-FITC and 5 μL PI for 30 min. the apoptosis of cells were measured by BD FACS Calibur and data were analyzed by Cell-Quest.
EdU incorporation assay
Cells were seeded into 6-well plates and incubated with serum-free medium for 2 h. Subsequently, cells were transfected with Lipo3000 and plasmids for 72 h. Then, cells were cultured with 10 μM Edu for 4 h and washed with PBS for three times. After fixation with 4% polyoxymethylene, the Edu assays were detected according to the manuscript. Photos were taken and analyzed by Olympus BX51 light microscope (Olympus, Japan) at a magnification of ×400 with an OlympusDP70 digital imaging system (Olympus, Japan).
EPC tube formation
Cells were seeded into 6-well plates and incubated with serum-free medium for 2 h. Subsequently, cell were transfected with Lipo3000 and plasmids for 72 h. Cells (1×106 cells/ml) were seeded into a 24-well plate pre-coated with 100 µl Matrigel for 8 h. tube formation were detected using an inverted light microscope (magnification, ×200).
Statistical analysis
Data are presented as mean ± standard deviation (SD) from at least six independent experiments. Significant differences among groups were compared by using ANOVA with P < 0.05 being considered statistically significant.
Results
Expression of CtBP2 and the number of microvessels increased in esophageal squamous cell carcinoma (ESCC)
CtBP2 expression has been reported to be up-regulated in ESCC [17]. To confirm this research, we examined expression of CtBP2 in tumor tissues and adjacent non-tumor tissues from 20 ESCC tissue specimens. As shown in Figure 1A, we found that expression of CtBP2 was dramatically increased in tumor tissues, compared with adjacent non-tumor tissues. Immunohistochemical staining also showed that CtBP2 was highly expressed in tumor tissues (Figure 1B). Moreover, the numbers of vessels in ESCC were higher in tumor tissues than in adjacent non-tumor tissues (Figure 1C), indicating that there may exist a correlation between CtBP2 and the number of vessels in ESCC.
Figure 1.
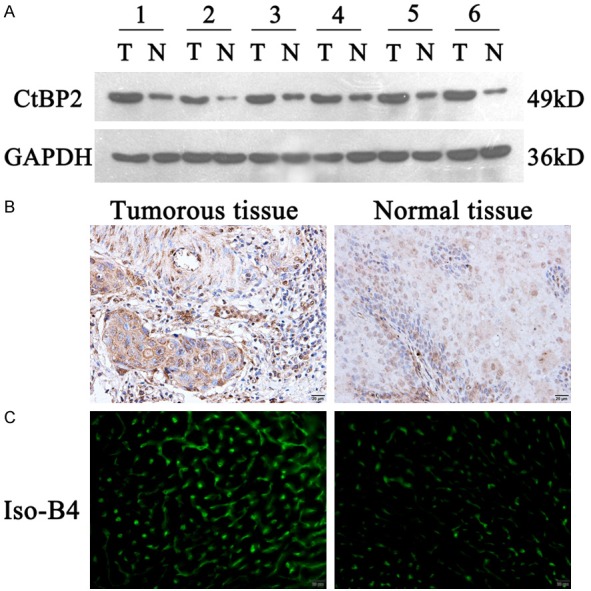
Comparison of CtBP2 and the number of microvessels in tumorous and para-carcinoma tissue. A: The expression of CtBP2 was observed by Immunohistochemical assay. Paraffin-embedded ESCC tumorous and para-carcinoma tissue sections were stained with CtBP2 antibody. The CtBP2 expression was recorded using an inverted microscope (magnification 400×). B: The preparation and culture of esophageal squamous cell were described in Materials and Methods Section. The proteins were extracted with lysis buffer. The level of CtBP2 were analyzed by 10% SDS-PAGE, β-actin was used as the internal control. C: The sections of ESCC tumorous and para-carcinoma tissue were stained with Iso-B4. The numbers of microvascular were recorded using an inverted microscope (magnification 400×).
