Abstract
The glycerol-3-phosphate acyltransferase mitochondrial gene (GPAM) variant has been associated with serum lipid levels in the Eurpean ancestry, but little is known about such association in Chinese populations. The aim of the present study was to investigate the relationship between the GPAM rs1129555 single nucleotide polymorphism (SNP) and several environment factors with blood lipid profiles in the Guangxi Maonan and Han populations. A total of 720 individuals of Maonan nationality and 780 participants of Han nationality were randomly selected from our previous stratified randomized samples. Genotyping of the rs1129555 SNP was carried out using the polymerase chain reaction-restriction fragment length polymorphism technique, and then confirmed by direct sequencing. The frequencies of C and T alleles were 72.85% and 27.15% in Maonan, and 65.19% and 34.81% in Han (P < 0.001); respectively. The frequencies of CC, CT, and TT genotypes were 51.53%, 42.36%, and 5.97% in Maonan, and 43.08%, 44.23%, and 12.69% in Han populations (P < 0.001). The T allele carriers had higher serum triglyceride (TG) in Han and higher low-density lipoprotein cholesterol (LDL-C) in both Maonan and Han than the T allele non-carriers (P < 0.05-0.01). Gender subgroup analyses showed that the T allele carriers had higher TG levels in Han males (P < 0.05) and higher LDL-C levels in Maonan males but not in famales (P < 0.01). Serum lipid parameters were also associated with several environmental factors (P < 0.05-0.001). These findings suggest that racial/ethnic- and/or gender-specific association occurs between the GPAM rs1129555 variant and serum lipid parameters in our study populations.
Keywords: Glycerol-3-phosphate acyltransferase mitochondrial, single nucleotide polymorphism, lipids, environmental factor
Introduction
Cornary heart disease (CHD) has been the largest threat to public health in developed contries [1]. CHD and its sequelae (angina, myocardial infarction, cardiac revascularization or transplantation) affect more than 16 million Americans and untold numbers of people world-wide [2]. In China, the CHD prevalence and mortality rate have been increasing in recent decades [3]. It is generally accepted that CHD is a multifactorial disease [4]. Reduction of known risk factors for CHD such as hyperlipidemia, hypertension, diabetes, heavy alcohol consumption, and smoking has been assessed in multiple clinical trials and is associated with 30% to 40% less clinical events such as myocardial infarction and chronic heart failure [5]. Dyslipidemia, particularly hypertriglyceridemia, hypercholesterolemia and low high-density lipoprotein cholesterol (HDL-C), is well-described independent predictor for atherosclerosis and CHD [6]. Low-density lipoprotein cholesterol (LDL-C) has been considered to be the major lipid risk factor and main target of lipid-lowering therapy in most national guidelines [7,8]. Lipid disorder is a common result that is determined by age, gender, genetic, ethnicity, environmental factors and their interactions [9]. Family studies based on twins suggest that in different populations, genetic factors contribute to about half of the variation in serum lipid profiles [10]. Although it has been estimated that up to 41% of interindividual variability may be determined by genetic factors [11], the genetic architecture of dyslipidemia and CHD remains largely undefined. Genome-wide association studies (GWASs) have identified that the common variants at loci together can explain about 10-12% of the variations in each lipid trait and rare variants with large individual effects may also contribute to the heritability of lipid traits [12]. Gentic variants with small effects can point to pathways and therapeutic targets that enable clinically-important changes in blood lipids. Therefore, understanding of the association of SNP and serum lipid levels may become a new approach for preventing and treatment of CHD.
In recent years, a multitude of new gene loci associated with blood lipid levels have been discovered by GWAS, the glycerol-3-phosphate acyltransferase mitochondrial gene (GPAM; also knows as: GPAT, GPAT1; Gene ID: 57678, HGNC ID: 24805, chromosomal location: 10q25.2) is one of the potential candidate genes that play an important role in serum lipid metabolism [13]. GPAM, is a member of protein family (pfam) 01553 family of glycerolipid acyltransferases, located in the outer mitochondrial membrane. GPAM is most highly expressed in liver and adipose tissue, but is also present in many other tissues including brain, kidney, heart, and adrenal gland. Liver is a major organ of regulating lipid metabolism, knockout and overexpression studies suggest that GPAM isoforms play a critical role in the development of hepatic steatosis and that steatosis initiated by overexpression of GPAM result in fatty liver, obesity, insulin resistance, and hyperlipidemia [14,15]. Gain-of-function and loss-of-function studies highlight the importance of GPAM in de novo triglyceride (TG) synthesis. The pathway of TG biosynthesis is remarkable for the number of enzymes that catalyze each step. For example, four independent GPAM isoforms, each encoded by a separate gene, catalyze the synthesis of lysophosphatidic acid from glycerol-3-phosphate and long-chain acyl-CoA. Coleman et al. showed that the esterification of glycerol-3-phosphate with a long-chain acyl-CoA was the initial step in the synthesis of phospholipids and TG [16]. Plasmid- and adenovirus-mediated overexpression of GPAM in Chinese hamster ovary and HEK293 cells and in primary rat hepatocytes increases TG content and [14C]oleate incorporation into TG 3- to 4-fold, whereas hepatic TG content variably affects very-low-density lipoprotein (VLDL) secretion [17,18]. More recently, research of human populations has showed that genetic polymorphsims in GPAM are significantly associated with plasma tatol cholesterol (TC) and LDL-C levels [13,19-21]. But the reproducibility and repeatability of this association has not been performed in Chinese populations. Genetic variation is known to have different magnitudes of effect in the different ethnic groups. Therefore, it may be acctractive to characterize the association between the rs1129555 variation and serum lipid levels in the Chinese populations.
China is a multiethnic country, with a culture of different ethnic groups having different content and features. The Han population accounts for the majority of the total population of our country. Maonan, on the other hand, is one of the minorities with a population of 107,166 according to the China’s fifth national census in 2000. They are mainly distributed in the Shangnan, Zhongnan, and Xianan townships of Huanjiang Maonan Autonomous County in Guangxi Zhuang Autonomous Region, which is situated in Southwestern China. The Maonan ethnic group has various lifestyles and different eating habits which may result in the effect of hereditary variation to be further modified. Several previous studies have showed that the genetic relationship between Maonan nationality and other minorities in Guangxi [22] was much closer than that between Maonan and Han nationalities [23]. Furthermore, they have their tradition of ethnic endogamy and consanguineous marriage to cousins of maternal side, suggesting that the genetic background of Maonan nationality may be less heterogeneous within the population. Taken together, we believed that the Maonan nationality has become a useful group for population genetic studies. Thus, the present study was to detect the association of the rs1129555 SNP and serum lipid levels in the Maonan and Han populations.
Materials and methods
Subjects
A total of 780 unrelated participants of Han (306 males, 39.23% and 474 females, 60.77%) and 720 unrelated subjects of Maonan (291 males, 40.42% and 429 females, 59.58%) were randomly selected from our previous stratified randomized samples [24]. All participants were agricultural workers living in Huanjiang Maonan Autonomous Region, Guangxi Zhuang Autonomous Region, People’s Republic of China. The age of the participants ranged from 25 to 80 years, with a mean age of 55.90±13.54 years in Han and 57.07±15.02 years in Maonan (P > 0.05), respectively. All study subjects were essentially healthy with no history of cardiovascular disease such as CHD and stroke, diabetes, hyper-or hypo-thyroids, and chronic renal disease. We excluded the subjects who had a history of taking medications known to affect serum lipid levels (lipid-lowering drugs such as statins or fibrates, beta blockers, diuretics, or hormones) before the blood sample was drawn. The present study was approved by the Ethics Committee of the First Affiliated Hospital, Guangxi Medical University (No. Lunshen-2014-KY-Guoji-001, Mar. 7, 2014). Informed consent was taken from all participants.
Epidemiological survey
The survey was carried out using internationally standardized methods, following a common protocol [25]. Information on demographics, socioeconomic status, and lifestyle factors was collected with standardized questionnaires. Alcohol consumption was categorized into groups of grams of alcohol per day: 0 (non-drinker), < 25 and ≥ 25. Smoking status was categorized into groups of cigarettes per day: 0 (non-smoker), < 20 and ≥ 20. Several parameters such as blood pressure, height, weight, waist circumference, and body mass index (BMI) were measured or calculated. The methods of measuring above parameters were referred to previous studies [26].
Biochemical measurements
A fasting venous blood sample of 5 mL was drawn from the participants. A part of the sample (2 mL) was collected into glass tubes and used to determine serum lipid levels, and another part (3 mL) was shifted to tubes with anticoagulants (4.80 g/L citric acid, 14.70 g/L glucose and 13.20 g/L tri-sodium citrate) and used to extract deoxyribonucleic acid (DNA). Measurements of serum TC, TG, HDL-C, and LDL-C levels in the samples were performed by enzymatic methods with commercially available kits (RANDOX Laboratories Ltd., Ardmore, Diamond Road, Crumlin Co. Antrim, United Kingdom, BT29 4QY; Daiichi Pure Chemicals Co., Ltd., Tokyo, Japan). Serum apolipoprotein (Apo) A1 and ApoB levels were measured by the immunoturbidimetric immunoassay using a commercial kit (RANDOX Laboratories Ltd.). All determinations were performed with an auto-analyzer (Type 7170A; Hitachi Ltd., Tokyo, Japan) in the Clinical Science Experiment Center of the First Affiliated Hospital, Guangxi Medical University [27].
