Abstract
Type 2 diabetes mellitus (T2DM) and diabetic nephropathy (DN) seriously impact the prognosis and survival of patients. Abundant studies suggest that single nucleotide polymorphisms (SNPs) within regulatory regions of miRNAs modulate progression of various diseases including DM and DN. However, evidence for exploring the mechanism in DM and DN are still insufficient. This study was performed to discuss the association of SNPs in the regulatory region of hsa-miR-34a with DM or DN susceptibility in the southwest Chinese Han population. Three SNPs (rs12128240, rs2666433, rs6577555) in miR-34a were analyzed in the T2DM patients with or without DN and normal controls. RT-qPCR was performed to measure expression of miR-34a. Real-time PCR was used for amplification of SNPs. MiR-34a was over-expressed in T2DM and DM patients. Both the distribution of GG genotypes and the G allele frequency of rs2666433 were significantly different between the T2DM and control groups (P=0.009, P=0.008 respectively). Univariate analysis with additive and recessive models for rs2666433 polymorphisms showed an association with DM (P=0.023; P=0.009 respectively), and a stronger association was found in an additive model (P=0.001) when compared with DN. After adjustment for clinical covariates, the GG genotype was still significantly different for the recessive model (OR 2.297; CI 1.031-5.121; P=0.042) in T2DM by multivariate analysis. In conclusion, our study demonstrates that a potential variant rs2666433 in the miR-34a regulatory region may significantly associate with the occurrence of T2DM.
Keywords: Diabetic nephropathy, polymorphism, type 2 diabetes
Introduction
Diabetes mellitus (DM) is a disease that is universally epidemic andis induced by a combination of genetic and environmental factors, which destroy both the insulin sensitivity and insulin secretion, and result in a damaged balance of intracorporal glucose [1]. The human population with diabetes is about 180 million in the world and Chinese adults with diabetes number more than 100 million [2]. It is estimated that the number of diabetics will increase worldwide to 366 million in 2030 [3]. Diabetic nephropathy (DN), one of the most common diabetic microangiopathies, is a leading cause of end-stage renal disease. Research has revealed that the diabetic nephropathy prevalence epidemiologically reaches about 30%-40% of hospitalized patients with diabetes [4]. Further research isurgently needed for better treatments for DM and DN. However, the mechanism of type 2 diabetes mellitus (T2DM) and DN is not fully clear.
MicroRNA (miRNA) is a class of endogenous, small (21 to 24 nucleotides in length), single-stranded, non-coding but functional RNAs. They are seen as the important regulatory factors that mainly regulate gene expression through posttranscriptional mechanisms. Abundant research has indicated that single-nucleotide polymorphisms (SNPs) within miRNAs are associated with the risk of various diseases, such as hepatocellular carcinoma [5], gastric adenocarcinoma [6], and so on. However, little has been known about the SNPs within miRNAs associated with T2DM and DN.
In this study, we found that miR-34a was significantly up-regulated in T2DM from the previous miRNA expression profiling studies [7-22]. Also, our result showed that miR-34a was over-expressed in DN patients by real-time PCR. Moreover, three SNPs (rs12128240 C>T, rs6577555 A>C, rs2666433 G>A) in the regulatory region of miR-34a were selected as the candidate SNPs by bioinformatics prediction. Furthermore, we found that the genotype frequency of the rs266433 polymorphism (P<0.05) was dramatically different in T2DM. The GG genotype was significantly different in the recessive model (OR 2.297; CI 1.031-5.121; P=0.042) by multivariate analysis. In univariate analysis, the additive model and the recessive model of rs2666433 were statistically different (P=0.023, P=0.009, respectively). We also show a connection with DN by the univariate analysis in the additive model (P=0.001). Therefore, our data indicate that miR-34a may participate in T2DM and DN, and a potential variant rs2666433 in miR-34a regulatory region may significantly associate with the occurrence of T2DM.
Material and methods
Ethics statement
This study was approved by the Ethics Committee of Chongqing Medical University, and was conducted in compliance with the Helsinki Declaration. All patients involved in this study consented to participate in the study and publication of the results.
