Abstract
It is well known that Helicobacter pylori (H. pylori) is not the only indigenous bacterium in the stomach, as numerous studies have revealed that the gastric microbiota contributes to the pathogenesis of gastric disease. However, the correlation between the gastric bacterial flora and gastritis is unclear. By comparing differences in viable gastric bacteria between a gastritis group and a healthy group, we examined the potential species related to chronic gastritis. We collected juice and mucosa samples from 103 consecutive patients and identified 81 species by culturing and matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS). The positive rates of Streptococcus and Neisseria were markedly higher in the gastritis group than those in the normal group, suggesting that certain bacterial species may play vital roles in the development of gastritis rather than acting as transient microbes. This finding can be applied to the diagnosis and treatment of chronic gastritis as evidence supporting non-Helicobacter pylori infection-related gastritis.
Keywords: Gastric microbiota, helicobacter pylori, gastritis, infection disease, matrix-assisted laser desorption ionization time-of-flight mass spectrometry
Introduction
Chronic inflammation is one of the most common gastric conditions worldwide due to bacterial infection, alcohol consumption and biliary reflux. Helicobacter pylori (H. pylori) infection is the most well-known risk factor [1]. However, infection with other gastric pathogens has been overlooked. The bacteria of gastritis patients must be reviewed, as they were once considered transient bacteria.
Recent studies have indicated that the gastric microbiota are entirely different from the microbiota of the oral cavity, throat, or other parts of the upper gastrointestinal tract [2]. The gastric microbiota may promote H. pylori-associated disease [3]. The interaction between H. pylori and the microbiota also influences gastric disease progression [4,5]. Candida albicans is an opportunistic pathogen that induces gastritis in patients taking antibiotics [6]. Streptococcus-related gastritis has been described in several case reports, but it was never investigated thoroughly [7,8]. Exploration of gastric pathogens is essential to gain insight into infectious diseases of the stomach.
The 5th Maastricht V/Florence Consensus [9] presented a workshop to discuss the role of the gastric microbiota in H. pylori-associated disease and H. pylori eradication effects on the microbiota. Although the evidence level was low and the grade of the recommendation was weak, maintaining the balance of the gastric ecosystem was reported to be crucial. Similar to pneumonia, chronic gastritis can also be induced by different pathogens. However, all the microbes cultured from the stomach have been considered transient bacteria, and no solid evidence is available regarding potential pathogens in the gastric microbiota. Marshall’s experiment [10] must be repeated to identify other bacteria in the stomachs of patients with chronic gastritis.
MALDI-TOF MS is an innovative tool that is easy to use, rapid, accurate, and cost-effective, and it has revolutionized the identification of highly pathogenic bacteria in clinical microbiology laboratories [11,12]. By immediately culturing gastric specimens and implementing MALDI-TOF MS in the hospital laboratory, we attempted to examine gastritis pathogens from 50 healthy subjects and 53 gastritis patients. Two types of medium were used to culture gastric juice and mucosa samples under three conditions. Four species, including H. pylori, were found to be significantly associated with chronic gastritis. Additionally, bacterial overgrowth and an abnormal pH were associated with the severity of gastritis. These species may facilitate the understanding of other infection factors that predispose individuals to gastritis.
Materials and methods
Overview of the study
From May 2015 to May 2016, 103 patients referred for endoscopy and histologic examination at Peking University Third Hospital were recruited for this research. Gastric juice and mucosa samples of the corpus and antrum were collected into sterile containers and were sent to the microbiology laboratory for immediate bacterial culture. All enrolled patients provided informed consent for clinical sample collection. This research was approved by the Medical Scientific Ethics Committees of Peking University Third Hospital (IRB00006761-2015172).
Patients and sample culturing
Of the participants enrolled in this study, 53 did not have gastritis and 50 had chronic gastritis. Individuals were recruited if they were 20 to 60 years old without severe complications or other systemic disease and normal lifestyle. Persons diagnosed with cancer, or those who had taken antibiotics or acid inhibitory drugs in recent months before the endoscopy examination were excluded. Mucosal biopsies and two 3-mm diameter ceramic beads were placed into a homogenizing pipe (Eppendorf Corporation, Germany) containing 400 µl of sterile normal saline. The auto homogenizer (Retsch Corporation, Germany) was set to 30 times per second to produce tissue homogenate. According to the pH of the gastric juice, an appropriate volume of 10-100 µl of tissue homogenate and gastric juice were incubated on agar plates. All samples were cultured on Columbia blood agar (Oxoid LTD, Basingstoke, Hampshire, England) and Helicobacter pylori-selective culture medium (Oxoid LTD, Basingstoke, Hampshire, England) for 1, 5 and 3 days under aerobic, anaerobic and microaerophilic conditions respectively. Colony-forming units (CFU) were detected by morphology and microscopy examination.
