Abstract
The present study aimed to investigate abnormal fetal cardiac pathological injury with exposure to a hypoxic environment during pregnancy. Pregnant hypoxia sheep models were prepared using artificial cabins. Sixteen pregnant sheep were randomly divided into three groups: normal group (NG), mild hypoxia group (MHG), and severe hypoxia group (SHG). The degree of SpO2, SaO2, breathing and heart rate, CO, and MDA were determined. HE staining, SEM, and TEM were used to evaluate the pathological changes of fetal myocardium. iTRAQ-based quantitative proteomics was employed to identify differentially expressed proteins. At day 30 and 90, the levels of SpO2 and SaO2 in SHG and MHG sheep met the diagnostic criteria. Ninety days after modeling, a dramatic reduction of breathing rate and increase of heart rate was observed in MHG and SHG comparing with that in NG (P < 0.05). At day 120, CO and MDA increased significantly in SHG and MHG than those in NG (P < 0.05), also in SHG compared with that in MHG (P < 0.05). HE staining and electron microscopy scanning showed that the myocardium in SHG and MHG had different levels of tissue edema and abnormal distribution of mitochondrion compared to NG. Eighty differentially expressed proteins were identified in fetal cardiac tissues. In conclusion, different degrees of pregnant hypoxia sheep models were successfully established. Prenatal hypoxia can cause serious damage to heart development of fetal sheep, as well as lead to the significant changes of hypoxia-associated proteins expression, which may be used to the further investigation for clinical applications.
Keywords: Prenatal hypoxia, fetal sheep, heart development, proteomics analysis
Introduction
All over the world, more than 140 million people settle in high altitude environments at > 2500 m. Of them, about 80 million live in Asia, and 35 million live in the Andean mountains [1]. High altitude regions are of thin air and low oxygen availability. As we known, oxygen content is critical for cell survival and embryo development [2]. Camm et al. reported that when Wistar dams undergo a prolonged hypoxic condition, the PaO2 level will drop to 40 mmHg, which can decrease the PaO2 content of the fetus to 10~12 mmHg, and further result in fetal hypoxia [3]. Therefore, in humans, fetal hypoxia is one of the most common suboptimal conditions in complicated pregnancy, specially, high-altitude pregnancy offers one of the prevalent forms of sustained fetal hypoxia. Hypoxic environments not only contribute to the redistribution of blood flow and oxygen molecule, but also trigger a series of abnormal responses including hypoxic stress and pathological changes of multiple tissues or organs, finally enlarge the risks of fetal growth restriction and dysplasia [3]. Studies of highland populations have proven that decreased birth weight and adverse cardiovascular alterations can occur in babies born at a high altitude in contrast with those born at sea level [4,5]. Meanwhile, prenatal hypoxia also increases the possibility of suffering from multiple chronic adult diseases, such as coronary atherosclerotic heart disease, hypertension, diabetes, etc. [6].
Cardiogenesis is started at the formation of the embryonic heart. After the heart has emerged, its volume growth rises rapidly during fetal development. Oxygen concentration plays a vital regulatory role in the process of heart formation and maturation. Compared with the adult heart, the fetal heart is more liable to hypoxia-induced cell death because of its strong ability to increase glycolytic flux [7], which means that the fetus has a great tendency to congenital heart defects with exposure to a hypoxic environment induration of pregnancy. Congenital heart defects, a common hypoxia-induced cardiopathy in high-altitude populations, is characterized by the cardiac structural change and myocardial microstructures damage, which can lead to the occurrence of diversified congenital heart diseases or cardiomyopathy. Relative studies reported the prevalence rate of congenital heart defects at sea level is 0.8-1.2% [8], but the value rise to 2.5-2.8% at high altitude [5]. Emerging epidemiological statistical analyses and related studies indirectly supported that there is a close relationship between congenital heart defects and prenatal hypoxia [9,10].
Despite numerous investigations have focused on the role of hypoxia in fetal heart development, thus far, the exact pathophysiological impact and potential mechanisms have not be completely understood due to the lack of successful animal models. In this study, we summarize the data associated with hypoxia to confirm the successful establishment of highland hypoxia model, and then characterize myocardial development and its pathological structural changes of fetal sheep. Next, we performed proteomics analysis to identify the hypoxia-independent proteins and the relative pathways in fetal hearts. The objective of this study was to gain knowledge of the relationship between prenatal hypoxia and cardiogenesis, as well as the possible molecular mechanisms, which may provide the important theoretical basis for the prevention and treatment of congenital heart diseases.
Material and methods
Animals
Thirty healthy pregnant sheep weighting 25~35 kg were provided by the laboratory animal center of Urumqi General Hospital in Lanzhou Military Region. The animals were randomly divided into three groups (10 sheep for each) consisting of a: low altitude control group (normal group, NG), mild hypoxia exposure group (mild hypoxia group, MHG), and severe hypoxia exposure group (severe hypoxia group, SHG). NG sheep were kept in the artificial cabin of plain environment at sea level with 20°C temperature, 11.0 g/m3 relative humidity, 260 g/m3 oxygen content, and 100.0 kPa pressure. MHG and SHG sheep were respectively maintained in the cabins of high altitude environment at 3000 m (temperature of 5°C, humidity of 3.68 g/m3, oxygen content of 206 g/m3, pressure of 67.7 kPa) and 5000 m (temperature of -6°C, humidity of 1.77 g/m3, oxygen content of 166 g/m3, pressure of 54.1 kPa) above sea level, and the cabins were gradually increased to the high altitude with 10 m/s speed. All animals were given free access to water and food. The levels of SpO2 (percutaneous oxygen saturation) and SaO2 (arterial oxygen saturation) were detected to confirm whether different degrees of hypoxia models of pregnant sheep were established successfully. MHG-sheep and SHG-sheep were required to meet the following requirements: 30 < SpO2 < 60 mmHg and 60% < SaO2 < 90%; SpO2 < 30 mmHg and SaO2 < 60%, respectively [11]. All tests were performed at the second trimester. The experimental procedures were in accordance with the “Guide for the Care and Use of Laboratory Animals of China”.
Measurement of life indicators
At day 90 of experiments, the sheep in different groups were anesthetized by an intraperitoneal injection of chloral hydrate (400 pl/100 g), then thoracotomy was done and fresh liver tissues were extracted. 0.1~0.2 g liver tissues were mixed with 0.01 mol/l cold phosphate buffer in a ratio of 1:20, and then homogenized at the temperature of 0~4°C. The content of CO (carbonyl groups) was determined using the spectrophotometric method [12], and the fluorometric thiobarbituric acid reactive substances (TBARS) method was conducted to measure MDA (malondialdehyde) level. Also, the breathing rate and heart rate of parental sheep were observed and recorded. The arterial blood samples were collected and assayed for the level of SpO2 and SaO2 within 2 hours at the time point of 30, 90, 120 d.
Echocardiography
Echocardiography was performed using a Visualsonics Series 2100 high-resolution imaging system with a 38 MHz Microscan transducer probe. Briefly, pregnant sheep from all groups were anesthetized using sevoflurane by inhalation, and then were placed on the inspection table after removing abdominal hairs for echocardiography examination. Fetal sheep were also examined by echocardiography to further confirm whether the cardiac malformation occurred or not according to previous articles [13,14].
