Abstract
Although chemotherapy has significantly improved the prognosis of patients with cervical cancer, many patients exhibit selective responses to chemotherapy. This study aimed to identify genes associated with the sensitivity of cervical cancer cells to paclitaxel. SiHa cells were transfected with lentiCRISPR (genome-scale CRISPR knockout library v.2) and then treated with paclitaxel or vehicle for 14 days. Subsequently, we screened for candidate genes whose loss resulted in sensitivity to paclitaxel. We obtained 374 candidates, some of which were consistent with those reported in previous studies, including ABCC9, IL37, EIF3C, AKT1S1, and several members of the PPP gene family (PPP1R7, PPP2R5B, PPP1R7, and PPP1R11). Importantly, our findings highlighted the newly identified genes that could provide novel insights into the mechanisms of paclitaxel sensitivity in cervical cancer. Cas9 technology provides a reliable method for the exploration of more effective combination chemotherapies for patients at the gene level.
Keywords: Paclitaxel sensitivity, cervical cancer, CRISPR-Cas9 technology
Introduction
Cervical cancer is the second most common cancer in women worldwide [1]. The primary therapeutic options include surgery, radiotherapy, and chemotherapy. However, these methods cannot always prevent metastasis and recurrence of cancer, thereby affecting the prognosis. One of the primary causes of treatment failure is poor chemotherapy sensitivity. Chemotherapy is also important in patients after surgery, particularly in those who show risk factors for tumor recurrence and metastasis [2,3]. Moreover, neoadjuvant chemotherapy has been applied to patients with locally advanced cervical cancer to improve surgical outcomes [4]. However, many patients exhibit selective responses to chemotherapy.
In patients with cervical cancer, paclitaxel (PTX) is a commonly used chemotherapeutic agent [5], and the mechanisms affecting PTX sensitivity in cervical cancer have been studied. Tissue culture studies have consistently shown that ABCC1 and ABCG2, belonging to the ABC family, are involved in reduce the sensitivity of multidrug chemotherapy, including that to PTX [6-9]. Zhang reported that nuclear localization of β-catenin is associated with chemotherapy sensitivity in human cervical squamous cell carcinoma [10], and several studies have described the roles of apoptosis-associated proteins (Bax, Bad, and the BCL family) in PTX therapeutic effect [11].
Although mechanisms mediating chemosensitivity in cervical cancer have been extensively studied, they are still poorly understood. Therefore, this study aimed to systematically screen genes related to PTX sensitivity using a genome-scale CRISPR-Cas9 screen in human SiHa cells.
Materials and methods
Cell culture
SiHa cells were purchased from American Type Culture Collection (ATCC; Manassas, VA, USA) and cultured in RPMI-1640 medium supplemented with 10% fetal bovine serum (FBS; Sigma, St. Louis, MO, USA) and 1% penicillin/streptomycin (HyClone, Logan, UT, USA). Cells were incubated at 37°C with 5% CO2. The cells were subjected to short tandem repeat classification recommended by ATCC.
LentiCRISPR construction
CRISPR-Pool KOUT products from GeneChem were prepared using the GeCKO plasmid library constructed by the Zhang Feng laboratory at the Broad Institute. We used the Zhang Feng (GeCKO v.2) plasmid library, which included 123,411 single-guide RNAs (sgRNAs) that targeted 19,050 protein-coding genes (six sgRNAs per gene), 1,864 microRNAs (four sgRNAs per microRNA), and approximately 1,000 nontargeting control sgRNAs.
Briefly, for each virus, the lentiviral plasmid and packaging constructs (pHelper 1.0 and pHelper 2.0) were cotransfected into HEK293T cells grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% FBS using a transfection reagent developed by GeneChem. The transfection mixture and medium were discarded after 6 h incubation, and 10 mL phosphate-buffered saline was added to wash the sample once. After 6 h, the medium was changed to D10 medium, i.e., DMEM (Hyclone, SH30022.01B Life Technologies) supplemented with 10% FBS (Shanghai Micronas Biochemicals Co., Ltd., Shanghai, China), followed by incubation in the presence of 5% CO2 at 37°C for 60 h. Next, 1% bovine serum albumin (Sigma) was added to improve virus stability.