Blocking CtBP2 expression inhibits vascular endothelial cell angiogenesis
To detect function of CtBP2 in tumor angiogenesis, we overexpressed or inhibited CtBP2 expression in ECA109 cells in vitro. As shown in Figure 2A, expression of CtBP2 was dramatically increased after transfection of the plasmid flag-CtBP2, and transfection with si-CtBP2 inhibited the level of CtBP2. Moreover, CtBP2 over-expression significantly promoted the number of vessels, shown with CD31 expression. Data also confirmed that blocking CtBP2 expression could down-regulate CD31 in ECA109 cells. A previous study identified tube formation as closely related to CD31 [18]. As shown in Figure 2B, we found that overexpression of CtBP2 in ECA109 cells further enhanced tube formation, and knockout of CtBP2 significantly blunted tube formation compared with control group.
Figure 2.
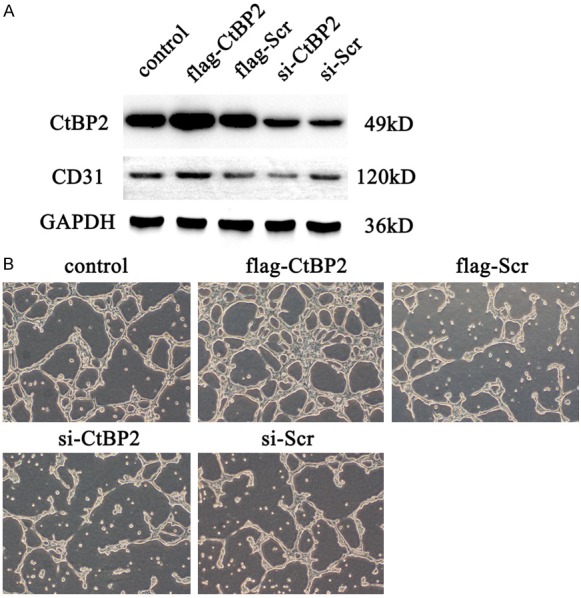
Effect of CtBP2 in angiogenesis. The preparation and culture of esophageal squamous cell is described in the Materials and Methods Section. Cells were transfected with Lipo3000 and plasmids. A: The expression of CtBP2 and CD31 was measured by Western blot. B: 1×106 cells were seeded in 6-well plated per-coating with Matrigel. Photos were taken at 8 h by an inverted microscope (magnification 200×).
Blocking CtBP2 expression decreases proliferation and migration in vascular endothelial cells while increasing apoptosis
The effect of CtBP2 on endothelial cell proliferation, apoptosis, and migration was identified with or without CtBP2 expression. As shown in Figure 3A and 3B, over-expressing CtBP2 in ECA109 cells could promote endothelium proliferation, whereas the proliferation was significantly inhibited under CtBP2 blocking, compared with control groups. Moreover, up-regulation CtBP2 in ECA109 dramatically reduced apoptosis in endothelial cells, and CtBP2 inhibition also increased the level of apoptosis in endothelium (Figure 3C, 3D). Wound healing assays showed that over-expressing CtBP2 in ECA109 cells enhanced endothelial cell migration, while inhibiting CtBP2 expression in ECA109 cells could suppress cell migration (Figure 3E, 3F). Taken together, these results certify that CtBP2 participates in the development of angiogenesis, and acts as an angiogenesis promoter in vitro.
Figure 3.
