Abstract
Background: Many studies have shown that differentially expressed circular RNA (circRNA) in plasma can serve as biomarkers in non-invasive detection of cancers during screening. However, the clinical significance of plasma circRNA in the diagnosis of colorectal cancer (CRC) is still not clear. Therefore, we examined expression of hsa_circ_0007534 in plasma to verify whether it can be utilized to diagnose and monitor CRC in routine clinical practice. Methods: 112 CRC patients and 46 healthy controls were recruited to participate in our study. The levels of hsa_circ_0007534 in plasma samples and tumor tissues were identified by real-time quantitative polymerase chain reaction (RT-qPCR). The diagnostic value was evaluated using receiver operating characteristics (ROC) curves and the area under the ROC curves (AUC). The Kaplan-Meier survival curve was used to evaluate whether the expression level of hsa_circ_0007534 was associated with overall survival rate. Results: Compared with the healthy control group, hsa_circ_0007534 expression was significantly increased in plasma from CRC patients. Increased hsa_circ_0007534 expression level in plasma was associated with progression of clinical classifications, metastatic phenotype, and poor differentiation in CRC patients. ROC analysis showed that hsa_circ_0007534 could distinguish CRC patients from healthy controls with high AUC (0.780), sensitivity (0.92) and specificity (0.522). Finally, high hsa_circ_0007534 expression was positively correlated with poor prognosis in CRC patients. Conclusion: All of the results suggest that hsa_circ_0007534 may be a potential cancer marker of patients with CRC and may associate with poor prognosis.
Keywords: Colorectal cancer, circular RNA, biomarker, diagnosis, prognosis
Introduction
Colorectal cancer (CRC) is the fourth most common malignant gastrointestinal cancer and accounts for approximately 9% of all cancer cases. The incidence of CRC is more than 376,000 and corresponded to about 191,000 deaths in China by 2015 [1]. In the past few decades, colonoscopy has been the dominant and preferred screening modality for CRC worldwide, and surgical resection is the ultimate therapeutic treatment [2]. However, current methods of screening have been far from adequate in building a complete early-warning system and prognostic value for CRC patients. Previous studies suggested that early diagnosis and treatment have improved patient outcomes, which largely depended on the stage of the tumor at the time of detection, and for CRC patients with advanced stage disease, prognosis remained quite poor [3-5]. In addition, prognostic markers can facilitate stratification of CRC patients in that it is beneficial to monitor tumor recurrence and metastasis [6]. Therefore, exploring novel and specific biomarkers is an imperative need for improving the current diagnostic and prognostic tools for CRC patients.
Circular RNA (circRNA) is a class of endogenous non-coding RNA and exists ubiquitously in the cytoplasm of eukaryotic cells [7,8]. CircRNA is generally formed by back-splicing with high stability, covalently closed continuous loop, without 5’ to 3’ polarity and polyadenylated tail, endowing them with a stable structure to counteract RNA exonucleolytic digestion [9,10]. CircRNAs perform a wide variety of biological functions in eukaryotic cells primarily through competing endogenous RNAs (ceRNAs) or miRNA sponges [11]. Recently, emerging evidence has revealed that circRNAs are stably present in body fluid and can serve as novel non-invasive biomarkers in various diseases, including arthritis [12], pre-eclampsia [13] and malignancies [14,15]. It is established that abnormally expressed circRNAs are strongly linked to the progression and development of CRC [16-18]. Hsa_circ_0003906 is dramatically downregulated in CRC tumor tissues and significantly correlates with lymphatic metastasis and poor differentiation, suggesting that hsa_circ_0003906 may serve as a potential biomarker for the diagnosis of CRC [19]. To the best of our knowledge, there is little literature focused on the blood-based non-invasive detection of CRC.