Based on the cell state, supernatant from 293T cells was collected 24 and 48 h after transfection and stored at 4°C. The fluid was then suction-filtered and passed through a tangential flow filtration system. The sample was further purified using an AKTA ion-exchange chromatography system, packaged into ultrafiltration centrifuge tubes, and centrifuged at 4°C and 5500 rpm. The duration of each centrifuge cycle was adjusted according to the rate of vom Virus concentration, until the target volume was achieved. Through these methods, we designed a single lentiviral vector to deliver Cas9 sgRNA and a puromycin selection marker to the target cells (lentiCRISPR).
Cytotoxicity assay
We used a Cell Counting Kit-8 (CCK-8) assay to assess cell viability. Cells were seeded in 96-well plates at a density of 2 × 103 cells/well. Next, 10 μL of CCK-8 solution was added into each well at the indicated time. After incubation for 4 h at 37°C in a humidified atmosphere containing 5% CO2, the optical density (OD) value was recorded at 450 nm using a microplate reader (Bio-Rad, Richmond, VA, USA). The experiment was performed in triplicate.
Cell count assays
Cells were plated at a density of 20% and then treated with the target drugs. When the control cells reached a density of approximately 80%, cell counting was performed using a Cell counter (Nexcelom, Bioscience LLC, Lawrence, MA, USA). Cells were counted at least four times for each treatment at each time.
Statistical analysis
Calculation of the half-maximal inhibitory concentration (IC50) was performed using GraphPadPrism5 Software, and gene ontology (GO) terms and pathway enrichment analysis were evaluated using Fisher’s exact tests (P < 0.05). Gene screening was performed based on the fold change (FC) and the Bayes factor.
Results
Multiplicity of infection
In the pre-experiment, the lentivirus multiplicity of infection (MOI) was measured and determined by the functional MOI value (Table 1; Figure 1). When the MOI was 0.5 or 1, both the groups showed functional MOIs close to 0.4. Thus, to ensure that cells were only infected with one virus, we used an MOI of 0.5 for the actual experiment.
Table 1.
Functional MOI values for different lentivirus MOIs
| MOI | CCK8 OD450 (no puromycin) | CCK8 OD450 (plus puromycin) | Functional MOI |
|---|---|---|---|
| 0 | 3.656 | 0.0081 | 2.2% |
| 0.125 | 3.632 | 0.349 | 9.5% |
| 0.25 | 3.617 | 0.728 | 19.9% |
| 0.5 | 3.660 | 1.197 | 32.7% |
| 1 | 3.661 | 1.684 | 46.1% |
| 2 | 3.660 | 2.353 | 64.3% |
Figure 1.
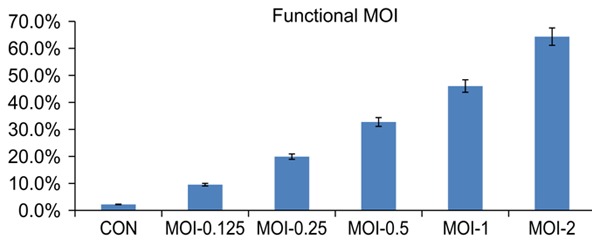
Lentivirus multiplicity of infection pre-experiment. Functional multiplicity of infection (MOI) was determined by measuring the absorbance at 450 nm.
Determination of the PTX concentration
Next, we evaluated the optimal concentration of PTX for treating the cells. The concentration on a semi-logarithmic plot was found to be 8.789 ng/mL. We then evaluated the appropriate concentration for achieving growth inhibition of SiHa cells. Our results indicated that 3 ng/mL was the optimal concentration (Table 2; Figure 2).
Table 2.
Cell growth conditions under different drug concentrations
| Drug concentration (ng/mL) | Multiplicity of infection | Cell inhibition rate (%) |
|---|---|---|
| 0 | 1438.2428 | 0.00 |
| 0.5 | 1348.0105 | 6.27 |
| 1.0 | 1233.936 | 14.21 |
| 2.0 | 1102.5219 | 23.34 |
| 4.0 | 372.2468 | 74.12 |
| 6.0 | 0 | 100.00 |
Figure 2.
