Abstract
To explore the correlation of microRNA (miRNA) profiles with disease risk and severity in patients with allergic rhinitis (AR). miRNA expression profiles in nasal mucosa samples from 8 AR patients and 8 matched non-atopic controls were detected by microarray. Twelve differentially expressed miRNAs (DEMs) in microarray analysis were further validated in nasal mucosa samples from 48 AR patients and 50 controls by qPCR assay. Individual nasal symptom score (INSS) and total nasal symptom score (TNSS) were used to evaluated the disease severity of AR. AR patients could be distinguished from controls according to the principal component analysis (PCA) plot analysis in the microarray, and 27 down-regulated and 51 up-regulated DEMs were identified by volcano plot. qPCR validation disclosed that miR-126-5p, miR-19a-5p and miR-26a-5p expression was up-regulated in AR patients compared with controls. Multivariate logistic regression displayed that miR-126-5p, miR-19a-5p and miR-26a-5p are independent predictive factors for AR risk, and receiver operating characteristic (ROC) analysis exhibited that the combination of miR-126-5p, miR-19a-5p and miR-26a-5p predicts the risk of AR with a high area under curve (AUC) of 0.866 (95% CI: 0.797-0.936). In addition, expression of miR-126-5p, miR-19a-5p and miR-181c-3p were positively correlated with TNSS. Therefore, miRNA profiles distinguish AR patients from controls and the combination of miR-126-5p, miR-19a-5p and miR-26a-5p could serve as novel biomarker for AR risk.
Keywords: MicroRNAs, profile, allergic rhinitis, risk, disease severity
Introduction
Allergic rhinitis (AR) is an immunoglobulin (Ig) E-mediated disorder triggered by exposure of nasal mucosa to allergens [1]. Four hundredmillion people suffer from AR worldwide and the prevalence of AR is still rising due to changes in the socioeconomic environment (industrialization and air pollution) [2,3]. The symptoms of AR seriously affect the patient’s life, work and study, especially in children and adolescents [3,4]. AR patients mostly present with nasal rhinorrhea, itching, sneezing and congestion while a considerable proportion of AR patients are misdiagnosed as non-inflammatory rhinopathy (also known as vasomotor rhinitis) and non-allergic chronic rhinosinusitis [5]. Misdiagnoses lead to delayed timing of treatment in clinical practice and thus, biomarkers for predicting the risk of AR and monitoring disease progression in AR patients are much needed so as to provide an accurate diagnosis as well as timely treatment.
microRNAs (miRNAs), a family of small noncoding RNAs with 21-25 nucleotides, act mainly as suppressors of gene expression by translational repression and/or mRNA degradation by binding to the 30 untranslated regions (30 UTRs) of their target mRNAs [6]. miRNA shave recently been recognized as pivotal for immune cell differentiation and immune regulation, and their dysregulation has been reported to correlate with many allergic diseases. miRNAs mediateimmune responses such as activating eosinophil proliferation and regulate the signal transduction sensitivity of the T cell antigen receptor and Th1/Th2 imbalance [7,8]. In addition, increasing evidence supports miRNA expressionas altered in AR patients and dysregulated miRNAs have been reported to be correlated with the development of AR [9-11], such as up-regulated miRNA-155 expression innasal mucosa of AR patients compared with healthy controls. Modulation of the IL-13 pathway in human macrophages and B and T lymphocytes influences AR progression [10]. Although several miRNAs have been reported to be dysregulated in AR, little is known about the entire miRNA expression profile in AR patients in regardto potential correlation with disease risk and severity.
Thus, this study aimed to explore differences in miRNA profiles in nasal mucosa between AR patients and non-atopic patients, and to explore the correlation of microRNA profiles with disease risk and severity in patients with AR.
Materials and methods
Participants
EightAR patients in the Central Hospital of Wuhan, Tongji Medical College, Huazhong University of Science and Technology from Oct 2016 to Nov 2016 were consecutively enrolled in Stage I of this case-control study. The inclusion criteria were: Diagnosis as AR according to the criteria of the Allergic Rhinitis and Its Impact on Asthma (ARIA) guideline (2010); Age less than 60 years. Patients with the following conditions were excluded: complicated with bronchial asthma, chronic rhinosinusitis, nasal polyposis, excessive septal deviation; current smokers; chronic renal disease or hepatobiliary disease; history of immunological disease, solid cancers or malignant hematological diseases. In addition, 8 age and gender matched non-atopic patients with obstructive snoring undergoing adenoid surgery were recruited as controls. Patients with chronic obstructive pulmonary disease (COPD), asthma, immunological disease, infection, renal or hepatic dysfunction, solid cancers or malignant hematological diseases were excluded from the study.
Apart from the 8 pair of AR and controls enrolled in Stage I, another 40 AR patients and 42 controls were recruited in the Central Hospital of Wuhan, Tongji Medical College, Huazhong University of Science and Technology between Dem 2016 and Jun 2017 in Stage II of this casecontrol study. The inclusion and exclusion criteria were the same as in Stage I. Thus, in total 48 AR patients and 50 controls were included in the analysis in Stage II.
Ethical approval was obtained from the Ethics Committee Boards of The Central Hospital of Wuhan, Tongji Medical College, Huazhong University of Science and Technology. Informed consent was acquired from all participants.
Sample collection
Nasal mucosa sample from the inferior turbinate was obtained from all AR patients and controls and subsequently stored in liquid nitrogen under microarray detection or quantitative real-time polymerase chain reaction (qRT-PCR).
Microarray in stage I
Total RNA was extracted from nasal mucosa sample of 8 AR patients and 8 controls in Stage I using TRIzol regent (Invitrogen, USA) according to the manufacturer’s instructions. 500 ng total RNA was used for miRNA microarray which was done according to the protocol of the manufacturer’s (LC Sciences, USA). In addition, the chips were marked by biotin-labelled DNA molecule and hybridized, and then washed on the GeneChip Fluidic Station 450 platform.