Study subjects
In this study, three independent groups were included to discuss the relationship between the SNPs of the miR-34a and the risk for T2DM and DN. The test group included 135 subjects undergoing T2DM more than one year (urinary albumin excretion rate (UAER)<20 µg/min) (T2DM group), 30 diabetes with DN (UAER: 20-200 µg/min) (DN group), and 117 healthy subjects having no any other chronic illness, excluding hepatic, renal, thyroid, or any other metabolic diseases, were enrolled as the control group (CON group). All the subjects were from the Han ethnic group aged from 30 to 75 years, and consecutively gathered from the Department of Endocrinology and Department of Nephrology at the first affiliated hospital of Chongqing Medical University from January 2014 to January 2016. The study design was approved by the hospital ethics review committee. Each eligible participant gave a written informed consent prior to participation and were interviewed to provide data regarding age, sex, ethnicity, duration of diabetes, glycosylated hemoglobin, fasting blood-glucose (FBG), 2-hour post-meal blood glucose (2 h PG), glycated hemoglobin A1c(HbA1c) and urinary albumin excretion rate (UARE) status [23].
Search strategies and selection of miRNA
To found out the miRNAs related to T2DM, the terms of “diabetes”, “miRNA”, “expression” and “profile” were searched in PubMed for miRNA expression profiling studies correlative with T2DM between 2010 and 6 June 2016. The process was conducted as previously described [23]. The final studies must meet the criteria: (1) They are miRNA expression profiling studies carried out on the blood from non-diabetes and T2DM; (2) They used miRNA expression arrays.
Total RNA isolation
Venous blood samples (6 ml) on empty stomach in the early morning were collected and transferred into PreAnalytiXPAXgeneTM blood RNA tubes (Qiagen, Valencia, CA, USA). Total RNA from human plasma was extracted by Trizol reagent (Invitrogen) according to the manufacturer’s instructions.
Real-time reverse transcription (RT)-PCR
PrimeScriptTM RT reagent Kit with gDNA Eraser (TaKaRa Bio Inc, Tokyo, Japan) was used for semi-quantitative determination for the expression of miR-34a according to the m anufacturer’s instructions. Briefly, 0.8 µg of total RNA was reverse transcribed to complementary DNA (cDNA) using miRNA-specific stem-loop RT primers (Supplementary Table 1), 5× g DNA Eraser Buffer, gDNA Eraser, PrimeScript RT Enzyme Mix I, RT Primer Mix, 5xPrimeScript Buffer, and RNase Free dH2O in the Bio-RAD T100TM Thermal Cycler (Applied Bio systems)under the following conditions: 42°C for 15 min, 85°C for 5 s. The expression of miR-34a was determined using the obtained cDNA, SYBR® Premix Ex TaqII (TliRNaseH Plus), Primer (10 µM), RT products, dH2O in the Bio-RADC-FX96TM Real-Time system under the following conditions: 95°C for 3 min followed by 40 cycles of 95°C for 30 s, and 60°C for 30 s. The relative expression was normalized to the expression of RNU48.
Candidate SNPs selection
The upstream 1.4 kb and downstream 578 bp of the 5’ transcription start site of the miR-34a and the regulatory activity had been validated in other research [24]. Next, 202 SNPs within the hsa-miR-34a were found in public SNP database (http://www.ncbi.nlm.nih.gov/SNP). 1000 Genomes were applied to pick out the SNPs in Han population with minor allele frequency (MAF)>0.05. Eight SNPs (rs70985597, rs17393158, rs12128240, rs6577555, rs2666433, rs59453084, rs147829974 and rs371949727) were finally included. But only three SNPs (rs12128240, rs2666433, rs6577555) were in the regulatory region of miR-34a. Thus these three SNPs were chosen for further research. TFSEARCHver1.3 software was used to predict transcription factor binding sites of these three SNPs and detail information was shown in Supplementary Table 2.
Total DNA isolation
2 ml venous blood from each subject on empty stomach in the early morning was collected. Genomic DNA was extracted from each whole-blood samples using TIANamp Blood DNA Kit (TIANGE), according to the manufacturers’ instructions. DNA purity and concentration were measured through NanoDrop 2000 (Thermo Scientific Inc) and the DNA was stored at -80°C until use.
PCR analyses and genotyping
PCR primers were designed by Primer Premier 5.0 and blasted from NCBI. Details were shown in Supplementary Table 1. PCR was conducted in 25 µl volumes including Premix Taq 12.5 µl, PCR primers mixture (10 µmol/µl) 2 µl, DNA 0.1 µg, and ddH2O. The PCR program was initiated as the following conditions: denaturation at 94°C for 3 min, followed by 34 cycles of 94°C for 30 s, annealing at 49°C for 30 s for rs6577555, 51°C for 30 s for rs2666433, 56°C for 30 s for rs12128240, and extension at 72°C for 50 s. A final extension step was at 72°C for 5 min. All the PCR products were sent for sequencing (Thermo Fisher Scientific Inc, Rockford, IL, USA). The genotype was analyzed by Chromas software.