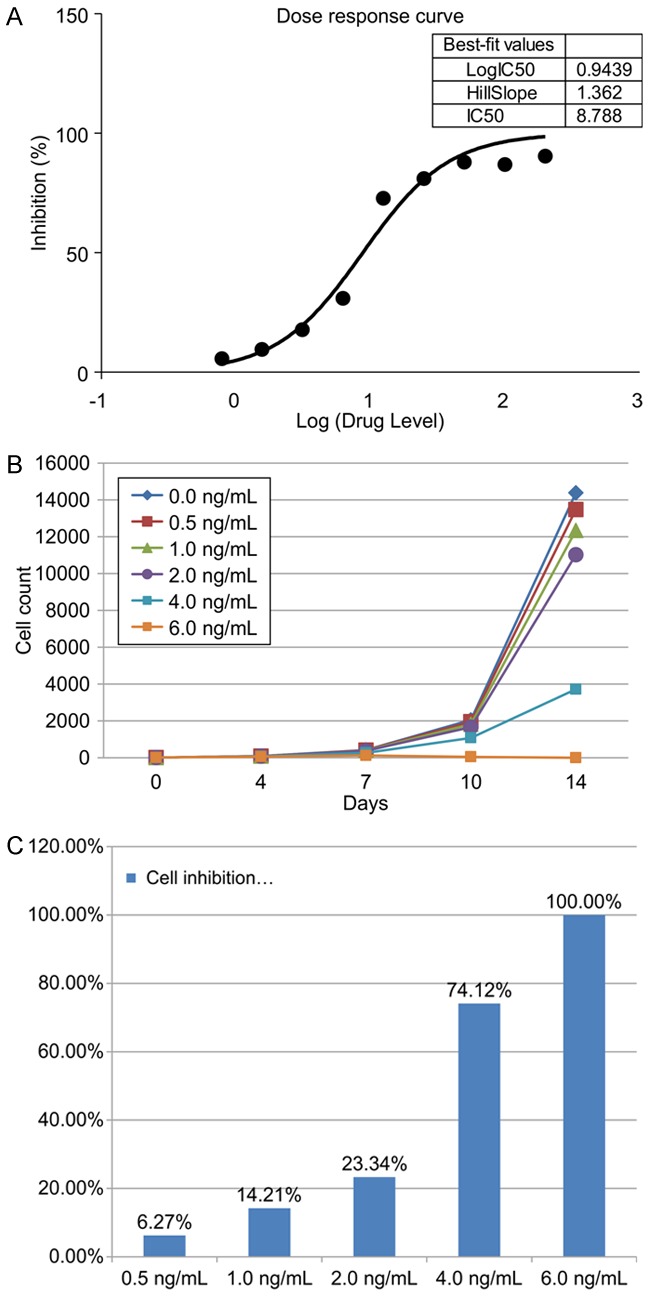
Dose response to paclitaxel. A. The IC50 of paclitaxel (PTX) was calculated as 8.789 ng/mL. B. Increasing concentrations of PTX were added to SiHa cells, and the cells were counted on days 0, 4, 10, and 14. C. Untreated cells were set as the baseline.
Screening of candidate genes whose loss contributed to sensitivity to PTX
To identify genes whose loss induced sensitivity to PTX, the cells were treated with PTX or the vehicle dimethylsulfoxide for 0-14 days and then subjected to high-throughput sequencing (Figure 3).
Figure 3.