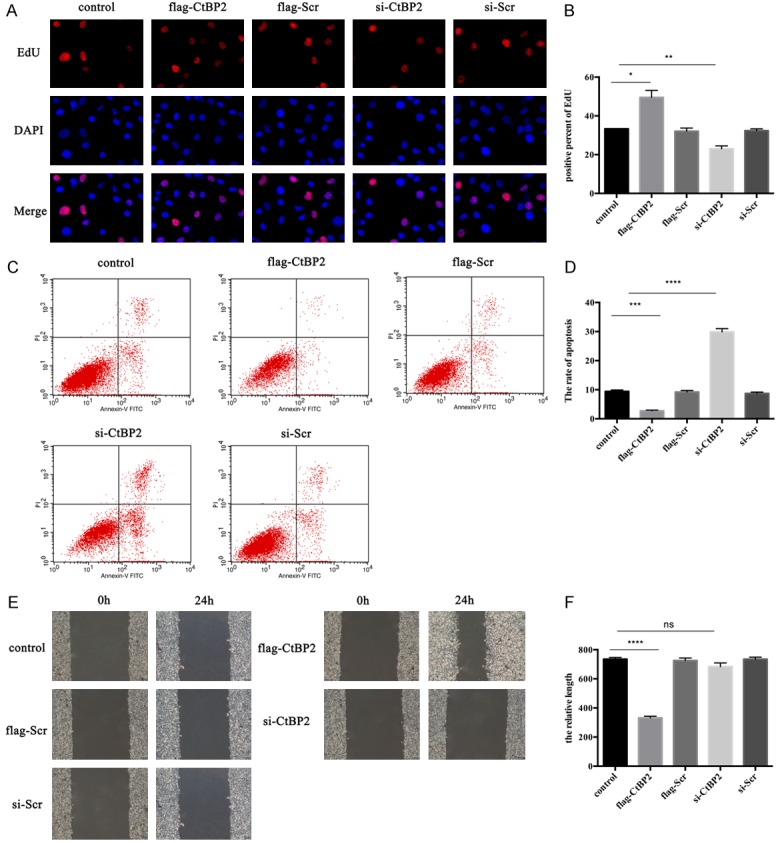
Effect of CtBP2 in proliferation, apoptosis, and migration. The preparation and culture of esophageal squamous cell was described in the Materials and Methods Section. Cells were transfected with Lipo3000 and plasmids. A, B: The proliferation of cells was observed by EdU assay, with the pictures recorded by an inverted microscope (magnification 400×). C, D: Cells were cultured with annexin-V FITC/PI for 30 min, then the apoptosis of cells were analyzed by flow cytometry. E, F: Migration were measured by wound healing assay. The wound healing lengths were recorded at 0 h and 24 h.
Reduced CtBP2 expression in ECA109 decreases the release of angiogenetic factors
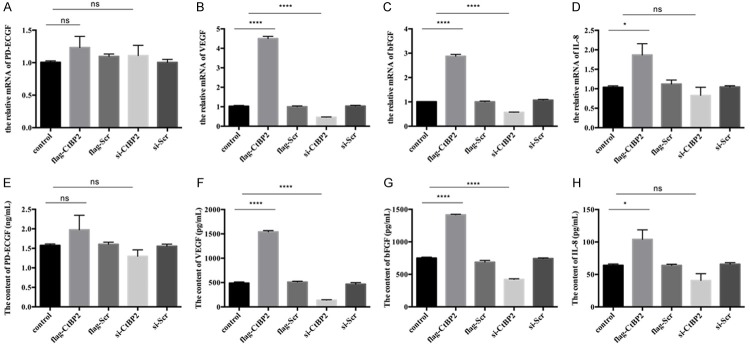
Since CtBP2 could promote angiogenesis in endothelium, we examined the relative angiogenetic factors in endothelial cells to further validate our data. The release and mRNA level of VEGF, platelet derived endothelial cell growth factor (PD-ECGF), basic fibroblast growth factor (bFGF) and interleukin (IL)-8 in endothelium were examined by ELISA and real-time qPCR. As shown in Figure 4A-D, except PD-ECGF, the mRNA level of VEGF, bFGF, and IL-8 were dramatically increased with CtBP2 insult, whereas CtBP2 inhibition could decrease the mRNA level of VEGF, bFGF, and IL-8. Furthermore, we collected the supernatant of endothelial cells with or without CtBP2 stimulation. ELISA assay found that the content of VEGF, bFGF, and IL-8 was also up-regulated when CtBP2 was overexpressed, while blocking expression of CtBP2 reduced the content of VEGF, bFGF, and IL-8. The content of PD-ECGF didn’t change with or without CtBP2 stimulation. These findings suggest that CtBP2 overexpression in ECA109 promotes the release of VEGF, bFGF, and IL-8 to induce angiogenesis.
Figure 4.