Hsa_circ_0007534 is located in chr17: 61869771-61877977, the spliced sequence length is 400 nt, and its associated-gene symbol is DDX42 (DEAD-box helicase 42; circBase database, http://www.circbase.org/). Based on our previous results, we first verified that hsa_circ_0007534 was significantly upregulated in CRC cell lines and tumor tissues. Then, we hypothesized that hsa_circ_0007534 might be released into circulation during CRC initiation and could be utilized to detect and monitor CRC. To validate the hypothesis, first, the expression levels of hsa_circ_0007534 in plasma needed to be addressed; second, we performed a correlation analysis between plasma and tumor tissues hsa_circ_0007534 level to confirm the accuracy of a blood-based tumor marker; third, the receiver operating characteristics (ROC) curves and the area under the ROC curves (AUC) were calculated to investigate the clinical diagnostic significance of hsa_circ_0007534; fourth, Kaplan-Meier survival curve was used to evaluate whether the expression level of hsa_circ_0007534 was associated with overall survival rate.
Material and methods
Patients and specimens
112 CRC patients and 46 healthy controls were recruited from the Cancer Hospital of China Medical University (Shenyang, China). 112 pairs of tumor tissue and matched adjacent non-tumorous tissues were collected from CRC patients who had undergone surgery at the Cancer Hospital of China Medical University (Shenyang, China) between January 2014 and June 2016. None of the patients were subjected to preoperative radiotherapy or chemotherapy and diagnosed with CRC based on histopathological evaluation. All collected tissue samples were immediately stored in liquid nitrogen. 10 mL of blood samples from preoperative and postoperative CRC patients were collected with ethylenediaminetetraacetic acid (EDTA)-containing tubes (Becton, Dickinson and Company), plasma samples were separated immediately, as described previously [20], and stored at -80°C in an ultra-low temperature refrigerator. Human samples were obtained with written informed consent from all of the patients. The study was approved by the Ethics Committee of the Department of Colorectal Surgery, Cancer Hospital of China Medical University (Shenyang, China).
Total RNA extraction
RNA extraction from tumor tissue, non-tumorous tissue, and plasma was performed using TRIzol Reagent (Invitrogen, Carlsbad, CA, USA) and preserved at -80°C until the time of usage. RNA concentration was measured using NanoDrop ND-2000 (Thermo Fisher Scientific, Wilmington, DE, USA).
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Moloney Murine Leukemia Virus reverse transcriptase (Promega Corporation, Madison, WI, USA) was used to synthesize cDNA. Divergent primers were designed to amplify the head-to-tail splicing of circRNA using an ABI7300 System (Applied Biosystems, Foster City, CA, USA) with SYBR Select Master Mix (Applied Biosystems). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was utilized to normalize the expression of the circRNAs. The PCR primers were used in this study as follows: hsa_circ_0007534, forward 5’-GTGACGGAAATCCAATTGCACC-3’ and reverse 5’-ATGGAATTGCTGGCGAGTTG-3’; GAPDH, forward 5’-GCACCGTCAAGCTGAGAAC-3’ and reverse 5’-TGGTGAAGACGCCAGTGGA-3’. The relative expression levels of hsa_circ_0007534 were calculated using the 2-ΔΔCt method [21].
Statistical analysis
Data were presented as the mean ± standard deviation (SD) for each group. All statistical analyses were performed using PRISM version 7.0 (GraphPad Software, Inc., La Jolla, CA, USA) and SPSS version 17.0 (SPSS, Inc., Chicago, IL, USA). Student’s t-test was used to analyze the two-group differences. Inter-group differences were analyzed by one-way analysis of variance, followed by a post hoc Tukey’s test for multiple comparisons. Pearson χ2 tests were used to evaluate differences in the clinical characteristics between the two groups. Receiver operating characteristic (ROC) curves and the area under the ROC curve (AUC) were used to assess the ability of using plasma hsa_circ_0007534 as a diagnostic tool for CRC patients. Spearman’s Rank analysis was used to identify the correlation of hsa_circ_0007534 levels between plasma and tumor tissues. A Bland-Altman plot (difference plot) was used to analyze the agreement of hsa_circ_0007534 levels between plasma and tumor tissues. Survival analysis was performed using the Kaplan-Meier method with the log-rank test applied for comparison. P < 0.05 was considered to indicate a statistically significant difference.