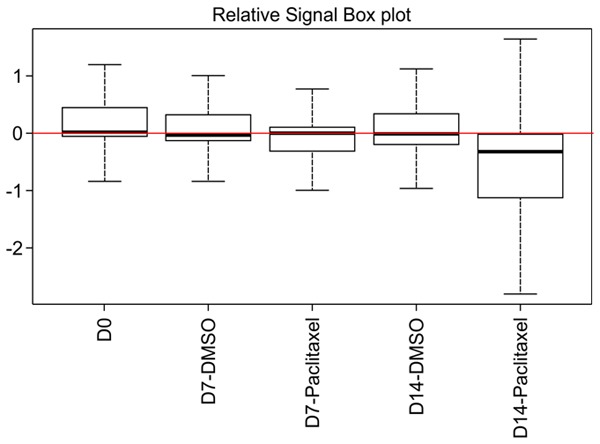
Boxplot showing the distribution of sgRNA frequencies using high-throughput sequencing. The Y-axis corresponds to the relative log2 counts per million (CPM) for each cell sample at different times. The red line represents the average relative log2, and the box extends from the first to the third quartile, with the whiskers denoting 1.5 times the interquartile range.
Compared with vehicle-treated cells, the sgRNA frequency distribution in PTX-treated cells changed under PTX-selected pressure in two opposite directions, enrichment or depletion. We selected genes whose loss sensitized SiHa cells to PTX by satisfying the filter criteria (Table 3). A total of 101 deleted genes were selected after 7 days, and 374 genes were selected after 14 days. The FC was determined by comparing the frequencies at 7 and 14 days with that at 0 days. The Bayes factor was also calculated for each gene [12].
Table 3.
Candidate gene screening conditions
| # | Screening method | Day 7 versus day 0 | Day 14 versus day 0 |
|---|---|---|---|
| 1 | |FCDMSO| < 2 & FCPaclitaxel ≤ -2 | 101 | 374 |
| 2 | BFDMSO < 0 & BFPaclitaxel > 2 | ||
| Total | 461 | ||
GO and KEGG pathway analyses
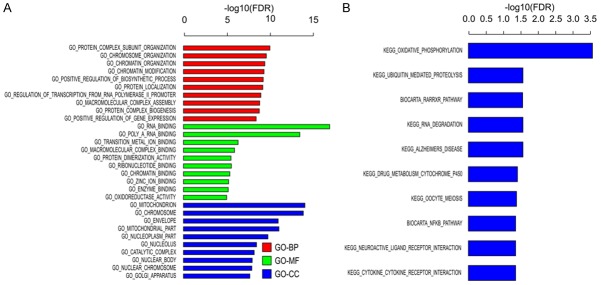
Using GO analysis, we identified the functions of the target genes. The functions were ranked by gene set enrichment analysis [13] (F-test, Bonferroni-corrected P < 0.05; Figure 4A, 4B). The results revealed that the deleted genes play a role in NF-κB signaling. It also revealed some pathways, such as oxidative phosphorylation, ubiquitin-mediated proteolysis, RARRXR pathway, and cytokine receptor interaction, which have not been studied before.
Figure 4.
GO and KEGG pathway analyses. The KEGG database and GO terms were used to analyze the pathways involved in PTX activity (F-test, Bonferroni-corrected P < 0.05).
Discussion
In this study, we used the Cas9 system to explore the genes associated with sensitivity to PTX in cervical cancer for the first time. Among the depleted genes under selective pressure, we identified ABCC9, which belongs to the ABC transporter gene family, related to multidrug transport. We also selected representative genes previously shown to participate in regulating cancer cell reactivity to chemotherapy.
Of the genes obtained as hits on our screen, IL-37 has been reported to be involved in the phosphatidylinositol 3-kinase (PI3K)/Akt pathway [14]. Moreover, EIF3C and PPP2R5B play a role in the mammalian target of rapamycin (mTOR) pathway [15-17]. mTOR, the downstream target of the PI3K/Akt pathway, is critical for tumor cell proliferation, growth, differentiation, metabolism and apoptosis [18]. Recent studies have shown that human papilloma virus oncogenes E6/E7/E5 activate the PI3K/Akt/mTOR signaling pathway, which modulates tumorigenesis and progression [19]. Moreover, high mTOR expression is associated with chemoresistance in cervical cancer [20]. Other studies have shown that Akt and mTORC1 signaling mediates resistance to PTX by protecting cancer cells from death/apoptosis [21,22]. Furthermore, previous reports have shown that PI3K, Akt and mTOR inhibitors upregulate sensitivity to various conventional chemotherapeutic agents [23-25]. These results explain the mechanisms and pathways through which the loss of IL-37 increased PTX sensitivity in our study. We also identified AKT1S1, a component of mTORC1 [26], as a gene related to drug sensitivity, further highlighting the role of the mTOR pathway in affecting PTX sensitivity.
