Abstract
There are two commonly accepted methods for detecting microsatellite status. One is to detect amplified microsatellite loci by polymerase chain reaction (PCR) and the other is to detect mismatch repair gene (MMR) protein expression by immunohistochemistry (IHC). PCR detection is considered to be accurate in clinical operations while IHC is widely used due to ease of operation and lesser expense. In order to compare IHC with PCR in detecting microsatellite status in colorectal carcinoma, a total of 569 samples of colorectal carcinoma resection were collected in the Department of Pathology, Nanjing Drum Tower Hospital, between June 2014 and June 2017. In all samples, IHC and PCR was used to detect microsatellite status and the consistency of results between the two methods was compared. We found that 48 cases of microsatellite instability (MSI) were detected by PCR including 37 cases of microsatellite instability high (MSI-H), 11 cases of microsatellite instability low (MSI-L), and 521 cases of MSS. MSI accounted for 8.44% of all cases and MSI-H accounted for 6.50%. IHC results of the 569 patients showed that 69 cases were deficient mismatch repair (dMMR) and 500 cases were proficient mismatch repair (pMMR). dMMR accounted for 12.13% of all cases. Loss expression of PMS2 protein was the most common while MSH6 was rare. The coincidence rate of the two methods for detecting microsatellite states was 91.92%. IHC and the PCR method had high consistency in microsatellite status. Compared with PCR, the IHC method is more economical and more convenient for clinical operations. When the 4 repair proteins were without deficiency detected by IHC, it could be diagnosed as MSS/MSI-L and further PCR was not necessary. When any repair protein was found to be deficient, PCR detection was needed to determine whether MSI existed. Our conclusion will save a lot of time and costs in clinical work.
Keywords: Immunohistochemistry (IHC), polymerase chain reaction (PCR), colorectal carcinoma, microsatellite instability (MSI), mismatch repair gene (MMR)
Introduction
Colorectal carcinoma is a common gastrointestinal carcinoma. Its pathogenesis is complex and it can be divided into hereditary and sporadic colorectal carcinoma. Lynch syndrome is the most important etiology that relates to hereditary colorectal carcinoma. Incidence of colorectal carcinoma in East Asia is on the rise. In China, there were 376,300 new cases of colorectal carcinoma and 191,000 death cases in 2015 [1]. Incidence and mortality of colorectal carcinoma in Hong Kong ranks second in malignant tumors. In colorectal carcinoma, microsatellite instability (MSI) exists, caused by insertion or deletion of a microsatellite in a tumor that leads to change in microsatellite length, with new microsatellite alleles emerging. MSI correlates with development, prognosis, efficacy, and inheritance of colorectal carcinoma [2-9]. Recent studies have shown that MSI is instructive for anti-PD-1 immunotherapy. The 0RR in dMMR and pMMR groups was 40% and 0%, respectively. DCR in the two groups was 78% and 11%, respectively, both with significant differences [10].
In previous studies, MSI was presented in both hereditary and sporadic colorectal carcinomas. Approximately 15% to 24.3% of sporadic colorectal carcinoma in Western countries presented MSI [11-14] with 7.75% to 13% in China, which was close to South Korea but lower than Western population [15-18]. Different incidences of MSI were likely due to different genetic backgrounds and testing techniques. In hereditary nonpolyposis colorectal carcinoma, limited reports from China showed the same result as Western countries, as MSI could be detected in 80 to 90% of patients [19,20]. It is evident that detection of MSI in hereditary colorectal carcinoma is essential.