Results
Expression of hsa_circ_0007534 in tumor tissues and plasma from CRC patients
RT-qPCR analysis of tumor tissue and non-tumorous adjacent tissue from 112 CRC patients showed that 78 of these cases (69.6%) exhibited upregulation of hsa_circ_0007534 levels in tumor tissues (Figure 1A). To validate whether differentially expressed hsa_circ_0007534 in plasma could serve as a potential biomarker for CRC detection, RT-qPCR was also performed on plasma from 112 CRC patients and 46 healthy controls. Consistent with the results using tumor tissue, we found that the levels of hsa_circ_0007534 in the plasma from CRC patients were significantly increased compared with those of the healthy control (Figure 1B). Increased hsa_circ_0007534 expression level in plasma was associated with the progression of T classifications (Figure 1C), N classifications (Figure 1D), metastatic phenotype (Figure 1E) and poor differentiation (Figure 1F) in CRC patients. These findings suggest a link between upregulation of hsa_circ_0007534 and the aggressive characteristics of CRC.
Figure 1.
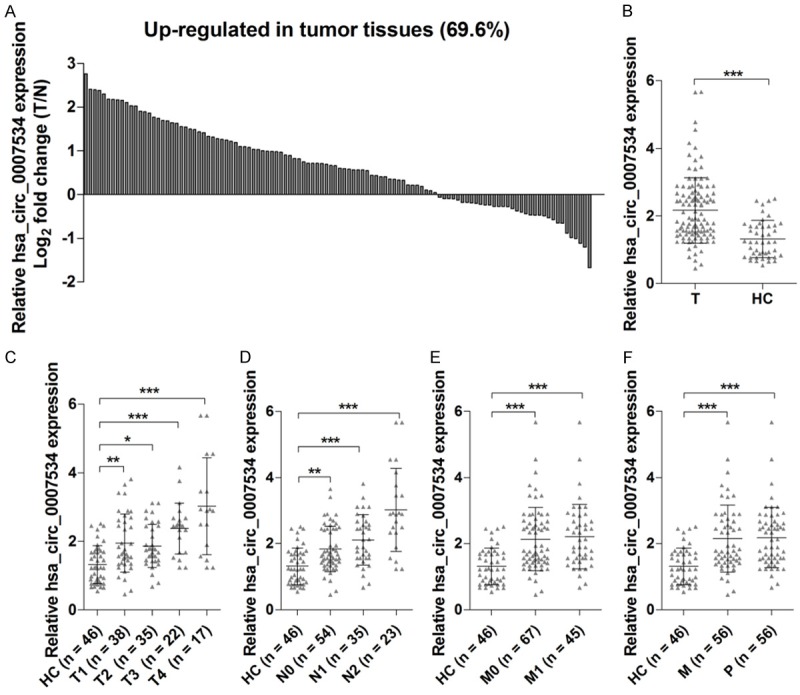
Expression of hsa_circ_0007534 was upregulated in tumor tissues from CRC patients. Hsa_circ_0007534 expression was measured in 112 pairs of tumor tissue and matched adjacent non-tumorous tissues from CRC patients, and the fold change was calculated (A). Hsa_circ_0007534 expression was measured in plasma from 112 CRC patients and 46 healthy controls (B). Hsa_circ_0007534 expression levels were measured with T classifications (C), N classifications (D), metastatic phenotype (E) and poor differentiation (F) in CRC patients. T, tumor tissues; N, non-tumorous tissues; HC, healthy control; M, moderate; P, poor. *P < 0.05, **P < 0.01, ***P < 0.001.
Correlation between clinical parameters and plasma hsa_circ_0007534
To verify whether hsa_circ_0007534 can serve as a biomarker for predicting clinical outcomes in CRC patients, we performed the correlation analysis between clinical parameters and plasma hsa_circ_0007534 levels. 112 CRC patients were divided into 2 groups: a high expression group (n = 72) and a low expression group (n = 40). The levels of hsa_circ_0007534 in plasma had no significant relationship to age and gender (Table 1). In addition, the high hsa_circ_0007534 expression group exhibited a significantly higher incidence of poor differentiation, distant metastasis, and clinical classifications than the low hsa_circ_0007534 expression group (Table 1). Previous studies have reported that both poor differentiation and metastatic phenotype are the independent risk factors for CRC patient survival [19]. Our results indicate that hsa_circ_0007534 as an oncogene can promote CRC progression and lead to a poor prognosis.
Table 1.