DNA amplification and genotyping
Genomic DNA of the samples was extracted from peripheral blood leucocytes according to the phenol-chloroform method [28]. The extracted DNA was stored at 4°C until analysis. Genotyping of the GPAM rs1129555 SNP was performed by polymerase chain reaction and restriction fragment length polymorphism (PCR-RFLP). PCR amplification was performed using 5’-AGAGAGGAGGGAAGTTGTGCA-3’ and 5’-TAACCCAGCATTGCCCAAAC-3’ (Sangon, Shanghai, People’s Republic of China) as the forward and reverse primer pairs, respectively. Each 25 μL PCR reaction mixture consisted of 2.0 μL genomic DNA, 1.0 μL each primer (10 μmol/L), 12.5 μL of 2 × Taq PCR Master mix (constituent: 0.1 U Taq polymerase/μL, 500 μM dNTP each and PCR buffer.), and 8.5 μL of ddH2O (DNase/RNase-free). PCR was performed with an initialization step of 95°C for 5 min, followed by 30 s denaturing at 95°C, 30 s of annealing at 58°C and 30 s of elongation at 72°C for 33 cycles. The amplification was completed by a final extension at 72°C for 7 min. Following electrophoresis on a 2.0% agarose gel with 0.5 µg/mL ethidium bromide, the amplification products were visualized under ultraviolet light. Subsequently, each restriction enzyme reaction was performed with 5.0 μL amplified DNA, 8.8 μL nuclease-free water, 1.0 μL of 10 × buffer solution and 0.2 μL BstMAI restriction enzyme in a total volume of 15 µL digested at 55°C overnight. After restriction enzyme digestion of the amplified DNA, genotypes were identified by electrophoresis on 2% ethidium-bromide stained agarose gels and visualized with UV illumination. Genotypes were scored by an experienced reader blinded to the epidemiological and serum lipid results. Six samples (CC, CT, and TT genotypes in two; respectively) detected by the PCR-RFLP were also confirmed by direct sequencing with an ABI Prism 3100 (Applied Biosystems) in Shanghai Sangon Biological Engineering Technology & Services Co., Ltd., People’s Republic of China.
Diagnostic criteria
The normal values of serum TC, TG, HDL-C, LDL-C, ApoA1, ApoB levels and the ApoA1/ApoB ratio in our Clinical Science Experiment Center were 3.10-5.17, 0.56-1.70, 1.16-1.42, 2.70-3.10 mmol/L, 1.20-1.60, 0.80-1.05 g/L and 1.00-2.50, respectively. The individuals with TC > 5.17 mmol/L and/or TG > 1.70 mmol/L were defined as hyperlipidaemic [29]. Hypertension diagnosis standard is according to the cirteria of 1999 and 2003 World Health Organization-International Society of Hypertension Guidelines for the management of hypertension [30]. The diagnostic criteria of overweight and obesity were according to the Cooperative Meta analysis Group of China Obesity Task Force. Normal weight, overweight and obesity were defined as a BMI < 24, 24-28 and > 28 kg/m2, respectively.
Statistical analyses
Statistical analyses were performed with the statistical software package SPSS 22.0 (SPSS Inc., Chicago, Illinois). Quantitative variables were presented as mean ± standard deviation (serum TG levels were presented as median and interquartile ranges). Allele frequency was determined via direct counting, and the Hardy-Weinberg equilibrium was verified with the standard goodness-of-fit test. The genotype distribution between the two ethnic groups was analyzed by the Chi-square test. General characteristics between two ethnic groups were compared by the Student’s unpaired t-test. Association between genotypes and serum lipid parameters was tested by covariance analysis (ANCOVA). Gender, age, BMI, blood pressure, alcohol consumption, and cigarette smoking were adjusted for the statistical analysis. Multivariate linear regression analyses with stepwise modeling were used to determine the correlation between the genotypes (CC = 1, CT = 2, TT = 3) and several environmental factors with serum lipid levels in males and females of Han and Maonan populations. Two-sided P values < 0.05 were considered statistically significant.
Results
General characteristics and serum lipid levels
Table 1 shows the general characteristics and serum lipid parameters between the Han and Maonan populations. The levels of serum HDL-C and ApoA1 were higher in Han than in Maonan (P < 0.01), whereas the percentages of cigarette smoking, alcohol consumption, and the levels of systolic blood pressure, pulse pressure, body waist circumference, serum TG were lower in Han than in Maonan (P < 0.05-0.001). There were no significant differences in the gender ratio, age structure, height, weight, BMI, diastolic blood pressure, glucose, serum TC, LDL-C, ApoB levels and the ratio of ApoA1 to ApoB between the two ethnic groups (P > 0.05 for all).
Table 1.
Comparison of demographic, lifestyle characteristics, and serum lipid levels between the Han and Maonan populations
| Parameter | Han | Maonan | t (x2) | P |
|---|---|---|---|---|
| Number | 780 | 720 | ||
| Male/female | 306/474 | 291/429 | 0.220 | 0.639 |
| Age (years) | 55.90±13.54 | 57.07±15.02 | -1.578 | 0.115 |
| Height (cm) | 154.25±7.90 | 153.87±8.05 | 0.908 | 0.364 |
| Weight (kg) | 53.40±9.18 | 53.17±10.72 | 0.443 | 0.658 |
| Body mass index (kg/m2) | 22.39±3.25 | 22.34±3.63 | 0.325 | 0.745 |
| Waist circumference (cm) | 75.53±8.14 | 76.60±9.25 | -2.360 | 0.018 |
| Smoking status [n (%)] | ||||
| Non-smoker | 593 (76.03) | 538 (74.72) | ||
| ≤ 20 cigarettes/day | 166 (21.28) | 142 (19.72) | ||
| > 20 cigarettes/day | 21 (2.69) | 40 (5.56) | 8.076 | 0.018 |
| Alcohol consumption [n (%)] | ||||
| Non-drinker | 625 (80.13) | 511 (70.97) | ||
| ≤ 25 g/day | 83 (10.64) | 110 (15.28) | ||
| > 25 g/day | 72 (9.23) | 99 (13.75) | 17.108 | 0.000 |
| Systolic blood pressure (mmHg) | 130.37±19.43 | 135.57±23.80 | -4.589 | 0.000 |
| Diastolic blood pressure (mmHg) | 82.01±11.51 | 83.20±12.44 | -1.911 | 0.056 |
| Pulse pressure (mmHg) | 48.36±15.73 | 52.37±17.44 | -4.639 | 0.000 |
| Glucose (mmol/L) | 6.29±1.96 | 6.19±1.43 | 1.133 | 0.257 |
| Total cholesterol (mmol/L) | 5.00±1.02 | 4.99±0.94 | 0.156 | 0.876 |
| Triglyceride (mmol/L) | 1.14 (0.51) | 1.28 (0.52) | -3.917 | 0.000 |
| HDL-C (mmol/L) | 1.70±0.46 | 1.58±0.39 | 5.128 | 0.000 |
| LDL-C (mmol/L) | 2.87±0.75 | 2.93±0.66 | -1.552 | 0.121 |
| ApoA1 (g/L) | 1.33±0.23 | 1.36±0.21 | -3.101 | 0.002 |
| ApoB (g/L) | 0.86±0.20 | 0.88±0.18 | -1.665 | 0.096 |
| ApoA1/ApoB | 1.63±0.53 | 1.62±0.44 | 0.282 | 0.778 |
HDL-C, high-density lipoprotein cholesterol; LDL-C, low-density lipoprotein cholesterol; Apo, apolipoprotein. The value of triglyceride was presented as median (interquartile range), the difference between the two ethnic groups was determined by the Wilcoxon-Mann-Whitney test.
Results of electrophoresis and genotyping
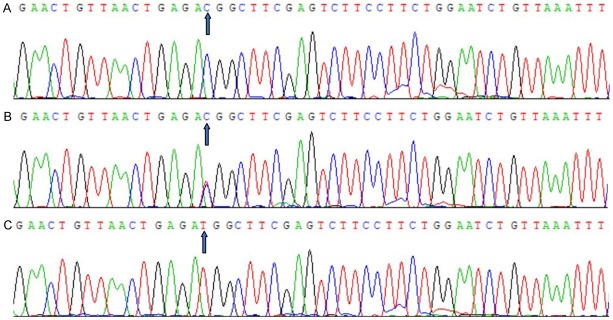
After the genomic DNA of the samples was amplified using PCR and visualized with 2% agarose gel electrophoresis, the products of 393 bp nucleotide sequences were observed in all samples (Figure 1). The genotypes identified were termed according to the presence (C allele) or absence (T allele) of the enzyme restriction sites. Thus, the CC genotype is homozygous for the presence of the site (bands at 307- and 86-bp), the CT genotype is heterozygous for the presence and absence of the site (bands at 393-, 307- and 86-bp) and the TT genotype is homozygous for the absence of the site (bands at 393 bp; Figure 2). The CC, CT and TT genotypes detected by PCR-RFLP were also confirmed by direct sequencing (Figure 3), respectively.