At present, there are two methods to detect the stability of microsatellites. One is to detect the amplified microsatellite loci by PCR. Commonly used detection markers are BAT25, BAT26, D5S346, D2S123, and D17S250, as recommended by the National Carcinoma Institute. The state of MSI is determined by comparing the shift of the markers in tumor tissue and normal tissue. High level of instability is gauged by tumors with a shift in at least two markers. At least 30 percent of interpretable markers were classified as having high levels of microsatellite instability (MSI-H), in accordance with international criteria. A low level of microsatellite instability (MSI-L) was defined as a shift in only one dinucleotide marker. Tumors without any shift in markers were categorized as microsatellite stable (MSS) tumors. Another method is to determine microsatellite status by immunohistochemistry (IHC) detection of proteins encoded relating to DNA mismatch repair genes (MMR) including MLH1, MSH2, MSH6, and PMS2. Deficient mismatch repair (dMMR) was defined by the presence of either MSI-H or by loss of MLH1, MSH2, MSH6, or PMS2 protein expression, as outlined above. Proficient mismatch repair (pMMR) was defined by presence of either MSS/MSI-L (i.e., instability at <30% of loci screened) or by intact MMR protein expression [21]. MSI-L has been shown to be biologically similar to tumors exhibiting MSS at all loci tested and these two molecular phenotypes can be grouped together. Detection of MSI status by PCR is the earliest established molecular detection to identify MSI in colorectal carcinoma and is considered to be the gold standard for detecting MSI [22]. PCR shortcomings include a long experimental period, high laboratory conditions, and high cost. At present, with continuous understanding of the mechanism of MSI and popularity of commercial monoclonal antibody of MMR proteins, the simple IHC method is more often being applied for MSI screening in colorectal carcinoma. It has been reported that IHC detection of MSI had similar results with PCR [23,24].
Materials and methods
Materials
Patients and tissues
Five hundred sixty-nine surgical resection samples pathologically diagnosed as colorectal carcinoma were collected from the Department of Pathology, Nanjing Drum Tower Hospital, between June 2014 and June 2017. All samples satisfied the following criteria: (1) sporadic colon or rectal cancer confirmed by pathological diagnosis, (2) no preoperative therapy, including preoperative radiotherapy, in rectal cancer patients, and (3) received radical resection or a palliative operation. Exclusion criteria were (1) tumors in the appendix and anal canal; (2) second primary tumor out of colorectal; (3) in situ carcinoma (high-grade intraepithelial neoplasia). This study was approved by the Ethics Committee of Nanjing Drum Tower Hospital. Written informed consent was obtained from each individual.
Reagents
Anti-MLH1, mouse monoclonal, ES05, 1:100 dilution, Leica/Novocastra, UK; anti-PSM2, rabbit monoclonal, EP51, 1:100 dilution, Epitomics, USA; anti-MSH2, mouse monoclonal, FE11, 1:100 dilution, Dako, Denmark; anti-MSH6, rabbit monoclonal, EP49, 1:150 dilution, Epitomics, USA; MSI MIX 1, 140 μL, Yuanqi Bio-Pharmaceutical CO, Shanghai, China; MIX 2, 200 μL, Yuanqi Bio-Pharmaceutical CO, Shanghai, China; PCR Premix (containing AmpliTaq Gold DNA Polymerase buffer, magnesium chloride and dNTPs), Yuanqi Bio-Pharmaceutical CO, Shanghai, China.
Methods
PCR-capillary electrophoresis detection for MSI
DNA was extracted from formalin-fixed paraffin-embedded tissue with QIAamp Tissue kit (Qiagen), according to manufacturer instructions. MSI detection kit was used to amplify the mutations of BAT25, BAT26, D5S346, D2S123, and D17S250. DNA samples from tumor tissues and normal tissues were amplified in a 20 μL volume containing 100 ng of DNA, 1 μmol/L of dye-labeled forward and unlabeled reverse primers, 200 μmol/L of deoxynucleotide, 1.5 mmol/L of MgCl2, and 0.75 U of Taq DNA polymerase. PCR was performed under the following conditions: denaturation at 95°C for 5 minutes, 35 cycles of denaturation at 95°C for 30 seconds, annealing at 53°C for 30 seconds, and extension at 72°C for 30 seconds. Final extension was at 72°C for 10 minutes. PCR product was analyzed by a genetic analyzer (Applied Biosystems 3500, ABI). Raw data were analyzed using GeneMapper 4.1 software. In accordance with National Cancer Institute (NCI) guidelines, MSI at ≥ 2 loci was defined as MSI high (MSI-H) (Figure 1B), instability at a single locus was defined as MSI low (MSI-L), and no instability at any of the loci tested was defined as microsatellite stable (MSS) (Figure 1A). Because extensive data indicate that tumors with low frequency are biologically similar to those exhibiting MSS, these two molecular phenotypes were grouped as MSS.
Figure 1.