Correlation between hsa_circ_0007534 levels and clinical parameters
| Characteristics | Hsa_circ_0007534 expression | P-value | |
|---|---|---|---|
|
| |||
| Low | High | ||
| Age | 0.734 | ||
| ≤ 60 | 33 | 17 | |
| > 60 | 39 | 23 | |
| Gender | 0.134 | ||
| Male | 40 | 28 | |
| Female | 32 | 12 | |
| T classification | < 0.001 | ||
| I-II | 58 | 15 | |
| III-IV | 14 | 25 | |
| N classification | 0.037 | ||
| 0 | 40 | 14 | |
| 1-2 | 32 | 26 | |
| Distant metastasis | 0.047 | ||
| No | 48 | 19 | |
| Yes | 24 | 21 | |
| Differentiation | < 0.001 | ||
| Well-moderate | 50 | 6 | |
| Poor | 22 | 34 | |
Correlation of hsa_circ_0007534 expression between plasma and tumor tissues
To further confirm the reliability of a blood-based marker for CRC diagnosis, we performed a correlation analysis of hsa_circ_0007534 expression between plasma samples and tumor tissues from CRC patients. Hsa_circ_0007534 expression levels obtained from plasma samples and tumor tissues were strongly correlated (r = 0.505, P < 0.001; mean differences = -0.551 ± 0.913; Figure 2A and 2B). To investigate whether tumor resection can regulate plasma hsa_circ_0007534 levels, RT-qPCR was carried out to investigate the expression of hsa_circ_0007534 in plasma from CRC patients before tumor resection and 14 days after tumor resection. As expected, the data showed that plasma levels of hsa_circ_0007534 significantly declined 14 days after tumor resection compared with levels before surgery (Figure 2C). Furthermore, the stability of hsa_circ_0007534 in plasma was investigated and the results showed that hsa_circ_0007534 was relatively stable within nine hours at room temperature. However, with the extension of incubation time, the expression levels of hsa_circ_0007534 significantly decreased compared with the control group (Figure 2D). These results suggest that hsa_circ_0007534 is relatively stable in plasma samples and can effectively reflect the actual yield of hsa_circ_0007534 in tumor tissues. Therefore, this evidence strongly demonstrates that plasma-based hsa_circ_0007534 can serve as a tumor marker for CRC screening.
Figure 2.
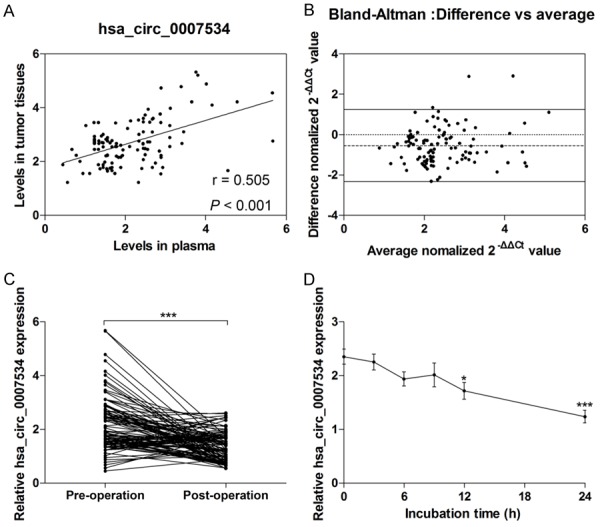
The correlation of hsa_circ_0007534 expression between plasma and tumor tissues. Spearman’s Rank analysis was used to identify the correlation of hsa_circ_0007534 levels between plasma and tumor tissues (A). Bland-Altman plots of the difference between plasma and tumor tissues were shown (B). Horizontal imaginary lines in the middle represent the mean difference, and upper and lower solid lines represent the limits of agreement (95% confidence intervals. Hsa_circ_0007534 expression levels were measured in the plasma from pre-operative and post-operative CRC patients (C). The expression of hsa_circ_0007534 was measured in plasma under room temperature with different exposure duration (D). *P < 0.05, ***P < 0.001.