Statistical analysis
SPSS software version 19.0 was performed for Statistical analyses. Hardy-Weinberg equilibrium of the genotypes for each SNP was evaluated using a Chi-square test for goodness of fit. The results for normally distributed continuous variables were shown as mean ± standard deviation (SD). Student’s t test was employed to compare age, gender, fasting blood-glucose (FBG) and 2-hour post-meal blood glucose (2 h PG) between CON group and T2DM group or DN group. The differences of diabetes duration and HbA1c between DM group and DN group were also analyzed by t-test. One-way ANOVAor Chi-square was used to compare allele frequencies and genotype distribution among three subject groups. The associations of allele and genotype were calculated as the odds ratio (OR) and its 95% confidence intervals (CI). Univariate analysis of allele or genotype frequencies between cases and controls for additive, dominant and recessive models were performed with X2 analysis. Multivariate analysis, adjusting for age, FBG and 2 h PG, were performed with binary logistic regression analysis. Statistical significance was accepted at P<0.05.
Results
Intimate connection between miR-34a and diabetes mellitus
After searching PubMed, eighteen eligible studies were chosen to discuss the differentially expressed miRNA in T2DM. The detailed characteristics of these studies areshown in Supplementary Table 3. Sixteen miRNAs were significantly increased in expression and eight miRNAs had reduced expression in T2DM compared with normal controls. These results were consistent with at least two other studies. MiR-34a was one of the significantly increased miRNAs in these studies. Furthermore, it was found that miR-34a could regulate mesangial proliferation and glomerular hypertrophy, which shows a tight link with the development of diabetes mellitus and diabetic nephropathy [25]. Moreover, the significantly up-regulatedmiR-34a had commonality among blood, liver, and pancreas in T2DM [23]. Furthermore, we also measured the expression of miR-34a in healthy persons, T2DM, and DN patients, and the results were consistent with other research studies (Figure 1).
Figure 1.
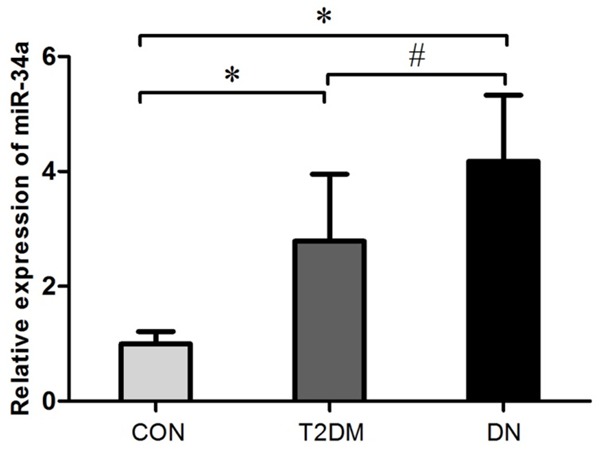
Expression of miR-34a by real-time PCR. The expression of miR-34a was tested by real-time RT-PCR in DN, T2DM patients, and normal controls. Expression of miR-34a was increased in DM and DN compared with the CON group. Also, the expression of miR-34a in the DN group was higher than that in T2DM group. *P<0.05, vs CON group; #P<0.05, vs T2DM group.
Characteristics of clinical data
The clinical characteristics of the 130 T2DM patients, 30 DM patients with DN, and 117 controls applied in the study are summarized in Table 1. No statistically significant difference in terms of the frequent distribution of sex among the three groups or the duration of diabetes. HbA1c was not significantly different between the T2DM group and DN group either. Both the average age of T2DM patients and the DN subjects were significantly largerthan control subjects and the FBG and 2 h PG in T2DM and DN groups were significantly higher than in the CON group. UAER in DN group was alsohigher than in the T2DM group.
Table 1.