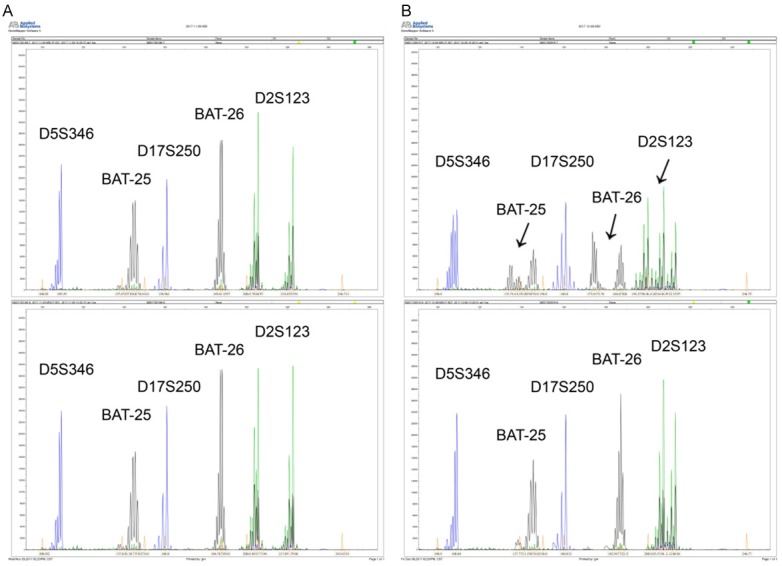
Results of paired normal and colorectal cancer DNA tissues tested for MSI using Bethesda recommended 5 markers by PCR. A. MSS patient. B. MSI-H patient. Black arrows indicate the mutation sites.
Tumor MMR protein expression detected by IHC
Specimens of colorectal carcinoma were fixed in 10% neutral formalin for 24-48 hours. After routine dehydration, fixation, paraffin embedding, and a series of routine IHC procedures such as dewaxing, antigen repair, staining, dehydration, and mounting were performed (primary antibodies inferred in the above were anti-MLH1, anti-PSM2, anti-MSH2, and anti-MSH6). Adjacent normal tissue from each sample served as positive controls. Immunohistochemical staining results are interpresented, according to the literature (Figure 2). Two independent observers carried out immunohistochemistry analysis, both observers were blinded to any prior information regarding clinical or pathological characteristics of the cases. If there was discrepancy between analyses performed by the two observers, the slides were reinvestigated by both investigators using a multi-headed microscope and a consensus was reached.
Figure 2.
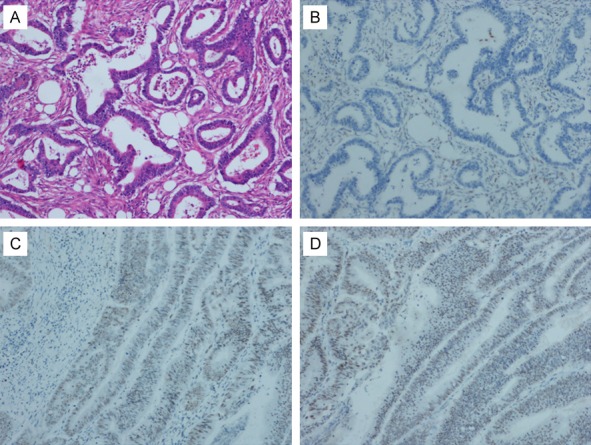
Immunohistochemical expression of MLH1 and PMS2 and HE staining in colorectal cancer tissue (SP ×100). A. HE staining in colorectal cancer tissue. B. Negative expression of MLH1 in colorectal cancer tissue. C. Positive expression of MLH1 in colorectal cancer tissue. D. Positive expression of PMS2 in colorectal cancer tissue.
Statistics
Data were analyzed by Statistical Package for the Social Sciences for Windows version 19 (SPSS Inc., Chicago, IL, USA). Consistency of the two methods was analyzed by Kappa consistency test. The significance level was set at P<0.05.
Results
PCR detection for microsatellite status
Of the 569 patients with colorectal carcinoma that underwent PCR detection (Figure 3A), 521 were MSS, accounting for 91.56%, 48 were MSI, accounting for 8.44%, of which 37 were MSI-H and 11 were MSI-L, accounting for 6.50% and 1.93%, respectively.
Figure 3.