Histology and ultrastructural analysis
The heart tissues extracted from fetal sheep were washed, frozen, and stored at -80°C. The frozen sections were cut into 6 um thickness, and stained with hematoxylin and eosin (HE) for morphological evaluation by a light microscopy (Olympus, CX23RTFS). Transmission electron microscopy (TEM) and scanning electron microscopy (SEM) were applied to determine the ultrastructural integrity of cardiac tissues slices. Tissues were fixed with 2.5% buffered glutaraldehyde, post fixed with 1% osmium tetroxide, dehydrated in ascending grades of ethyl alcohol, and immersed in propylene-epon mixture, embedded in epoxy, then stained with 1% uranyl acetate and 1% lead citrate for TEM. Samples for SEM were prepared in a similar manner, in addition to the slices were critically point dried and sputter coated with gold. TEM samples were assayed by a JEOL 1200EX (Welwyn Garden City, UK), and SEM samples were detected using a JEOL 35 SEM at 15 kV.
Protein extraction
All samples were obtained from the heart tissues of fetal sheep with approval of the Institutional Ethics Committee. The collected samples were homogenized in lysis buffer (4% SDS, 1 mM DTT, 150 mM Tris-HCl pH 8.0, protease inhibitor), incubated in boiling water for 5 minutes, and then sonicated on ice. The crude extract was then boiled in a water bath again and centrifuged at 25°C for 10 min. Total protein was quantified using the BCA protein assay reagent (Beyotime), and then stored at -20°C.
Digestion and iTRAQ labeling
Protein digestion was conducted according to FASP procedure [15]. Briefly, a total of 200 μg proteins from each sample were incubated with trypsin (Promega, Madison, WI, USA) overnight at 37°C. After digestion, the peptides were dried and collected by vacuum centrifugation, then quantification was done by UV light spectral density. Labeling was performed following the manufacturer’s manual for the iTRAQ (Applied Biosystems, Foster City, CA USA). Normal group samples (100 mg) were labeled with tags 113 and 114 at room temperature for 1 h with the following groups: mild hypoxia samples were labeled with tags 115, 116, and 117 and severe hypoxia samples were labeled with tags 118, 119, and 121.
Strong cationic-exchange (SCX) chromatography separation
The labeled peptides were pooled, dried, and fractionated by an SCX chromatography (LC-20AB HPLC pumpsystem, Shimadzu Corporation, Kyoto, Japan). Briefly, each of the peptide mixture was reconstituted in 2 mL buffer A (10 mM KH2PO4 in 25% of ACN, pH 2.7), loaded into a PolySulfoethyl column (4.6 × 100 mm, 5 μm, 200 Å, PolyLCInc, Maryland, USA). The peptides were eluted at 1 mL/min with a gradient of 0%-10% buffer B (10 mM KH2PO4, pH 3.0, 500 mM KCl, 25% acetonitrile) for 2 min, 10% to 20% buffer B for 25 min followed by 20% to 45% buffer B for 5 min and 50% to 100% buffer B for 5 min. The UV absorbance at 214 nm was monitored when the fractions were collected. Fractions were then combined into 10 pools, and desalted on Empore SPEC18 Cartridges (Sigma, Santa Clara, CA, USA). Each fraction was concentrated by vacuum centrifugation and reconstituted in 40 μL of 0.1% formic acid for LC-MS/MS analysis.
LC-electrospray Ionization (ESI)-MS/MS analysis by Q exactive
Experiments were carried out on an Easy nLC 1000 system coupled to Q Exactive mass spectrometer (Proxeon Biosystems, now Thermo Fisher Scientific). In brief, 5 μg peptide mixture was loaded onto a Zorbax 300SB C18-reversed phase column (Agilent Technologies, Wilmington, DE, USA) for 20 min in buffer A (0.1% formic acid), then separated with a linear gradient of buffer B (80% acetonitrile and 0.1% formic acid) at 250 nl/min controlled by IntelliFlow technology over 140 min. MS data was acquired using the 10 most abundant precursor ions from the survey scan (300-1800 m/z) for HCD fragmentation. Target values were obtained based on predictive automatic gain control (pAGC). Dynamic exclusion duration was 60 s. Survey scans were acquired at a resolution of 70000 at m/z 200 and resolution for HCD spectra was set to 17500 at m/z 200. Normalized collision energy was 30 eV and the underfill ratio was set as 0.1%. The instrument was operated with peptide recognition mode enabled. The whole workflow of iTRAQ was exhibited as Figure 2A.
Figure 2.
Work flow of iTRAQ (A) and protein purity verified with SDS-PAGE (B).
Data analysis
Protein identification and iTRAQ quantification were performed as previously described by a MASCOT engine (Matrix Science) embedded into Proteome Discoverer 1.4 (Thermo Electron, San Jose, CA) [16]. Briefly, protein identification was performed with peptide mass tolerance at 20 ppm, MS/ MS tolerance at 0.1 Da, enzyme of trypsin, number of missed cleavage up to 2, fixed modification of carbamidomethyl (C), iTRAQ 8plex (K), iTRAQ8plex (N-term), variable modification of oxidation (M), and FDR ≤ 0.01. For iTRAQ data, to be identified as being significantly differentially changed, a protein must pass the test with P < 0.05 and a ratio fold change > 1.3 or < 0.77 must be identified in at least two of the three biological replicates. These limits were selected according to previous studies using iTRAQ reagents with some modifications [17].
Bioinformatics
The molecular functions of identified proteins were classified based on their gene ontology annotations and biological functions. Hierarchical clustering of proteins abundance was conducted by Heat mapper server (http://www.heatmapper.ca) [18]. Gene Ontology (GO) analyses (http://www.geneontology.org) were performed according to a previous report [19]. KEGG pathway database (http://www.genome.jp/kegg/pathway.html) was used to analysis the metabolic pathways of the identified proteins. Protein-protein interaction networks were processed by the publicly available program STRING (http://string-db.org/).
Statistical analysis
Statistical analysis was conducted using SPSS 16.0 software. All data are presented as mean ± SD (standard deviation). Student’s t-test was performed to make comparisons between two groups. One-way analysis of variance (ANOVA) and SNK-q test were applied to analyze the differences between groups. P < 0.05 was considered as the significant difference.
Results
Establishment of the prenatal hypoxic sheep model
To confirm whether the hypoxia models of pregnant sheep were established successfully, we determined the levels of SpO2 and SaO2, as well as the breathing rate and heart rate (Table 1). Compared with normal group, SpO2 and SaO2 of mild hypoxia- and severe hypoxia-sheep were both diminished remarkably from the 30th day (P < 0.05, P < 0.01), whereas the levels in SHG was much lower than that in MHG (P < 0.05). At day 90 of the experiment, a dramatically reduction of breathing rate was observed in MHG and SHG comparing with NG (P < 0.05, P < 0.01), while the heart rate was significantly increased (P < 0.05). According to the diagnostic criteria of hypoxia [11], we successfully established the mild hypoxic sheep model at 30th day, as well as severe hypoxic sheep model at 90th day.
Table 1.