Clinical characteristics of all subjects
| CON (n=117) | T2DM (n=130) | DN (n=30) | ||
|---|---|---|---|---|
| Age | 45.7±7.3 | 58.7±11.2a | 63.2±7.3a | |
| Sex | Male | 55 | 62 | 16 |
| Female | 62 | 68 | 14 | |
| Diabetes duration (years) | - | 6.4±5.5 | 10.9±5.7 | |
| FBG (mmol/l) | 5.1±0.8 | 9.7±6.6a | 9.8±1.6a | |
| 2hPG (mmol/l) | 9.4±1.0 | 12.7±1.3a | 14.5±0.8a | |
| HbA1c (mmol/l) | - | 9.0±2.4 | 9.9±2.8 | |
| UAER (µg/min) | - | <20 | 109.2±53.5b | |
P<0.01, vs CON;
P<0.01, vs T2DM.
Genotype of SNPs
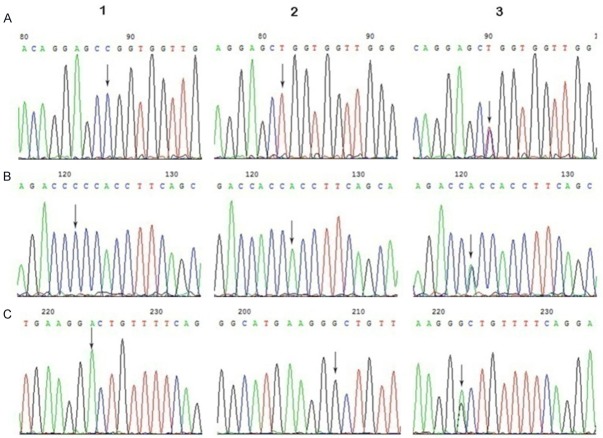
The products of PCR were all analyzed by sequencing (Figure 2). The allelic and genotype data of these three SNPs were all in Hardy-Weinberg equilibrium (P>0.05). The allelic or genotypic distributions and the associations among cases, control group, and the three SNPs areshown in Table 2.
Figure 2.
The genotypes of miR-34a by sequencing. A: rs12128240-F, 1-C, 2-T, 3-CT; B: rs6577555-F, 1-C, 2-A, 3-AC; C: rs2666433-R, 1-A, 2-G, 3-AG.
Table 2.
Association between miRNA SNPs and presence of diabetes mellitus and diabetic nephropathy
| SNP (gene) | Genotypes/allele | CON number (%) | DM number (%) | DN number (%) | DM vs CON | DN vs CON | ||||
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| P | P a | ORa (95% CI) | P | P a | ORa (95% CI) | |||||
| rs12128240 | CC | 74 (63.2) | 86 (66.2) | 18 (60) | 0.633 | - | 0.743 | - | ||
| CT | 40 (34.2) | 39 (30) | 10 (33.3) | 0.481 | 0.226 | 0.665 (0.344-1.287) | 0.930 | 0.068 | 0.035 (0.001-1.286) | |
| TT | 3 (2.6) | 5 (3.8) | 2 (6.7) | 0.725 | 0.630 | 1.560 (0.256-9.487) | 0.269 | 0.063 | 0.034 (0.001-1.205) | |
| T | 46 (19.7) | 49 (18.8) | 14 (23.3) | 0.819 | 0.437 | 0.642 (0.210-1.964) | 0.529 | 0.426 | 1.550 (0.526-4.564) | |
| rs6577555 | AA | 12 (10.3) | 14 (10.8) | 3 (10) | 0.896 | - | 0.967 | - | ||
| AC | 42 (35.9) | 46 (35.4) | 9 (30) | 0.933 | 0.520 | 0.519 (0.071-3.814) | 0.545 | 0.508 | 0.520 (0.075-3.617) | |
| CC | 63 (53.8) | 70 (53.8) | 18 (60) | 1 | 0.695 | 0.685 (0.103-4.551) | 0.545 | 0.959 | 1.047 (0.177-6.214) | |
| C | 168 (71.8) | 186 (71.5) | 45 (75) | 0.950 | 0.858 | 0.917 (0.355-2.369) | 0.620 | 0.288 | 0.023 (0.000-23.916) | |
| rs2666433 | AA | 5 (4.3) | 12 (9.2) | 2 (6.7) | 0.124 | - | 0.583 | - | ||
| AG | 43 (36.7) | 63 (48.5) | 8 (26.7) | 0.063 | 0.940 | 0.902 (0.062-13.114) | 0.301 | 0.991 | 1.015 (0.086-12.047) | |
| GG | 69 (59.0) | 55 (42.3) | 20 (66.6) | <0.05 | 0.484 | 0.381 (0.026-5.679) | 0.442 | 0.642 | 0.704 (0.161-3.086) | |
| G | 181 (78.4) | 173 (66.5) | 48 (80) | <0.05 | 0.274 | 0.586 (0.225-1.527) | 0.659 | 0.778 | 0.852 (0.279-2.601) | |
ORs and P-values were adjusted for age, FBG, 2 h PG in a logistic regression model.