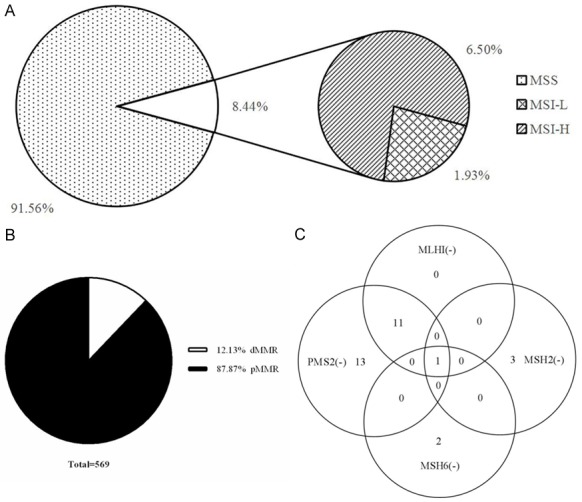
Results of immunohistochemistry (IHC) and polymerase chain reaction(PCR) in detecting microsatellite status in colorectal carcinoma. A. PCR detection for microsatellite status. B. IHC detection for microsatellite status. C. Expression of MMR protein in 37 cases of MSI-H colorectal cancer which were determined by PCR.
IHC detection for microsatellite status
Of the 569 cases receiving IHC staining of MMR proteins (Figure 3B), there were 69 cases of dMMR and the remaining 500 cases were pMMR. Incidence of dMMR and pMMR was 12.13% and 87.87%, respectively.
Comparison of IHC and PCR detection
PCR is shown in Table 1 and Figure 3C. Except for one case in which MLH1, MSH2, MSH6, and PMS2 expression was deficient, expression of MMR protein was positive in the other 36 cases of MSI-H. It is worth noting that there were 7 cases determined as MSI-H by PCR while all of the MMR proteins were positively expressed, meaning that these 7 cases should be judged as MSS/MSI-L by IHC. There were even two cases expressing moderate/strong positive of all four proteins. Only one of the cases was negative for both PMS2 and MSH6. Among 532 cases of MSI-L/MSS determined by PCR, 493 were pMMR and the rest were dMMR.
Table 1.
Expression of mismatch repair gene proteins in 37 cases of MSI-H colorectal carcinoma determined by PCR (n)
| MMR | - | + | 2+/3+ |
|---|---|---|---|
| MLH1 | 12 | 17 | 8 |
| MSH2 | 4 | 10 | 23 |
| MSH6 | 3 | 8 | 2 |
| PMS2 | 25 | 7 | 5 |
Note: PCR, polymerase chain reaction; MSI-H, microsatellite instability high; MMR, mismatch repair gene.
Sensitivity and specificity of IHC for detection of MSI-H were 81.08% and 92.67%, respectively. Positive predictive value and negative predictive value was 43.48% and 98.60%, if using PCR method as the gold standard for clinical detection of microsatellite status (Table 2). Coincidence rate of the two methods was 91.92% (523/569), with good consistency (Kappa = 0.526).
Table 2.
Comparison between IHC and PCR of microsatellite status in 569 cases of colorectal carcinoma (n)
| IHC | PCR | Total | Kappa consistency test | |
|---|---|---|---|---|
|
| ||||
| MSI-H | MSI-L/MSS | |||
| MSI-H | 30 | 39 | 69 | 0.526 |
| MSI-L/MSS | 7 | 493 | 500 | P<0.001 |
| Total | 37 | 532 | 569 | |
Note: PCR, polymerase chain reaction; IHC, immunohistochemistry; MSS, microsatellite stable; MSI-L, microsatellite instability low; MSI-H, microsatellite instability high.
There were 500 cases of pMMR (MSI-L/MSS) detected by IHC and just 7 cases were judged as MSI-H by PCR. The error rate was only 1.40%. There were 69 patients with dMMR (MSI-H), of which 39 were judged as MSI-L/MSS by PCR, accounting for 56.52%. The error rate was very high.
Expression of MMR proteins in colorectal carcinoma of MSI-H determined by PCR
In 37 cases of colorectal carcinoma with MSI-H determined by PCR, the rate of expression loss of MMR was 81.08% (30/37) (Figure 4). The vast majority of occurrences with dMMR were due to inactivation of PMS2 (67.57% (25/37)), similar to previous reports [24-26]. MSH2 and MSH6 account for a much smaller percentage (10.81% and 8.11) while MLH1 accounted for 32.43%. Negative staining was not found in the 7 MSI-H cases.