SpO2, SaO2, breathing and heart rate in pregnant sheep of 3 groups at different time points during the exposure to hypoxia or normoxia (x ± s, n=8)
| Group | SpO2 (mmHg) | SaO2 (%) | Breathing (/min) | Heart rate (/min) | ||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| 30 d | 90 d | 120 d | 30 d | 90 d | 120 d | 90 d | 90 d | |
| Normal | 78 ± 5 | 81 ± 4 | 86 ± 2 | 95 ± 5 | 94 ± 3 | 96 ± 4 | 19 ± 2 | 72 ± 4 |
| Mild hypoxia | 58 ± 4a | 54 ± 5a | 44 ± 2a | 76 ± 4a | 64 ± 5a | 73 ± 2a | 15 ± 1a | 78 ± 3a |
| Severe hypoxia | 32 ± 4a,b | 30 ± 2a,b | 26 ± 4a,b | 54 ± 3a,b | 42 ± 4a,b | 40 ± 3a,b | 12 ± 2a,b | 79 ± 3a |
P < 0.01, compared with normal;
P < 0.01, compared with severe hypoxia.
Prenatal hypoxia aggravated the heart damage of fetal sheep
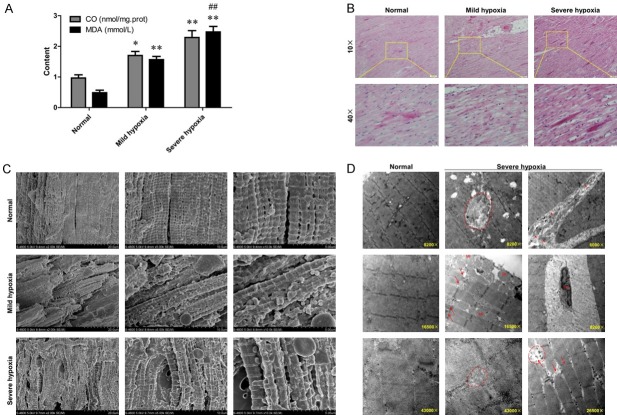
Echocardiography was performed to explore the effect of hypoxia on cardiovascular structure of parental sheep, and no obvious abnormalities were found in both pregnant sheep and fetal sheep. Also, the cardiac hemodynamics did not manifest any abnormality based on the images of CDFI (Color Doppler). Specific results are shown in our previous published article [20]. Furthermore, compared with NG, the levels of CO and MDA were significantly increased in both MHG (P < 0.05, P < 0.01) and SHG sheep (P < 0.01, P < 0.01), simultaneously, MDA level in SHG were much higher than that in MHG (P < 0.01), implying that hypoxia would aggravate oxidative damage of heart tissues (Figure 1A). Next, H&E staining was performed to provide a global histology and morphology representation of myocardium. Compared with NG, the MHG showed much worse morphology, with myocardial fibers being disorderly arranged with large spaces, and myocardial cells appeared rounded or irregular. When the hypoxia environment deteriorated, except for the aggravation of above damage, some other significant changes also presented, such as rupture of the myofilaments and agglomerationof the cardiomyocytes (Figure 1B). Simultaneously, SEM and TEM were carried out to assess ultrastructural integrity of heart tissues exposed to different hypoxia conditions. SEM revealed the disrupted myocardium and disorganized mitochondria in hypoxia treated-sheep. The normal group demonstrated a good ultrastructural morphology, characterized by well-organized myocardial tissues, intact mitochondria, and all myofilaments were coplanar. However, myocardial tissues from hypoxia-treated sheep were in disorder with dilated space, and myofilaments were on different planes. Particularly, myocardial fibers stimulated by severe hypoxia appeared to be disrupted and then redistributed. Magnifying the specimen, some mitochondria presented abnormally dispersed and clumped, which suggested a negative effect of hypoxia on the metabolism process of myocardial tissues (Figure 1C). Figure 1D exhibited evident ultrastructural changes in the organization of microfilaments simulated by normal conditions or severe hypoxia based on the following reasons: 1) no evident difference of TEM images between mild hypoxia and severe hypoxia group; 2) obvious characteristic lesions were occurred in severe hypoxia-treated myocardium. Clearly, the heart samples in NG showed a dense matrix, visible fine cristae and Z lines, and arranged in lines between the microfilaments. However, severe hypoxia treatment contributed to significant changes in myocardium, such as the disorganization of microfilaments with the loss of Z lines, degeneration of microfibers and mitochondria, increase of the collagen fibers in the intracellular spaces, the slightly swollen of mitochondria, and the indentation of nuclear envelop. The above outcomes suggested that prenatal hypoxia can lead to an obvious damage of fetal heart in a dependent manner.
Figure 1.
Prenatal hypoxic sheep showed different oxidative stress and their fetal hearts had different histomorphological characteristics. A. The levels of carbonyl groups (CO) and malondialdehyde (MDA) in prenatal hypoxic sheep; *P < 0.05, **P < 0.01, ##P < 0.01. B. Representative photomicrographs of fetal sheep myocardial tissues sections stained by hematoxylin and eosin (HE). C. Representative scanning electron microscopy (SEM) micrographs of fetal sheep myocardial tissues sections. D. Representative transmission electron microscopy (TEM) micrographs of fetal sheep myocardial tissues sections.
Identification of differentially expressed proteins under the three hypoxia conditions
Figure 2B provided the result of SDS-PAGE analysis. Obviously, the eight protein samples showed a complete parallelism, and the corresponding bands could be subsequently visualized, suggesting a very good extraction. Therefore, the purified samples were appropriate for LC/MS/MS. In tissue analysis, a total of 16679 peptides (Table S1) and 2506 proteins (Table S2) were quantified. Compared with NG, 113 proteins were upregulated and 70 proteins were downregulated in MHG, respectively; 47 proteins were upregulated and 54 proteins were downregulated in SHG (Table 2). Compared with MHG, SHG showed 80 upregulated proteins and 117 downregulated proteins (Table 2). To access the proteins with gradient change (up- or down-regulated proteins), we used more than 1.3-fold or less than 0.77-fold (severe hypoxia vs. normal condition), and more than 1-fold or less than 1-fold (severe hypoxia vs. mild hypoxia) cut off to define the proteins that had significantly expression difference with the degree of hypoxia in a dependent manner. Finally, a total of 80 differentially expressed proteins with 41 upregulated and 39 downregulated proteins were identified (Tables 2, S3), of which, the top 26 proteins were based on a threshold of > 1.3-fold and P value < 0.05 for severe hypoxia-induced protein expression levels compared with mild hypoxia, or P value < 0.05 of mild hypoxia relative to normal condition were shown in Table 3.
Table 2.
The number of differentially expressed proteins in hearts fetal sheep that exposed to mild hypoxia or severe hypoxia (multiple comparison, P < 0.05 and ratio > 1.3 or < 0.77)
| Comparison | Up | Down | Differentially expressed |
|---|---|---|---|
| Mild hypoxia vs. Normal | 113 | 70 | 183 |
| Severe hypoxia vs. Normal | 47 | 54 | 101 |
| Severe hypoxia vs. Mild hypoxia | 80 | 117 | 197 |
| Severe hypoxia vs. Mild hypoxia vs. Normal (Gradient) | 41 | 39 | 80 |
Table 3.