The diagnostic value of hsa_circ_0007534 in CRC patients
To further estimate the diagnostic value of hsa_circ_0007534 in CRC patients, ROC curves and the area under the ROC curves (AUC) were performed in all of the participants, including 112 CRC patients and 46 healthy controls. The ROC curves showed that hsa_circ_0007534 was a strong distinguishing mark between the CRC patients and healthy controls, with an AUC of 0.780 (95% CI: 0.703-0.857; P < 0.001; Figure 3A and Table 2). The sensitivity and specificity were 0.920 and 0.522, respectively. The fundamental aim of this study was to diagnose CRC patients at the early stage. Therefore, the ROC curves were performed in different clinical stages. The AUC for stage I (Figure 3B), II (Figure 3C), III (Figure 3D) or IV (Figure 3E) was 0.721, 0.739, 0.871 or 0.890, respectively. Although hsa_circ_0007534 had the highest diagnostic value for CRC patients with stage IV, detecting plasma hsa_circ_0007534 might be helpful for differentiating stage I CRC patients from healthy control.
Figure 3.
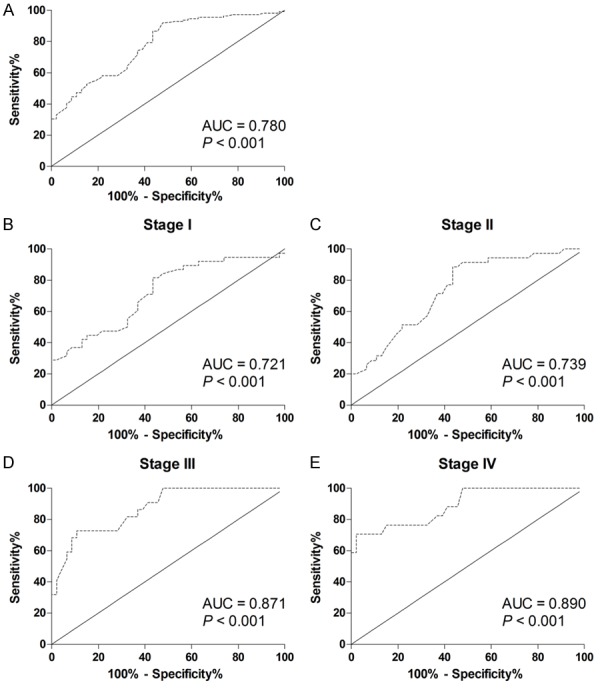
Evaluation of hsa_circ_0007534 for CRC detection. Receiver operating characteristics (ROC) curves were drawn with the data of plasma hsa_circ_0007534 levels from 112 CRC patients and 46 healthy controls (A). ROC curves were drawn based on stage I patients and 46 healthy controls (B). ROC curves were drawn based on stage II patients and 46 healthy controls (C). ROC curves were drawn based on stage III patients and 46 healthy controls (D). ROC curves were drawn based on stage IV patients and 46 healthy controls (E).
Table 2.
Clinical diagnostic significance of hsa_circ_0007534 in CRC patients
| AUC | P-value | 95% CI | Sensitivity | Specificity | Youden index | Cut-off | ||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| Lower | Upper | |||||||
| Stage I-IV | 0.780 | < 0.001 | 0.703 | 0.857 | 0.920 | 0.522 | 0.442 | 1.225 |
| Stage I | 0.721 | < 0.001 | 0.611 | 0.831 | 0.816 | 0.565 | 0.381 | 1.290 |
| Stage II | 0.739 | < 0.001 | 0.631 | 0.846 | 0.886 | 0.565 | 0.451 | 1.290 |
| Stage III | 0.871 | < 0.001 | 0.785 | 0.957 | 0.727 | 0.891 | 0.618 | 2.060 |
| Stage IV | 0.890 | < 0.001 | 0.797 | 0.983 | 0.706 | 0.978 | 0.684 | 2.470 |
The prognostic value of hsa_circ_0007534 in CRC patients
Kaplan-Meier analysis was performed to evaluate whether plasma hsa_circ_0007534 levels can predict a CRC prognosis. CRC patients were segregated into a hsa_circ_0007534 high expression group and a low expression group according to Log2FC ≥ 1. CRC patients with high hsa_circ_0007534 levels had a significantly poorer prognosis than that of patients with low hsa_circ_0007534 levels (P = 0.048; Figure 4).
Figure 4.