Figure 4.

Expression of MMR protein in 37 cases of MSI-H colorectal cancer determined by PCR. White: negative expressive, gray: weak positive expression or “+”, black: strong positive expression or “++/+++”.
Expression of MMR protein in colorectal carcinoma of MSI-L/MSS determined by PCR
In 11 cases of colorectal carcinoma determined as MSI-L by PCR, there were eight cases expressing positive of all four proteins, showing the same results with PCR. There were still 3 cases found with negative staining but they were dMMR. The error rate was 27.27%. Among 521 cases of colorectal carcinoma detected as MSS by PCR, 485 were judged as pMMR by IHC, consistent with results of PCR. However, there were also 36 cases of dMMR and the error rate was 6.91%.
Therefore, in 532 cases of MSS and MSI-L determined by PCR, there were 39 cases determined as MSI-H by IHC. The total error rate was 7.33%. Among them, 9 of the cases were negative for MLH1, 5 for MSH2, 3 for MSH6, 27 for PMS2. Only 1 case was negative for both MSH2 and MSH6, 1 case for both MLH1 and MSH6, and 2 cases for both MLH1 and PMS2. We did not find any cases negative for both PMS2 and MSH6, nor with more than 2 deletions of MMR protein expression.
Discussion
Colorectal carcinoma remains an important risk factor for human life and health. Microsatellite status has had a great impact on its chemotherapy and immunotherapy [2-10], which is of great significance in determination of treatment, judgment of prognosis, and screening of family genetic diseases. In our experiment, PCR and IHC were used to detect microsatellite status of colorectal carcinoma. Results show that the coincidence rate was 91.92%, which was high in consistency. In our study, MSI cases accounted for approximately 8.44%-12.13% of all 569 cases, lower than that reported in Western populations [11-14] but close to reports from Korea. This may be related to differences with race, genetics, and lifestyle [15-18].
Our study results showed that deletion rate of PMS2 was the highest among four MMR while MSH6 was the lowest. We saw that negative staining for PMS2 protein had higher sensitivity but lower specificity for detecting microsatellite status, while MSH6 showed the opposite.
With improvement of molecular biological experiments, accuracy and convenience of microsatellite detection has been greatly enhanced but with the sensitivity and specificity of IHC detection, 100% is still difficult to reach. It has been reported that detection of MSH6 mismatch repair protein expression in vitro or MSH6 mutant mice may be regarded as MSS by PCR [27-29] and even 14/18 (78%) cases were regarded as MSS or MSI-L when MSH6 expression was abnormal [30]. The results of our study also demonstrated that 39 out of 532 MSI-L/MSS cases were regarded as dMMR by IHC and 7 out of 37 MSI-H cases were regarded as pMMR by IHC. The reasons for different results between the two detection methods may include the following aspects: (1) PCR detection showed that some of the genes that affected microsatellite status were mutated but the antigenic determinant of expression production was not undermined. Although the production was not functional, IHC result was still positive, which may lead to false negatives; (2) Due to MMR protein’s functional redundancy, it may not be enough to lead to occurrence of MSI-H when individual MMR protein was missing; (3) The time of tissue fixation and staining will also affect IHC results during the process; (4) Sensitivity of IHC is also dependent on its antibody type and other types of MMR genes can also lead to dMMR besides MLH1, MSH2, MSH6, and PMS2.
Because IHC detection of microsatellite status has the advantages of being a simple operation, short time of operation, low cost, low requirement of experimental instruments, and display of each repair protein, it has high application value. It can be used as a first-line screening method for detecting microsatellite status in colorectal carcinoma. However, Vasen et al. [31] indicated that IHC could not completely replace PCR method in determining microsatellite status until other dMMR genes were elucidated. Our study shows 500 cases of pMMR detected by IHC. Just 7 cases were judged as MSI-H by PCR and the error rate was only 1.40%. There were 69 patients with dMMR (MSI-H), 39 were judged as MSI-L/MSS by PCR, accounting for 56.52%. The error rate was very high. Therefore, we believe that when the 4 repair proteins were without deficiency detected by immunohistochemistry, it could be diagnosed as MSS/MSI-L and further PCR was not necessary. But when any repair protein was found to be deficient, PCR detection was needed to determine whether microsatellite instability existed. Our conclusion will save a lot of time and cost for clinical work.