The top 26 differentially expressed proteins in the fetal heart tissues of the sheep exposed to hypoxia
| Differentially expression | Gene name | Severe/Mild | Mild/Normal | Severe/Normal | |||
|---|---|---|---|---|---|---|---|
|
| |||||||
| Ratio | T test | Ratio | T test | Ratio | T test | ||
| Upregulated | SURF1 | 2.27440 | 0.00945 | 1.01100 | 0.59218 | 2.29941 | 0.03539 |
| PHYH | 1.83916 | 0.24122 | 1.37521 | 0.03986 | 2.52923 | 0.25186 | |
| CCS | 1.63985 | 0.00244 | 1.01762 | 0.71198 | 1.66875 | 0.01432 | |
| SKOR2 | 1.51264 | 0.00355 | 1.18486 | 0.02225 | 1.79226 | 0.00717 | |
| REN | 1.50684 | 0.00018 | 1.27150 | 0.03053 | 1.91595 | 0.00071 | |
| W5Q9A6 | 1.50557 | 0.01366 | 1.18016 | 0.21017 | 1.77681 | 0.01248 | |
| TSPAN17 | 1.43757 | 0.00854 | 1.03390 | 0.58952 | 1.48630 | 0.02273 | |
| ACTR1B | 1.41908 | 0.01250 | 1.16014 | 0.31777 | 1.64633 | 0.04005 | |
| GUCY2F | 1.38522 | 0.02842 | 1.06556 | 0.55657 | 1.47604 | 0.04131 | |
| LOC101115423 | 1.37638 | 0.56495 | 1.36951 | 0.03605 | 1.88497 | 0.46592 | |
| DSCR3 | 1.36059 | 0.01065 | 1.03826 | 0.52394 | 1.41265 | 0.02394 | |
| IGHM | 1.35870 | 0.00482 | 1.18352 | 0.00974 | 1.60804 | 0.00742 | |
| APOBEC3F | 1.31595 | 0.00027 | 1.09504 | 0.00418 | 1.44101 | 0.00132 | |
| GPD1L | 1.30613 | 0.00213 | 1.06665 | 0.06008 | 1.39319 | 0.00756 | |
| Downregulated | RTN3 | 0.75898 | 0.01218 | 0.98075 | 0.49804 | 0.74437 | 0.03207 |
| SCN3B | 0.75803 | 0.00146 | 0.95341 | 0.01356 | 0.72272 | 0.00497 | |
| EXOC8 | 0.74039 | 0.01855 | 0.91427 | 0.25967 | 0.67692 | 0.02973 | |
| PGK2 | 0.72730 | 0.05809 | 0.73456 | 0.01284 | 0.53424 | 0.01367 | |
| TNFAIP8 | 0.72274 | 0.04213 | 0.78152 | 0.07815 | 0.56483 | 0.04068 | |
| ACOT4 | 0.68085 | 0.01129 | 0.99989 | 0.99896 | 0.68078 | 0.04264 | |
| CPED1 | 0.67732 | 0.33791 | 0.51392 | 0.01448 | 0.34809 | 0.05065 | |
| TMEM14C | 0.63955 | 0.00131 | 0.91158 | 0.15693 | 0.58300 | 0.00233 | |
| MPDZ | 0.54306 | 0.15814 | 0.91208 | 0.80293 | 0.49531 | 0.02003 | |
| COL28A1 | 0.45984 | 0.02078 | 0.86182 | 0.40641 | 0.39630 | 0.04065 | |
| TTN | 0.31632 | 0.00011 | 0.57689 | 0.00197 | 0.18248 | 0.00016 | |
| HBB | 0.09148 | 0.00007 | 0.95938 | 0.60434 | 0.08776 | 0.00012 | |
NOTE: The genes in bold font met the criterion that Severe/Mild T test P < 0.05, and Mild/Normal Ration > 1.3 or Mild/Normal T test P < 0.05.
Cluster analysis
To explore the complex potential interdependence of the proteins and visualize their expression patterns in response to hypoxia treatment, 80 differential proteins were further delivered to hierarchical clustering analysis via Cluster 3.0 software. Finally, two major clusters were obtained in the cluster tree (Figure 3). Almost all of the hypoxia-responsive proteins exhibited a coordinated change in different oxygenated environment, and their levels were declined significantly under hypoxia conditions, indicating an inhibitory effect of hypoxia on protein metabolism.
Figure 3.
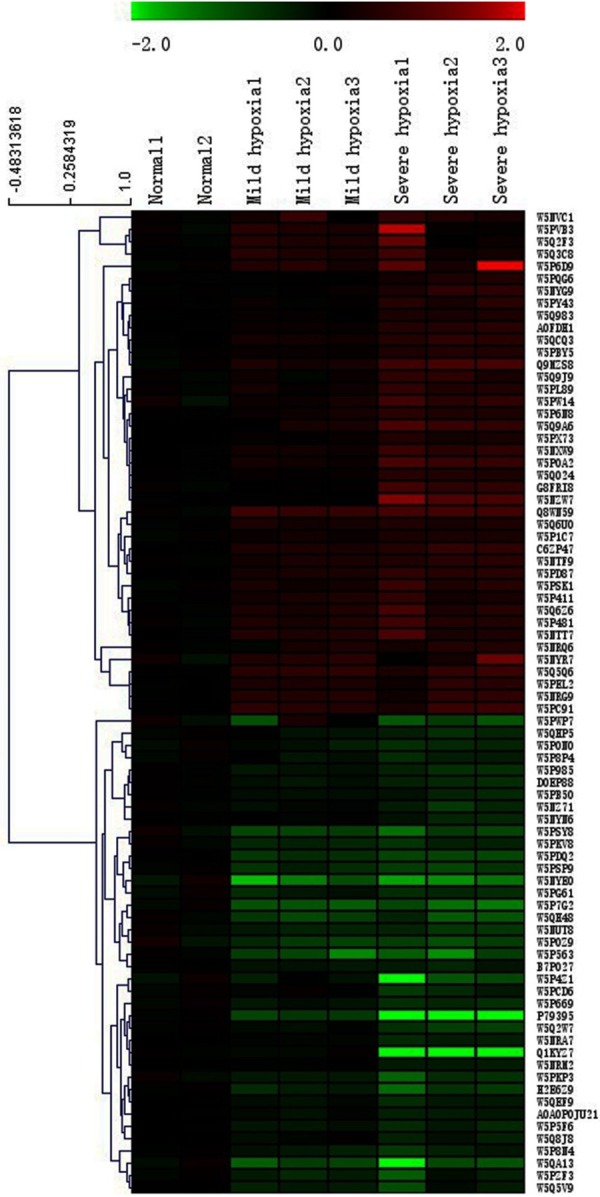
Hierarchical clustering analysis of eighty differentially expressed proteins. The identity of each protein can be found in Table S3. Each column represents an experimental condition, each row represents a gene, red indicates up-regulation, and green indicates down-regulation. Means are average values of the three independent replicates. NOTE: The genes in bold font met the following criterion: Severe/Normal > 1.3 or < 0.77, meanwhile, Severe/Mild > 1.0 or < 1.0.