Identification of isolates
Various forms of bacterial colonies were identified using the direct transfer approach [13] of the Bruker IVD MALDI (Matrix Assisted Laser Desorption Ionization) Biotyper (Bruker, Germany) according to the standard of operation based on the instruction of the facility [14]. A single colony was transferred to the MALDI target plate using an inoculating loop and smeared gently as a thin film within a spot on the plate. The dyed film was overlain with formic acid and then dyed again. One µl of IVD was added to the HCCA matrix. The film was allowed to dry at ambient temperature. The sample location on the plate was assessed and recorded. The plate was placed in the ionization chamber of the mass spectrometer and the MSP identification IVD method was selected.
A laser was applied to the spots subjected to analysis to desorb and ionize microbial and matrix molecules on the target plate. The cloud of ionized molecules was directed into the TOF mass analyzer, toward a detector. Lighter molecules travel faster, followed by progressively heavier analytes. A mass spectrum was generated, representing the number of ions hitting the detector over time [15]. The details of the parameter settings of the operation are available in the brochure (https://www.bruker.com/fileadmin/user_upload/8-PDF-Docs/Separations_MassSpectrometry/Literature/Brochures/1840190_MBT_Clinical_IVD_brochure_10-2015_eBook.pdf).
Assessment scores were applied to evaluate the quality of identification. A score of ≥ 2.000 indicated reliable identification at the species level, a score of 1.700 to 1.999 indicated reliable identification at the genus level, and a score of < 1.700 indicated unreliable identification [14].
Unreliably identified stains were determined by 16SrDNA sequencing via mass spectrometry. All bacterial colonies that were not identified were subcultured and collected for DNA extraction. Full-length 16 s rDNA was amplified with 1492R (5’-GGTTACCTTGTTACGACTT-3’) and 27F (5’-AGAGTTTGATCCTFFCTCAG-3’) primers. The PCR conditions were as follows: one cycle at 95°C for 5 min, 35 cycles at 94°C for 30 s, 50°C for 45 s and 72°C for 2 min, and a final extension step at 72°C for 10 min [2]. Then, 1500-bp PCR products were purified from the agarose gel and sent to BGI Company for sequencing. A Fastq file was uploaded to the NICB website (http://www.ncbi.nlm.nih.gov/BLAST/) for blasting. The identities of the isolates were determined based on the highest scores. Sequences with a percentage similarity of 97% or higher were assigned to a species category.
Statistical analysis
The data analysis was performed using SPSS (version 20.0). Continuous variables are described as the mean plus the stander error and counting variables are shown as numbers and ratios. Correlations between continuous variables were assessed using Pearson correlation coefficients. Wilcoxon signed rank tests were used to compare continuous variables. Patient characteristics and positive rates of certain bacteria were analyzed by Fisher’s test or the chi-squared (X2) method as appropriate. Odds ratios are summarized as asymptotic standard errors and 95% confidence intervals (95% CI). A P value ≤0.05 indicated a significant difference.
Results
One hundred three patients were included for culturing of gastric mucosa and gastric juice. According to the histopathology test of gastric biopsy tissues, 53 patients had chronic gastritis and 50 patients were normal. Therefore, the recruited cases were divided into a gastritis group and a control group. The patients in the chronic gastritis group presented with concomitant hemorrhage, atrophy, intestinal metaplasia or hyperplasia. The healthy group included 50 subjects with mild superficial gastritis and served as the control group. The two groups were matched according to mean age, gender, and endoscopy status (Table 1). The positive rate of mucosal bacterial growth was higher in the gastritis group (86.7%) than that in the normal group (48%), with a significant difference between the groups (P < 0.001) even after excluding H. pylori infection data (P=0.014).
Table 1.