Associations between SNPs and subjects
To further analyze the associations of the three SNPs with T2DM and DN, we compared the control group with DM and DN independently. As shown in Table 2, the distribution of GG genotypes of rs2666433 was significantly different between the DM group and control group (P=0.009). Furthermore, the G allele frequency of rs2666433 was also significantly higher in DM group than that in control group (P=0.008). However, there were no statistical differences in the frequencies of rs12128240 and rs6577555 in the control compared with DM and control compared with DN regardless of adjusting for age, FBG, 2 h PG or not. Therefore, these results show that rs2666433 of miR-34a may play an important role in T2DM.
Univariate analysis with additive and recessive models for rs2666433 polymorphisms showed an association with DM (P=0.023; P=0.009 respectively). Also, a strong association was found for rs2666433 and DN in an additive model (P=0.001). After adjustment for clinical covariates, the GG genotype was still significantly different in T2DM for the recessive model (OR 2.297; CI 1.031-5.121; P=0.042) by multivariate analysis. But there was no association between rs2666433 and DN after adjustment for clinical covariates. The lack of association might be explained by the influence of confounding variables. Therefore, our data show the variant GG genotype of miR-34a rs2666433 was positively associated with significantly increased risk of T2DM. Details areshown in Table 3.
Table 3.
Association of SNPs with DM and DN
| SNP (gene) | Model | DM case versus control | DN case versus control | ||
|
|
|
||||
| Univariate | X2 | P value | X2 | P value | |
|
| |||||
| rs12128240 | Additive | 0.730 | 0.694 | 1.227 | 0.541 |
| Dominant | 0.228 | 0.633 | 0.108 | 0.743 | |
| Recessive | 0.323 | 0.570 | 1.223 | 0.269 | |
| rs6577555 | Additive | 0.020 | 0.990 | 0.405 | 0.817 |
| Dominant | 0.017 | 0.896 | 0.002 | 0.967 | |
| Recessive | 0.000 | 1.000 | 0.366 | 0.545 | |
| rs2666433 | Additive | 7.573 | 0.023 | 15.058 | 0.001 |
| Dominant | 2.361 | 0.124 | 0.302 | 0.583 | |
| Recessive | 6.842 | 0.009 | 0.591 | 0.442 | |
|
| |||||
| Multivariate | ORa (95% CI) | P a | ORa (95% CI) | P a | |
|
| |||||
| rs12128240 | Additive | 1.560 (0.256-9.487) | 0.630 | 0.034 (0.001-1.205) | 0.068 |
| Dominant | 0.894 (0.393-2.033) | 0.789 | 0.761 (0.239-2.426) | 0.644 | |
| Recessive | 0.087 (0.010-0.742) | 0.026 | 6.252 (0.230-169.644) | 0.276 | |
| rs6577555 | Additive | 0.685 (0.103-4.551) | 0.695 | 1.047 (0.177-6.214) | 0.959 |
| Dominant | 0.989 (0.450-2.171) | 0.977 | 1.219 (0.218-6.806) | 0.822 | |
| Recessive | 0.853 (0.226-3.218) | 0.815 | 1.664 (0.516-5.359) | 0.394 | |
| rs2666433 | Additive | 0.381 (0.026-5.679) | 0.484 | 0.704 (0.161-3.086) | 0.642 |
| Dominant | 0.550 (0.110-2.756) | 0.467 | 0.391 (0.043-3.555) | 0.404 | |
| Recessive | 2.297 (1.031-5.121) | 0.042 | 0.891 (0.270-2.936) | 0.849 | |
Univariate and multivariate analysis for additive, dominant, and recessive models are presented.
Adjusted for age, FBG, 2 h PG. P<0.05 in bold.