GO analysis and KEGG pathway
Eighty differentially expressed proteins were annotated to 208 GO function entries according to the whole genome background. Second-level GO terms were used to classify proteins based on their involvement in three main categories: biological process (68 proteins), cellular component (73 proteins), and molecular function (67 proteins) (Table S3). Proteins belonging to each category are shown in Figure 4A. The most populated biological process was translation, to which 3 proteins were assigned. The top three cellular components were located on the lysosomal membrane (4 proteins), integral component of membrane (14 proteins), and Golgi apparatus (3 proteins), and the top three molecular functions were chromatin binding (3 proteins), identical protein binding (10 proteins), and zinc ion binding (5 proteins). Different proteins function together and cooperate to integrated biochemical reactions, a KEGG pathway-based analysis was conducted to confirm the pathways that would be potentially affected by differential protein expression in hypoxic myocardia. The top three pathways identified were the glycerophospholipid metabolism (3 proteins, P < 0.01), the protein digestion and absorption (3 proteins, P < 0.01), and the relaxin signaling pathway (3 proteins, P < 0.01), which were displayed in Table S3 and Figure 4B. Several important genes, such as collagen alpha-2(I) chain (COL1A2), and collagen alpha-1(I) chain (COL1A1), are involved in the process of infibrillar forming collagen, could be classified as part of the protein digestion and absorption. The expression of the majority of these proteins was significantly regulated in hypoxia-treated heart tissues compared with the normal tissues.
Figure 4.
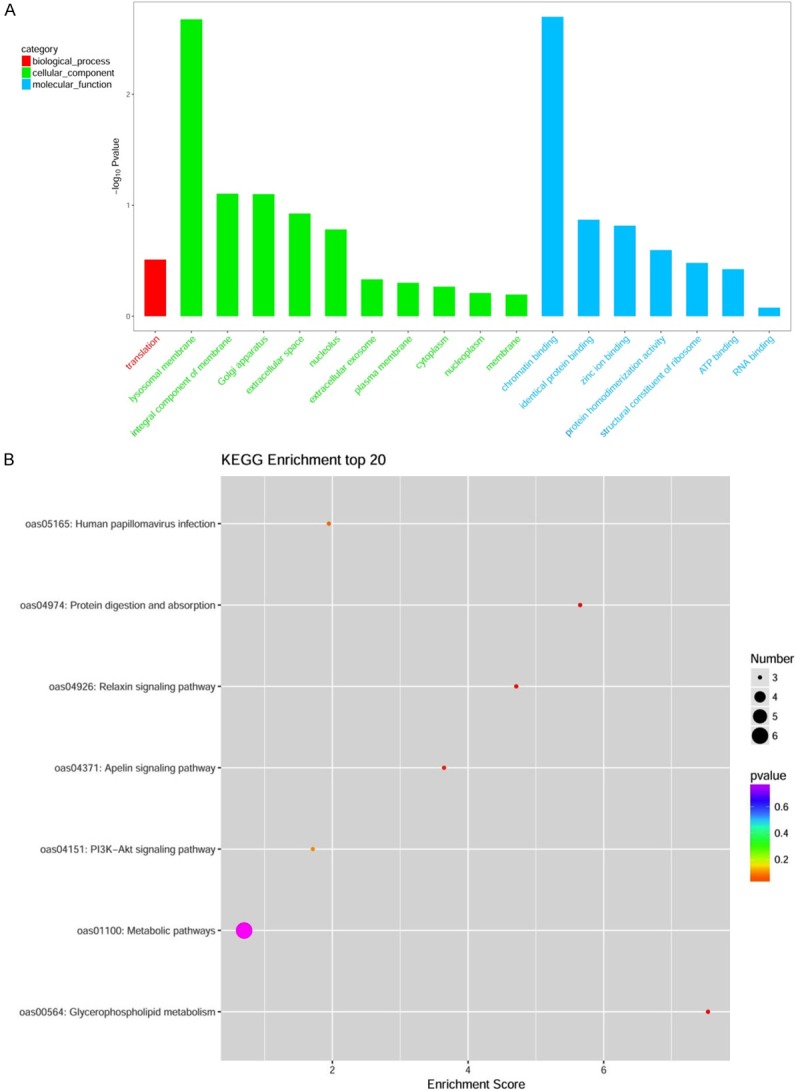
Enriched Gene Ontology (A) and the scatter plot for KEGG enrichment result (B).
Analysis of protein-protein interactions
Proteins in living cells do not serve as single entities, but rather function together in the context of networks. Next, to investigate how hypoxia stimulation exerted an influence on heart development through protein-protein interactions, we constructed a protein interaction network diagram for the 80 gradient differentially expressed proteins us-ing Pathway Studio software. Eight separate interaction networks were predicted in the network (Figure 5). 100851509 (an uncharacterized protein), LOC781493 (an uncharacterized protein), and collagen alpha-2(I) chain (COL1A2) were found to be actively interacted with collagen alpha-1(I) chain (COL1A1). One notable interacted protein group is composed of 60S ribosomal protein L36a (ENSBTAG00000027610), 60S ribosomal protein L13 (PRL13), 39S ribosomal protein L24 (MRPL24), 39S ribosomal protein L45 (MRPL45), WD repeat-containing protein 82 (TWF2), alpha-centractin (ACTR1A), tubulin beta-4A chain (TUBB4A), tubulin-folding cofactor B (TBCB), PABPC1L2A (an uncharacterized protein), PGK1 (Phosphoglycerate kinase 1), GPD1L (Glycerol-3-phosphate dehydrogenase), and Glycerol-3-phosphate dehydrogenase [NAD(+)], (GPD1). ATP6 (ATP synthase subunit α) was a key protein in another network and interacted with SURF1 (an uncharacterized protein), transmembrane protein 70 (TMEM70), V-type proton ATPase subunit d 1 (ATP6V0D1), and ATP synthase subunit d (ATP5H). Furthermore, there were five pairs of interactive protein-species. In this network, the main two pairs were beta-A globin chain HBB and hemoglobin subunit alpha (ENSBTAG00000026417), spectrin alpha chain (SPTA1), and band 3 anion transport protein (SLC4A1), ENSBTAG00000032603 (an uncharacterized protein), and glutathione reductase (GSR), DNA excision repair protein (ERCC6), and inositol-3-phosphate synthase 1 (ISYNA), mimecan precursor (OGN) and serine/threonine-protein kinase (MST4), respectively. Despite these predicted interaction networks required to be further verified, they have offered a narrow pool of protein-protein interactions in fetal heart development in responding to highland hypoxia environment for our further researches.
Figure 5.