Demographic characteristics of the study group and the control group
| No. (%) | P value | ||
|---|---|---|---|
|
| |||
| Gastritis (n=53) | Normal (n=50) | ||
| Males/Females | 26/27 | 1 | 0.924 |
| Mean age (SD) | 44.32 (10.84) | 43.16 (11.33) | 0.597 |
| H. pylori-positive | 22 | 2 | < 0.001 |
| Mean pH value | 2.7 (1.32) | 2.49 (1.21) | 0.403 |
| Bile reflux | 13 (24.5) | 18 (36) | 0.205 |
| Gastric juice growth | 33 (62.3) | 27 (54) | 0.395 |
| Mucosal growth | 46 (86.8) | 24 (48) | < 0.001 |
| Mucosal growth (no H. pylori) | 38 (71.7) | 24 (48) | 0.014 |
Gastric microbiota
Eighty-one species were isolated from all the gastric mucosa and juice samples, of which 56 of which were positive in the gastric juice samples, 53 were positive in the gastric mucosa samples, and 28 were positive in both gastric juice and mucosal biopsies. All the isolated gastric species are listed in Tables S1 and S2. As shown in Table 2, Helicobacter pylori (23.3%), Rothia mucilaginosa (23.3%) and Neisseria flavescens (22.3%) were common in the mucosa. In the gastric juice, Candida albicans (14.6%), Streptococcus Oralis (9.7%) and Rothia mucilaginosa (8.7%) were common, as shown in Table 3.
Table 2.
Gastric mucosal bacteria
| Mucosal bacteria | Aerobic | Microaerobic | Selective | Anaerobic | Colonized patients No. (%) | Gastritis No. (%) | Normal No. (%) | P value |
|---|---|---|---|---|---|---|---|---|
| Rothia mucilaginosa | 22 | 11 | 1 | 3 | 24 (23.3) | 9 (17.0) | 15 (30) | 0.188 |
| Helicobacter pylori | 0 | 10 | 24 | 0 | 24 (23.3) | 22 (41.5) | 2 (4.0) | < 0.001 |
| Neisseria flavescens | 16 | 15 | 15 | 0 | 23 (22.3) | 16 (30.2) | 7 (14) | 0.049 |
| Streptococcus mitis | 13 | 8 | 0 | 10 | 18 (17.5) | 14 (26.4) | 4 (8) | 0.014 |
| Streptococcus salivarius | 9 | 5 | 2 | 9 | 16 (15.5) | 10 (18.9) | 6 (12) | 0.336 |
| Streptococcus oralis | 9 | 5 | 0 | 11 | 16 (15.5) | 11 (20.8) | 5 (10) | 0.132 |
| Neisseria perflava | 6 | 3 | 5 | 0 | 11 (10.7) | 10 (18.9) | 1 (2) | 0.006 |
| Streptococcus parasanguis | 5 | 1 | 1 | 2 | 8 (7.8) | 5 (9.4) | 3 (6) | 0.716 |
| Rothia dentocariosa | 1 | 6 | 0 | 5 | 8 (7.8) | 4 (7.5) | 4 (8) | >0.999 |
| Staphylococcus aureus | 3 | 6 | 0 | 0 | 7 (6.8) | 2 (3.8) | 5 (10) | 0.261 |
Table 3.
Gastric juice culturable bacteria
| Juice species | Aerobic | Microaeroic | Selective | Anaerobic | Colonized patients No. (%) | Gastitis No. (n=53) | Normal No. (n=50) | P value |
|---|---|---|---|---|---|---|---|---|
| Candida albicans | 9 | 11 | 12 | 9 | 15 (14.6) | 9 | 6 | 0.337 |
| Streptococcus oralis | 8 | 4 | 0 | 4 | 10 (9.7) | 1 | 9 | 0.017 |
| Rothia mucilaginosa | 5 | 6 | 0 | 1 | 9 (8.7) | 0 | 9 | 0.003 |
| Neisseria flavescens | 4 | 5 | 5 | 1 | 7(6.8) | 2 | 5 | 0.438 |
| Streptococcus mitis | 1 | 2 | 1 | 6 | 6 (5.8) | 2 | 4 | 0.679 |
| Neisseria perflava | 6 | 3 | 3 | 0 | 6 (5.8) | 0 | 6 | 0.027 |
| Staphylococcus aureus | 5 | 4 | 0 | 3 | 6 (5.8) | 4 | 2 | 0.428 |
| Streptococcus salivarius | 1 | 2 | 1 | 4 | 5 (4.8) | 1 | 4 | 0.364 |
| Ralstonia pickettii | 1 | 3 | 0 | 1 | 5 (4.8) | 2 | 3 | >0.999 |
Gastritis and microbiota
Positive rates of Helicobacter pylori (P < 0.001), Streptococcus mitis (P=0.014), Neisseria flavescens (P=0.049) and Neisseria perflava (P=0.006) in the mucosa of the chronic gastritis group were significantly higher than those in the mucosa of the healthy group (Table 2). The odds ratios (ORs) of these potential pathogens are shown in Figures 1 and 2: Helicobacter pylori (OR=17.03, [95% CI, 3.74-77.59]), Streptococcus mitis (OR=4.13, [95% CI, 1.26-13.57]), Neisseria flavescens (OR=2.66, [95% CI, 1.00-7.16]), and Neisseria perflava (OR=11.40, [95% CI, 1.40-92.69]). The positive mucosal rates of Neisseria (41.5% vs. 16% P=0.009) and Streptococcus (50.9% vs. 28% P=0.017) were significantly different between the chronic gastritis group and the normal group.