Figure 1.
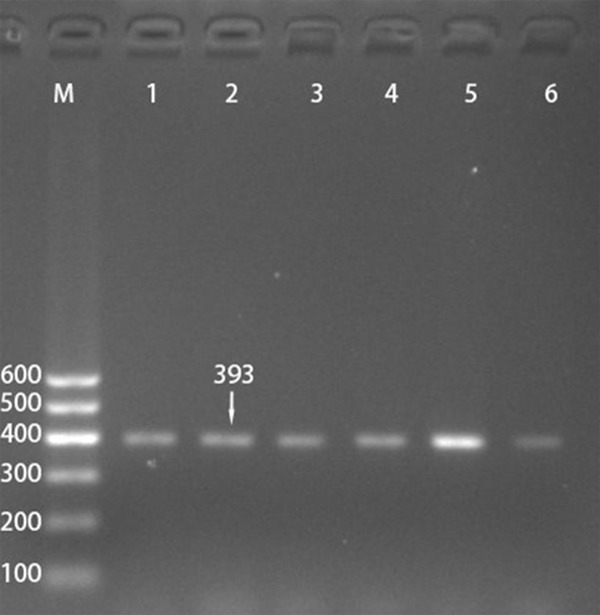
Electrophoresis of polymerase chain reaction products of the samples. Lane M is the 100 bp marker ladder; lanes 1-6 are samples, the 393 bp bands are the target genes.
Figure 2.
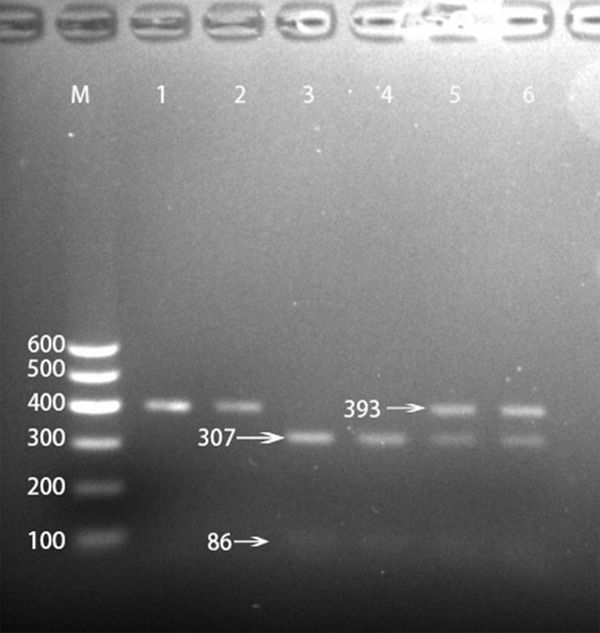
Genotyping of the GPAM rs1129555 SNP. Lane M, 100 bp marker ladder; lanes 1 and 2, TT genotype (393-bp); lanes 3 and 4, CC genotype (307- and 86-bp); and lanes 5 and 6, CT genotype (393-, 307- and 86-bp).
Figure 3.
Nucleotide sequence of the GPAM rs1129555 SNP. A. CC genotype; B. CT genotype; C. TT genotype.
Genotypic and allelic frequencies
The genotypic and allelic frequencies of the GPAM rs1129555 SNP are shown in Table 2. The genotype and allele frequencies were significantly different between the Han and Maonan populations (CC, 43.08% vs. 51.53%; CT, 44.23% vs. 42.36%; TT, 12.69% vs. 5.97%; P = 0.000; C, 65.19% vs. 72.85%; T, 34.81% vs. 27.15%; P = 0.000). Gender subgroup analysis showed that there were no differences in the genotypic and allele frequencies between males and females in the two ethnic groups (P > 0.05 for each).
Table 2.
Comparison of the genotype and allele frequencies of GPAM rs1129555 SNP in the Han and Maonan populations [n (%)]
| Group | n | Genotype | Allele | |||
|---|---|---|---|---|---|---|
|
| ||||||
| CC | CT | TT | C | T | ||
| Han | 780 | 336 (43.08) | 345 (44.23) | 99 (12.69) | 1017 (65.19) | 543 (34.81) |
| Maonan | 720 | 372 (51.53) | 305 (42.36) | 43 (5.97) | 1049 (72.85) | 391 (27.15) |
| x2 | 23.836 | 20.465 | ||||
| P | 0.000 | 0.000 | ||||
| Han | ||||||
| Male | 306 | 138 (45.10) | 125 (40.85) | 43 (14.05) | 404 (65.52) | 211 (34.48) |
| Female | 474 | 198 (41.77) | 220 (46.41) | 56 (11.81) | 616 (64.98) | 332 (35.02) |
| x2 | 2.513 | 0.048 | ||||
| P | 0.285 | 0.826 | ||||
| Maonan | ||||||
| Male | 291 | 158 (54.30) | 115 (39.52) | 18 (6.19) | 431 (74.05) | 151 (25.95) |
| Female | 429 | 214 (49.89) | 190 (44.29) | 25 (5.83) | 618 (72.03) | 240 (27.97) |
| x2 | 1.525 | 0.720 | ||||
| P | 0.467 | 0.396 | ||||
Genotypes and serum lipid levels
Tables 3 and 4 describe the association between genotypes and serum lipid levels. Serum LDL-C levels were different among the three genotypes in both ethnic groups (P < 0.05). Serum TG levels were different among the three genotypes in Han but not in Maonan (P < 0.05), the T allele carriers had higher TG and LDL-C levels than the T allele non-carriers (P < 0.05). Gender subgroup analysis showed that the TG levels in Han males and LDL-C levels in Maonan males were associated with the genotypes (P < 0.05-0.01).
Table 3.
Comparison of the genotype and serum lipid levels in the Han and Maonan populations
| Genotype | n | TC (mmol/L) | TG (mmol/L) | HDL-C (mmol/L) | LDL-C (mmol/L) | ApoA1 (g/L) | ApoB (g/L) | ApoA1/ApoB |
|---|---|---|---|---|---|---|---|---|
| Han | ||||||||
| CC | 335 | 4.85±1.17 | 1.03 (0.29) | 1.76±0.74 | 2.71±0.75 | 1.35±0.23 | 0.85±0.24 | 1.72±0.77 |
| CT | 345 | 5.00±1.01 | 1.10 (0.45) | 1.71±0.42 | 2.87±0.77 | 1.34±0.24 | 0.85±0.19 | 1.65±0.47 |
| TT | 99 | 5.04±0.98 | 1.16 (0.61) | 1.66±0.39 | 2.93±0.73 | 1.31±0.22 | 0.88±0.20 | 1.58±0.50 |
| F | 1.376 | 7.786 | 1.905 | 3.274 | 1.938 | 1.510 | 2.908 | |
| P | 0.253 | 0.020 | 0.150 | 0.038 | 0.145 | 0.221 | 0.055 | |
| Maonan | ||||||||
| CC | 372 | 4.76±1.05 | 1.16 (0.41) | 1.64±0.31 | 2.75±0.62 | 1.39±0.19 | 0.84±0.17 | 1.70±0.40 |
| CT | 305 | 5.00±0.96 | 1.28 (0.54) | 1.60±0.39 | 2.87±0.71 | 1.37±0.22 | 0.89±0.19 | 1.64±0.49 |
| TT | 43 | 5.01±0.91 | 1.25 (0.52) | 1.56±0.38 | 2.99±0.62 | 1.35±0.20 | 0.88±0.18 | 1.60±0.39 |
| F | 1.421 | 3.303 | 1.191 | 4.308 | 1.589 | 1.043 | 1.546 | |
| P | 0.242 | 0.192 | 0.305 | 0.014 | 0.205 | 0.353 | 0.214 |
TC, total cholesterol; TG, triglyceride; HDL-C, high-density lipoprotein cholesterol; LDL-C, low-density lipoprotein cholesterol; ApoA1, apolipoprotein A1; ApoB, apolipoprotein B; ApoA1/ApoB, the ratio of apolipoprotein A1 to apolipoprotein B. The value of TG was presented as median (interquartile range). The difference between the genotypes was determined by the Kruskal-Wallis test.
Table 4.