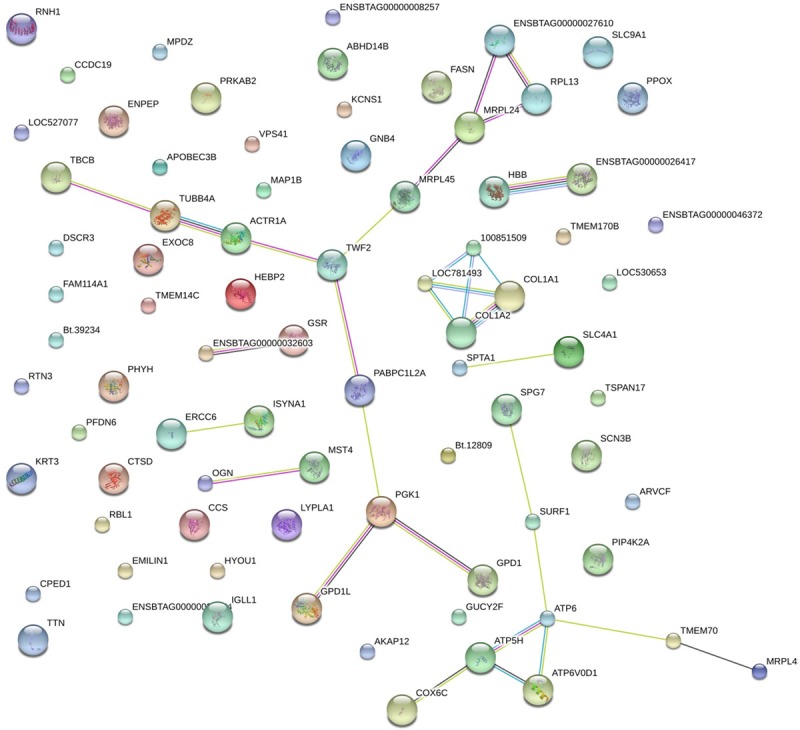
Protein-protein interaction network analysis of eighty gradient differential proteins. The interaction network as analyzed using the Pathway Studio software. In this network, nodes represent proteins, lines represent functional associations between proteins. Notes: red lines, the presence of fusion evidence; the green line, the neighborhood evidence; the blue line, the co-occurrence evidence; the purple line, the experimental evidence; the yellow line, text mining.
Discussion
Medically, high altitude area typically refers to elevations over at 3000 m, where there is thin air and hypothermia-hypoxia. The heart development of an early embryo is highly sensitive to environmental factors, and these development processes comprise cell proliferation, differentiation, and apoptosis. An observation by Camm et al. has made it clear that the PaO2 level of pregnant Wistar dams will drop to 40 mmHg when they were raised in a hypoxic condition for a long time, then the PaO2 concentration of the fetus will decrease to 10~12 mmHg and fetal hypoxia will occur [3]. Accordingly, a relative study has reported that pregnancy in a hypoxic environment can induce fetal to be attacked by various underlying diseases, such as coronary atherosclerotic heart disease, hypertension, diabetes, and so on [6]. Specially, congenital heart defects are a common hypoxia-induced cardiopathy, with the exact pathogenesis being a subject of much interest in recently years. Most converging evidence supports that the formation of congenital heart defect is a complex process involving the gene-environment interaction [21,22]. However, the specific pathogenic factor and potential mechanism remains largely unknown.
Currently, very limited research concerning the role of high-altitude hypoxia on fetal heart development is effectively performed due to the following reasons: 1) the field study is hard to conduct under highland environment; 2) a relatively simple and meaningful animal model has not been successfully established. In present study, we successfully prepared varying degrees of prenatal hypoxia sheep models via repeated adjusting the temperature, humidity, pressure, and oxygen content of artificial cabin to furthest simulate the low-oxygen environment of plateau region. After 90 days of experiments, the pregnant sheep developed symptoms of reduced breathing rate and increased heart rates in MHG and SHG by contrast with NG, meanwhile, the SpO2 and SaO2 levels were also markedly altered from the 30th day. Our observations in part were in good agreement with previous reports and further confirmed that hypoxia treatment can regulate the vital signs and behaviors of animals [23,24]. Reactive oxygen species (ROS) and reactive nitrogen species (RNS) are produced from multiple some metabolic pathways, which are the main factors for causing protein oxidation [25]. Previous study showed that, ROS and RNS are easy to react with intracellular macromolecular substances, then contribute to the extensive pathological injury of cellular structure [6]. For this reason, the extent of oxidative damage can be assessed by the free radicals-mediated proteins and lipid oxidation products [12]. At day 90 of the experiment, the levels of CO (the biomarker of oxidative damage in liver) and MDA (the biomarker of cellular oxidative damage) were remarkably improved in both mild-treated and severe hypoxia-treated sheep comparing with NG sheep, specially, MDA level in SHG were much higher than that in MHG (P < 0.01), which indicated that high-altitude hypoxia induces the oxidative stress response of animals, as well as aggravates the oxidative damage of proteins and lipids.
Previous investigation showed that hypoxia stimulation exerts an important impact on myocardial tissues [26], which can promote the proliferation of cardiac fibroblasts and destroy the cytoskeleton of myocardial microtubules, then prolong the open time of L-type calcium channel and cause the intracellular calcium overload via inducing calcium inward current. Sim-ultaneously, the highly expressed inflammatory cytokines, matrix metalloproteinases, anaerobic inductance, and hypoxia-inducible factor-1α in hypoxic myocardium would lead to myocardial damage. In our study, images of HE staining and SEM detection showed that the heart damages caused by prenatal hypoxia exposure is mainly reflected in two aspects: 1) damage to heart tissue microstructure, including the disorganization of microfilaments with the loss of Z lines, the degeneration of microfibers, the increase of the collagen fibers in the intracellular spaces, and the indentation of nuclear envelop; 2) the damage to the energy metabolism of myocardium, for example, the mitochondria on myocardial surface were arranged disorderly and degenerated, even, some mitochondria appeared to be obvious aggregated and obvious swollen. If above status continues for an unusually long time, the heart motor function will be destroyed by disorganizing the contractility and rhythm. Finally, a series of the heart pathological changes would break out, such as cardiac hypertrophy, cardiac enlargement, myocardial fibrosis, and arrhythmia.
Prior our research, several studies have provided proteomic data for injury of myocardial tissues induced by some harmful stimulation. For example, a pilot study performed by Xu et al. [27] investigated the proteomic differential expression of cardiomyocyte after hypoxia preconditioning, and finally identified 12 decreased proteins and 6 highly expressed proteins. Through the establishment of a myocardium injury model induced by restraint stress in rats, Gong et al. [28] found that there were 10 differentially expressed proteins involved in the stress-induced injury to myocardium. However, to our knowledge, not much attention has been paid to the proteomics analysis of fetal heart tissues exposure to prenatal hypoxia thus far. In present study, a total of 80 differentially expressed proteins containing 41 upregulated and 39 downregulated proteins were identified with the degree of hypoxia in a dependent manner. Superoxide dismutase [Cu-Zn] (SOD1) and cathepsin D are the typical upregulated proteins. As we have known, SODs represent the first line of defense against oxidative stress. Large amounts of data suggest that SOD1 overexpression may protect some organisms against oxidative stress conditions and extend their life span [29,30]. In this study, hypoxia treatment exacerbates the oxidative damage of myocardial mitochondria, thus further contributes to the high expression of SOD1. Cathepsin D play a promoter in the degradation of cardiac mitochondria, which is generally considered as the mitochondrial destruction characteristic of ischemic injury in myocardial tissues [31]. Therefore, hypoxia induced-mitochondrial damage is inevitable to be accompanied with upregulated cathepsin D.