Effect of CtBP2 on PD-ECGF, VEGF, bFGF, and IL-8. The preparation and culture of esophageal squamous cell were described in Materials and Methods Section. Cells were transfected with Lipo3000 and plasmids. A-D: Total RNA were extracted by TRIzol and the cDNA were synthesized according to the manuscript. The expression of PD-ECGF, VEGF, bFGF, and IL-8 were analyzed by real-time qPCR. E-H: The supernatants were collected and certificated at 12,000 rpm for 10 min. The contents of PD-ECGF, VEGF, bFGF, and IL-8 were measured by ELISA.
CtBP2 activates notch by CtBP2-Tel complex
A previous study certified that Notch signaling participates in the development of angiogenesis [10]. Moreover, Tel, a translocation ets leukemia gene, also has a role in the process of angiogenesis [12]. Evidence found that mice lacking Tel exhibit defects in angiogenesis [19]. To confirm whether CtBP2 activates Notch signaling through Tel, we carried out co-immunoprecipitation experiments. As shown in Figure 5A, Tel was found in the CtBP2 immunoprecipitates from cells co-transfected with flag-CtBP2, whereas it was detected at low levels in the cells transfected with Tel alone. No Tel was detected by the Myc antibody when IgG was used as an immunoprecipitation reagent in the same experiment (Figure 5B). Moreover, endogenous Tel and CtBP2 were co-immunoprecipitated using a CtBP2 antibody and IgG, when used as the control (Figure 5C). Finally, co-localization of Tel and CtBP2 was detected by confocal microscopy. As detected in Figure 5D, CtBP2 was present in the nucleus, and Tel was enriched in some areas, which were co-localized with CtBP2 as well. These results identify that CtBP2 interacts with Tel.
Figure 5.
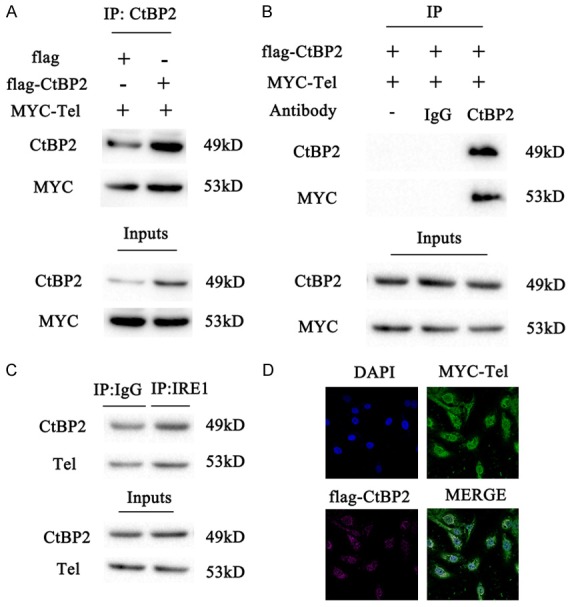
Tel interacts with CtBP2. A: The lysates of ECA109 cells co-transfected with HA-CtBP2/MYC-Tel or HA-EMPTY/ MYC-Tel were immunoprecipitated with an CtBP2 antibody and analyzed by Western blot with antibodies against CtBP2 and Tel. B: The lysates of ECA109 cells co-transfected with HA- CtBP2/MYC-Tel were immunoprecipitated with IgG, CtBP2 antibody, or without antibody, respectively. The immunoprecipitated proteins were analyzed by Western blot with antibodies against CtBP2 and Tel. C: ECA109 cell lysates were immunoprecipitated with an CtBP2 antibody or normal IgG (control), followed by Western blot with antibodies against CtBP2 and Tel. D: Confocal microscopy images were obtained from ECA109 cells co-transfected with HA-CtBP2 (purple) and MYC-Tel (green); the nuclei were imaged by DAPI staining (blue). Bar, 10 “m.