Comparison of the genotypes and serum lipid levels between males and females in the Han and Maonan populations
| Ethnic/Genotype | n | TC (mmol/L) | TG (mmol/L) | HDL-C (mmol/L) | LDL-C (mmol/L) | ApoA1 (g/L) | ApoB (g/L) | ApoA1/ApoB |
|---|---|---|---|---|---|---|---|---|
| Han/male | ||||||||
| CC | 138 | 5.00±1.32 | 1.03 (0.29) | 1.74±0.44 | 2.79±0.81 | 1.36±0.26 | 0.92±0.27 | 1.58±0.45 |
| CT | 125 | 5.09±0.95 | 1.12 (0.56) | 1.67±0.45 | 2.88±0.75 | 1.35±0.28 | 0.88±0.20 | 1.60±0.46 |
| TT | 43 | 5.16±0.96 | 1.32 (0.79) | 1.59±0.37 | 2.96±0.68 | 1.30±0.24 | 0.91±0.19 | 1.49±0.45 |
| F | 0.421 | 8.685 | 2.756 | 0.993 | 1.940 | 0.859 | 2.177 | |
| P | 0.657 | 0.013 | 0.065 | 0.372 | 0.145 | 0.425 | 0.115 | |
| Han/female | ||||||||
| CC | 198 | 4.73±1.05 | 1.02 (0.34) | 1.78±0.92 | 2.64±0.70 | 1.34±0.19 | 0.80±0.20 | 1.82±0.94 |
| CT | 220 | 4.94±1.03 | 1.09 (0.44) | 1.73±0.40 | 2.85±0.79 | 1.33±0.22 | 0.83±0.18 | 1.68±0.47 |
| TT | 56 | 4.97±0.99 | 1.11 (0.47) | 1.72±0.40 | 2.90±0.76 | 1.32±0.21 | 0.85±0.21 | 1.65±0.52 |
| F | 1.179 | 2.054 | 0.305 | 2.511 | 0.350 | 1.375 | 1.998 | |
| P | 0.308 | 0.358 | 0.737 | 0.082 | 0.705 | 0.254 | 0.137 | |
| Maonan/male | ||||||||
| CC | 158 | 4.69±0.90 | 1.15 (0.28) | 1.61±0.31 | 2.71±0.58 | 1.34±0.21 | 0.80±0.16 | 1.73±0.39 |
| CT | 115 | 4.98±0.95 | 1.25 (0.76) | 1.53±0.38 | 2.76±0.68 | 1.36±0.24 | 0.89±0.21 | 1.63±0.61 |
| TT | 18 | 4.99±0.89 | 1.38 (0.57) | 1.55±0.43 | 2.99±0.58 | 1.33±0.19 | 0.89±0.17 | 1.56±0.38 |
| F | 0.908 | 1.917 | 0.339 | 5.463 | 0.448 | 2.132 | 1.458 | |
| P | 0.405 | 0.384 | 0.713 | 0.005 | 0.639 | 0.120 | 0.234 | |
| Maonan/female | ||||||||
| CC | 214 | 4.81±1.16 | 1.16 (0.49) | 1.67±0.31 | 2.78±0.66 | 1.42±0.18 | 0.87±0.18 | 1.69±0.42 |
| CT | 190 | 5.01±0.97 | 1.20 (0.47) | 1.64±0.40 | 2.94±0.71 | 1.38±0.20 | 0.88±0.19 | 1.64±0.40 |
| TT | 25 | 5.03±0.92 | 1.26 (0.39) | 1.57±0.35 | 2.98±0.64 | 1.36±0.20 | 0.87±0.18 | 1.63±0.39 |
| F | 0.585 | 2.818 | 2.042 | 1.125 | 1.356 | 0.156 | 0.277 | |
| P | 0.557 | 0.244 | 0.131 | 0.326 | 0.259 | 0.855 | 0.758 |
TC, total cholesterol; TG, triglyceride; HDL-C, high-density lipoprotein cholesterol; LDL-C, low-density lipoprotein cholesterol; ApoA1, apolipoprotein A1; ApoB, apolipoprotein B; ApoA1/ApoB, the ratio of apolipoprotein A1 to apolipoprotein B. The value of TG was presented as median (interquartile range). The difference between the genotypes was determined by the Kruskal-Wallis test.
Relative factors for serum lipid parameters
The risk factors for serum lipid parameters in Maonan and Han are shown in Tables 5 and 6. Multiple linear regression analysis showed that serum LDL-C levels in both Maonan and Han populations; TG, LDL-C, in Han and LDL-C levels in Maonan were correlated with the genotypes of the GPAM rs1129555 SNP (P < 0.05-0.01; Table 5). As shown in Table 6, when the correlation between serum lipid parameters and the genotypes was analyzed according to gender, the TG levels were assocated with genotypes in Han males and serum LDL-C levels in Maonan males were correlated with the genotypes (P < 0.05-0.01 for all; Table 6). Blood lipid phenotypes were also correlated with several environmental factors such as age, gender, weight, waist circumference, alcohol consumption, and cigarette smoking, and traditional cardiovascular risk factors such as BMI, fasting blood glucose, and blood pressure levels in males and females of both ethnics (P < 0.05-0.001; Tables 5 and 6).
Table 5.
Relationship between serum lipid parameters and relative factors in the Han and Maonan populations
| Lipid | Risk factor | B | Std.error | Beta | t | P |
|---|---|---|---|---|---|---|
| Han and Maonan | ||||||
| TC | Waist circumference | 0.019 | 0.005 | 0.165 | 3.903 | 0.000 |
| Diastolic blood pressure | 0.008 | 0.002 | 0.093 | 3.438 | 0.001 | |
| Glucose | 0.042 | 0.015 | 0.072 | 2.735 | 0.006 | |
| TG | Alocohol consumption | 0.164 | 0.075 | 0.065 | 2.185 | 0.029 |
| Cigarette smoking | 0.428 | 0.101 | 0.134 | 4.247 | 0.000 | |
| Distolic blood pressure | 0.010 | 0.003 | 0.079 | 3.021 | 0.003 | |
| Waist cirumference | 0.041 | 0.007 | 0.230 | 5.637 | 0.000 | |
| Glucose | 0.096 | 0.023 | 0.106 | 4.164 | 0.000 | |
| HDL-C | Ethnic group | -0.099 | 0.0262 | -0.114 | -4.488 | 0.000 |
| Gender | 0.131 | 0.034 | 0.148 | 3.809 | 0.000 | |
| Cigarette smoking | 0.075 | 0.028 | 0.084 | 2.635 | 0.009 | |
| Alocohol consumption | 0.089 | 0.021 | 0.128 | 4.210 | 0.000 | |
| Waist circumference | -0.008 | 0.002 | -0.159 | -3.843 | 0.000 | |
| LDL-C | Waist circumference | 0.012 | 0.003 | 0.150 | 3.533 | 0.000 |
| Genotype | 0.011 | 0.004 | 0.124 | 3.296 | 0.001 | |
| Distolic blood pressure | 0.004 | 0.002 | 0.065 | 2.393 | 0.017 | |
| Glucose | 0.022 | 0.011 | 0.053 | 1.980 | 0.048 | |
| ApoA1 | Alcohol consumption | 0.076 | 0.011 | 0.217 | 7.077 | 0.000 |
| Ethnic group | 0.036 | 0.011 | 0.081 | 3.161 | 0.002 | |
| Cigarette smoking | 0.069 | 0.014 | 0.154 | 4.784 | 0.000 | |
| Gender | 0.087 | 0.018 | 0.193 | 4.940 | 0.000 | |
| ApoB | Waist circumference | 0.005 | 0.001 | 0.229 | 5.617 | 0.000 |
| Glucose | 0.010 | 0.003 | 0.094 | 3.661 | 0.000 | |
| Age | 0.001 | 0.000 | 0.107 | 3.639 | 0.000 | |
| Distolic blood pressure | 0.001 | 0.000 | 0.061 | 2.346 | 0.019 | |
| ApoA1/ApoB | Gender | 0.182 | 0.038 | 0.182 | 4.769 | 0.000 |
| Age | -0.004 | 0.001 | -0.130 | -4.398 | 0.000 | |
| Alcohol consumption | 0.097 | 0.023 | 0.124 | 4.141 | 0.000 | |
| Cigarette smoking | 0.105 | 0.031 | 0.105 | 3.330 | 0.001 | |
| Height | 0.018 | 0.007 | 0.287 | 2.396 | 0.017 | |
| Weight | -0.022 | 0.010 | -0.440 | -2.157 | 0.023 | |
| Waist circumference | -0.010 | 0.002 | -0.185 | -4.532 | 0.000 | |
| Glucose | -0.015 | 0.007 | -0.052 | -2.056 | 0.040 | |
| Han | ||||||
| TC | Waist circumference | 0.002 | 0.008 | 0.178 | 2.898 | 0004 |
| Glucose | 0.071 | 0.019 | 0.136 | 3.750 | 0.000 | |
| Distolic blood pressure | 0.012 | 0.003 | 0.139 | 3.810 | 0.000 | |
| TG | Age | -0.014 | 0.005 | -0.109 | -2.726 | 0.007 |
| Cigarette smoking | 0.471 | 0.152 | 0.135 | 3.098 | 0.002 | |
| Waist circumference | 0.057 | 0.013 | 0.269 | 4.423 | 0.000 | |
| Glucose | 0.105 | 0.032 | 0.119 | 3.327 | 0.001 | |
| Genotype | 0.354 | 0.154 | 0.103 | 2.297 | 0.022 | |
| HDL-C | Gender | 0.139 | 0.052 | 0.145 | 2.658 | 0.008 |
| Cigarette smoking | 0.113 | 0.042 | 0.121 | 2.708 | 0.007 | |
| Alcohol consumption | 0.087 | 0.032 | 0.117 | 2.715 | 0.007 | |
| LDL-C | Gender | -0.214 | 0.085 | -0.138 | -2.523 | 0.012 |