Data processing of microarray, differentially expressed miRNAs (DEMs) and enrichment analysis
In consideration of the essential background of each chip, the overall signal strength varies among chips, which results in error of the miRNA expression calculation, thus in order to normalize the raw data of each chip, the Robust Multichip Average (RMA) was performed. The RMA was an algorithm used for creating an expression matrix from Affymetrix data, and the raw intensity values were background corrected, log2 transformed, and subsequently quantiles were normalized using the RMA method. For the access of the expression measure for each probe set on each array, a linear model was performed for the normalized data.
DEMs were identified between AR patients and controls via the limma package in R software Version 3.2.5 (MathSoft, USA). MiRNAs with the thresholds of Benjamini-Hochberg adjusted P < 0.05 and |log2 (fold change)|>1 were identified as DEMs. The annotation of DEMs was regulated by miRNA enrichment analysis in annotation (miEAA) database, including Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database. Fisher’s exact test was used to distinguish overrepresented miRNA-related items for the enrichment analysis of DEMs and its precursors.
qRT-PCR validation in stage II
In stage II, qPCR assay was used to detect the relative expression of the top 12 significant DEMs that were identified in Stage I by microarray. Total RNA was extracted from nasal mucosa sample of 48 AR patients and 50 controls by TRIzol reagent (Invitrogen, USA) in Stage II, which was then reverse transcribed into cDNAs by One Step Primer Script miRNA cDNA Synthesis Kit (Takara, China). SYBR Premix Ex Taq kit (Takara, China) was then usedto detect candidate miRNAs expression, while the data of candidate miRNAs were normalized according to the expression of U6. The final results were analyzed using SDS 1.4 software according to the 2-ΔΔCt method.
AR severity assessment
Individual nasal symptom score (INSS) was evaluated in AR patients after enrollment. The scoring consists of nasal rhinorrhea, sneezing, itching and congestion, and each item was scored as 0 = no symptoms, 1 = mild symptoms, 2 = moderate symptoms, and 3 = severe symptoms. Subsequently total nasal symptom score (TNSS) was calculated by adding nasal rhinorrhea, sneezing, itching, and congestion scores together.
Statistics
Statistical analysis was performed by SPSS 21.0 (IBM, USA), GraphPad Prism 6 (GraphPad Software, USA) and R software Version 3.2.5 (MathSoft, USA). Data are presented as mean ± standard deviation, median and (25th-75th) or count (percentage). Comparison of baseline characteristics was determined by t test or Chi-square test. The comparison of validated miRNAs in stage II was determined by Wilcoxon rank-sum test. Univariate logistic regression model was performed to analyze the factors for predicting AR risk, while factors with a P value no more than 0.1 were further analyzed by multivariate logistic regression analysis. Receiver operating characteristic (ROC) curve was used to assess the predictive value of validated miRNAs affecting AR risk. P value < 0.05 was considered statistically significant.
Results
Characteristics of AR patients and controls in stage I of microarray assay
The overall demographic and clinical characteristics of AR patients (n = 8) and controls (n = 8) in Stage I arelisted in Table 1. No differences were observed in age as well as gender between AR patients and controls (P = 0.955 and P = 0.614, respectively). Among the AR patients, the disease duration was 76 (59-114) months, and the TNSS score was 8.38 ± 0.16, while INSS scores were as follows: nasal rhinorrhea 2.00 ± 0.18, itching 2.13 ± 0.25, sneezing 2.25 ± 0.13 and congestion 2.00 ± 0.27.
Table 1.
Characteristics of AR patients and controls in Stage I of microarray assay
| Parameters | AR patients (N = 8) | Controls (N = 8) | P Value |
|---|---|---|---|
| Age (years) | 20.88 ± 5.79 | 21.13 ± 10.75 | 0.955 |
| Female (n/%) | 5 (62.5) | 4 (50.0) | 0.614 |
| Disease Duration (months) | 76 (59-114) | - | - |
| INSS score | - | - | |
| Nasal rhinorrhea | 2.00 ± 0.18 | - | - |
| Itching | 2.13 ± 0.25 | - | - |
| Sneezing | 2.25 ± 0.13 | - | - |
| Congestion | 2.00 ± 0.27 | - | - |
| TNSS score | 8.38 ± 0.16 | - | - |
Data arepresented as mean value ± standard deviation, median (quartile 25th-75th) or count (%). Comparison was determined by t test or Chi-square test. P value < 0.05 was considered statistically significant. AR, allergic rhinitis; INSS, individual nasal symptom score; TNSS, total nasal symptom score.
DEM analysis and enrichment analysis
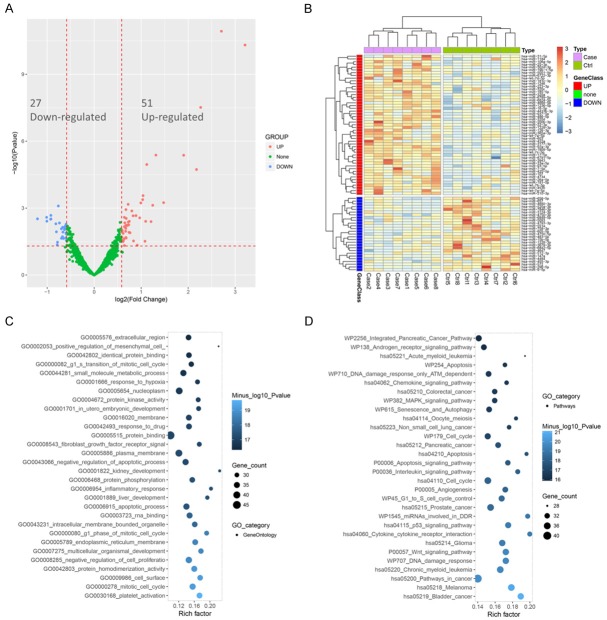
The PCA plot analysis was performed to evaluate the difference of miRNA aggregate between two groups, which showed that miRNA expression can clearly distinguish AR patients from controls in comp_2 axe, while in comp_1 axe (Figure 1A) and comp_3 axe (Figure 1B), the differences of miRNA expression between AR patients and controls were not found. Furthermore, as presented in volcano plot (Figure 2A), 27 down-regulated miRNAs and 51 up-regulated miRNAs were detected in AR patients and the dysregulated miRNAs analyzed by heat mapshowed that AR patients and controls could be differentiated by the expression of up-regulated miRNAs and down-regulated miRNAs (Figure 2B).