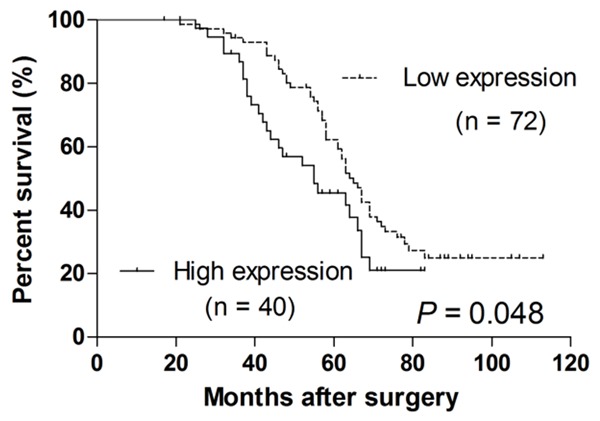
The prognostic value of hsa_circ_0007534 in CRC patients. Kaplan-Meier survival curve was used to evaluate whether hsa_circ_0007534 expression levels were associated with overall survival rate in patients with CRC.
Discussion
Accumulating evidence suggests that cell-free nucleic acids are detectable in plasma and serum of cancer patients and can serve as biomarkers for cancer diagnosis, including esophageal squamous cell carcinoma [22], bladder cancer [23], epithelial ovarian cancer [24] and lung cancer [25]. Although numerous studies have focused on DNA, mRNA, microRNAs, and long non-coding RNA as potential tumor markers for cancer screening [26], plasma circRNAs as diagnostic and prognostic markers for CRC have seldom been reported.
In our study, CRC-related circRNA was screened based on different expression profiling between CRC tumorous tissues and adjacent non-tumorous tissues than in our previous study, which was not published. Hsa_circ_0007534 was identified and significantly upregulated in CRC tumorous tissues, and then we further measured its expression level in plasma from CRC patients and healthy subjects. Our findings suggest that the levels of hsa_circ_0007534 are significantly upregulated in plasma from CRC patients compared with the healthy control group, indicating strong evidence that hsa_circ_0007534 can be detected in plasma and can serve as a diagnostic marker for CRC.
Blood-based nucleic acids are considered to be unstable, which is attributed to the high level of RNase activity in plasma. Increased plasma RNase activity has been confirmed in patients with malignancies [27]. To verify the stability of hsa_circ_0007534 in plasma from CRC patients, we performed an experiment in that plasma samples were stored at room temperature for different times. The results demonstrated that hsa_circ_0007534 was relatively stable existing at room temperature for nine hours. The precise mechanism of circRNAs resistance to endogenous RNase may be closely related to its closed continuous cyclic structure [14]. Therefore, hsa_circ_0007534 serving as a promising CRC biomarker is reasonable in routine clinical practice.
One of the major goals of the current study was to diagnose CRC patients at the earliest stage possible, which may increase success in clinical treatment and may be beneficial in improving overall survival [28]. Therefore, the diagnostic value of hsa_circ_0007534 in different clinical stages was investigated in all of subjects, including 112 CRC patients and 46 healthy controls. The ROC curves illustrated strong diagnostic value (AUC = 0.780; 95% CI: 0.703-0.857; P < 0.001) for CRC patients from all of the subjects. Interestingly, the diagnostic value of hsa_circ_0007534 could distinguish stage I patients from healthy control with an AUC of 0.721 (95% CI: 0.611-0.831; P < 0.001), which is similar in stage I-IV patients. In general, hsa_circ_0007534 provided high diagnostic power for the detection of CRC in patients and plasma hsa_circ_0007534 could serve as a promising tumor marker for the early diagnosis of CRC.
The distant metastasis and TNM stages are the most important parameters in the evaluation of the prognosis of patients with CRC [29]. Zhuo, et al. showed that the downregulation of circRNA0003906 levels significantly correlated with lymphatic metastasis and poor differentiation in CRC patients [19]. Moreover, Wang, et al. indicated that hsa_circ_001988 was significantly correlated with differentiation and perineural invasion in CRC patients [30]. In our study, we found that there was a significant relationship between high expression levels of hsa_circ_0007534 in plasma from CRC patients and clinical classifications, metastatic phenotype, poor differentiation and poor prognosis. However, the number of samples was limited, which could have affected our results.