| Glucose | 0.041 | 0.014 | 0.107 | 2.895 | 0.004 | |
| Genotype | 0.016 | 0.008 | 0.118 | 2.001 | 0.046 | |
| ApoA1 | Gender | 0.064 | 0.025 | 0.133 | 2.514 | 0.012 |
| Age | -0.002 | 0.001 | -0.116 | -2.903 | 0.004 | |
| Cigarette smoking | 0.093 | 0.020 | 0.201 | 4.616 | 0.000 | |
| Alcohol consumption | 0.092 | 0.016 | 0.246 | 5.897 | 0.000 | |
| Weight | -0.014 | 0.007 | -0.554 | -2.074 | 0.038 | |
| ApoB | Gender | -0.059 | 0.022 | -0.143 | -2.696 | 0.007 |
| Waist circumference | 0.004 | 0.002 | 0.168 | 2.779 | 0.006 | |
| Glucose | 0.015 | 0.004 | 0.143 | 4.207 | 0.000 | |
| ApoA1/ApoB | Gender | 0.229 | 0.058 | 0.209 | 3.934 | 0.000 |
| Age | -0.006 | 0.002 | -0.143 | -3.583 | 0.000 | |
| Cigarette smoking | 0.151 | 0.047 | 0.141 | 3.252 | 0.001 | |
| Alcohol consumption | 0.091 | 0.036 | 0.106 | 2.525 | 0.012 | |
| Maonan | ||||||
| TC | Age | 0.007 | 0.004 | 0.108 | 2.008 | 0.045 |
| Waist circumference | 0.023 | 0.007 | 0.203 | 3.041 | 0.002 | |
| TG | Waist circumference | 0.030 | 0.006 | 0.299 | 4.764 | 0.000 |
| HDL-C | Gender | 0.148 | 0.048 | 0.174 | 3.112 | 0.002 |
| Height | 0.019 | 0.009 | 0.407 | 2.159 | 0.031 | |
| Waist circumference | -0.006 | 0.003 | -0.146 | -2.251 | 0.025 | |
| LDL-C | Waist circumference | 0.019 | 0.005 | 0.246 | 3.731 | 0.000 |
| Genotype | 0.020 | 0.005 | 0.260 | 3.970 | 0.000 | |
| ApoA1 | Gender | 0.094 | 0.027 | 0.202 | 3.519 | 0.000 |
| ApoB | Age | 0.002 | 0.001 | 0.154 | 3.037 | 0.003 |
| Waist circumference | 0.007 | 0.001 | 0.360 | 5.698 | 0.000 | |
| ApoA1/ApoB | Waist circumference | -0.015 | 0.003 | -0.350 | -5.546 | 0.000 |
TC, total cholesterol; TG, triglyceride; HDL-C, high-density lipoprotein cholesterol; LDL-C, low-density lipoprotein cholesterol; ApoA1, apolipoprotein A1; ApoB, apolipoprotein B; ApoA1/ApoB, the ratio of apolipoprotein A1 to apolipoprotein B; B, unstandardized coefficient; Beta, standardized coefficient.
Table 6.
Relationship between serum lipid parameters and relative factors in the males and females of the Han and Maonan populations
| Lipid | Risk factor | B | Std.error | Beta | t | P |
|---|---|---|---|---|---|---|
| Han/male | ||||||
| TC | Waist circumference | 0.027 | 0.012 | 0.226 | 2.216 | 0.027 |
| TG | Genotype | 0.554 | 0.225 | 0.142 | 2.421 | 0.016 |
| Age | -0.027 | 0.012 | -0.163 | -2.333 | 0.020 | |
| Cigarette smoking | 0.460 | 0.226 | 0.120 | 2.036 | 0.043 | |
| Waist circumference | 0.090 | 0.028 | 0.322 | 3.214 | 0.001 | |
| Glucose | 0.177 | 0.083 | 0.127 | 2.138 | 0.033 | |
| HDL-C | Cigarette smoking | 0.094 | 0.040 | 0.137 | 2.349 | 0.019 |
| Alcohol consumption | 0.086 | 0.030 | 0.167 | 2.856 | 0.005 | |
| LDL-C | Age | -0.008 | 0.004 | -0.158 | -2.188 | 0.030 |
| Cigarette smoking | -0.189 | 0.073 | -0.158 | -2.558 | 0.010 | |
| ApoA1 | Cigarette smoking | 0.080 | 0.024 | 0.189 | 3.381 | 0.001 |
| Alcohol consumption | 0.095 | 0.018 | 0.296 | 5.297 | 0.000 | |
| Weight | -0.021 | 0.010 | -0.748 | -2.190 | 0.029 | |
| ApoB | Diastolic blood pressure | 0.003 | 0.001 | 0.173 | 2.627 | 0.009 |
| Glucose | 0.018 | 0.007 | 0.149 | 2.475 | 0.014 | |
| ApoA1/ApoB | Cigarette smoking | 0.118 | 0.043 | 0.157 | 2.760 | 0.006 |
| Alcohol consumption | 0.098 | 0.032 | 0.171 | 3.018 | 0.003 | |
| Han/female | ||||||
| TC | Glucose | 0.078 | 0.022 | 0.162 | 3.563 | 0.000 |
| TG | Waist circumference | 0.029 | 0.012 | 0.186 | 2.359 | 0.019 |
| Diastolic blood pressure | 0.015 | 0.005 | 0.138 | 2.879 | 0.004 | |
| Glucose | 0.075 | 0.025 | 0.136 | 2.955 | 0.003 | |
| HDL-C | Waist circumference | -0.009 | 0.003 | -0.145 | -3.031 | 0.003 |
| LDL-C | Age | 0.010 | 0.003 | 0.176 | 3,231 | 0.001 |
| Glucose | 0.039 | 0.017 | 0.108 | 2.352 | 0.019 | |
| ApoA1 | Age | -0.003 | 0.001 | -0.163 | -2.939 | 0.003 |
| Cigarette smoking | 0.197 | 0.061 | 0.157 | 3.211 | 0.011 | |
| Body mass index | -0.084 | 0.036 | -1.174 | -2.330 | 0.020 | |
| Height | -0.022 | 0.010 | -0.641 | -2.109 | 0.035 | |
| ApoB | Age | 0.002 | 0.001 | 0.165 | 3.105 | 0.002 |
| Cigarette smoking | -0.122 | 0.054 | -0.106 | -2.273 | 0.023 | |
| Glucose | 0.012 | 0.004 | 0.135 | 3.010 | 0.003 | |
| ApoA1/ApoB | Age | -0.010 | 0.002 | -0.235 | -4.369 | 0.000 |
| Cigarette smoking | 0.555 | 0.158 | 0.166 | 3.504 | 0.001 | |
| Maonan/male | ||||||
| TC | Height | -0.125 | 0.062 | -0.917 | -2.010 | 0.045 |
| Weight | 0.179 | 0.083 | 1.983 | 2.144 | 0.033 | |
| TG | Alcohol consumption | 0.272 | 0.136 | 0.117 | 2.044 | 0.046 |
| Waist circumference | 0.044 | 0.021 | 0.216 | 2.130 | 0.034 | |
| Glucose | 0.214 | 0.073 | 0.166 | 2.920 | 0.004 | |
| HDL-C | Waist circumference | -0.017 | 0.004 | -0.395 | -3.856 | 0.000 |
| Alcohol consumption | 0.115 | 0.030 | 0.230 | 3.896 | 0.000 | |
| LDL-C | Alcohol consumption | -0.098 | 0.049 | -0.123 | -1.985 | 0.048 |
| Genotype | 0.120 | 0.029 | 0.243 | 4.100 | 0.000 | |
| ApoA1 | Waist circumference | -0.005 | 0.002 | -0.231 | -2.206 | 0.028 |
| Alcohol consumption | 0.074 | 0.016 | 0.286 | 4.749 | 0.000 | |
| ApoB | Weight | 0.037 | 0.016 | 1.805 | 2.083 | 0.038 |
| Age | 0.002 | 0.001 | 0.158 | 2.248 | 0.025 | |
| Glucose | 0.019 | 0.007 | 0.145 | 2.620 | 0.009 | |
| ApoA1/ApoB | Waist circumference | -0.012 | 0.005 | -0.227 | -2.246 | 0.026 |
| Maonan/female | ||||||
| TC | Age | 0.011 | 0.004 | 0.162 | 2.771 | 0.006 |
| Waist circumference | 0.023 | 0.008 | 0.186 | 2.739 | 0.006 | |
| TG | Waist circumference | 0.028 | 0.006 | 0.316 | 4.944 | 0.000 |
| HDL-C | Height | 0.022 | 0.011 | 0.305 | 2.033 | 0.043 |
| Waist circumference | -0.006 | 0.003 | -0.139 | -2.072 | 0.039 | |
| LDL-C | Age | 0.007 | 0.003 | 0.154 | 2.680 | 0.008 |
| Waist circumference | 0.018 | 0.005 | 0.226 | 3.381 | 0.001 | |
| ApoA1 | Waist circumference | -0.003 | 0.002 | -0.146 | -2.146 | 0.032 |
| ApoB | Age | 0.003 | 0.001 | 0.209 | 3.793 | 0.000 |
| Waist circumference | 0.008 | 0.001 | 0.354 | 5.528 | 0.000 | |
| Pulse pressure | 0.002 | 0.001 | 0.187 | 3.716 | 0.000 | |
| ApoA1/ApoB | Age | -0.004 | 0.001 | -0.139 | -2.497 | 0.013 |
| Waist circumference | -0.016 | 0.003 | -0.350 | -5.436 | 0.000 | |
| Systolic blood pressure | -0.003 | 0.001 | -0.175 | -2.616 | 0.009 |
TC, total cholesterol; TG, triglyceride; HDL-C, high-density lipoprotein cholesterol; LDL-C, low-density lipoprotein cholesterol; ApoA1, apolipoprotein A1; ApoB, apolipoprotein B; ApoA1/ApoB, the ratio of apolipoprotein A1 to apolipoprotein B; B, unstandardized coefficient; Beta, standardized coefficien.