Figure 1.
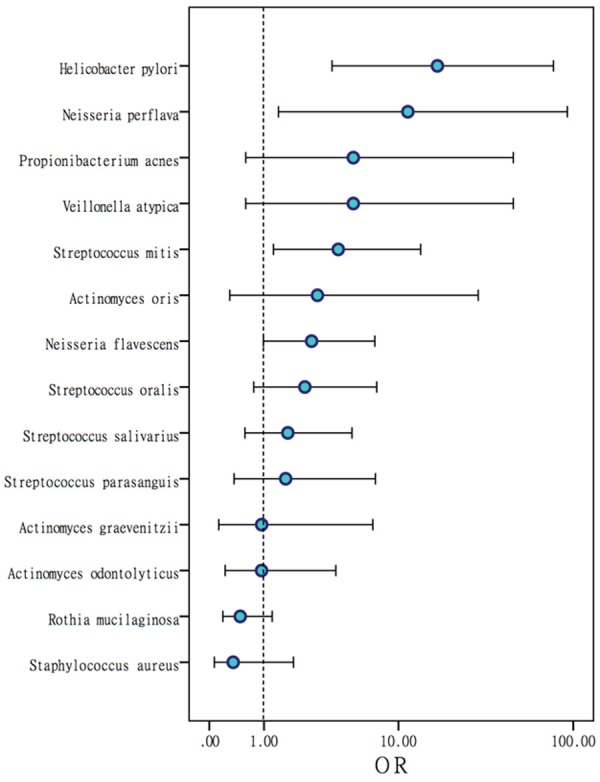
Odds ratios (ORs) of colonized gastric mucosal bacteria. OR values show the risk of gastritis. An OR value greater than 1 reflects a high risk of gastritis.
Figure 2.
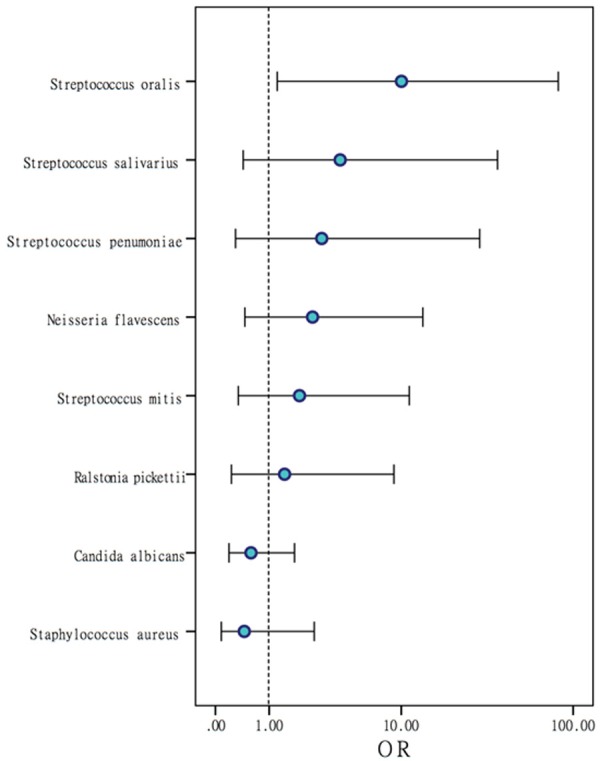
Odds ratios (ORs) of positive gastric juice bacteria. Species with a greater OR value are associated with a higher risk of gastritis.