Next, we conducted GO analysis and KEGG pathway analysis to further confirm the functions of 80 differentially expressed proteins and the relative protein pathways. In GO analysis, the top three cellular components were lysosomal membrane (4 proteins), integral component of membrane (14 proteins), and Golgi apparatus (3 proteins). Lysosomal membrane damage has been turned out to be the main contributing factor of myocardial infarction followed by necrosis, which can be destabilized under the impact oxidative stress [32]. The golgi apparatus is a key organelle in cell metabolism, which is involved in modifying, sorting, and packaging macromolecules for cell secretion or use. One study supported that the golgi apparatus not only participated in the process of oxidative stress, but also plays a more important role than mitochondria [33]. Zinc ion binding is one of the top three molecular functions. Hypoxia can result in increased superoxide anions and the decrease of Zinc ion, meanwhile, Zinc deficiency causes oxidative damage to proteins, lipids and DNA in rat myocardium, then contribute to the apoptosis of cardiomyocytes [34,35]. The relaxin signaling pathway is one of the top three pathways identified from the KEGG database analysis. Relaxin, as a peptide hormone, has been proved to significantly decrease the production of MDA in the ischemic-re-perfusion rat hearts [36]. Furthermore, relaxin also acts at multiple levels in the cascade of events leading to myocardial damage, which refer to the endothelial dysfunction [37], generation of oxygen-derived free radicals by these cells [38], neutrophil accumulation in the myocardium [38], platelet and mast cell activation [39], and calcium overload that eventually leads to myocardial cell damage and death [40]. Our data was in part in agreement with a previous report suggesting relaxin is involved in the myocardial injury [41].
In current study, the different degrees of hypoxic pregnancy in the sheep model were successfully established for simulating high altitude hypoxia environment by using artificial cabin. Our results reveal the effect of hypoxia exposure during pregnancy on the congenital heart injury of fetal sheep, and preliminarily identify the pathological changes in heart tissues. Furthermore, systematic proteomic analysis of fetal heart tissues generated the hypoxia-associated differential proteins, which may lay a theoretical basis for the pathogenesis and intervention of prenatal hypoxia-induced diseases.
Acknowledgements
This study was supported by Regional Project of National Natural Science Foundation of China (Grant No. 81560291).
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Penaloza D, Ariasstella J. The heart and pulmonary circulation at high altitudes: healthy highlanders and chronic mountain sickness. Circulation. 2007;115:1132–1146. doi: 10.1161/CIRCULATIONAHA.106.624544. [DOI] [PubMed] [Google Scholar]
- 2.Powers DE, Millman JR, Huang RB, Colton CK. Effects of oxygen on mouse embryonic stem cell growth, phenotype retention, and cellular energetics. Biotechnol Bioeng. 2008;101:241–54. doi: 10.1002/bit.21986. [DOI] [PubMed] [Google Scholar]
- 3.Camm EJ, Hansell JA, Kane AD, Herrera EA, Lewis C, Wong S, Morrell NW, Giussani DA. Partial contributions of developmental hypoxia and undernutrition to prenatal alterations in somatic growth and cardiovascular structure and function. Am J Obstet Gynecol. 2010;203:24–34. doi: 10.1016/j.ajog.2010.06.046. [DOI] [PubMed] [Google Scholar]
- 4.Hartinger S, Tapia V, Carrillo C, Bejarano L, Gonzales GF. Birth weight at high altitudes in Peru. Int J Gynaecol Obstet. 2006;93:275–281. doi: 10.1016/j.ijgo.2006.02.023. [DOI] [PubMed] [Google Scholar]
- 5.Huicho L, Niermeyer S. Cardiopulmonary pathology among children resident at high altitude in Tintaya, Peru: a cross-sectional study. High Alt Med Biol. 2006;7:168–179. doi: 10.1089/ham.2006.7.168. [DOI] [PubMed] [Google Scholar]
- 6.Kadiiska MB, Peddada S, Herbert RA, Basu S, Hensley K, Jones DP, Hatch GE, Mason RP. Biomarkers of oxidative stress study VI. Endogenous plasma antioxidants fail as useful biomarkers of endotoxin-induced oxidative stress. Free Radic Biol Med. 2015;81:100–106. doi: 10.1016/j.freeradbiomed.2015.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ascuitto RJ, Ross-Ascuitto NT. Substrate metabolism in the developing heart. Semin Perinatol. 1996;20:542–63. doi: 10.1016/s0146-0005(96)80068-1. [DOI] [PubMed] [Google Scholar]
- 8.Ferencz C, Neill CA. Cardiovascular malformations: prevalence at livebirth. 1992 [Google Scholar]
- 9.Monroymuñoz IE, Pérezhernández N, Vargasalarcón G, Ortizsan JG, Buendíahernández A, Calderóncolmenero J, Ramírezmarroquín S, Cervantessalazar JL, Curicuri P, Martínezrodríguez N. [Changing the paradigm of congenital heart disease: from the anatomy to the molecular etiology] . Gac Med Mex. 2013;149:212–219. [PubMed] [Google Scholar]
- 10.Zaffran S, Kelly RG. New developments in the second heart field. Differentiation. 2012;84:17–24. doi: 10.1016/j.diff.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 11.Jie HU, Hua LI, Jiao MK, Duan HY Echocardiography DO. Reproduction of the model of plateau hypoxia in different degrees in gestational sheep. Medical Journal of Chinese Peoples Liberation Army. 2015;40:358–361. [Google Scholar]
- 12.Parraguez VH, Urquieta B, Pérez L, Castellaro G, De los Reyes M, Torres-Rovira L, Aguadomartínez A, Astiz S, González-Bulnes A. Fertility in a high-altitude environment is compromised by luteal dysfunction: the relative roles of hypoxia and oxidative stress. Reprod Biol Endocrinol. 2013;11:24. doi: 10.1186/1477-7827-11-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chunhua X. Color doppler combined three dimensional ultrasound in diagnosis of fetal distress in uterus. Chinese Journal of Ultrasound in Medicine. 2013;29:158–160. [Google Scholar]
- 14.Zhihua C, Yicheng W, Ruimin Y, Hongyu J, Bing L, Likun W, Liwei Z. Prediction of fetal hypoxia using resistant parameter ratio of fetal renal artery to middle cerebral artery combined with tei index in hypertensive disorders complicating pregnancy. Chinese Journal of Ultrasound in Medicine. 2015;31:38–40. [Google Scholar]
- 15.Umezawa T, Sugiyama N, Takahashi F, Anderson JC, Ishihama Y, Peck SC, Shinozaki K. Genetics and phosphoproteomics reveal a protein phosphorylation network in the abscisic acid signaling pathway in Arabidopsis thaliana. Sci Signal. 2013;6:rs8. doi: 10.1126/scisignal.2003509. [DOI] [PubMed] [Google Scholar]