Discussion
China has experienced a considerable increase in the prevalence of CHD over the past years [31]. Disorders of lipid metabolism plays an important role in the pathogenesis and development of atherosclerosis and CHD. It is widely accepted that genetic variants and interactions with environmental factors have a great impact on serum lipid levels. Numerous studies have indicated that ~40-60% of variation in serum lipid profiles was genetically determined [11]. The Han nationality is the largest ethnic group and widely live in 2/3 regions of China, whereas the Maonan nationality is a peculiar mountainous minority in Guangxi. Maonan people mainly engage in agriculture and are good at raising beef cattle and preparing the bamboo hat. Special geographical and folk customs formed a cooking culture with distinct characteristics. Corn and rice are the main food for them and sweet potatoes, pumpkin, and sorghum are an important supplement. In addition, the individuals of Maonan nationality are more inclined to eat pickles, sauerkraut and spicy and sour dishes. Maonan nationality preserves their custom of ethnic intermarriages, with parents arranging marriages being common. They have their culture of consanguineous marriage to cousins of maternal side, intermarriage with Han or Zhuang people seldom occurs. In summary, owing to their unique diet habits, lifestyle, and endogamy customs, we speculate that the hereditary characteristics of lipid metabolism-realted genes in Maonan ethnic group may be different from those in Han Chinese.
The genotypic and allelic frequencies of the rs1129555 SNP in diverse ethnic groups are significantly different, which can be found on the International HapMap project data-base. The frequencies of C and T alleles were 70.34% and 34.81% in European; 74.17% and 31.71% in Yoruba; 80% and 20% in Japanese; 68.29% and 31.71% in Chinese Han in Beijing. In the present study, we showed that the frequencies of C and T alleles were 72.85% and 27.15% in Maonan, and 65.19% and 34.81% in Han (P < 0.001), which were in close proximity to those of Chinese Han Beijing; the T allele frequency of the rs1129555 SNP was lower in Maonan than in Han. The distribution of TT and CT genotypes was also different in two ethnic groups (P < 0.001). Gender subgroup analysis showed that there were no conspicuous differences in the genotypic and allelic frequencies between males and females in the Maonan and Han populations. These findings suggest that the genotype and allele frequencies of the rs1129555 SNP in GPAM may exhibit a racial/ethnic specificity.
Several previous GWASs reported that genetic polymorphism in GPAM was significantly associated with plasma lipid levels. A study of common and rare lipid-associated risk loci has indentified that the GPAM rs1129555 SNP is associated with serum LDL-C concentrations in European population [19]. A meta-analysis of the mechanisms and genetic determinant regulating human lipid disease showed that the minor allele of the rs1129555 SNP was related with higher blood LDL-C and TC levels [32]. In the current study, we found that the rs1129555 SNP was significant associated with multiple serum lipid parameters in the Maonan and Han populaitons. The T allele carriers had higher TG in Han and higher LDL-C in both ethnic groups than the T allele non-carriers. Gender subgroup analyses showed that the T allele carriers had higher TG levels in Han males and higher LDL-C levels in Maonan males but not in famales in both ethnics. These experimental results indicate that the association between the GPAM 1129555 SNP and serum lipid levels may have racial/ethnic and/or sex specificity. GPAM has been considered to be the rate-limiting step in the pathway of glycerolipid synthesis and to regulate fatty acid flux through the pathway, which plays a key role in lipid biosynthesis. Animal models and in vitro studies showed that GPAM is upregulated transcriptionally by sterol regulatory element-binding protein-1c (SREBP-1c) and downregulated acutely by AMP-activated protein kinase, consistent with a role in TG synthesis [33]. Combined with our experimental results, we speculate that the GPAM rs1129555 mutation may act in the rate-limiting step of glycerolipid synthesis and bring about the cascade of events in dyslipidemia. However, the biogical role and function of the rs1129555 SNP in lipid metabolism need to be further investigation.
The influence extents of genetic and environmental factors on serum lipid levels remain a controversial issue, but several environmental factors such as low-carbohydrate and high-fat dietary patterns, obesity, hypertension, diabetes and unhealthy lifestyle have been associated with serum lipid levels [34]. In the present study, multivariate linear regression analysis showed that serum lipid parameters were correlated to age, gender, weight, waist circumference, BMI, blood pressure, fasting blood glucose levels, alcohol consumption, and cigarette smoking in both ethnic groups. These results suggest that environmental factors and their interactions with a hereditary component also play a critical role in determining serum lipid levels in our study populations. Moreover, the percentages of subjects who consumed alcohol and smoked cigarettes were higher in Maonan than in Han. Moderate alcohol intake has been showed to reduce cardiovascular events, and the beneficial effects of alcohol on CHD have been ascribed to the increase in HDL-C and ApoA1 levels [35]. Long-term alcohol abuse can cause liver damage, hypertriglyceridemia, hypertension and serious cardiovascular lesions. In a previous meta-analysis, 30 g of alcohol daily was associated with a plasma TG increase of 5.69 mg/dl, wheres alcohol intake of 60 g/day increased the TG levels by about 0.19 mg/dL per 1 gram of alcohol consumed [36]. The influence of alcohol on blood lipid metabolism seems to be different among males and famales. A previous study of Turks found that the levels of LDL-C as well as ApoB and TG were increased in male drinkers, while females had decreased TG and no change in LDL-C or ApoB with alcohol [37]. Another research study indicated that the effects of alcohol consumptionon on LDL-C levels appear to vary by specific patient types or patterns of alcohol intake, and sex as well as genetic variants [38]. Smoking has been strongly implicated as a risk factor for dyslipidemia, arteriosclerosis, chronic obstructive pulmonary disease, and lung cancer. Cigarettes contain a large number of oxidants and many adverse effects of smoking result from oxidative damage to cirtical biologic substances. A related study indicated that oxidation of LDL by cigarette smoking may contribute a causative link between cigarette smoking and atherogenesis [39]. Additionally, according to the results of a meta analysis based on 7,256 subjects, smoking increased TG by 13 mg/dl (0.15 mmol/L) and decreased HDL-C by 3.5 mg/dl (0.09 mmol/L) with every 20 cigarettes smoked [40]. It is well accepted that dietary patterns are strongly related with serum lipid profiles and the prevalence of dyslipidemia. The people of Maonan nationality like to eat cold foods along with acidic and spicy dishes, so acidic meat and pickled vegetables are among their most popular dishes, which contain abundant sodium salt. This preference of high-carbohydrates and salt diet may be related to the higher TG, LDL-C levels, and waist circumference in Maonan than in Han populations. Numerous studies have indentified that dietary intake of saturated and trans-fat raises blood cholesterol concentrations and CVD risk [41,42]. Diet and relative weight could account for up to 6% of the variability in serum cholesterol levels and every 1% decrease in energy consumed as dietary saturated fatty acid, TC decreased by 0.056 mmol/L and LDL-C by 0.05 mmol/L [43]. To summarize, the mutual effects of different eating habits, lifestyles, and environmental factors probably further modify the association of genetic variations and serum lipid levels in our study populations.
Limitations
There are several potential limitations in our study. First, the sample size is relatively small compared to many GWASs and replication studies, further studies with larger sample sizes are needed to confirm our results. Second, we were not able to alleviate the effect of diet and several environmental factors during the statistical analysis. Third, although we have detected effects of GPAM rs1129555 SNP on serum lipid levels in our study, there are still many lipid-related SNPs and the interaction of SNP-SNP and/or SNP-environmental factors. Thus, further studies on biological functions of the rs1129555 variation and interactions of gene-environment are necessary.
Conclusions
In conclusion, the genotype and allele frequencies of the rs1129555 SNP were significantly different between the Han and Maonan populations. The minor T allele carriers have more unfavorable serum lipid profiles than the T allele non-carriers in both ethnic groups. These findings suggest that the association between the GPAM rs1129555 variant and serum lipid levels might have racial/ethnic- and/or sex- specificity.
Acknowledgements
This study was supported by the National Natural Science Foundation of China (No. 81460169).
Disclosure of conflict of interest
None.