The findings would be more interesting if the diagnostic and prognostic value of hsa_circ_0007534 were stablished in a larger sample size. Taken together, hsa_circ_0007534 may be potentially associated with the CRC pathogenesis and can serve as a potential biomarker for diagnosis and prognosis.
Acknowledgements
This study was supported by the National Natural Science Foundation of China (Grant No. 81402367), “Liaoning BaiQianWan Talents Program” (Grant No. [2017] No. B44), Liaoning Clinical Research Center for Colorectal Cancer (Grant No. 2015225005) and Clinical Capability Construction Project for Liaoning Provincial Hospitals (Grant No. LNCCC-D44-2015).
Disclosure of conflict of interest
None.
References
- 1.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 2.Sun Y, Shen S, Tang H, Xiang J, Peng Y, Tang A, Li N, Zhou W, Wang Z, Zhang D, Xiang B, Ge J, Li G, Wu M, Li X. miR-429 identified by dynamic transcriptome analysis as a new candidate biomarker for colorectal cancer prognosis. OMICS. 2014;18:54–64. doi: 10.1089/omi.2012.0132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu L, Li M, Wang M, Yan D, Feng G, An G. The expression of microRNA-375 in plasma and tissue is matched in human colorectal cancer. BMC Cancer. 2014;14:714. doi: 10.1186/1471-2407-14-714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Soerjomataram I, Thong MS, Ezzati M, Lamont EB, Nusselder WJ, van de Poll-Franse LV. Most colorectal cancer survivors live a large proportion of their remaining life in good health. Cancer Causes Control. 2012;23:1421–1428. doi: 10.1007/s10552-012-0010-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bosch LJ, Carvalho B, Fijneman RJ, Jimenez CR, Pinedo HM, van Engeland M, Meijer GA. Molecular tests for colorectal cancer screening. Clin Colorectal Cancer. 2011;10:8–23. doi: 10.3816/CCC.2011.n.002. [DOI] [PubMed] [Google Scholar]
- 6.Perez-Carbonell L, Sinicrope FA, Alberts SR, Oberg AL, Balaguer F, Castells A, Boland CR, Goel A. MiR-320e is a novel prognostic biomarker in colorectal cancer. Br J Cancer. 2015;113:83–90. doi: 10.1038/bjc.2015.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9:e1003777. doi: 10.1371/journal.pgen.1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, Loewer A, Ziebold U, Landthaler M, Kocks C, le Noble F, Rajewsky N. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 9.Vicens Q, Westhof E. Biogenesis of circular RNAs. Cell. 2014;159:13–14. doi: 10.1016/j.cell.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 10.Hentze MW, Preiss T. Circular RNAs: splicing’s enigma variations. EMBO J. 2013;32:923–925. doi: 10.1038/emboj.2013.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 12.Ouyang Q, Wu J, Jiang Z, Zhao J, Wang R, Lou A, Zhu D, Shi GP, Yang M. Microarray expression profile of circular RNAs in peripheral blood mononuclear cells from rheumatoid arthritis patients. Cell Physiol Biochem. 2017;42:651–659. doi: 10.1159/000477883. [DOI] [PubMed] [Google Scholar]
- 13.Zhang YG, Yang HL, Long Y, Li WL. Circular RNA in blood corpuscles combined with plasma protein factor for early prediction of preeclampsia. BJOG. 2016;123:2113–2118. doi: 10.1111/1471-0528.13897. [DOI] [PubMed] [Google Scholar]
- 14.Meng S, Zhou H, Feng Z, Xu Z, Tang Y, Li P, Wu M. CircRNA: functions and properties of a novel potential biomarker for cancer. Mol Cancer. 2017;16:94. doi: 10.1186/s12943-017-0663-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yao JT, Zhao SH, Liu QP, Lv MQ, Zhou DX, Liao ZJ, Nan KJ. Over-expression of circRNA_100876 in non-small cell lung cancer and its prognostic value. Pathol Res Pract. 2017;213:453–456. doi: 10.1016/j.prp.2017.02.011. [DOI] [PubMed] [Google Scholar]