Figure 1.
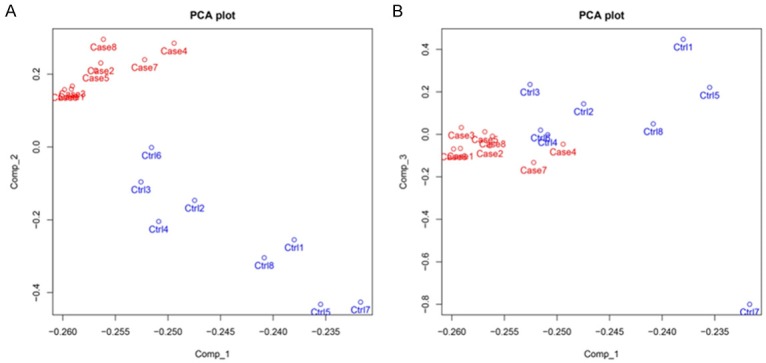
The principal component analysis (PCA) plot. The Principal Components Analysis (PCA) plot showed that in comp_2 axe (A), AR patients can be distinguished from controls via miRNA profiles, whiledifferences of miRNA expressionbetween AR patients and controls were not found in comp_1 axe (A) as well as comp_3 axe (B). The PCA plot analysis of the 8 pairs of samples in microarray was used to distinguish AR patients and controls (Comp_1, Comp_2 and Comp_3 stand for three principal components respectively).
Figure 2.
Differential analysis of DEMs and Enrichment analysis. Volcano plot displayed 27 down-regulated miRNAs and 51 up-regulated DEMs (A) and heat map analysis showed that AR patients could be distinguished from controls according to dysregulated DEMs (B). Enrichment analysis showed that the DEMs were mostly associated with inflammatory responses (C) and pathways (chemokines, MAPK and ILs) related to regulation of inflammatory disease (D). DEMs was compared by R package limma, Benjamini as well as Hochberg procedure was carried out to adjust P values, and clinical significance defined as a difference of 2.0 folds (absolute (log2 (fold change))>1.0). Fisher’s exact test was used to distinguish overrepresented miRNA-related items for the enrichment analysis of DEMs and its precursors.
The enrichment analysis of DEMs consisted of two areas, including GO (Figure 2C) and KEGG pathway (Figure 2D), which showed that the DEMs were mainly associated with inflammatory responses and pathways mediated by chemokines, MAPK and ILs.
Top 12 DEMs between AR patients and controls in microarray
The top 12 DEMs were selected in AR patients according to their P values (Table 2), which consisted of 9 up-regulated miRNAs, miR-1908-5p, miR-126-5p, miR-92a-3p, miR-125a-5p, miR-19a-5p, miR-26a-5p, miR-106a-5p, miR-181c-3p, and miR-3177-3p, and 3 down-regulated miRNAs, miR-572, miR-1228-3p, and miR-483-5p.
Table 2.
Top 12 DEMs between AR patients and controls in microarray
| DEMs | LogFC | AveExpr | P Value | Trend |
|---|---|---|---|---|
| miR-1908-5p | 2.708755 | 4.308389 | 1.17E-11 | UP |
| miR-126-5p | 3.21136 | 4.562552 | 4.90E-11 | UP |
| miR-92a-3p | 2.266234 | 3.53371 | 3.07E-08 | UP |
| miR-572 | -1.20753 | 1.494931 | 4.18E-06 | DOWN |
| miR-1228-3p | -0.91754 | 1.178168 | 4.28E-06 | DOWN |
| miR-483-5p | -0.9257 | 1.357245 | 1.11E-05 | DOWN |
| miR-125a-5p | 2.177425 | 3.772401 | 1.84E-05 | UP |
| miR-19a-5p | 1.039185 | 1.351904 | 0.000276 | UP |
| miR-26a-5p | 1.032444 | 1.02019 | 0.000417 | UP |
| miR-106a-5p | 1.477438 | 2.025603 | 0.000566 | UP |
| miR-181c-3p | 0.992658 | 0.714471 | 0.000582 | UP |
| miR-3177-3p | 0.723816 | 0.699096 | 0.00067 | UP |
Top 12 DEMs were selected according to P value. Comparison was determined by limma package in R software. AR, allergic rhinitis; DEMs, differentially expressed miRNAs; LogFC, log2 (fold change); AveExpr, average of expression level; Trend: UP, Up-regulated; Down, Down-regulated.
Characteristics of AR patients and controls in stage II
The overall demographic and clinical characteristics of AR patients (n = 48) and controls (n = 50) in Stage II are shown in Table 3. There was no difference in terms of age (P = 0.266) and gender distribution (P = 0.552) between the two groups. Among the AR patients, the disease duration was 76 (61-118) months, and the TNSS score was 8.02 ± 0.23, while INSS scores were as follows: nasal rhinorrhea 1.90 ± 0.10, itching 1.85 ± 0.08, sneezing 2.31 ± 0.10 and congestion 1.96 ± 0.10.
Table 3.
Characteristics of AR patients and controls in stage II
| Parameters | AR patients (N = 48) | Controls (N = 50) | P Value |
|---|---|---|---|
| Age (years) | 23.25 ± 6.90 | 25.14 ± 8.38 | 0.266 |
| Female (n/%) | 24 (50.0) | 28 (56.0) | 0.552 |
| Disease Duration (months) | 79 (61-118) | - | - |
| INSS score | - | - | |
| Nasal rhinorrhea | 1.90 ± 0.10 | - | - |
| Itching | 1.85 ± 0.08 | - | - |
| Sneezing | 2.31 ± 0.10 | - | - |
| Congestion | 1.96 ± 0.10 | - | - |
| TNSS score | 8.02 ± 0.23 | - | - |
Data arepresented as mean value ± standard deviation, median (quartile 25th-75th) or count (%). Comparison weredetermined by t test or Chi-square test. P value < 0.05 was considered statistically significant. AR, allergic rhinitis; INSS, individual nasal symptom score; TNSS, total nasal symptom score.