References
- 1.Roberts R, Stewart AF. Genes and coronary artery disease: where are we? J Am Coll Cardiol. 2012;60:1715–1721. doi: 10.1016/j.jacc.2011.12.062. [DOI] [PubMed] [Google Scholar]
- 2.Dorn GW, Cresci S. Genome-wide association studies of coronary artery disease and heart failure: where are we going. Pharmacogenomics. 2009;10:213–223. doi: 10.2217/14622416.10.2.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ye X, Peng L, Kan H, Wang W, Geng F, Mu Z, Zhou J, Yang D. Acute effects of particulate air pollution on the incidence of coronary heart disease in Shanghai, China. PLoS One. 2016;11:e0151119. doi: 10.1371/journal.pone.0151119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Y, Du F, Zhao H, Yu X, Liu J, Xiao Y, Lu C, Li X, Wang Y, Wang B, Niu W. Synergistic association between two alcohol metabolism relevant genes and coronary artery disease among Chinese hypertensive patients. PLoS One. 2014;9:e103161. doi: 10.1371/journal.pone.0103161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wald NJ, Law MR. A strategy to reduce cardiovascular disease by more than 80% BMJ. 2004;326:1419. doi: 10.1136/bmj.326.7404.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qian DF, Fan GL, Chen P, He DC, Fan JD, Feng C, Zhu PG, Zhou ZH, Liao YH. Risk factors for hyperuricemia in active and retired employees underwent physical examination. Zhonghua Xin Xue Guan Bing Za Zhi. 2013;41:60–64. [PubMed] [Google Scholar]
- 7.Vaverkova H. LDL-C or apoB as the best target for reducing coronary heart disease: should apoB be implemented into clinical practice. Clin Lipidol. 2011;6:35–48. [Google Scholar]
- 8.Graham I, Dan A, Borch-Johnsen K, Boysen G, Burell G, Cifkova R, Dallongeville J, Backer GD, Ebrahim S, Gjelsvik B. European guidelines on cardiovascular disease prevention in clinical practice: executive summary. Fourth joint task force of the European society of cardiology and other societies on cardiovascular disease prevention in clinical practice. Eur Heart J. 2007;28:2375–2414. doi: 10.1093/eurheartj/ehm316. [DOI] [PubMed] [Google Scholar]
- 9.Ordovás JM, Robertson R, Cléirigh EN. Genegene and gene-environment interactions defining lipid-related traits. Curr Opin Lipidol. 2011;22:129–136. doi: 10.1097/MOL.0b013e32834477a9. [DOI] [PubMed] [Google Scholar]
- 10.Pilia G, Chen WM, Scuteri A, Orrú M, Albai G, Dei M, Lai S, Usala G, Lai M, Loi P. Heritability of cardiovascular and personality traits in 6,148 Sardinians. PLoS Genet. 2006;2:e132. doi: 10.1371/journal.pgen.0020132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Draisma HH, Reijmers TH, Meulman JJ, van der Greef J, Hankemeier T, Boomsma DI. Hierarchical clustering analysis of blood plasma lipidomics profiles from mono- and dizygotic twin families. Eur J Hum Genet. 2013;21:95–101. doi: 10.1038/ejhg.2012.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kathiresan S, Willer CJ, Peloso GM, Demissie S, Musunuru K, Schadt EE, Kaplan L, Bennett D, Li Y, Tanaka T, Voight BF, Bonnycastle LL, Jackson AU, Crawford G, Surti A, Guiducci C, Burtt NP, Parish S, Clarke R, Zelenika D, Kubalanza KA, Morken MA, Scott LJ, Stringham HM, Galan P, Swift AJ, Kuusisto J, Bergman RN, Sundvall J, Laakso M, Ferrucci L, Scheet P, Sanna S, Uda M, Yang Q, Lunetta KL, Dupuis J, de Bakker PI, O’Donnell CJ, Chambers JC, Kooner JS, Hercberg S, Meneton P, Lakatta EG, Scuteri A, Schlessinger D, Tuomilehto J, Collins FS, Groop L, Altshuler D, Collins R, Lathrop GM, Melander O, Salomaa V, Peltonen L, Orho-Melander M, Ordovas JM, Boehnke M, Abecasis GR, Mohlke KL, Cupples LA. Common variants at 30 loci contribute to polygenic dyslipidemia. Nat Genet. 2009;41:56–65. doi: 10.1038/ng.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, Koseki M, Pirruccello JP, Ripatti S, Chasman DI, Willer CJ, Johansen CT, Fouchier SW, Isaacs A, Peloso GM, Barbalic M, Ricketts SL, Bis JC, Aulchenko YS, Thorleifsson G, Feitosa MF, Chambers J, Orho-Melander M, Melander O, Johnson T, Li X, Guo X, Li M, Shin CY, Jin GM, Jin KY, Lee JY, Park T, Kim K, Sim X, Twee-Hee OR, Croteau-Chonka DC, Lange LA, Smith JD, Song K, Hua ZJ, Yuan X, Luan J, Lamina C, Ziegler A, Zhang W, Zee RY, Wright AF, Witteman JC, Wilson JF, Willemsen G, Wichmann HE, Whitfield JB, Waterworth DM, Wareham NJ, Waeber G, Vollenweider P, Voight BF, Vitart V, Uitterlinden AG, Uda M, Tuomilehto J, Thompson JR, Tanaka T, Surakka I, Stringham HM, Spector TD, Soranzo N, Smit JH, Sinisalo J, Silander K, Sijbrands EJ, Scuteri A, Scott J, Schlessinger D, Sanna S, Salomaa V, Saharinen J, Sabatti C, Ruokonen A, Rudan I, Rose LM, Roberts R, Rieder M, Psaty BM, Pramstaller PP, Pichler I, Perola M, Penninx BW, Pedersen NL, Pattaro C, Parker AN, Pare G, Oostra BA, O’Donnell CJ, Nieminen MS, Nickerson DA, Montgomery GW, Meitinger T, McPherson R, McCarthy MI, McArdle W, Masson D, Martin NG, Marroni F, Mangino M, Magnusson PK, Lucas G, Luben R, Loos RJ, Lokki ML, Lettre G, Langenberg C, Launer LJ, Lakatta EG, Laaksonen R, Kyvik KO, Kronenberg F, König IR, Khaw KT, Kaprio J, Kaplan LM, Johansson A, Jarvelin MR, Janssens AC, Ingelsson E, Igl W, Kees HG, Hottenga JJ, Hofman A, Hicks AA, Hengstenberg C, Heid IM, Hayward C, Havulinna AS, Hastie ND, Harris TB, Haritunians T, Hall AS, Gyllensten U, Guiducci C, Groop LC, Gonzalez E, Gieger C, Freimer NB, Ferrucci L, Erdmann J, Elliott P, Ejebe KG, Döring A, Dominiczak AF, Demissie S, Deloukas P, de Geus EJ, de Faire U, Crawford G, Collins FS, Chen YD, Caulfield MJ, Campbell H, Burtt NP, Bonnycastle LL, Boomsma DI, Boekholdt SM, Bergman RN, Barroso I, Bandinelli S, Ballantyne CM, Assimes TL, Quertermous T, Altshuler D, Seielstad M, Wong TY, Tai ES, Feranil AB, Kuzawa CW, Adair LS, Taylor HA, Borecki IB, Gabriel SB, Wilson JG, Holm H, Thorsteinsdottir U, Gudnason V, Krauss RM, Mohlke KL, Ordovas JM, Munroe PB, Kooner JS, Tall AR, Hegele RA, Kastelein JJ, Schadt EE, Rotter JI, Boerwinkle E, Strachan DP, Mooser V, Stefansson K, Reilly MP, Samani NJ, Schunkert H, Cupples LA, Sandhu MS, Ridker PM, Rader DJ, van Duijn CM, Peltonen L, Abecasis GR, Boehnke M, Kathiresan S. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gimeno RE, Cao J. Thematic review series: glycerolipids. Mammalian glycerol-3-phosphate acyltransferases: new genes for an old activity. J Lipid Res. 2008;49:2079–88. doi: 10.1194/jlr.R800013-JLR200. [DOI] [PubMed] [Google Scholar]
- 15.Karlsson E, Wang SQ, Coleman R, Beck M. Glycerol-3-phosphate acyltransferase 1 is essential for the immune response to infection with coxsackievirus B3 in mice. J Nutr. 2009;139:779–783. doi: 10.3945/jn.108.101683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coleman RA, Lee DP. Enzymes of triacylglycerol synthesis and their regulation. Prog Lipid Res. 2004;43:134–176. doi: 10.1016/s0163-7827(03)00051-1. [DOI] [PubMed] [Google Scholar]
- 17.Igal RA, Wang S, Gonzalez-Baró M, Coleman RA. Mitochondrial glycerol phosphate acyltransferase directs the incorporation of exogenous fatty acids into triacylglycerol. J Biol Chem. 2001;276:42205–42212. doi: 10.1074/jbc.M103386200. [DOI] [PubMed] [Google Scholar]
- 18.Lewin TM, Wang S, Nagle CA, Van Horn CG, Coleman RA. Mitochondrial glycerol-3-phosphate acyltransferase-1 directs the metabolic fate of exogenous fatty acids in hepatocytes. Am J Physiol Endocrinol Metab. 2005;288:E835. doi: 10.1152/ajpendo.00300.2004. [DOI] [PubMed] [Google Scholar]