MAPK37 belongs to the MAP3K family, which is the dominant member of the mitogen-activated protein kinase (MAPK) signaling pathway [27]. In addition, some of the identified deleted genes, PPP1R7, PPP2R5B, PPP1R7 and PPP1R11, belong to the phosphoserine/threonine phosphatase (PPP) family and have been proven to regulate the MAPK signaling pathway [17,28,29]. MAPK pathways are activated by diverse extracellular and intracellular stimuli, including peptide growth factors, cytokines, hormones, and various cellular stressors, such as oxidative stress and endoplasmic reticulum stress. These signaling pathways regulate various cellular activities, including proliferation, differentiation, survival and death [30]. The anticancer effects of PTX can be partly attributed to apoptosis, which is mediated by multipolar spindles. Thus, apoptosis contributes to PTX sensitivity, and many studies have shown that changes in the apoptosis pathway result in relative sensitivity or resistance to PTX [31,32]. The MAPK pathway plays an important role in apoptosis [33] and mediates the variability PTX sensitivity [34]. So, loss of these genes contributes to PTX sensitivity.
The mTOR signaling pathway and the MAPK pathway have a common target: 4EBP. However, these two routes have opposing roles in determining PTX therapeutic efficacy (Figure 5).
Figure 5.
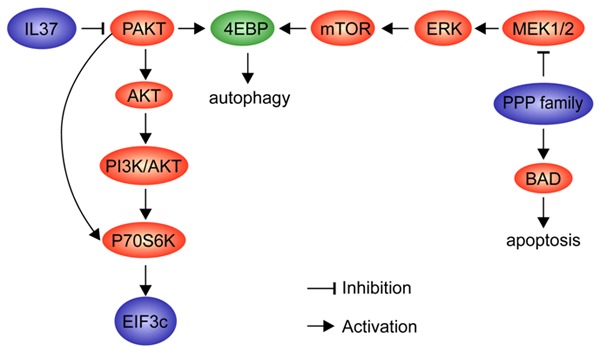
Gene interaction diagram. The screening hits in our study are marked in blue. The crossing target is marked in green. The pathways influence the drug sensitivity through apoptosis and autophagy.
Cas9 technology provides a reliable method for exploring the essential factors affecting the degree of PTX sensitivity at the gene level. In most traditional in vivo studies, RNA interference (RNAi) is the main method for gene knockout. However, in this study GeCKO screening was shown to provide an effective alternative toRNAi. First, RNAi reduced protein expression at the RNA level, whereas GeCKO targeted DNA. Cas9 could be programmed to induce a loss-of-function allele at specific genomic loci [19,35,36], which yielded high screening sensitivity. Second, the RNAi knockout zone only applied to transcripts, whereas Cas9 could target the entire gene range, including promoters, enhancers, introns and intergenic regions. Moreover, in previous studies, cancer cells associated with chemoresistance were routinely derived from patients who appeared chemoresistant in the clinical setting or from cell lines treated with chemotherapeutic agents for several days [37,38], and changes in gene expression were then analyzed. In contrast, in our study, the Cas9 system provided genome-wide knockout in chemo-naïve cells within a short period of time, avoiding additional gene mutations acquired during therapeutic intervention and prolonged culture. These factors support the reliability of the Cas9 system.
In summary, we identified novel candidate genes associated with sensitivity to PTX in cervical cancer using CRISPR-Cas9 to induce their deletion. However, there are some limitations to this study. This was an in vitro study, and the dosage and frequency of PTX are different from those in the clinical setting. Therefore, moving ahead, we need to conduct large sample clinical trials to develop related drugs considered more effective therapeutic options for patients with cervical cancer.
