Abstract
Melanoma is a malignant skin tumor and has a poor cure rate because of its high metastatic potential. Overexpression of long non-coding (lnc) RNA PANDAR has been observed in several kinds of cancer, but the function of PANDAR on melanoma is still unclear. Therefore, this study was to explore the mechanism of PANDAR on the occurrence and progression in malignant melanoma. We detected expression of PANDAR in malignant melanoma tissues and cell lines by qRT-PCR and analyzed correlation of PANDAR expression with the patients’ prognosis. Furthermore, we investigated the effects of PANDAR on cell viability, migration, invasion, tumorigenesis, and epithelial-mesenchymal transition (EMT) using CCK-8, Transwell, and nude mouse subcutaneous tumor formation model assays and Western blotting analysis, respectively. From the results, we discovered that the PANDAR expression is strikingly upregulated in melanoma tissues compared with paired-adjacent non-tumorous tissues and elevated PANDAR is positively correlated with short overall survival time. The results also demonstrate that knockdown of PANDAR inhibits cell viability, migration, invasion, tumorigenesis, and EMT, whereas overexpression of PANDAR gave opposite results by promoting cell viability, migration, invasion, tumorigenesis, and EMT of melanoma cells. These new findings all illustrate that PANDAR might play a pivotal oncogenic role in the occurrence and development of melanoma, and PANDAR might promote melanoma cell invasion through regulating EMT, providing a potential diagnostic and therapeutic target for melanoma.
Keywords: Long non-coding RNA PANDAR, melanoma, invasion, EMT
Introduction
Melanoma, derived from pigment cells, is responsible for the majority of skin cancer related deaths worldwide, as well as the most lethal cutaneous neoplasm [1,2]. Currently, estimated new cases of melanoma are 160,000, and there are about 48,000 deaths caused by melanoma each year [3], supporting the significant threat posed to human health. Until now, there have been three primary pharmacological therapeutic options for patients with metastatic melanoma: (1) immunotherapy; (2) targeted therapy, focusing on the mutational status of melanoma cells; and (3) conventional chemotherapy, if the first two options are not suitable or available [4-6]. Although advanced improvements have been made in treatment options, unfortunately the incidence of melanoma is rising worldwide and metastatic melanoma is still associated with high rates of mortality and poor prognosis. Once metastasis occurs in patients with melanoma, the 5-year survival rate is 16% [7]. Therefore, the identifying possible molecular mechanisms of melanoma metastasis and tumorigenesis and researching biomarkers for early diagnosis and the effective therapeutic strategies for melanoma are urgently needed.
Long non-coding RNAs (lncRNA) represent a highly heterogeneous group of RNAs, which have an extensive variability in their cellular effects, and their molecular influences. They can be classified by their length (>200 nt) and by their lack of a functional open reading frame, meaning they encompass less than 100 amino acids [8-10]. However, more and more evidence has revealed that lncRNAs play irreplaceable roles in various pathophysiological processes and are frequently dysregulated in many cancers including melanoma [11-14]. LncRNAs may act as oncogenes or tumor suppressor genes, with a specifically role in cancer metastasis and progression [15]. To date, many lncRNAs have been studied in melanomas such as lncRNAs HOXA11-AS [16], SPRY4-IT1 [17], HEIH [18], CCAT1 [19] and PVT1 [20], where increased expression and pro-oncogenic function was observed. Furthermore, lncRNAs GAS5 [21], NKILA [22] are decreased expression and exert a tumor suppressing function in melanoma. But there are still lots of lncRNAs that need to be explored and analyzed, providing the possibility for delivering them as therapeutic targets for melanoma.