Discussion
From previous miRNA expression profiling studies, sixteen up-regulated and eight down-regulated miRNAs were found in T2DM by meta-analysis. Also, miR-34a was one of the up-regulated miRNAs, and we focused on miR-34a because it was the only one related to T2DM and DN. MiR-34a is involved in several biological pathways, including diabetes mellitus. In non-alcoholic fatty liver disease (NAFLD), miR-34a caninhibit very low-density lipoprotein secretion and promote liver steatosis and hypolipidemia in a HNF 4α-dependent manner [26]. Knockdown of miR-34a can reduce apoptosis in the β-cell line MIN6 [27] and protect β cells from cytokine-induced cell death [28]. Kong L et al. explored the significance of miR-34a in the pathogenesis of T2DM [16] and the expression of miR-34a was increased in islets of the non-obese diabetic mice during the development of pre-diabetic insulin [29]. These discoveries suggested that miR-34a can beinvolved in the progression of diabetes mellitus. Furthermore, by directly targeting GAS1, down-regulation of miR-34a inhibited mesangial proliferation and glomerular hypertrophy in early DN [29]. Furthermore, our real-time PCR results showed that miR-34a was over-expressed in T2DM and DN patients. Therefore, thesedata suggested that miR-34a might play a critical role in the development of T2DM and DN.
Vast research has revealed SNPs within miRNAs are associated with the risk of various diseases, including T2DM and DN. But the SNPs within in miR-34a relating with T2DM or DN were not known. In our study, we found that the SNP rs2666433, which located within the regulation region of the hsa-miR-34a, presented a tight relationship with T2DM. The GG genotype of rs2666433 was found to be significantly associated with T2DM in univariate and multivariate analysis, which might strongly indicate the significant association of GG genotype with the risk of T2DM. Although, rs2666433 only showed a significantly association with DN by univariate analysis, and there was no relationship between rs2666433 and DN after adjusting by the age, FBG and 2 h PG by multivariate analysis. Thesedata might suggest the contribution of the environmental factors in the development of DN. Therefore, our results imply that miR-34a rs2666433 variants may affect the incidence of T2DM.
Similar to most of the association studies, there are some potential limitations of our study. First, it will benecessary to enlarge the size of studied groups, especially for DN group in this study. Although our study has considered some confounding variables, some weak elements and associations may have been missed. Furthermore, our study was only processed in the Han ethnic population and the polymorphism of DM and DN might differ between ethnic populations. Additional studies with more samples in other populations should be conducted.
In conclusion, our results suggest that miR-34a is over-expressed in both T2DM and DN patients, and indicate that rs2666433 A/G in miR-34a is associated with an increased risk of T2DM.
Acknowledgements
This study was supported by the National Natural Science Foundation of China (81570747).
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Prentki M, Nolan CJ. Islet beta cell failure in type 2 diabetes. J Clin Invest. 2006;116:1802–1812. doi: 10.1172/JCI29103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang W, Lu J, Weng J, Jia W, Ji L, Xiao J, Shan Z, Liu J, Tian H, Ji Q, Zhu D, Ge J, Lin L, Chen L, Guo X, Zhao Z, Li Q, Zhou Z, Shan G, He J China National Diabetes and Metabolic Disorders Study Group. Prevalence of diabetes among men and women in China. N Engl J Med. 2010;362:1090–1101. doi: 10.1056/NEJMoa0908292. [DOI] [PubMed] [Google Scholar]
- 3.Wild S, Roglic G, Green A, Sicree R, King H. Global prevalence of diabetes: estimates for the year 2000 and projections for 2030. Diabetes Care. 2004;27:1047–1053. doi: 10.2337/diacare.27.5.1047. [DOI] [PubMed] [Google Scholar]