Acknowledgements
This study was supported by grants from Beijing Municipal Science and Technology Commission (D131100005313009) and the Special Program for Development of Clinical Medicine of Beijing Municipal Administration of Hospitals (ZYLX201705).
Disclosure of conflict of interest
None.
References
- 1.Pisani P, Parkin DM, Bray F, Ferlay J. Estimates of the worldwide mortality from 25 cancers in 1990. Int J Cancer. 1999;83:18–29. doi: 10.1002/(sici)1097-0215(19990924)83:1<18::aid-ijc5>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 2.Seidman AD, Berry D, Cirrincione C, Harris L, Muss H, Marcom PK, Gipson G, Burstein H, Lake D, Shapiro CL. Randomized phase III trial of weekly compared with every-3-weeks paclitaxel for metastatic breast cancer, with trastuzumab for all HER-2 overexpressors and random assignment to trastuzumab or not in HER-2 nonoverexpressors: final results of cancer and leukemia group B protocol 9840. J. Clin. Oncol. 2008;26:1642–1649. doi: 10.1200/JCO.2007.11.6699. [DOI] [PubMed] [Google Scholar]
- 3.Sparano JA, Wang M, Martino S, Jones V, Perez EA, Saphner T, Wolff AC, Sledge GW Jr, Wood WC, Davidson NE. Weekly paclitaxel in the adjuvant treatment of breast cancer. N Engl J Med. 2008;358:1663–1671. doi: 10.1056/NEJMoa0707056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mori T, Hosokawa K, Sawada M, Kuroboshi H, Tatsumi H, Koshiba H, Okubo T, Kitawaki J. Neoadjuvant weekly carboplatin and paclitaxel followed by radical hysterectomy for locally advanced cervical cancer: long-term results. Int J Gynecol Cancer. 2010;20:611–616. doi: 10.1111/IGC.0b013e3181d80aa9. [DOI] [PubMed] [Google Scholar]
- 5.Saito I, Kitagawa R, Fukuda H, Shibata T, Katsumata N, Konishi I, Yoshikawa H, Kamura T. A phase III trial of paclitaxel plus carboplatin versus paclitaxel plus cisplatin in stage IVB, persistent or recurrent cervical cancer: gynecologic cancer study group/Japan clinical oncology group study (JCOG0505) Jap J Clin Oncol. 2009;40:90–93. doi: 10.1093/jjco/hyp117. [DOI] [PubMed] [Google Scholar]
- 6.Gottesman MM, Pastan I. Biochemistry of multidrug resistance mediated by the multidrug transporter. Annu Rev Biochem. 1993;62:385–427. doi: 10.1146/annurev.bi.62.070193.002125. [DOI] [PubMed] [Google Scholar]
- 7.Juliano RL, Ling V. A surface glycoprotein modulating drug permeability in Chinese hamster ovary cell mutants. Biochim Biophys Acta. 1976;455:152–162. doi: 10.1016/0005-2736(76)90160-7. [DOI] [PubMed] [Google Scholar]
- 8.Szakács G, Paterson JK, Ludwig JA, Booth-Genthe C, Gottesman MM. Targeting multidrug resistance in cancer. Nature Rev Drug Discov. 2006;5:219–234. doi: 10.1038/nrd1984. [DOI] [PubMed] [Google Scholar]
- 9.Ueda K, Cardarelli C, Gottesman MM, Pastan I. Expression of a full-length cDNA for the human “MDR1” gene confers resistance to colchicine, doxorubicin, and vinblastine. Proc Natl Acad Sci U S A. 1987;84:3004–3008. doi: 10.1073/pnas.84.9.3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Y, Liu B, Zhao Q, Hou T, Huang X. Nuclear localizaiton of β-catenin is associated with poor survival and chemo-/radioresistance in human cervical squamous cell cancer. Int J Clin Exp Pathol. 2014;7:3908. [PMC free article] [PubMed] [Google Scholar]