- 16.Xie H, Ren X, Xin S, Lan X, Lu G, Lin Y, Yang S, Zeng Z, Liao W, Ding YQ, Liang L. Emerging roles of circRNA_001569 targeting miR-145 in the proliferation and invasion of colorectal cancer. Oncotarget. 2016;7:26680–26691. doi: 10.18632/oncotarget.8589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hsiao KY, Lin YC, Gupta SK, Chang N, Yen L, Sun HS, Tsai SJ. Noncoding effects of circular RNA CCDC66 promote colon cancer growth and metastasis. Cancer Res. 2017;77:2339–2350. doi: 10.1158/0008-5472.CAN-16-1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo JN, Li J, Zhu CL, Feng WT, Shao JX, Wan L, Huang MD, He JD. Comprehensive profile of differentially expressed circular RNAs reveals that hsa_circ_0000069 is upregulated and promotes cell proliferation, migration, and invasion in colorectal cancer. Onco Targets Ther. 2016;9:7451–7458. doi: 10.2147/OTT.S123220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhuo F, Lin H, Chen Z, Huang Z, Hu J. The expression profile and clinical significance of circRNA0003906 in colorectal cancer. Onco Targets Ther. 2017;10:5187–5193. doi: 10.2147/OTT.S147378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou X, Wen W, Shan X, Zhu W, Xu J, Guo R, Cheng W, Wang F, Qi LW, Chen Y, Huang Z, Wang T, Zhu D, Liu P, Shu Y. A six-microRNA panel in plasma was identified as a potential biomarker for lung adenocarcinoma diagnosis. Oncotarget. 2017;8:6513–6525. doi: 10.18632/oncotarget.14311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 22.Tong YS, Wang XW, Zhou XL, Liu ZH, Yang TX, Shi WH, Xie HW, Lv J, Wu QQ, Cao XF. Identification of the long non-coding RNA POU3F3 in plasma as a novel biomarker for diagnosis of esophageal squamous cell carcinoma. Mol Cancer. 2015;14:3. doi: 10.1186/1476-4598-14-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Du M, Shi D, Yuan L, Li P, Chu H, Qin C, Yin C, Zhang Z, Wang M. Circulating miR-497 and miR-663b in plasma are potential novel biomarkers for bladder cancer. Sci Rep. 2015;5:10437. doi: 10.1038/srep10437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zuberi M, Mir R, Das J, Ahmad I, Javid J, Yadav P, Masroor M, Ahmad S, Ray PC, Saxena A. Expression of serum miR-200a, miR-200b, and miR-200c as candidate biomarkers in epithelial ovarian cancer and their association with clinicopathological features. Clin Transl Oncol. 2015;17:779–787. doi: 10.1007/s12094-015-1303-1. [DOI] [PubMed] [Google Scholar]
- 25.Chen J, Wang R, Zhang K, Chen LB. Long non-coding RNAs in non-small cell lung cancer as biomarkers and therapeutic targets. J Cell Mol Med. 2014;18:2425–2436. doi: 10.1111/jcmm.12431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schwarzenbach H, Hoon DS, Pantel K. Cellfree nucleic acids as biomarkers in cancer patients. Nat Rev Cancer. 2011;11:426–437. doi: 10.1038/nrc3066. [DOI] [PubMed] [Google Scholar]
- 27.Tsui NB, Ng EK, Lo YM. Stability of endogenous and added RNA in blood specimens, serum, and plasma. Clin Chem. 2002;48:1647–1653. [PubMed] [Google Scholar]
- 28.Shah R, Jones E, Vidart V, Kuppen PJ, Conti JA, Francis NK. Biomarkers for early detection of colorectal cancer and polyps: systematic review. Cancer Epidemiol Biomarkers Prev. 2014;23:1712–1728. doi: 10.1158/1055-9965.EPI-14-0412. [DOI] [PubMed] [Google Scholar]
- 29.Shao Y, Li J, Lu R, Li T, Yang Y, Xiao B, Guo J. Global circular RNA expression profile of human gastric cancer and its clinical significance. Cancer Med. 2017;6:1173–1180. doi: 10.1002/cam4.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang X, Zhang Y, Huang L, Zhang J, Pan F, Li B, Yan Y, Jia B, Liu H, Li S, Zheng W. Decreased expression of hsa_circ_001988 in colorectal cancer and its clinical significances. Int J Clin Exp Pathol. 2015;8:16020–16025. [PMC free article] [PubMed] [Google Scholar]