- 19.Johansen CT, Wang J, Lanktree MB, McIntyre AD, Ban MR, Martins RA, Kennedy BA, Hassell RG, Visser ME, Schwartz SM, Voight BF, Elosua R, Salomaa V, O’Donnell CJ, Dallinga-Thie GM, Anand SS, Yusuf S, Huff MW, Kathiresan S, Cao H, Hegele RA. An increased burden of common and rare lipid-associated risk alleles contributes to the phenotypic spectrum of hypertriglyceridemia. Arterioscler Thromb Vasc Biol. 2011;31:1916–1926. doi: 10.1161/ATVBAHA.111.226365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tikkanen E, Tuovinen T, Widén E, Lehtimäki T, Viikari J, Kähönen M, Peltonen L, Raitakari OT, Ripatti S. Association of known loci with lipid levels among children and prediction of dyslipidemia in adults. Circ Cardiovasc Genet. 2011;4:673–80. doi: 10.1161/CIRCGENETICS.111.960369. [DOI] [PubMed] [Google Scholar]
- 21.Qi Q, Liang L, Doria A, Hu FB, Qi L. Genetic predisposition to dyslipidemia and type 2 diabetes risk in two prospective cohorts. Diabetes. 2012;61:745–752. doi: 10.2337/db11-1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deng Q, Lin X, Gong J, Zhou L, Li S, Deng X, Luo G, Xie X. Genetic relationships among four minorities in Guangxi revealed by analysis of 15 STRs. J Genet Genomics. 2007;34:1072–1079. doi: 10.1016/S1673-8527(07)60122-2. [DOI] [PubMed] [Google Scholar]
- 23.Wang Y, Aung LHH, Tan JY, Yin RX, Hu XJ, Long XJ, Wu DF, Miao L, Yang DZ, Pan SL. Prevalence of dyslipidemia and its risk factors in the Chinese Maonan and Han populations. International Journal of Clinical & Experimental Medicine. 2016 [Google Scholar]
- 24.Lin QZ, Yin RX, Guo T, Wu J, Sun JQ, Shen SW, Shi GY, Wu JZ, Liu CW, Pan SL. Association of the ST3GAL4 rs11220462 polymorphism and serum lipid levels in the Mulao and Han populations. Lipids Health Dis. 2014;13:123–123. doi: 10.1186/1476-511X-13-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.An epidemiological study of cardiovascular and cardiopulmonary disease risk factors in four populations in the People’s Republic of China. Baseline report from the P.R.C.-U.S.A. collaborative study. People’s Republic of China--United states cardiovascular and cardiopulmonary epidemiology research group. Circulation. 1992;85:1083–1096. doi: 10.1161/01.cir.85.3.1083. [DOI] [PubMed] [Google Scholar]
- 26.Guo T, Yin RX, Li H, Wang YM, Wu JZ, Yang DZ. Association of the Trp316Ser variant (rs1801690) near the apolipoprotein H (beta2glycoprotein-I) gene and serum lipid levels. Int J Clin Exp Pathol. 2015;8:7291–7304. [PMC free article] [PubMed] [Google Scholar]
- 27.Miao L, Yin RX, Wu JZ, Yang S, Lin WX, Pan SL. The SRGAP2 SNPs, their haplotypes and G × E interactions on serum lipid traits. Sci Rep. 2017;7:11626. doi: 10.1038/s41598-017-10950-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tao G, Yin RX, Yuan B, Nie RJ, Xia C, Pan SL. Association of the SPT2 chromatin protein domain containing 1 gene rs17579600 polymorphism and serum lipid traits. Int J Clin Exp Pathol. 2015;8:12995–13010. [PMC free article] [PubMed] [Google Scholar]
- 29.Ramazauskiene V, Petkeviciene J, Klumbiene J, Kriaucioniene V, Sakytė E. Diet and serum lipids: changes over socio-economic transition period in Lithuanian rural population. BMC Public Health. 2011;11:447. doi: 10.1186/1471-2458-11-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Whitworth JA. 2003 World Health Organization (WHO)/International Society of Hypertension (ISH) statement on management of hypertension. J Hypertens. 2003;21:1983–1992. doi: 10.1097/00004872-200311000-00002. [DOI] [PubMed] [Google Scholar]
- 31.Joanne F, Huo Y, Ji L, Zhao D, Dylan B, Meng HJ, Susan S, Hu D. Unique and varied contributions of traditional CVD risk factors: a systematic literature review of CAD risk factors in China. Clin Med Insights Cardiol. 2013;7:59–86. doi: 10.4137/CMC.S10225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Calandra S, Tarugi P, Speedy HE, Dean AF, Bertolini S, Shoulders CC. Mechanisms and genetic determinants regulating sterol absorption, circulating LDL levels, and sterol elimination: implications for classificationand disease risk. J Lipid Res. 2011;52:1885–926. doi: 10.1194/jlr.R017855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ehara T, Kamei Y, Takahashi M, Yuan X, Kanai S, Tamura E, Tanaka M, Yamazaki T, Miura S, Ezaki O, Suganami T, Okano M, Ogawa Y. Role of DNA methylation in the regulation of lipogenic glycerol-3-phosphate acyltransferase 1 gene expression in the mouse neonatal liver. Diabetes. 2012;61:2442–2450. doi: 10.2337/db11-1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ruixing Y, Qiming F, Dezhai Y, Shuquan L, Weixiong L, Shangling P, Hai W, Yongzhong Y, Feng H, Shuming Q. Comparison of demography, diet, lifestyle, and serum lipid levels between the Guangxi Bai Ku Yao and Han populations. J Lipid Res. 2007;48:2673–81. doi: 10.1194/jlr.M700335-JLR200. [DOI] [PubMed] [Google Scholar]
- 35.Matsumoto C, Miedema MD, Ofman P, Gaziano JM, Sesso HD. An expanding knowledge of the mechanisms and effects of alcohol consumption on cardiovascular disease. J Cardiopulm Rehabil Prev. 2014;34:159–71. doi: 10.1097/HCR.0000000000000042. [DOI] [PubMed] [Google Scholar]
- 36.Perissinotto E, Buja A, Maggi S, Enzi G, Manzato E, Scafato E, Mastrangelo G, Frigo AC, Coin A, Crepaldi G, Sergi G ILSA Working Group. Alcohol consumption and cardiovascular risk factors in older lifelong wine drinkers: the Italian longitudinal study on aging. Nutr Metab Cardiovasc Dis. 2010;20:647–655. doi: 10.1016/j.numecd.2009.05.014. [DOI] [PubMed] [Google Scholar]
- 37.Onat A, Hergenc G, Dursunoglu D, Ordu S, Can G, Bulur S, Yüksel H. Associations of alcohol consumption with blood pressure, lipoproteins, and subclinical inflammation among Turks. Alcohol. 2008;42:593–601. doi: 10.1016/j.alcohol.2008.06.007. [DOI] [PubMed] [Google Scholar]
- 38.Brinton EA. Effects of ethanol intake on lipoproteins and atherosclerosis. Curr Opin Lipidol. 2010;21:346–51. doi: 10.1097/MOL.0b013e32833c1f41. [DOI] [PubMed] [Google Scholar]
- 39.Morrow JD, Frei B, Longmire AW, Gaziano JM, Lynch SM, Shyr Y, Strauss WE, Oates JA, Roberts LJ 2nd. Increase in circulating products of lipid peroxidation (F2-isoprostanes) in smokers. Smoking as a cause of oxidative damage. N Engl J Med. 1995;332:1198–203. doi: 10.1056/NEJM199505043321804. [DOI] [PubMed] [Google Scholar]
- 40.Rao CS, Subash YE. The effect of chronic tobacco smoking and chewing on the lipid profile. J Clin Diagn Res. 2013;7:31–34. doi: 10.7860/JCDR/2012/5086.2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Valente EA, Sheehy ME, Avila JJ, Gutierres JA, Delmonico MJ, Lofgren IE. The effect of the addition of resistance training to a dietary education intervention on apolipoproteins and diet quality in overweight and obese older adults. Clin Interv Aging. 2011;6:235–241. doi: 10.2147/CIA.S23583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Solá R, Fitó M, Estruch R, Salas-Salvadó J, Corella D, de La Torre R, Muñoz MA, López-Sabater Mdel C, Martínez-González MA, Arós F, Ruiz-Gutierrez V, Fiol M, Casals E, Wärnberg J, Buil-Cosiales P, Ros E, Konstantinidou V, Lapetra J, Serra-Majem L, Covas MI. Effect of a traditional Mediterranean diet on apolipoproteins B, A-I, and their ratio: a randomized, controlled trial. Atherosclerosis. 2011;218:174–80. doi: 10.1016/j.atherosclerosis.2011.04.026. [DOI] [PubMed] [Google Scholar]
- 43.Bermudez OI, Velez-Carrasco W, Schaefer EJ, Tucker KL. Dietary and plasma lipid, lipoprotein, and apolipoprotein profiles among elderly Hispanics and non-Hispanics and their association with diabetes. Am J Clin Nutr. 2002;76:1214–1221. doi: 10.1093/ajcn/76.6.1214. [DOI] [PubMed] [Google Scholar]