Discussion
CtBP2 has been identified to be a vital tumor promoter, which participates in the development of breast cancer [20], prostate cancer [21], neuroblastoma [22], and human esophageal squamous cell carcinoma [15]. A previous study found that LncRNA NEAT1 regulated cell progression in ESCC through the miR-129/CtBP2 axis, which leads to a better research of mechanism of ESCC [23]. Moreover, another investigation identified that CtBP2 promoted ESCC migration in a CCNH/CDK7-dependent manner [16]. However, the effect of CtBP2 on angiogenesis under ESCC is still unknown.
In the present study, we confirmed that the expression of CtBP2 is significantly increased during ESCC, compared with adjacent non-tumorous tissue specimens. Immunostaining showed CtBP2 expression is inversely associated with the number of microvessels in ESCC. The expression of CD31, a microvessel marker, was also up-regulated after overexpression CtBP2 as detected by Western blot, as well as CtBP2. Tube formation analysis demonstrated that CtBP2 could promote the formation of microvessels, and that angiogenesis was inhibited without CtBP2. From these results, we recognize the strong association between CtBP2 and angiogenesis in ESCC. However, the mechanism of CtBP2 in angiogenesis under ESCC is still unclear.
We also clarified the function of CtBP2 on endothelial proliferation, migration, and apoptosis. Edu analysis showed CtBP2 promoted the proliferation of endothelium, whereas proliferation was inhibited without CtBP2. Furthermore, CtBP2 was identified as an apoptosis inhibitor in ESCC cell lines. Flow analysis found that CtBP2 significantly suppressed endothelial apoptosis in vitro. Transwell invasion assays also demonstrated endothelial invasion was promoted with CtBP2 overexpression. Furthermore, the level of VEGF, bFGF, and IL-8, which were tumor angiogenesis controllers [24], were highly expressed under CtBP2 insult through ELISA assay. Taken together, our results suggested that CtBP2 could promote angiogenesis in ESCC to aggravate the occurrence of ESCC.
We noticed that CtBP2 is directly combined with Tel in ECA109 cells. Previous research found Tel was a vital factor during tumor angiogenesis [25]. Accumulating evidence also demonstrated that lacking Tel in embryonic stem cells leaded to die with defective angiogenesis [19]. In the present study, we identified CtBP2, as a target of Tel in ESCC, in agreement with previous work in endothelial sprouting [26]. Co-immunoprecipitation and confocal analysis also showed that CtBP2 combined with Tel. These results indicate that Tel is a functional target of CtBP2 in regulating angiogenesis during the development of ESCC.
Conclusions
In conclusion, our study identified that CtBP2 inhibition could suppress esophageal squamous cell carcinoma via decreasing angiogenesis.
Acknowledgements
This work was supported by 2016 Science and Technology Guiding Project of Nantong Municipal Bureau of Science and Technology (Project number: GJZ16072).
Disclosure of conflict of interest
None.
References
- 1.Pennathur A, Gibson MK, Jobe BA, Luketich JD. Oesophageal carcinoma. Lancet. 2013;381:400–412. doi: 10.1016/S0140-6736(12)60643-6. [DOI] [PubMed] [Google Scholar]