Comparison of 12 DEMs expression between AR group and control group in stage II
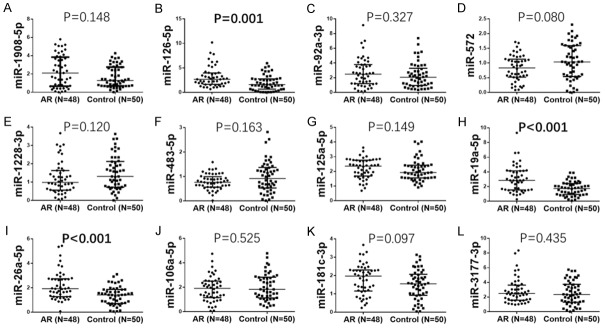
In stage II, miR-126-5p (P = 0.001) (Figure 3B), miR-19a-5p (P < 0.001) (Figure 3H) and miR-26a-5p (P < 0.001) (Figure 3I) were remarkably up-regulated in AR patients compared with controls. No difference between expression of miR-1908-5p (P = 0.148) (Figure 3A), miR-92a-3p (P = 0.327) (Figure 3C), miR-572 (P = 0.080) (Figure 3D), miR-1228-3p (P = 0.120) (Figure 3E), miR-483-5p (P = 0.163) (Figure 3F), miR-125a-5p (P = 0.149) (Figure 3G), miR-106a-5p (P = 0.525) (Figure 3J), miR-181c-3p (P = 0.097) (Figure 3K) or miR-3177-3p (P = 0.435) (Figure 3L) was found between AR patients compared to controls.
Figure 3.
Relative expression of 12 DEMs in stage II. The relative expressions of 12 DEMs were assessed by qPCR in the stage II, which displayed that relative levels of miR-126-5p (B), miR-19a-5p (H) and miR-26a-5p (I) were markedly up regulated in AR patients. No difference concerning the relative expressionof miR-1908-5p (A), miR-92a-3p (C), miR-572 (D), miR-1228-3p (E), miR-483-5p (F), miR-125a-5p (G), miR-106a-5p (J), miR-181c-3p (K) or miR-3177-3p (L) was found between AR patients and controls. Comparison between two groups was determined by Wilcoxon rank-sum test. P < 0.05 was considered significant.
Predictive value of validated miRNAs for AR risk
In the univariate logistic regression model, expression of miR-1908-5p (P = 0.041), miR-126-5p (P = 0.003), miR-19a-5p (P < 0.001) and miR-26a-5p (P < 0.001) were positively correlated with risk of AR, while expression of miR-572 (P = 0.036) was negatively associated with AR risk. All factors with P value ≤0.1 were included in the multivariate logistic regression, which showed that higher expression of miR-126-5p (P = 0.024), miR-19a-5p (P < 0.001), and miR-26a-5p (P = 0.002) is independently associated with increased risk of AR (Table 4).
Table 4.
Predictive value of validated miRNAs for AR risk
| Parameters | Univariate logistic regression | Multivariate logistic regression | ||||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| P value | OR | 95% CI | P value | OR | 95% CI | |||
|
|
|
|||||||
| Lower | Higher | Lower | Higher | |||||
| miR-1908-5p | 0.041 | 1.335 | 1.012 | 1.761 | 0.074 | 1.472 | 0.963 | 2.247 |
| miR-126-5p | 0.003 | 1.478 | 1.137 | 1.922 | 0.024 | 1.562 | 1.059 | 2.304 |
| miR-92a-3p | 0.300 | 1.124 | 0.901 | 1.402 | - | - | - | - |
| miR-572 | 0.036 | 0.435 | 0.200 | 0.946 | 0.052 | 0.310 | 0.095 | 1.011 |
| miR-1228-3p | 0.139 | 0.710 | 0.451 | 1.118 | - | - | - | - |
| miR-483-5p | 0.057 | 0.436 | 0.185 | 1.026 | 0.158 | 0.398 | 0.111 | 1.430 |
| miR-125a-5p | 0.417 | 1.264 | 0.718 | 2.226 | - | - | - | - |
| miR-19a-5p | < 0.001 | 2.172 | 1.472 | 3.206 | < 0.001 | 3.375 | 1.820 | 6.259 |
| miR-26a-5p | < 0.001 | 2.654 | 1.543 | 4.565 | 0.002 | 3.393 | 1.542 | 7.465 |
| miR-106a-5p | 0.478 | 0.873 | 0.600 | 1.271 | - | - | - | - |
| miR-181c-3p | 0.074 | 1.577 | 0.958 | 2.596 | 0.251 | 1.529 | 0.741 | 3.153 |
| miR-3177-3p | 0.342 | 1.125 | 0.882 | 1.436 | - | - | - | - |
Data arepresented as P value, OR (odds ratio) and 95% CI (confidence interval). Factors affecting AR risk were determined by univariate logistic regression analysis, while all factors with P value no more than 0.1 were further detected by multivariate logistic regression analysis. P Value < 0.05 was considered statistically significant. AR, allergic rhinitis.
The correlation of miRNA expression and the risk of AR
ROC curves were performed to evaluate the diagnostic value of miRNAs which could predict AR risk independently in the multivariate logistic models. As shown in Figure 4, the area under curve (AUC) of individual miR-126-5p, miR-19a-5p and miR-26a-5p were 0.685 (95% CI: 0.581-0.790) (Figure 4A), 0.742 (95% CI: 0.644-0.840) (Figure 4B) and 0.719 (95% CI: 0.619-0.819) (Figure 4C), respectively, which indicate that miR-126-5p, miR-19a-5p, and miR-26a-5p could be good predictors for risk of AR separately. When combining the expression of miR-126-5p, miR-19a-5p, and miR-26a-5p, the ROC curve showed a high AUC of 0.866 (95% CI: 0.797-0.936) with good sensitivity and specificity (89.6% and 70%, respectively) at best cut-off point which yielded the maximum value of sensitivity plus specificity (Figure 4D), suggesting that the combination of the three miRNAs presented with a great value for predicting the risk of AR.