Cultures of gastric juice showed notable differences in the positive rates of Streptococcus oralis (P=0.017), Rothia mucilaginosa (P=0.003) and Neisseria perflava (P=0.027) between the gastritis and normal groups (Table 3). Similar to the mucosal culture results, the positive rates of certain bacterial species were higher in the gastritis group than those in the control group.
Symbiosis
Bacterial overgrowth (> 105 CFU/L) of 16 species was found in 17 gastric juice samples, and overgrowth (> 105 CFU/g) of 8 species was observed in 22 mucosal samples. Bacterial overgrowth in gastric juice (P < 0.001) and mucosa (P=0.009) was significantly associated with an abnormal pH value of gastric juice (pH > 4), which was significantly correlated with chronic gastritis (P=0.036). Sixteen mucosal samples from 24 Helicobacter pylori-positive selective agars were colonized by non-Helicobacter pylori bacterial flora, including Streptococcus, Neisseria, Ralstonia and Staphylococcus, suggesting that other bacteria live in the gastric mucosa concurrently with Helicobacter pylori.
Discussion
This study revealed that Streptococcus, Neisseria and H. pylori were significantly associated with chronic gastritis. As a form of infectious disease, chronic gastritis can be caused by pathogens other than H. pylori. Other potential pathogens in the stomach may include a range of bacterial species such as Mycobacterium and Enterococcus [1]. Evidence-based data regarding other etiologies of chronic gastritis are limited, although several cases studies have reported that Streptococcus spp. are associated with acute gastritis [16,17]. Additionally, the clinical diagnosis and treatment of chronic gastritis generally focus on H. pylori infection.
H. pylori is a well-known bacterial species in the stomach that can survive among hundreds of species in gastric acid [18]. However, the H. pylori sequence can be detected in 60% of conventionally negative samples [19,20]. Therefore, H. pylori is considered a normal microbe that is beneficial to its host for a certain period. Blaser et al [21] investigated a series of possible disorders that may be caused by H. pylori loss. Eradicating H. pylori is controversial and requires long-term observational and clinical research.
The latest consensus on the management of H. pylori discussed the gastric microbiota in terms of its composition as determined by sequencing. Bik et al [22] found 128 species in the stomach and invalidated the concept of H. pylori as the only gastric colonizer. Schulz identified 687 phylotypes from 24 individuals by RNA amplification and sequencing [23]. However, comparisons of the gastric microbiota between gastritis and non-gastritis subjects have been neglected. In our study, specimens from 103 individuals with or without gastritis were cultured for live bacterial detection. We identified 81 species in samples from healthy subjects and gastritis patients by MLDI-TOF. Delgado et al [2] isolated 16 species from mucosal and gastric samples of healthy human stomachs; the dominant genus was entirely different for the various community sources of the samples.
The prevalence of non-H. pylori pathogens in the stomach is associated with the pH value of gastric juice. A pH value > 4 reflects a risk of gastric flora overgrowth [24]. Rosen et al [25] reported that acid suppression resulted in gastric bacterial overgrowth of genera including Staphylococcus and Streptococcus. We observed the same phenomenon. In addition, we found that Streptococcus could inhibit the growth of H. pylori on culture medium. Khosravi et al [26] reported that Streptococcus mitis induced the conversion of H. pylori to coccid cells and growth inhibition. Therefore, we can infer that the gastric microbiota is the key to restricting the overgrowth of H. pylori or other pathogens.
Gastritis is similar to pneumonia and nephritis in that it can be induced by many kinds of bacteria. In addition to H. pylori, Streptococcus and Neisseria may also contribute to the pathogenesis of gastritis and even gastric cancer [27]. To our best knowledge, our study is the first to show the relationship between the risk of gastritis and infection with non-H. pylori bacteria. Our results are consistent with current reports on H. pylori-related disease. Unlike Marshall et al [10], we identified all cultivable colonies, including H. pylori, on the available medium, which could eventually lead to the identification of other pathogens. Until now, no studies have clearly linked specific gastric microbiomes besides H. pylori to an increased risk of gastritis. Future work should focus on the pathogenic effects of Streptococcus and Neisseria.
Acknowledgements
We are grateful for the generous help from the staff of the Department of Laboratory Medicine with the process of bacterial identification in this study. We also wish to thank the Medical Central Lab staff of Peking University Third Hospital for their support. This work was supported by the National Natural Science Foundation of China Grant 81672410 and the National Technology R&D Program in the 12th Five-Year Plan of China (Grant 2012BAI06B02).