- 4.Reutens AT, Atkins RC. Epidemiology of diabetic nephropathy. Contrib Nephrol. 2011;170:1–7. doi: 10.1159/000324934. [DOI] [PubMed] [Google Scholar]
- 5.Yu G, Xiao Q, Ma XP, Chen X, Shi Z, Zhang LY, Chen H, Zhang P, Ding DL, Huang HX, Saiyin H, Chen TY, Lu PX, Wang NJ, Yu H, Sun J, Conran C, Zheng SL, Xu J, Yu L, Jiang DK. miR-492G>C polymorphism (rs2289030) is associated with overall survival of hepatocellular carcinoma patients. Tumour Biol. 2016;37:8961–8972. doi: 10.1007/s13277-015-4752-9. [DOI] [PubMed] [Google Scholar]
- 6.Wang S, Lv C, Jin H, Xu M, Kang M, Chu H, Tong N, Wu D, Zhu H, Gong W, Zhao Q, Tao G, Zhou J, Zhang Z, Wang M. A common genetic variation in the promoter of miR-107 is associated with gastric adenocarcinoma susceptibility and survival. Mutat Res. 2014;769:35–41. doi: 10.1016/j.mrfmmm.2014.07.002. [DOI] [PubMed] [Google Scholar]
- 7.Wang X, Sundquist J, Zoller B, Memon AA, Palmer K, Sundquist K, Bennet L. Determination of 14 circulating microRNAs in Swedes and Iraqis with and without diabetes mellitus type 2. PLoS One. 2014;9:e86792. doi: 10.1371/journal.pone.0086792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spinetti G, Fortunato O, Caporali A, Shantikumar S, Marchetti M, Meloni M, Descamps B, Floris I, Sangalli E, Vono R, Faglia E, Specchia C, Pintus G, Madeddu P, Emanueli C. MicroRNA-15a and microRNA-16 impair human circulating proangiogenic cell functions and are increased in the proangiogenic cells and serum of patients with critical limb ischemia. Circ Res. 2013;112:335–346. doi: 10.1161/CIRCRESAHA.111.300418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zampetaki A, Kiechl S, Drozdov I, Willeit P, Mayr U, Prokopi M, Mayr A, Weger S, Oberhollenzer F, Bonora E, Shah A, Willeit J, Mayr M. Plasma microRNA profiling reveals loss of endothelial miR-126 and other microRNAs in type 2 diabetes. Circ Res. 2010;107:810–817. doi: 10.1161/CIRCRESAHA.110.226357. [DOI] [PubMed] [Google Scholar]
- 10.Zhang T, Lv C, Li L, Chen S, Liu S, Wang C, Su B. Plasma miR-126 is a potential biomarker for early prediction of type 2 diabetes mellitus in susceptible individuals. Biomed Res Int. 2013;2013:761617. doi: 10.1155/2013/761617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Balasubramanyam M, Aravind S, Gokulakrishnan K, Prabu P, Sathishkumar C, Ranjani H, Mohan V. Impaired miR-146a expression links subclinical inflammation and insulin resistance in Type 2 diabetes. Mol Cell Biochem. 2011;351:197–205. doi: 10.1007/s11010-011-0727-3. [DOI] [PubMed] [Google Scholar]
- 12.Corral-Fernandez NE, Salgado-Bustamante M, Martinez-Leija ME, Cortez-Espinosa N, Garcia-Hernandez MH, Reynaga-Hernandez E, Quezada-Calvillo R, Portales-Perez DP. Dysregulated miR-155 expression in peripheral blood mononuclear cells from patients with type 2 diabetes. Exp Clin Endocrinol Diabetes. 2013;121:347–353. doi: 10.1055/s-0033-1341516. [DOI] [PubMed] [Google Scholar]
- 13.Karolina DS, Armugam A, Tavintharan S, Wong MT, Lim SC, Sum CF, Jeyaseelan K. MicroRNA 144 impairs insulin signaling by inhibiting the expression of insulin receptor substrate 1 in type 2 diabetes mellitus. PLoS One. 2011;6:e22839. doi: 10.1371/journal.pone.0022839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karolina DS, Tavintharan S, Armugam A, Sepramaniam S, Pek SL, Wong MT, Lim SC, Sum CF, Jeyaseelan K. Circulating miRNA profiles in patients with metabolic syndrome. J Clin Endocrinol Metab. 2012;97:E2271–2276. doi: 10.1210/jc.2012-1996. [DOI] [PubMed] [Google Scholar]
- 15.Kong L, Han WX, Jiang XY, Jin XU, Zhang X, Qiu LI, Guan QB, Gao L, Zhao JJ. Expression and clinical significance of peripheral miR-34a during the onset of type 2 diabetes. Journal of Shandong University. 2010;5:368–373. [Google Scholar]
- 16.Kong L, Zhu J, Han W, Jiang X, Xu M, Zhao Y, Dong Q, Pang Z, Guan Q, Gao L, Zhao J, Zhao L. Significance of serum microRNAs in pre-diabetes and newly diagnosed type 2 diabetes: a clinical study. Acta Diabetol. 2011;48:61–69. doi: 10.1007/s00592-010-0226-0. [DOI] [PubMed] [Google Scholar]