- 16.Hu X, Li N, Wu L, Li C, Li C, Zhang L, Liu T, Wang W. Quantitative iTRAQ-based proteomic analysis of phosphoproteins and ABA-regulatedphosphoproteins in maize leaves under osmotic stress. Sci Rep. 2015;5:15626. doi: 10.1038/srep15626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ren Y, Hao P, Dutta B, Cheow ES, Sim KH, Gan CS, Lim SK, Sze SK. Hypoxia modulates A431 cellular pathways association to tumor radioresistance and enhanced migration revealed by comprehensive proteomic and functional studies. Mol Cell Proteomics. 2013;12:485–98. doi: 10.1074/mcp.M112.018325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang Y, Bu D, Zhao X, Sun P, Wang J, Zhou L. Proteomic analysis of cow, yak, buffalo, goat and camel milk whey proteins: quantitative differential expression patterns. J Proteome Res. 2013;12:1660–7. doi: 10.1021/pr301001m. [DOI] [PubMed] [Google Scholar]
- 19.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The gene ontology consortium. Nat Genet. 2000;25:25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu J, Li H, Niu M, Sun Y, Li Y. Correlation analysis of sheep fetal cariac pathological injury with hypoxia environment induration of pregnancy: experimental study. Chinese Journal of Ultrasound in Medicine. 2016;32:1041–1044. [Google Scholar]
- 21.Hobbs CA, Cleves MA, Karim MA, Zhao W, Macleod SL. Maternal folate-related gene environment interactions and congenital heart defects. Obstet Gynecol. 2010;116:316–22. doi: 10.1097/AOG.0b013e3181e80979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang X, Li P, Chen S, Xi L, Guo Y, Guo A, Sun K. Influence of genes and the environment in familial congenital heart defects. Mol Med Rep. 2014;9:695–700. doi: 10.3892/mmr.2013.1847. [DOI] [PubMed] [Google Scholar]
- 23.Seet RC, Lee CY, Loke WM, Huang SH, Huang H, Looi WF, Chew ES, Quek AM, Lim EC, Halliwell B. Biomarkers of oxidative damage in cigarette smokers: which biomarkers might reflect acute versus chronic oxidative stress? Free Radic Biol Med. 2011;50:1787–1793. doi: 10.1016/j.freeradbiomed.2011.03.019. [DOI] [PubMed] [Google Scholar]
- 24.Nishimura R, Okuda K. Hypoxia is important for establishing vascularization during corpus luteum formation in cattle. J Reprod Dev. 2010;56:110–116. doi: 10.1262/jrd.09-162e. [DOI] [PubMed] [Google Scholar]
- 25.Yan-Bo LI, Zhou W, Yong-Bo YU, Duan JC, Guo CX, Sun ZW. Cytotoxicity and oxidative damage effect of silica nanoparticles on vascular endothelial cells. Journal of Jilin University. 2014;40:476–481. [Google Scholar]
- 26.Anderson JD, Honigman B. The effect of altitude-induced hypoxia on heart disease: do acute, intermittent, and chronic exposures provide cardioprotection? High Alt Med Biol. 2011;12:45–55. doi: 10.1089/ham.2010.1021. [DOI] [PubMed] [Google Scholar]
- 27.Xu FF, Sun S, Liu XH. Pilot Study of proteomic changes of cardiomyocyte induced by hypoxia preconditioning. Acta Chimica Sinica. 2006;64:543–550. [Google Scholar]
- 28.Gong J, Wu S, Qian L. Proteomics analysis on stressed myocardium injury-related proteins. Chinese Journal of Applied Physiology. 2005;21:171–174. [PubMed] [Google Scholar]
- 29.Landis GN, Tower J. Superoxide dismutase evolution and life span regulation. Mech Ageing Dev. 2005;126:365–79. doi: 10.1016/j.mad.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 30.Parker JD, Parker KM, Sohal BH, Sohal RS, Keller L. Decreased expression of Cu-Zn superoxide dismutase 1 in ants with extreme lifespan. Proc Natl Acad Sci U S A. 2004;101:3486–9. doi: 10.1073/pnas.0400222101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hines JL, Ogunro EA, Ferguson AG, Lesch M. Cathepsin D catalyzed degradation of cardiac mitochondria. A study of enzyme inactivation and ultrastructure. J Mol Cell Cardiol. 1982;14:91–97. doi: 10.1016/0022-2828(82)90197-3. [DOI] [PubMed] [Google Scholar]
- 32.Pullaiah CP, Kumar GVN, Jyothsna K, Thyagaraju K, Nelson VK, Reddy GD. Rosa damascena Mill. L. attenuates myocardial lysosomal membrane destabilization in isoproterenol induced oxidative stress. Oriental Pharmacy and Experimental Medicine. 2017:1–8. [Google Scholar]
- 33.Jiang Z, Hu Z, Zeng L, Lu W, Zhang H, Li T, Xiao H. The role of the Golgi apparatus in oxidative stress: is this organelle less significant than mitochondria? Free Radic Biol Med. 2011;50:907–917. doi: 10.1016/j.freeradbiomed.2011.01.011. [DOI] [PubMed] [Google Scholar]
- 34.Nakajima H, Hangaishi M, Ishizaka N, Taguchi J, Igarashi R, Mizushima Y, Nagai R, Ohno M. Lecithinized copper, zinc-superoxide dismutase ameliorates ischemia-induced myocardial damage. Life Sci. 2001;69:935–44. doi: 10.1016/s0024-3205(01)01188-2. [DOI] [PubMed] [Google Scholar]
- 35.Oteiza PI, Olin KL, Fraga CG, Keen CL. Zinc deficiency causes oxidative damage to proteins, lipids and DNA in rat testes. J Nutr. 1995;125:823–829. doi: 10.1093/jn/125.4.823. [DOI] [PubMed] [Google Scholar]
- 36.Rao PS, Cohen MV, Mueller HS. Production of free radicals and lipid peroxides in early experimental myocardial ischemia. J Mol Cell Cardiol. 1983;15:713–716. doi: 10.1016/0022-2828(83)90260-2. [DOI] [PubMed] [Google Scholar]
- 37.VanBenthuysen KM, McMurtry IF, Horwitz LD. Reperfusion after acute coronary occlusion in dogs impairs endothelium-dependent relaxation to acetylcholine and augments contractile reactivity in vitro. J Clin Invest. 1987;79:265–274. doi: 10.1172/JCI112793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nichols WW, Mehta JL, Donnelly WH, Lawson D, Thompson L, Ter RM. Reduction in coronary vasodilator reserve following coronary occlusion and reperfusion in anesthetized dog: role of endothelium-derived relaxing factor, myocardial neutrophil infiltration and prostaglandins. J Mol Cell Cardiol. 1988;20:943–954. doi: 10.1016/s0022-2828(88)80148-2. [DOI] [PubMed] [Google Scholar]
- 39.May GR, Crook P, Moore PK, Page CP. The role of nitric oxide as an endogenous regulator of platelet and neutrophil activation within the pulmonary circulation of the rabbit. Br J Pharmacol. 1991;102:759–763. doi: 10.1111/j.1476-5381.1991.tb12246.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zimmerman AN, Hülsmann WC. Paradoxical influence of calcium Ions on the permeability of the cell membranes of the isolated rat heart. Nature. 1966;211:646–647. doi: 10.1038/211646a0. [DOI] [PubMed] [Google Scholar]
- 41.Bani D, Masini E, Bello MG, Bigazzi M, Sacchi TB. Relaxin protects against myocardial injury caused by ischemia and reperfusion in rat heart. Am J Pathol. 1998;152:1367–1376. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.