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Sugano K, Tack J, Kuipers EJ, Graham DY, El-Omar EM, Miura S, Haruma K, Asaka M, Uemura N, Malfertheiner P. Kyoto global consensus report on helicobacter pylori gastritis. Gut. 2015;64:1353–1367. doi: 10.1136/gutjnl-2015-309252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Delgado S, Cabrera-Rubio R, Mira A, Suarez A, Mayo B. Microbiological survey of the human gastric ecosystem using culturing and pyrosequencing methods. Microb Ecol. 2013;65:763–772. doi: 10.1007/s00248-013-0192-5. [DOI] [PubMed] [Google Scholar]
- 3.Lofgren JL, Whary MT, Ge Z, Muthupalani S, Taylor NS, Mobley M, Potter A, Varro A, Eibach D, Suerbaum S, Wang TC, Fox JG. Lack of commensal flora in helicobacter pylori-infected INS-GAS mice reduces gastritis and delays intraepithelial neoplasia. Gastroenterology. 2011;140:210–220. doi: 10.1053/j.gastro.2010.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wroblewski LE, Peek RM Jr. Helicobacter pylori, cancer, and the gastric microbiota. Adv Exp Med Biol. 2016;908:393–408. doi: 10.1007/978-3-319-41388-4_19. [DOI] [PubMed] [Google Scholar]
- 5.Engstrand L, Lindberg M. Helicobacter pylori and the gastric microbiota. Best Pract Res Clin Gastroenterol. 2013;27:39–45. doi: 10.1016/j.bpg.2013.03.016. [DOI] [PubMed] [Google Scholar]
- 6.Mason KL, Erb Downward JR, Falkowski NR, Young VB, Kao JY, Huffnagle GB. Interplay between the gastric bacterial microbiota and Candida albicans during postantibiotic recolonization and gastritis. Infect Immun. 2012;80:150–158. doi: 10.1128/IAI.05162-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sohn SH, Kim N, Jo HJ, Kim J, Park JH, Nam RH, Seok YJ, Kim YR, Lee DH. Analysis of gastric body microbiota by pyrosequencing: Possible role of bacteria other than helicobacter pylori in the gastric carcinogenesis. J Cancer Prev. 2017;22:115–125. doi: 10.15430/JCP.2017.22.2.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu G, Torres J, Hu N, Medrano-Guzman R, Herrera-Goepfert R, Humphrys MS, Wang L, Wang C, Ding T, Ravel J, Taylor PR, Abnet CC, Goldstein AM. Molecular characterization of the human stomach microbiota in gastric cancer patients. Front Cell Infect Microbiol. 2017;7:302. doi: 10.3389/fcimb.2017.00302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Malfertheiner P, Megraud F, O’Morain CA, Gisbert JP, Kuipers EJ, Axon AT, Bazzoli F, Gasbarrini A, Atherton J, Graham DY, Hunt R, Moayyedi P, Rokkas T, Rugge M, Selgrad M, Suerbaum S, Sugano K, El-Omar EM European Helicobacter and Microbiota Study Group and Consensus panel. Management of helicobacter pylori infection-the maastricht V/Florence consensus report. Gut. 2017;66:6–30. doi: 10.1136/gutjnl-2016-312288. [DOI] [PubMed] [Google Scholar]
- 10.Marshall BJ, Warren JR. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet. 1984;1:1311–1315. doi: 10.1016/s0140-6736(84)91816-6. [DOI] [PubMed] [Google Scholar]
- 11.Sauget M, Valot B, Bertrand X, Hocquet D. Can MALDI-TOF mass spectrometry reasonably type bacteria? Trends Microbiol. 2017;25:447–455. doi: 10.1016/j.tim.2016.12.006. [DOI] [PubMed] [Google Scholar]
- 12.Lasch P, Jacob D, Grunow R, Schwecke T, Doellinger J. Matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry (MS) for the identification of highly pathogenic bacteria. Trac-Trends in Analytical Chemistry. 2016;85:103–111. [Google Scholar]
- 13.Sauer S, Kliem M. Mass spectrometry tools for the classification and identification of bacteria. Nat Rev Microbiol. 2010;8:74–82. doi: 10.1038/nrmicro2243. [DOI] [PubMed] [Google Scholar]