- 11.Huang Y, Ray S, Reed JC, Ibrado AM, Tang C, Nawabi A, Bhalla K. Estrogen increases intracellular p26Bcl-2 to p21Bax ratios and inhibits taxol-induced apoptosis of human breast cancer MCF-7 cells. Breast Cancer Res Treat. 1997;42:73–81. doi: 10.1023/a:1005777219997. [DOI] [PubMed] [Google Scholar]
- 12.Hart T, Brown KR, Sircoulomb F, Rottapel R, Moffat J. Measuring error rates in genomic perturbation screens: gold standards for human functional genomics. Mol Syst Biol. 2014;10:733. doi: 10.15252/msb.20145216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2011;40:D109–D114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li TT, Zhu D, Mou T, Guo Z, Pu JL, Chen QS, Wei XF, Wu ZJ. IL-37 induces autophagy in hepatocellular carcinoma cells by inhibiting the PI3K/AKT/mTOR pathway. Mol Immunol. 2017;87:132–140. doi: 10.1016/j.molimm.2017.04.010. [DOI] [PubMed] [Google Scholar]
- 15.Zhao W, Li X, Wang J, Wang C, Jia Y, Yuan S, Huang Y, Shi Y, Tong Z. Decreasing eukaryotic initiation factor 3c (EIF3C) suppresses proliferation and stimulates apoptosis in breast cancer cell lines through mammalian target of rapamycin (mtor) pathway. Med Sci Monit. 2017;23:4182. doi: 10.12659/MSM.906389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giannios J, Ginopulos P. Pegylated liposomal anti-eIF3c shRNA-vinorelbine formulation (SEVINA-V) inhibits oncogenic protein translational initiation and oncogene addiction inducing apoptosis in HRBC chemoresistant to taxanes. Drug Chem Toxicol. 2007:1–14. [Google Scholar]
- 17.Janssens V, Goris J. Protein phosphatase 2A: a highly regulated family of serine/threonine phosphatases implicated in cell growth and signalling. Biochem J. 2001;353:417–439. doi: 10.1042/0264-6021:3530417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Polivka J, Janku F. Molecular targets for cancer therapy in the PI3K/AKT/mTOR pathway. Pharmacol Ther. 2014;142:164–175. doi: 10.1016/j.pharmthera.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 19.Zhang L, Wu J, Ling MT, Zhao L, Zhao KN. The role of the PI3K/Akt/mTOR signalling pathway in human cancers induced by infection with human papillomaviruses. Mol Cancer. 2015;14:87. doi: 10.1186/s12943-015-0361-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xie G, Wang Z, Chen Y, Zhang S, Feng L, Meng F, Yu Z. Dual blocking of PI3K and mTOR signaling by NVP-BEZ235 inhibits proliferation in cervical carcinoma cells and enhances therapeutic response. Cancer Lett. 2017;388:12–20. doi: 10.1016/j.canlet.2016.11.024. [DOI] [PubMed] [Google Scholar]
- 21.Bava SV, Puliappadamba VT, Deepti A, Nair A, Karunagaran D, Anto RJ. Sensitization of taxol-induced apoptosis by curcumin involves down-regulation of nuclear factor-κB and the serine/threonine kinase Akt and is independent of tubulin polymerization. J Biol Chem. 2005;280:6301–6308. doi: 10.1074/jbc.M410647200. [DOI] [PubMed] [Google Scholar]
- 22.Lin H, Lui W, Liu T, Chi C. Reversal of Taxol resistance in hepatoma by cyclosporin A: involvement of the PI-3 kinase-AKT 1 pathway. Br J Cancer. 2003;88:973–980. doi: 10.1038/sj.bjc.6600788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zitzmann K, von Rüden J, Brand S, Göke B, Lichtl J, Spöttl G, Auernhammer CJ. Compensatory activation of Akt in response to mTOR and Raf inhibitors-a rationale for dual-targeted therapy approaches in neuroendocrine tumor disease. Cancer Lett. 2010;295:100–109. doi: 10.1016/j.canlet.2010.02.018. [DOI] [PubMed] [Google Scholar]