- 2.Wang X, Li M, Wang Z, Han S, Tang X, Ge Y, Zhou L, Zhou C, Yuan Q, Yang M. Silencing of long noncoding RNA MALAT1 by miR-101 and miR-217 inhibits proliferation, migration, and invasion of esophageal squamous cell carcinoma cells. J Biol Chem. 2015;290:3925–3935. doi: 10.1074/jbc.M114.596866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bass AJ, Watanabe H, Mermel CH, Yu S, Perner S, Verhaak RG, Kim SY, Wardwell L, Tamayo P, Gat-Viks I, Ramos AH, Woo MS, Weir BA, Getz G, Beroukhim R, O’Kelly M, Dutt A, Rozenblatt-Rosen O, Dziunycz P, Komisarof J, Chirieac LR, Lafargue CJ, Scheble V, Wilbertz T, Ma C, Rao S, Nakagawa H, Stairs DB, Lin L, Giordano TJ, Wagner P, Minna JD, Gazdar AF, Zhu CQ, Brose MS, Cecconello I, Ribeiro U Jr, Marie SK, Dahl O, Shivdasani RA, Tsao MS, Rubin MA, Wong KK, Regev A, Hahn WC, Beer DG, Rustgi AK, Meyerson M. SOX2 is an amplified lineage survival oncogene in lung and esophageal squamous cell carcinomas. Nat Genet. 2009;41:1238–1242. doi: 10.1038/ng.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oshima Y, Yajima S, Yamazaki K, Matsushita K, Tagawa M, Shimada H. Angiogenesis-related factors are molecular targets for diagnosis and treatment of patients with esophageal carcinoma. Ann Thorac Cardiovasc Surg. 2010;16:389–393. [PubMed] [Google Scholar]
- 5.Phng LK, Gerhardt H. Angiogenesis: a team effort coordinated by Notch. Dev Cell. 2009;16:196–208. doi: 10.1016/j.devcel.2009.01.015. [DOI] [PubMed] [Google Scholar]
- 6.Gerhardt H. VEGF and endothelial guidance in angiogenic sprouting. Organogenesis. 2008;4:241–246. doi: 10.4161/org.4.4.7414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kleespies A, Bruns C, Jauch KW. Clinical significance of VEGF-A, -C and -D expression in esophageal malignancies. Onkologie. 2005;28:281–288. doi: 10.1159/000085198. [DOI] [PubMed] [Google Scholar]
- 8.Zhang L, Ma J, Han Y, Liu J, Zhou W, Hong L, Fan D. Targeted therapy in esophageal cancer. Expert Rev Gastroenterol Hepatol. 2016;10:595–604. doi: 10.1586/17474124.2016.1140036. [DOI] [PubMed] [Google Scholar]
- 9.Gerhardt H, Golding M, Fruttiger M, Ruhrberg C, Lundkvist A, Abramsson A, Jeltsch M, Mitchell C, Alitalo K, Shima D, Betsholtz C. VEGF guides angiogenic sprouting utilizing endothelial tip cell filopodia. J Cell Biol. 2003;161:1163–1177. doi: 10.1083/jcb.200302047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hellström M, Phng LK, Hofmann JJ, Wallgard E, Coultas L, Lindblom P, Alva J, Nilsson AK, Karlsson L, Gaiano N, Yoon K, Rossant J, Iruela-Arispe ML, Kalén M, Gerhardt H, Betsholtz C. Dll4 signalling through Notch1 regulates formation of tip cells during angiogenesis. Nature. 2007;445:776–780. doi: 10.1038/nature05571. [DOI] [PubMed] [Google Scholar]
- 11.Tammela T, Zarkada G, Wallgard E, Murtomäki A, Suchting S, Wirzenius M, Waltari M, Hellström M, Schomber T, Peltonen R, Freitas C, Duarte A, Isoniemi H, Laakkonen P, Christofori G, Ylä-Herttuala S, Shibuya M, Pytowski B, Eichmann A, Betsholtz C, Alitalo K. Blocking VEGFR-3 suppresses angiogenic sprouting and vascular network formation. Nature. 2008;454:656–660. doi: 10.1038/nature07083. [DOI] [PubMed] [Google Scholar]
- 12.Golub TR, Barker GF, Lovett M, Gilliland DG. Fusion of PDGF receptor beta to a novel ets-like gene, tel, in chronic myelomonocytic leukemia with t(5;12) chromosomal translocation. Cell. 1994;77:307–316. doi: 10.1016/0092-8674(94)90322-0. [DOI] [PubMed] [Google Scholar]
- 13.Wang LC, Kuo F, Fujiwara Y, Gilliland DG, Golub TR, Orkin SH. Yolk sac angiogenic defect and intra-embryonic apoptosis in mice lacking the Ets-related factor TEL. EMBO J. 1997;16:4374–4383. doi: 10.1093/emboj/16.14.4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stankiewicz TR, Gray JJ, Winter AN, Linseman DA. C-terminal binding proteins: central players in development and disease. Biomol Concepts. 2014;5:489–511. doi: 10.1515/bmc-2014-0027. [DOI] [PubMed] [Google Scholar]