In order to reduce false negatives and positive proportions detected by IHC method during our application, we strictly performed every step of the operation and results of IHC were confirmed by two experienced pathologists, independently. Errors caused by subjective factors should be avoided as much as possible. It is critical that we discover new genes that affect deletion of MMR proteins.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 81272741), the Project of Making Health Become Strong by Science and Teaching at the Health and Family Planning Commission in Jiangsu Province (QNRC2016042) and Nanjing Medical Science and Technique Development Foundation (Outstanding Youth Foundation, JQX14001). These funding sources had no role in the study design, data collection, data analysis, data interpretation, or writing of this report.
Disclosure of conflict of interest
None.
References
- 1.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 2.Collura A, Lagrange A, Svrcek M, Marisa L, Buhard O, Guilloux A, Wanherdrick K, Dorard C, Taieb A, Saget A, Loh M, Soong R, Zeps N, Platell C, Mews A, Iacopetta B, De Thonel A, Seigneuric R, Marcion G, Chapusot C, Lepage C, Bouvier AM, Gaub MP, Milano G, Selves J, Senet P, Delarue P, Arzouk H, Lacoste C, Coquelle A, Bengrine-Lefevre L, Tournigand C, Lefevre JH, Parc Y, Biard DS, Flejou JF, Garrido C, Duval A. Patients with colorectal tumors with microsatellite instability and large deletions in HSP110 T17 have improved response to 5-fluorouracil-based chemotherapy. Gastroenterology. 2014;146:401–11. e1. doi: 10.1053/j.gastro.2013.10.054. [DOI] [PubMed] [Google Scholar]
- 3.Goldstein J, Tran B, Ensor J, Gibbs P, Wong HL, Wong SF, Vilar E, Tie J, Broaddus R, Kopetz S, Desai J, Overman MJ. Multicenter retrospective analysis of metastatic colorectal cancer (CRC) with high-level microsatellite instability (MSI-H) Ann Oncol. 2014;25:1032–1038. doi: 10.1093/annonc/mdu100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mouradov D, Domingo E, Gibbs P, Jorissen RN, Li S, Soo PY, Lipton L, Desai J, Danielsen HE, Oukrif D, Novelli M, Yau C, Holmes CC, Jones IT, McLaughlin S, Molloy P, Hawkins NJ, Ward R, Midgely R, Kerr D, Tomlinson IP, Sieber OM. Survival in stage II/III colorectal cancer is independently predicted by chromosomal and microsatellite instability, but not by specific driver mutations. Am J Gastroenterol. 2013;108:1785–1793. doi: 10.1038/ajg.2013.292. [DOI] [PubMed] [Google Scholar]
- 5.Pogue-Geile K, Yothers G, Taniyama Y, Tanaka N, Gavin P, Colangelo L, Blackmon N, Lipchik C, Kim SR, Sharif S, Allegra C, Petrelli N, O’Connell MJ, Wolmark N, Paik S. Defective mismatch repair and benefit from bevacizumab for colon cancer: findings from NSABP C-08. J Natl Cancer Inst. 2013;105:989–992. doi: 10.1093/jnci/djt140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shin US, Cho SS, Moon SM, Park SH, Jee SH, Jung EJ, Hwang DY. Is microsatellite instability really a good prognostic factor of colorectal cancer? Ann Coloproctol. 2014;30:28–34. doi: 10.3393/ac.2014.30.1.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thomas ML, Hewett PJ, Ruszkiewicz AR, Moore JW. Clinicopathological predictors of benefit from adjuvant chemotherapy for stage C colorectal cancer: microsatellite unstable cases benefit. Asia Pac J Clin Oncol. 2015;11:343–351. doi: 10.1111/ajco.12411. [DOI] [PubMed] [Google Scholar]
- 8.Yang L, Sun Y, Huang XE, Yu DS, Zhou JN, Zhou X, Li DZ, Guan X. Carcinoma microsatellite instability status as a predictor of benefit from fluorouracil-based adjuvant chemotherapy for stage II rectal cancer. Asian Pac J Cancer Prev. 2015;16:1545–1551. doi: 10.7314/apjcp.2015.16.4.1545. [DOI] [PubMed] [Google Scholar]