- 24.Mukherjee B, Tomimatsu N, Amancherla K, Camacho CV, Pichamoorthy N, Burma S. The dual PI3K/mTOR inhibitor NVP-BEZ235 is a potent inhibitor of ATM-and DNA-PKCs-mediated DNA damage responses. Neoplasia. 2012;14:34–43. doi: 10.1593/neo.111512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Inoki K, Corradetti MN, Guan KL. Dysregulation of the TSC-mTOR pathway in human disease. Nat Genet. 2005;37:19–24. doi: 10.1038/ng1494. [DOI] [PubMed] [Google Scholar]
- 26.Kim EK, Choi EJ. Pathological roles of MAPK signaling pathways in human diseases. Biochim Biophys Acta. 2010;1802:396–405. doi: 10.1016/j.bbadis.2009.12.009. [DOI] [PubMed] [Google Scholar]
- 27.Cohen PT. Protein phosphatase 1-targeted in many directions. J Cell Sci. 2002;115:241–256. doi: 10.1242/jcs.115.2.241. [DOI] [PubMed] [Google Scholar]
- 28.Zolnierowicz S. Type 2A protein phosphatase, the complex regulator of numerous signaling pathways. Biochem Pharmacol. 2000;60:1225–1235. doi: 10.1016/s0006-2952(00)00424-x. [DOI] [PubMed] [Google Scholar]
- 29.Jankowsky JL, Fadale DJ, Anderson J, Xu GM, Gonzales V, Jenkins NA, Copeland NG, Lee MK, Younkin LH, Wagner SL. Mutant presenilins specifically elevate the levels of the 42 residue β-amyloid peptide in vivo: evidence for augmentation of a 42-specific γ secretase. Hum Mol Genet. 2003;13:159–170. doi: 10.1093/hmg/ddh019. [DOI] [PubMed] [Google Scholar]
- 30.Musa F, Alard A, David-West G, Curtin JP, Blank SV, Schneider RJ. Dual mTORC1/2 inhibition as a novel strategy for the resensitization and treatment of platinum-resistant ovarian cancer. Mol Cancer Ther. 2016;15:1557–1567. doi: 10.1158/1535-7163.MCT-15-0926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yusuf R, Duan Z, Lamendola D, Penson R, Seiden M. Paclitaxel resistance: molecular mechanisms and pharmacologic manipulation. Curr Cancer Drug Targets. 2003;3:1–19. doi: 10.2174/1568009033333754. [DOI] [PubMed] [Google Scholar]
- 32.Johnstone RW, Ruefli AA, Lowe SW. Apoptosis: a link between cancer genetics and chemotherapy. Cell. 2002;108:153–164. doi: 10.1016/s0092-8674(02)00625-6. [DOI] [PubMed] [Google Scholar]
- 33.Yin B, Morgan K, Hasz D, Mao Z, Largaespada D. Nf1 gene inactivation in acute myeloid leukemia cells confers cytarabine resistance through MAPK and mTOR pathways. Leukemia. 2006;20:151–151. doi: 10.1038/sj.leu.2404033. [DOI] [PubMed] [Google Scholar]
- 34.Liu L, Gao F, Ye Y, Chen Z, Dai Y, Zhao P, Guan G, Zhao M. Influence of HMGB1/MAPK/m-TOR signaling pathway on cell autophagy and chemotherapy resistance in K562 cells. J Cent South Univ. 2016;41:1016–1023. doi: 10.11817/j.issn.1672-7347.2016.10.002. [DOI] [PubMed] [Google Scholar]
- 35.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ho CM, Huang CJ, Huang SH, Chang SF, Cheng WF. Demethylation of HIN-1 reverses paclitaxel-resistance of ovarian clear cell carcinoma through the AKT-mTOR signaling pathway. BMC Cancer. 2015;15:789. doi: 10.1186/s12885-015-1744-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu X, Wang D, Liu H, Feng Y, Zhu T, Zhang L, Zhu B, Zhang Y. Knockdown of astrocyte elevated gene-1 (AEG-1) in cervical cancer cells decreases their invasiveness, epithelial to mesenchymal transition, and chemoresistance. Cell Cycle. 2014;13:1702–1707. doi: 10.4161/cc.28607. [DOI] [PMC free article] [PubMed] [Google Scholar]