- 15.Guan C, Shi H, Wang H, Zhang J, Ni W, Chen B, Hou S, Yang X, Shen A, Ni R. CtBP2 contributes to malignant development of human esophageal squamous cell carcinoma by regulation of p16INK4A. J Cell Biochem. 2013;114:1343–1354. doi: 10.1002/jcb.24475. [DOI] [PubMed] [Google Scholar]
- 16.Zhang J, Zhu J, Yang L, Guan C, Ni R, Wang Y, Ji L, Tian Y. Interaction with CCNH/CDK7 facilitates CtBP2 promoting esophageal squamous cell carcinoma (ESCC) metastasis via upregulating epithelial-mesenchymal transition (EMT) progression. Tumour Biol. 2015;36:6701–6714. doi: 10.1007/s13277-015-3354-x. [DOI] [PubMed] [Google Scholar]
- 17.Guan C, Shi H, Wang H, Zhang J, Ni W, Chen B, Hou S, Yang X, Shen A, Ni R. CtBP2 contributes to malignant development of human esophageal squamous cell carcinoma by regulation of p16INK4A. J Cell Biochem. 2013;114:1343–1354. doi: 10.1002/jcb.24475. [DOI] [PubMed] [Google Scholar]
- 18.Lee C, Liu A, Miranda-Ribera A, Hyun SW, Lillehoj EP, Cross AS, Passaniti A, Grimm PR, Kim BY, Welling PA, Madri JA, DeLisser HM, Goldblum SE. NEU1 sialidase regulates the sialylation state of CD31 and disrupts CD31-driven capillary-like tube formation in human lung microvascular endothelia. J Biol Chem. 2014;289:9121–9135. doi: 10.1074/jbc.M114.555888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang LC, Kuo F, Fujiwara Y, Gilliland DG, Golub TR, Orkin SH. Yolk sac angiogenic defect and intra-embryonic apoptosis in mice lacking the Ets-related factor TEL. EMBO J. 1997;16:4374–4383. doi: 10.1093/emboj/16.14.4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Birts CN, Harding R, Soosaipillai G, Halder T, Azim-Araghi A, Darley M, Cutress RI, Bateman AC, Blaydes JP. Expression of CtBP family protein isoforms in breast cancer and their role in chemoresistance. Biol Cell. 2010;103:1–19. doi: 10.1042/BC20100067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang C, Li S, Qiao B, Yang K, Liu R, Ma B, Liu Y, Zhang Z, Xu Y. CtBP2 overexpression is associated with tumorigenesis and poor clinical outcome of prostate cancer. Arch Med Sci. 2015;11:1318–1323. doi: 10.5114/aoms.2015.56359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nan J, Guan S, Jin X, Jian Z, Linshan F, Jun G. Down-regulation of C-terminal binding protein 2 (CtBP2) inhibits proliferation, migration, and invasion of human SHSY5Y cells in vitro. Neurosci Lett. 2017;647:104–109. doi: 10.1016/j.neulet.2017.02.006. [DOI] [PubMed] [Google Scholar]
- 23.Li Y, Chen D, Gao X, Li X, Shi G. LncRNA NEAT1 regulates cell viability and invasion in esophageal squamous cell carcinoma through the miR-129/CTBP2 axis. Dis Markers. 2017;2017:5314649. doi: 10.1155/2017/5314649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kitadai Y, Onogawa S, Kuwai T, Matsumura S, Hamada H, Ito M, Tanaka S, Yoshihara M, Chayama K. Angiogenic switch occurs during the precancerous stage of human esophageal squamous cell carcinoma. Oncol Rep. 2004;11:315–319. [PubMed] [Google Scholar]
- 25.Edel MJ. Analysis of the TEL protein during tumour angiogenesis. Anticancer Res. 1999;19:2945–2951. [PubMed] [Google Scholar]
- 26.Roukens MG, Alloul-Ramdhani M, Baan B, Kobayashi K, Peterson-Maduro J, van Dam H, Schulte-Merker S, Baker DA. Control of endothelial sprouting by a Tel-CtBP complex. Nat Cell Biol. 2010;12:933–942. doi: 10.1038/ncb2096. [DOI] [PubMed] [Google Scholar]