Figure 4.
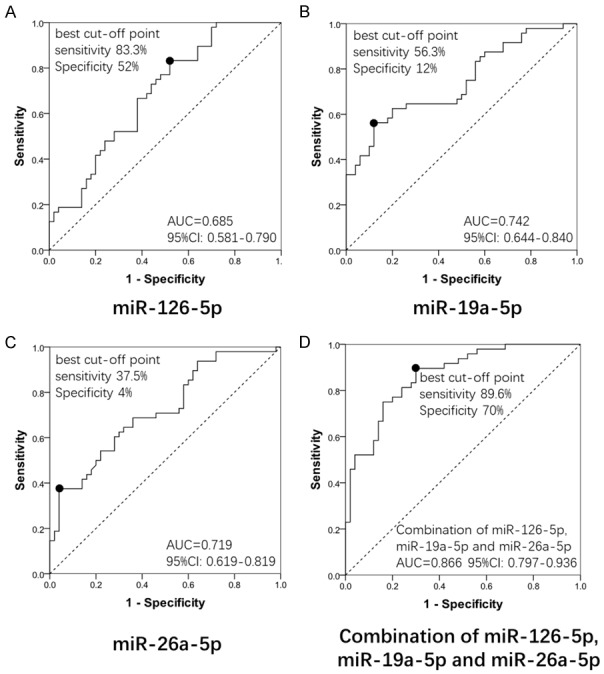
ROC curve analysis of 3 DEMs in AR patients. ROC curve analysis was performed to evaluate the diagnostic value of the independent predicting miRNAs for AR risk in the multivariate logistic models in AR patients. A. The area under curve (AUC) of miR-126-5p was 0.685 (95% CI: 0.581-0.790); B. The AUC of miR-19a-5p was 0.742 (95% CI: 0.644-0.840); C. miR-26a-5p was 0.719 (95% CI: 0.619-0.819); D. The AUC of combination of miR-126-5p, miR-19a-5p and miR-26a-5p was 0.866 (95% CI: 0.797-0.936) with good sensitivity and specificity (89.6% and 70%, respectively) at the best cut-off point.
Correlation of validated miRNAs with TNSS score
Disease severity of AR patients was evaluated by INSS and TNSS score. To explore the association of validated miRNA expression with disease severity of AR, Spearman tests were performed. The levels of miR-126-5p (P = 0.003) (Figure 5B), miR-19a-5p (P = 0.027) (Figure 5H) and miR-181c-3p (P = 0.005) (Figure 5K) in nasal mucosal were illustrated to be positively associated with TNSS score, while no correlation was found with TNSS score in other 9 DEMs (miR-1908-5p (P = 0.102) (Figure 5A), miR-92a-3p (P = 0.575) (Figure 5C), miR-572 (P = 0.241) (Figure 5D), miR-1228-3p (P = 0.991) (Figure 5E), miR-483-5p (P = 0.655) (Figure 5F), miR-125a-5p (P = 0.257) (Figure 5G), miR-26a-5p (P = 0.520) (Figure 5I), miR-106a-5p (P = 0.063) (Figure 5J) and miR-3177-3p (P = 0.945) (Figure 5L).
Figure 5.
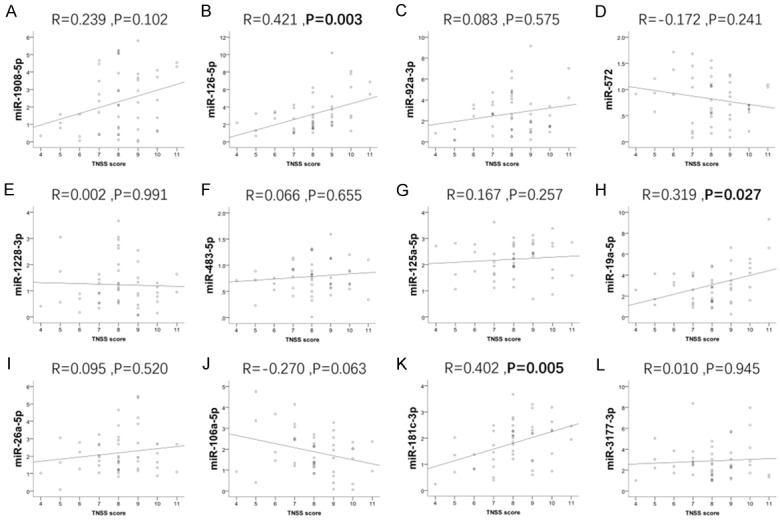
The associations of 12 DEMs with TNSS score. The expressions of miR-126-5p (B), miR-19a-5p (H) and miR-181c-3p (K) were positively correlated with TNSS score; while the correlations between another 9 DEMs (miR-1908-5p (A), miR-92a-3p (C), miR-572 (D), miR-1228-3p (E), miR-483-5p (F), miR-125a-5p (G), miR-26a-5p (I), miR-106a-5p (J) and miR-3177-3p (L)) and TNSS were not found. The association between 12 validated miRNAs and TNSS score was analyzed by Spearman test. P < 0.05 was considered statistically significant.
Correlation of validated miRNAs with INSS score
miR-126-5p expression was correlated with itching score (P = 0.033) and congestion score (P = 0.021). Additionally, the miR-19a-5p level was correlated with nasal rhinorrhea score (P = 0.024) and sneezing score (P = 0.038). For miR-181c-3p, a positive correlation was discovered with sneezing score (0.045) and congestion score (P = 0.008). No other correlation of other miRNAs with INSS score was found (Table 5).
Table 5.