- 14.Hu Y, He LH, Xiao D, Liu GD, Gu YX, Tao XX, Zhang JZ. Bacterial flora concurrent with Helicobacter pylori in the stomach of patients with upper gastrointestinal diseases. World J Gastroenterol. 2012;18:1257–1261. doi: 10.3748/wjg.v18.i11.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Patel R. MALDI-TOF MS for the diagnosis of infectious diseases. Clin Chem. 2015;61:100–111. doi: 10.1373/clinchem.2014.221770. [DOI] [PubMed] [Google Scholar]
- 16.Morimoto M, Tamura S, Hayakawa T, Yamanishi H, Nakamoto C, Nakamoto H, Ikebe T, Nakano Y, Fujimoto T. Phlegmonous gastritis associated with group A streptococcal toxic shock syndrome. Intern Med. 2014;53:2639–2642. doi: 10.2169/internalmedicine.53.2741. [DOI] [PubMed] [Google Scholar]
- 17.Odai T, Hibino T. The abdominal ultrasonographic appearance of acute phlegmonous gastritis. Kansenshogaku Zasshi. 2016;90:113–119. doi: 10.11150/kansenshogakuzasshi.90.113. [DOI] [PubMed] [Google Scholar]
- 18.Schulz C, Schutte K, Malfertheiner P. Helicobacter pylori and other gastric microbiota in gastroduodenal pathologies. Dig Dis. 2016;34:210–216. doi: 10.1159/000443353. [DOI] [PubMed] [Google Scholar]
- 19.Kim J, Kim N, Jo HJ, Park JH, Nam RH, Seok YJ, Kim YR, Kim JS, Kim JM, Kim JM, Lee DH, Jung HC. An appropriate cutoff value for determining the colonization of helicobacter pylori by the pyrosequencing method: comparison with conventional methods. Helicobacter. 2015;20:370–380. doi: 10.1111/hel.12214. [DOI] [PubMed] [Google Scholar]
- 20.Thorell K, Bengtsson-Palme J, Liu OH, Palacious Gonzales RV, Nookaew I, Rabeneck L, Paszat L, Graham DY, Nielsen J, Lundin SB, Sjoling A. In vivo analysis of the viable microbiota and Helicobacter pylori transcriptome in gastric infection and early stages of carcinogenesis. Infect Immun. 2017;85 doi: 10.1128/IAI.00031-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blaser MJ, Chen Y, Reibman J. Does helicobacter pylori protect against asthma and allergy? Gut. 2008;57:561–567. doi: 10.1136/gut.2007.133462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bik EM, Eckburg PB, Gill SR, Nelson KE, Purdom EA, Francois F, Perez-Perez G, Blaser MJ, Relman DA. Molecular analysis of the bacterial microbiota in the human stomach. Proc Natl Acad Sci U S A. 2006;103:732–737. doi: 10.1073/pnas.0506655103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schulz C, Schutte K, Koch N, Vilchez-Vargas R, Wos-Oxley ML, Oxley AP, Vital M, Malfertheiner P, Pieper DH. The active bacterial assemblages of the upper GI tract in individuals with and without helicobacter infection. Gut. 2018;67:216–225. doi: 10.1136/gutjnl-2016-312904. [DOI] [PubMed] [Google Scholar]
- 24.Sanduleanu S, Jonkers D, De Bruine A, Hameeteman W, Stockbrugger RW. Non-helicobacter pylori bacterial flora during acid-suppressive therapy: differential findings in gastric juice and gastric mucosa. Aliment Pharmacol Ther. 2001;15:379–388. doi: 10.1046/j.1365-2036.2001.00888.x. [DOI] [PubMed] [Google Scholar]
- 25.Rosen R, Amirault J, Liu HY, Mitchell P, Hu L, Khatwa U, Onderdonk A. Changes in gastric and lung microflora with acid suppression acid suppression and bacterial growth. JAMA Pediatr. 2014;168:932–937. doi: 10.1001/jamapediatrics.2014.696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khosravi Y, Dieye Y, Loke MF, Goh KL, Vadivelu J. Streptococcus mitis induces conversion of Helicobacter pylori to coccoid cells during co-culture in vitro. PLoS One. 2014;9:e112214. doi: 10.1371/journal.pone.0112214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Coker OO, Dai Z, Nie Y, Zhao G, Cao L, Nakatsu G, Wu WK, Wong SH, Chen Z, Sung JJY, Yu J. Mucosal microbiome dysbiosis in gastric carcinogenesis. Gut. 2017 doi: 10.1136/gutjnl-2017-314281. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