PANDAR (GenBank accession ID: 101154753), a novel lncRNA consisting of 1506 nucleotides in length, is located at 6p21.2 and was first reported by Hung et al. [23]. PANDAR can interact with the transcription factor NF-YA, which is linked to the expression of pro-apoptotic genes. In addition, lncRNA PANDAR is overexpressed in many cancers and serves as an oncogene. For example, in bladder cancer, Zhang et al. [24] found that lncRNA PANDAR promoted cell proliferation and migration. Ma et al. [25], Peng et al. [26], Min et al. [27] and Xu et al. [28] all revealed that LncRNA PANDAR was up-regulated and associated with poor prognosis in gastric cancer, hepatocellular carcinoma, colorectal cancer, and cholangiocarcinoma, respectively. But studies of the effect of lncRNA PANDAR on melanoma progression are rare. Therefore, the objective of the present study was to investigate the role of lncRNA promoter of CDKN1A antisense DNA damage activated RNA (PANDAR) in melanoma. We explored the expression of lncRNA PANDAR in melanoma and the correlation between lncRNA PANDAR expression and clinical outcome of melanoma patients. We assessed the roles of lncRNA PANDAR in melanoma cell proliferation, migration, invasion, tumorigenicity, and epithelial-mesenchymal transition (EMT) progress, and our results demonstrate molecular mechanisms underlying the roles of lncRNA PANDAR in melanoma.
Materials and methods
Tissue specimens and patient characterization
Tissue were collected from patients who underwent surgical resections at First Affiliated Hospital of Bengbu Medical College after obtaining informed consent and included 62 melanoma tissues and 24 benign nevi tissues (considered as controls). The specimens were immediately frozen in liquid nitrogen and then stored at -80°C for analysis. Overall survival (OS) time was calculated from the date of the initial surgery to death. This study was carried out in accordance with the approved guidelines by the Ethics Committee of First Affiliated Hospital of Bengbu Medical College.
Cell culture
The normal human skin HACAT cells and five human melanoma cells lines, including CHL-1, A375, SK-MEL-1, WM-35, and WM-115, were purchased from American Type Culture Collection (Manassas, VA, USA), and cultured in DMEM medium (GIBCO-BRL; Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS; Invitrogen) and 1% antibiotic/antimycotic solution in a humidified atmosphere containing 5% CO2 at 37°C.
Cell transfection
Plasmid vector (pcDNA-PANDAR and pcDNA3.1-NC) were prepared using DNA Midiprep kits (Qiagen, Hilden, Germany), and were transfected into melanoma cells. shRNAs targeting PANDAR (sh-PANDAR) and negative control shRNA (sh-NC) constructed by GenePharma (China) were transfected into melanoma cells, respectively, using Lipofectamine™ 3000 (Life Technologies, San Diego, CA, USA). 48 hours post-transfection, PANDAR expression levels were measured. PANDAR shRNA-1: GCAATCTACAACCTGTCTT, shRNA-2: GCCGATGCTCCCAGCTGAATA, shRNA-3: GCATAGCCATAGGTGATTAGA.
qRT-PCR assay
Total RNA was extracted using Trizol Reagent (Invitrogen). 2 ng of total RNA was reverse transcribed into cDNA using the GoScript Reverse Transcription (RT-PCR) System (Promega, Madison, WI, USA). Then, SYBR Premix Ex Taq (TaKaRa, Dalian, China) was used to conduct qRT-PCR on an ABI 7900 system (Applied Biosystems, Foster City, CA, USA). GAPDH was used as internal reference. The relative expression was analyzed by the 2-ΔΔCt method. The sequences of the primers are listed as follows: PANDAR (F): CCTGTTCGTCGATTCTGGCT; PANDAR (R): GTCTGGCCGTGAGATGTTTC; GAPDH (F): GAAGGTGAAGGTCGGAGT; GAPDH (R): GAAGATGGTGATGGGATTTC.
In vitro cell proliferation assays
Cell proliferation was evaluated using a Cell Counting Kit-8 (CCK-8; Dojindo, Kumamoto, Japan). The absorbance values of cells at various time points after transfection were measured using a microplate reader (Bio-Rad, Hercules, CA, USA). To perform colony formation assays, 400 cells were seeded per well in 6-well plates and cultured for two weeks. The colonies were fixed with 4% paraformaldehyde and stained with 0.1% crystal violet (Sigma, St Louis, MO, USA). Stained clones were observed under a microscope.