Correlation of validated miRNAs with INSS score
| Nasal rhinorrhea | Itching | Sneezing | Congestion | |
|---|---|---|---|---|
| miR-1908-5p | ||||
| Coefficient R | 0.137 | 0.063 | 0.167 | 0.275 |
| P value | 0.352 | 0.671 | 0.257 | 0.059 |
| miR-126-5p | ||||
| Coefficient R | 0.078 | 0.308 | 0.204 | 0.333 |
| P value | 0.597 | 0.033 | 0.163 | 0.021 |
| miR-92a-3p | ||||
| Coefficient R | -0.026 | 0.202 | -0.026 | 0.257 |
| P value | 0.861 | 0.168 | 0.859 | 0.078 |
| miR-572 | ||||
| Coefficient R | -0.079 | -0.012 | -0.085 | -0.146 |
| P value | 0.591 | 0.937 | 0.564 | 0.323 |
| miR-1228-3p | ||||
| Coefficient R | -0.110 | 0.042 | -0.048 | 0.188 |
| P value | 0.456 | 0.777 | 0.744 | 0.202 |
| miR-483-5p | ||||
| Coefficient R | -0.091 | 0.022 | 0.153 | 0.112 |
| P value | 0.538 | 0.880 | 0.298 | 0.449 |
| miR-125a-5p | ||||
| Coefficient R | 0.068 | 0.064 | 0.090 | 0.080 |
| P value | 0.646 | 0.667 | 0.545 | 0.587 |
| miR-19a-5p | ||||
| Coefficient R | 0.324 | 0.058 | 0.300 | 0.065 |
| P value | 0.024 | 0.697 | 0.038 | 0.660 |
| miR-26a-5p | ||||
| Coefficient R | 0.165 | 0.064 | 0.049 | -0.034 |
| P value | 0.262 | 0.664 | 0.739 | 0.818 |
| miR-106a-5p | ||||
| Coefficient R | -0.049 | 0.006 | -0.204 | -0.221 |
| P value | 0.742 | 0.970 | 0.165 | 0.132 |
| miR-181c-3p | ||||
| Coefficient R | 0.065 | 0.183 | 0.291 | 0.377 |
| P value | 0.663 | 0.213 | 0.045 | 0.008 |
| miR-3177-3p | ||||
| Coefficient R | 0.050 | -0.169 | 0.114 | 0.011 |
| P value | 0.735 | 0.249 | 0.441 | 0.941 |
Correlation between validated miRNAs and INSS score was analyzed by Spearman test. P < 0.05 was considered statistically significant. INSS, individual nasal symptom score.
Discussion
Our study demonstrates that (1) AR patients could be differentiated from controls according to miRNA profiles by PCA plot analysis, and 27 down-regulated miRNAs as well as 51 up-regulated miRNAs were identified by volcano map. (2) miR-126-5p, miR-19a-5p, and miR-26a-5p expression was shown to be remarkably up-regulated in AR patients compared with controls. Multivariate logistic regression displayed that miR-126-5p, miR-19a-5p, and miR-26a-5p are independent predictive factors for AR risk, and ROC curve displayed that the AUC of the combination of miR-126-5p, miR-19a-5p, and miR-26a-5p was 0.866 (95% CI: 0.797-0.936) with good sensitivity and specificity (89.6% and 70%, respectively). (3) The correlation of miRNAs with AR severity was also analyzed and the results indicate that miR-126-5p, miR-19a-5p, and miR-181c-3p arepositively associated with TNSS, which is a rating score of AR symptoms.
AR is a common disease characterized by chronic inflammation of the nasal mucosa, while unacceptable misdiagnosis of AR frequently occurs in clinical practice which results in delaying patients’ treatment and recovery [1,2,12]. Recently, accumulating studies have revealed that miRNAs are involved in the pathogenesis of AR in several ways such as regulation of mast cells, eosinophilic granulocytes, and T helper cell (Th)1/Th2 imbalance [8,13]. For instance, Xiao L et al. discovered that miR-133b appears to strongly attenuate pathological alterations, eosinophils and mast cells infiltration in nasal mucosa via up-regulating the levels of cytokines (TNF-α, IL-4, IL-5, and IFN-γ) and relieves allergic symptom, such as sneezing, in AR mice by inhibition of Nlrp3 inflammasome-meditated inflammation [13]. miRNA-135a is reported to down-regulate protein expression levels of GATA-3 and IL-4, and up-regulate the levels of T-bet and IFN-γ as well as correct the Th1/Th2 imbalance in the AR mice [8]. In addition, serum levels of miR-181a is found to be down-regulated in AR patients and is elucidated to have a negative correlation with IL-4/IL-5 and positive correlation with IFN-γ/IL-12, resulting in increasing disease severity in childhood of AR patients [14]. These indicate dysregulation of miRNAs is emerging as a crucial factor in the etiology and development of AR. However, altered miRNAs profiles in AR patients and the predicting value of the miRNAs for AR risk have not yet been fully described.
miR-126-5p, an intronic miRNA located within the Egfl7 gene, mediates mast cell degranulation, cytokine production in a mast cell-specific and activation dependent way, acting as a positive mediator of monocyte-mediated inflammatory responses [15,16]. A previous in vivo experiment showed that overexpression of miR126 suppressed Spred1 expression and enhanced extracellular signal-regulated kinase activity in MC9 mast cells, which was associated with elevated FcεRI-mediated cytokine production [15]. Furthermore, Huang J et al. illustrated that miR-126-5p could up-regulate phosphorylation of JNK protein and enhance inflammatory responses of monocytes to lipopolysaccharide stimulation by suppressing cylindromatosis in HIV patients [16]. In this present study, miR-126-5p was positively correlated with both risk and severity of AR, the probable explanation may be that miR-126-5p contributes to the process related to AR development by mediating mast cells and promotes inflammatory responses [15-17].