- 9.Gandhi JS, Goswami M, Sharma A, Tanwar P, Gupta G, Gupta N, Pasricha S, Mehta A, Singh S, Agarwal M, Gupta N. Clinical impact of mismatch repair protein testing on outcome of early staged colorectal carcinomas. J Gastrointest Cancer. 2017 doi: 10.1007/s12029-017-9954-5. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 10.Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, Skora AD, Luber BS, Azad NS, Laheru D, Biedrzycki B, Donehower RC, Zaheer A, Fisher GA, Crocenzi TS, Lee JJ, Duffy SM, Goldberg RM, de la Chapelle A, Koshiji M, Bhaijee F, Huebner T, Hruban RH, Wood LD, Cuka N, Pardoll DM, Papadopoulos N, Kinzler KW, Zhou S, Cornish TC, Taube JM, Anders RA, Eshleman JR, Vogelstein B, Diaz LA Jr. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med. 2015;372:2509–2520. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ghanipour L, Jirstrom K, Sundstrom M, Glimelius B, Birgisson H. Associations of defect mismatch repair genes with prognosis and heredity in sporadic colorectal cancer. Eur J Surg Oncol. 2017;43:311–321. doi: 10.1016/j.ejso.2016.10.013. [DOI] [PubMed] [Google Scholar]
- 12.Sinicrope FA, Foster NR, Yoon HH, Smyrk TC, Kim GP, Allegra CJ, Yothers G, Nikcevich DA, Sargent DJ. Association of obesity with DNA mismatch repair status and clinical outcome in patients with stage II or III colon carcinoma participating in NCCTG and NSABP adjuvant chemotherapy trials. J. Clin. Oncol. 2012;30:406–412. doi: 10.1200/JCO.2011.39.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vasovcak P, Pavlikova K, Sedlacek Z, Skapa P, Kouda M, Hoch J, Krepelova A. Molecular genetic analysis of 103 sporadic colorectal tumours in Czech patients. PLoS One. 2011;6:e24114. doi: 10.1371/journal.pone.0024114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abu Freha N, Leibovici Weissman Y, Fich A, Barnes Kedar I, Halpern M, Sztarkier I, Behar DM, Arbib Sneh O, Vilkin A, Baris HN, Gingold R, Lejbkowicz F, Niv Y, Goldberg Y, Levi Z. Constitutional mismatch repair deficiency and Lynch syndrome among consecutive Arab Bedouins with colorectal cancer in Israel. Fam Cancer. 2018;17:79–86. doi: 10.1007/s10689-017-0009-7. [DOI] [PubMed] [Google Scholar]
- 15.Jeon CH, Lee HI, Shin IH, Park JW. Genetic alterations of APC, K-ras, p53, MSI, and MAGE in Korean colorectal cancer patients. Int J Colorectal Dis. 2008;23:29–35. doi: 10.1007/s00384-007-0373-0. [DOI] [PubMed] [Google Scholar]
- 16.Ye JX, Liu Y, Qin Y, Zhong HH, Yi WN, Shi XY. KRAS and BRAF gene mutations and DNA mismatch repair status in Chinese colorectal carcinoma patients. World J Gastroenterol. 2015;21:1595–1605. doi: 10.3748/wjg.v21.i5.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peng J, Huang D, Poston G, Ma X, Wang R, Sheng W, Zhou X, Zhu X, Cai S. The molecular heterogeneity of sporadic colorectal cancer with different tumor sites in Chinese patients. Oncotarget. 2017;8:49076–49083. doi: 10.18632/oncotarget.16176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jung SB, Lee HI, Oh HK, Shin IH, Jeon CH. Clinico-pathologic parameters for prediction of microsatellite instability in colorectal cancer. Cancer Res Treat. 2012;44:179–186. doi: 10.4143/crt.2012.44.3.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pino MS, Chung DC. Application of molecular diagnostics for the detection of Lynch syndrome. Expert Rev Mol Diagn. 2010;10:651–665. doi: 10.1586/erm.10.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sheng JQ, Chan TL, Chan YW, Huang JS, Chen JG, Zhang MZ, Guo XL, Mu H, Chan AS, Li SR, Yuen ST, Leung SY. Microsatellite instability and novel mismatch repair gene mutations in northern Chinese population with hereditary non-polyposis colorectal cancer. Chin J Dig Dis. 2006;7:197–205. doi: 10.1111/j.1443-9573.2006.00269.x. [DOI] [PubMed] [Google Scholar]