- 17.Meng S, Cao JT, Zhang B, Zhou Q, Shen CX, Wang CQ. Downregulation of microRNA-126 in endothelial progenitor cells from diabetes patients, impairs their functional properties, via target gene Spred-1. J Mol Cell Cardiol. 2012;53:64–72. doi: 10.1016/j.yjmcc.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 18.Ortega FJ, Mercader JM, Moreno-Navarrete JM, Rovira O, Guerra E, Esteve E, Xifra G, Martinez C, Ricart W, Rieusset J, Rome S, Karczewska-Kupczewska M, Straczkowski M, Fernandez-Real JM. Profiling of circulating microRNAs reveals common microRNAs linked to type 2 diabetes that change with insulin sensitization. Diabetes Care. 2014;37:1375–1383. doi: 10.2337/dc13-1847. [DOI] [PubMed] [Google Scholar]
- 19.Pescador N, Perez-Barba M, Ibarra JM, Corbaton A, Martinez-Larrad MT, Serrano-Rios M. Serum circulating microRNA profiling for identification of potential type 2 diabetes and obesity biomarkers. PLoS One. 2013;8:e77251. doi: 10.1371/journal.pone.0077251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rong Y, Bao W, Shan Z, Liu J, Yu X, Xia S, Gao H, Wang X, Yao P, Hu FB, Liu L. Increased microRNA-146a levels in plasma of patients with newly diagnosed type 2 diabetes mellitus. PLoS One. 2013;8:e73272. doi: 10.1371/journal.pone.0073272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xavier DJ, Takahashi P, Evangelista AF, Foss-Freitas MC, Foss MC, Donadi EA, Passos GA, Sakamoto-Hojo ET. Assessment of DNA damage and mRNA/miRNA transcriptional expression profiles in hyperglycemic versus nonhyperglycemic patients with type 2 diabetes mellitus. Mutat Res. 2015;776:98–110. doi: 10.1016/j.mrfmmm.2015.01.016. [DOI] [PubMed] [Google Scholar]
- 22.Santovito D, De Nardis V, Marcantonio P, Mandolini C, Paganelli C, Vitale E, Buttitta F, Bucci M, Mezzetti A, Consoli A, Cipollone F. Plasma exosome microRNA profiling unravels a new potential modulator of adiponectin pathway in diabetes: effect of glycemic control. J Clin Endocrinol Metab. 2014;99:E1681–1685. doi: 10.1210/jc.2013-3843. [DOI] [PubMed] [Google Scholar]
- 23.Zhu H, Leung SW. Identification of microRNA biomarkers in type 2 diabetes: a meta-analysis of controlled profiling studies. Diabetologia. 2015;58:900–911. doi: 10.1007/s00125-015-3510-2. [DOI] [PubMed] [Google Scholar]
- 24.Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, Feldmann G, Yamakuchi M, Ferlito M, Lowenstein CJ, Arking DE, Beer MA, Maitra A, Mendell JT. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. 2007;26:745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang L, He S, Guo S, Xie W, Xin R, Yu H, Yang F, Qiu J, Zhang D, Zhou S, Zhang K. Downregulation of miR-34a alleviates mesangial proliferation in vitro and glomerular hypertrophy in early diabetic nephropathy mice by targeting GAS1. J Diabetes Complications. 2014;28:259–264. doi: 10.1016/j.jdiacomp.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu Y, Zalzala M, Xu J, Li Y, Yin L, Zhang Y. A metabolic stress-inducible miR-34a-HNF4alpha pathway regulates lipid and lipoprotein metabolism. Nat Commun. 2015;6:7466. doi: 10.1038/ncomms8466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roggli E, Britan A, Gattesco S, Lin-Marq N, Abderrahmani A, Meda P, Regazzi R. Involvement of microRNAs in the cytotoxic effects exerted by proinflammatory cytokines on pancreatic beta-cells. Diabetes. 2010;59:978–986. doi: 10.2337/db09-0881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Backe MB, Novotny GW, Christensen DP, Grunnet LG, Mandrup-Poulsen T. Altering beta-cell number through stable alteration of miR-21 and miR-34a expression. Islets. 2014;6:e27754. doi: 10.4161/isl.27754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rottiers V, Naar AM. MicroRNAs in metabolism and metabolic disorders. Nat Rev Mol Cell Biol. 2012;13:239–250. doi: 10.1038/nrm3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.