miR-19a, one of the members of the miR-17-92 cluster, has a strong regulatory effect on the immunity [18]. It has been reported that expression of miR-19a is higher in the peripheral B cells in AR patients, which is line with our study, and miR-19a facilitates AR development by mediating IL-4-suppressed IL-10 expression in B cells [19]. In their following study, they found that repressing expression of miR-19a in B cells could strengthen the effect of specific immunotherapy on Th2 response, which alleviated AR symptoms [19]. Coincidentally, another study by Geng XR et al. also found that the level of miR-19a is significantly higher in peripheral B cells from AR patients than that in B cells from healthy participants, and miR-19a mediates the allergen-specific immune response-decreased IL-10 expression in B cells in AR process [20]. In our study, miR-19b-5p was shown to be up-regulated in AR patients and it was an independent predictive factor for AR risk as well as correlated with the disease severity of AR, which could be explained by the destructive role of miR-19a in immune function discovered by the previous studies [19,20].
miR-26a-5p belongs to the miR-26 family, which is also reported to be involved in the pathogenesis of inflammation-related disease. For instance, miR-26a is reported to repress inflammatory responses likelyby promoting regulatory T cell responses or through NF-κB inhibition in chondrocytes, which results in the disease progression of both sepsis and systemic inflammatory response syndrome (SIRS) [21]. Several previous studies revealed that miR-26ais involved in inflammation through multiple pathways and it may enhance AKT pathway activation by PTEN suppression and promote cancer PTEN Signaling pathway [22], regulate endothelial cell dysfunction, apoptosis [23], and mediate Wnt and mitogen-activated protein kinase (MAPK) signaling pathways [24]. Those results suggest that miR-26a-5p could accelerate AR development, which might explain the higher level of miR-26a-5p in AR patients compared with controls in our study, however no association was found between miR-26a-5p expression and disease severity.
miR-181c-3p, one of the miR-181 family, has been demonstrated to play important role in regulating inflammatory response [25-27]. For instance, in a rat model, miR-181c suppressed TLR4 expression and subsequently reduced NF-κB/p65 activation to regulate inflammatory factor expression (TNF-α, IL-2, IL-8 and chemokine) as well as restricts burn-induced inflammation [25]. In our study, apositive correlation between miR-181c-3p level and disease severity of AR patients was found, which highlights the pivotal role of miR-181c-3p in regulating inflammation [25-27].
More importantly, the ROC curve showed that a combination of miR-126-5p, miR-19a-5p and miR-26a-5p expression predicts AR risk with a high AUC of 0.866 (95% CI: 0.797-0.936). The diagnostic value could be explained by miR-126-5p, miR-19a-5p and miR-26a-5p beinginvolved in the pathogenesis of AR via mediating mast cells, promoting inflammatory responses, and destructing immune function. No similar research has illustrated the combined predicting value of miR-126-5p, miR-19a-5p and miR-26a-5p for AR risk, which suggests that the combination of miR-126-5p, miR-19a-5p, and miR-26a-5p has a high potential for early diagnosis of AR as well as to be promising biomarkers for disease surveillance in AR patients.
Some limitations of our study include the following: (1) This case-control study enrolled 50 controls who were non-atopic patients with obstructive snoring undergoing adenoid surgery instead of health controls, and it resulted from nasal mucosa samples that were difficult and inappropriate to be obtained from healthy participants. Obstructive snoring did not share the same mechanism with AR, and it reduced the bias in this study. (2) Our study was only conducted in a single center and patients were enrolled from central China, which doesnot represent the general population of China. Thus multi-center, crossing regional studies are needed for more convincing results. (3) The sample size of this study was limited with 48 AR patients and 50 controls which needed to be enlarged in the future study.
In conclusion, miRNA profiles distinguish AR patients from controls, and the combination of miR-126-5p, miR-19a-5p and miR-26a-5p could serve as novel biomarker for AR risk.
Disclosure of conflict of interest
None.
References
- 1.Wheatley LM, Togias A. Clinical practice. Allergic rhinitis. N Engl J Med. 2015;372:456–463. doi: 10.1056/NEJMcp1412282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Greiner AN, Hellings PW, Rotiroti G, Scadding GK. Allergic rhinitis. Lancet. 2011;378:2112–2122. doi: 10.1016/S0140-6736(11)60130-X. [DOI] [PubMed] [Google Scholar]
- 3.Zhang Y, Zhang L. Prevalence of allergic rhinitis in china. Allergy Asthma Immunol Res. 2014;6:105–113. doi: 10.4168/aair.2014.6.2.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Luo X, Hong H, Tang J, Wu X, Lin Z, Ma R, Fan Y, Xu G, Liu D, Li H. Increased expression of miR-146a in children with allergic rhinitis after allergen-specific immunotherapy. Allergy Asthma Immunol Res. 2016;8:132–140. doi: 10.4168/aair.2016.8.2.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sin B, Togias A. Pathophysiology of allergic and nonallergic rhinitis. Proc Am Thorac Soc. 2011;8:106–114. doi: 10.1513/pats.201008-057RN. [DOI] [PubMed] [Google Scholar]