- 21.Sargent DJ, Marsoni S, Monges G, Thibodeau SN, Labianca R, Hamilton SR, French AJ, Kabat B, Foster NR, Torri V, Ribic C, Grothey A, Moore M, Zaniboni A, Seitz JF, Sinicrope F, Gallinger S. Defective mismatch repair as a predictive marker for lack of efficacy of fluorouracilbased adjuvant therapy in colon cancer. J. Clin. Oncol. 2010;28:3219–3226. doi: 10.1200/JCO.2009.27.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boland CR, Thibodeau SN, Hamilton SR, Sidransky D, Eshleman JR, Burt RW, Meltzer SJ, Rodriguez-Bigas MA, Fodde R, Ranzani GN, Srivastava S. A national cancer institute workshop on microsatellite instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998;58:5248–5257. [PubMed] [Google Scholar]
- 23.Amira AT, Mouna T, Ahlem B, Raoudha A, Majid BH, Amel H, Rachida Z, Nadia K. Immunohistochemical expression pattern of MMR protein can specifically identify patients with colorectal cancer microsatellite instability. Tumour Biol. 2014;35:6283–6291. doi: 10.1007/s13277-014-1831-2. [DOI] [PubMed] [Google Scholar]
- 24.Shia J. Immunohistochemistry versus microsatellite instability testing for screening colorectal cancer patients at risk for hereditary nonpolyposis colorectal cancer syndrome. Part I. The utility of immunohistochemistry. J Mol Diagn. 2008;10:293–300. doi: 10.2353/jmoldx.2008.080031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cicek MS, Lindor NM, Gallinger S, Bapat B, Hopper JL, Jenkins MA, Young J, Buchanan D, Walsh MD, Le Marchand L, Burnett T, Newcomb PA, Grady WM, Haile RW, Casey G, Plummer SJ, Krumroy LA, Baron JA, Thibodeau SN. Quality assessment and correlation of microsatellite instability and immunohistochemical markers among population- and clinicbased colorectal tumors results from the Colon Cancer family registry. J Mol Diagn. 2011;13:271–281. doi: 10.1016/j.jmoldx.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karahan B, Argon A, Yildirim M, Vardar E. Relationship between MLH-1, MSH-2, PMS-2, MSH-6 expression and clinicopathological features in colorectal cancer. Int J Clin Exp Pathol. 2015;8:4044–4053. [PMC free article] [PubMed] [Google Scholar]
- 27.Edelmann W, Yang K, Umar A, Heyer J, Lau K, Fan K, Liedtke W, Cohen PE, Kane MF, Lipford JR, Yu N, Crouse GF, Pollard JW, Kunkel T, Lipkin M, Kolodner R, Kucherlapati R. Mutation in the mismatch repair gene Msh6 causes cancer susceptibility. Cell. 1997;91:467–477. doi: 10.1016/s0092-8674(00)80433-x. [DOI] [PubMed] [Google Scholar]
- 28.Umar A, Risinger JI, Glaab WE, Tindall KR, Barrett JC, Kunkel TA. Functional overlap in mismatch repair by human MSH3 and MSH6. Genetics. 1998;148:1637–1646. doi: 10.1093/genetics/148.4.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamada NA, Castro A, Farber RA. Variation in the extent of microsatellite instability in human cell lines with defects in different mismatch repair genes. Mutagenesis. 2003;18:277–282. doi: 10.1093/mutage/18.3.277. [DOI] [PubMed] [Google Scholar]
- 30.Mojtahed A, Schrijver I, Ford JM, Longacre TA, Pai RK. A two-antibody mismatch repair protein immunohistochemistry screening approach for colorectal carcinomas, skin sebaceous tumors, and gynecologic tract carcinomas. Mod Pathol. 2011;24:1004–1014. doi: 10.1038/modpathol.2011.55. [DOI] [PubMed] [Google Scholar]
- 31.Vasen HF, Hendriks Y, de Jong AE, van Puijenbroek M, Tops C, Brocker-Vriends AH, Wijnen JT, Morreau H. Identification of HNPCC by molecular analysis of colorectal and endometrial tumors. Dis Markers. 2004;20:207–213. doi: 10.1155/2004/391039. [DOI] [PMC free article] [PubMed] [Google Scholar]