- 6.Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 7.Lu TX, Rothenberg ME. Diagnostic, functional, and therapeutic roles of microRNA in allergic diseases. J Allergy Clin Immunol. 2013;132:3–13. doi: 10.1016/j.jaci.2013.04.039. quiz 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luo Y, Deng Y, Tao Z, Chen S, Xiao B, Ren J, Chen Z, Han J, Kong Y, Xu Y, Deng M. Regulatory effect of microRNA-135a on the Th1/Th2 imbalance in a murine model of allergic rhinitis. Exp Ther Med. 2014;8:1105–1110. doi: 10.3892/etm.2014.1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu D, Zhang Z, Ke X, Kang H, Hong S. A functional variant of miRNA-149 confers risk for allergic rhinitis and comorbid asthma in Chinese children. Int J Immunogenet. 2017;44:62–70. doi: 10.1111/iji.12307. [DOI] [PubMed] [Google Scholar]
- 10.Suojalehto H, Lindstrom I, Majuri ML, Mitts C, Karjalainen J, Wolff H, Alenius H. Altered microRNA expression of nasal mucosa in longterm asthma and allergic rhinitis. Int Arch Allergy Immunol. 2014;163:168–178. doi: 10.1159/000358486. [DOI] [PubMed] [Google Scholar]
- 11.Wu G, Yang G, Zhang R, Xu G, Zhang L, Wen W, Lu J, Liu J, Yu Y. Altered microRNA expression profiles of extracellular vesicles in nasal mucus from patients with allergic rhinitis. Allergy Asthma Immunol Res. 2015;7:449–457. doi: 10.4168/aair.2015.7.5.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sin E, Anand P, Frieri M. A link: allergic rhinitis, Asthma & systemic lupus erythematosus. Autoimmun Rev. 2016;15:487–491. doi: 10.1016/j.autrev.2016.02.003. [DOI] [PubMed] [Google Scholar]
- 13.Xiao L, Jiang L, Hu Q, Li Y. MicroRNA-133b ameliorates allergic inflammation and symptom in murine model of allergic rhinitis by targeting Nlrp3. Cell Physiol Biochem. 2017;42:901–912. doi: 10.1159/000478645. [DOI] [PubMed] [Google Scholar]
- 14.Liu W, Zeng Q, Luo R. Correlation between serum osteopontin and miR-181a levels in allergic rhinitis children. Mediators Inflamm. 2016;2016:9471215. doi: 10.1155/2016/9471215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ishizaki T, Tamiya T, Taniguchi K, Morita R, Kato R, Okamoto F, Saeki K, Nomura M, Nojima Y, Yoshimura A. miR126 positively regulates mast cell proliferation and cytokine production through suppressing Spred1. Genes Cells. 2011;16:803–814. doi: 10.1111/j.1365-2443.2011.01529.x. [DOI] [PubMed] [Google Scholar]
- 16.Huang J, Zhu L, Qiu C, Xu X, Zhang L, Ding X, Liao Q, Xu J, Zhang X. MicroRNA miR-126-5p enhances the inflammatory responses of monocytes to lipopolysaccharide stimulation by suppressing cylindromatosis in chronic HIV-1 infection. J Virol. 2017;91:e02048–16. doi: 10.1128/JVI.02048-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shibayama Y, Kondo T, Ohya H, Fujisawa S, Teshima T, Iseki K. Upregulation of microRNA-126-5p is associated with drug resistance to cytarabine and poor prognosis in AML patients. Oncol Rep. 2015;33:2176–2182. doi: 10.3892/or.2015.3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haj-Salem I, Fakhfakh R, Berube JC, Jacques E, Plante S, Simard MJ, Bosse Y, Chakir J. MicroRNA-19a enhances proliferation of bronchial epithelial cells by targeting TGFbetaR2 gene in severe asthma. Allergy. 2015;70:212–219. doi: 10.1111/all.12551. [DOI] [PubMed] [Google Scholar]
- 19.Yu ZJ, Zeng L, Luo XQ, Geng XR, Xu R, Chen K, Yang G, Luo X, Liu ZQ, Liu ZG, Liu DB, Yang PC, Li HB. Vitamin D3 inhibits micro RNA-17-92 to promote specific immunotherapy in allergic rhinitis. Sci Rep. 2017;7:546. doi: 10.1038/s41598-017-00431-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geng XR, Qiu SQ, Yang LT, Liu ZQ, Yang G, Liu JQ, Zeng L, Li XX, Mo LH, Liu ZG, Yang PC. Allergen-specific immune response suppresses interleukin 10 expression in B cells via increasing micro-RNA-17-92 cluster. Cell Biochem Funct. 2016;34:449–454. doi: 10.1002/cbf.3207. [DOI] [PubMed] [Google Scholar]
- 21.Caserta S, Kern F, Cohen J, Drage S, Newbury SF, Llewelyn MJ. Circulating plasma microRNAs can differentiate human sepsis and systemic inflammatory response syndrome (SIRS) Sci Rep. 2016;6:28006. doi: 10.1038/srep28006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Capra E, Turri F, Lazzari B, Cremonesi P, Gliozzi TM, Fojadelli I, Stella A, Pizzi F. Small RNA sequencing of cryopreserved semen from single bull revealed altered miRNAs and piRNAs expression between High- and Low-motile sperm populations. BMC Genomics. 2017;18:14. doi: 10.1186/s12864-016-3394-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Silambarasan M, Tan JR, Karolina DS, Armugam A, Kaur C, Jeyaseelan K. MicroRNAs in hyperglycemia induced endothelial cell dysfunction. Int J Mol Sci. 2016;17:518. doi: 10.3390/ijms17040518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Enatescu VR, Papava I, Enatescu I, Antonescu M, Anghel A, Seclaman E, Sirbu IO, Marian C. Circulating plasma micro RNAs in patients with major depressive disorder treated with antidepressants: a pilot study. Psychiatry Investig. 2016;13:549–557. doi: 10.4306/pi.2016.13.5.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li X, Liu L, Yang J, Yu Y, Chai J, Wang L, Ma L, Yin H. Exosome derived from human umbilical cord mesenchymal stem cell mediates MiR-181c attenuating burn-induced excessive inflammation. EBioMedicine. 2016;8:72–82. doi: 10.1016/j.ebiom.2016.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang L, Li YJ, Wu XY, Hong Z, Wei WS. MicroRNA-181c negatively regulates the inflammatory response in oxygen-glucose-deprived microglia by targeting Toll-like receptor 4. J Neurochem. 2015;132:713–723. doi: 10.1111/jnc.13021. [DOI] [PubMed] [Google Scholar]
- 27.Hutchison ER, Kawamoto EM, Taub DD, Lal A, Abdelmohsen K, Zhang Y, Wood WH 3rd, Lehrmann E, Camandola S, Becker KG, Gorospe M, Mattson MP. Evidence for miR-181 involvement in neuroinflammatory responses of astrocytes. Glia. 2013;61:1018–1028. doi: 10.1002/glia.22483. [DOI] [PMC free article] [PubMed] [Google Scholar]