In vitro migration and invasion assays
Cell migration and invasion assays were performed in a 24-well transwell plate with 8-μm polyethylene terephthalate membrane filters (Costar, Corning, MA, USA). Cells in 500 μL of serum-free medium were added to the upper chambers, which contained either uncoated or Matrigel-coated membranes. Each lower chamber was filled with 500 μL medium with 10% FBS. After 24 h of incubation, the non-migrating cells on the upper sides of the filters were detached using cotton swabs. The cells located in the lower filters were fixed with 4% paraformaldehyde, and then stained with 0.1% crystal violet. Migrated or invaded cells were counted in five randomly chosen fields per well under a microscope.
In vivo tumorigenesis assay
Female athymic Balb/c nude mice (4-5 weeks of age, 18-20 g), obtained from Slac Laboratory Animal Co. Ltd. (Shanghai, China), were randomized to the control or experimental group (6 mice/group). Melanoma cells stably expressing sh-PANDAR or sh-NC were implanted subcutaneously into the back of each nude mouse. Tumors were measured with calipers every 3 days and the volume was calculated by the formula V = 0.5 × length × width2. Over a five-week period, all mice were euthanized, and the tumors were excised and weighed. All experimental procedures were approved by the animal care and ethics committee of First Affiliated Hospital of Bengbu Medical College.
Western blot analysis
Cells were lysed with RIPA buffer supplemented with protease inhibitors. Protein concentration was measured using a BCA protein assay (Thermo Scientific, Rockford, USA). Proteins were separated using SDS-PAGE electrophoresis and transferred to PVDF membranes. The membranes were blocked and then probed with primary antibodies against E-cadherin, vimentin (Abcam, MA, USA), N-cadherin, and β-actin (Cell Signaling Technology, Danvers, USA). After incubation with a secondary antibody, the blots were visualized by enhanced chemiluminescence (Millipore, Billerica, MA). β-actin was used as a loading control.
Statistical analysis
All statistical analyses were performed using SPSS version 18.0 software (SPSS Inc., Chicago, IL, USA) and Graphpad Prism (version 6.01) software (GraphPad Software, Inc., La Jolla, CA, USA). Each experiment was performed in triplicate, and repeated at least three times. The data are expressed as the mean ± SD. Kaplan-Meier plots and log-rank tests were used for survival analysis. P<0.05 was considered to be statistically significant.
Results
Upregulated PANDAR predicts poor clinical outcome in patients
Here, we examined the expression profiles of PANDAR in 62 melanoma tissues and 24 benign nevi tissues. We observed that, as shown in Figure 1A, PANDAR was significant highly expressed in the melanoma tissues than in benign nevi tissues. Also, the expression of PANDAR was remarkably higher in the melanoma cells than in the normal human skin HACAT cells (Figure 1B).
Figure 1.
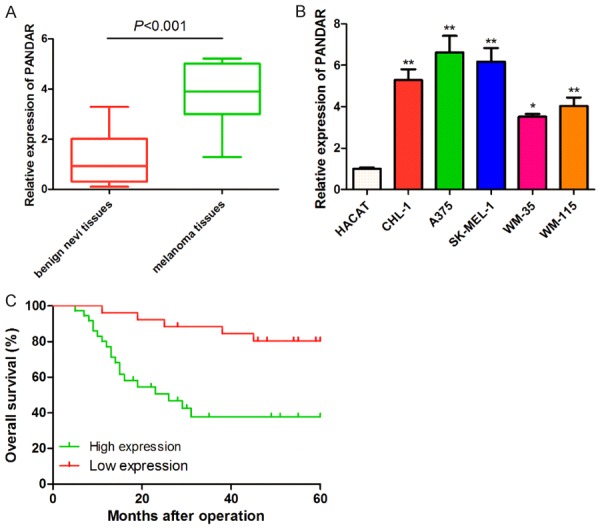
Upregulated PANDAR predicts poor clinical outcome in patients with melanoma. A: Expression of PANDAR in melanoma tissues and benign nevi tissues was detected by qRT-PCR analysis. B: Expression of PANDAR in melanoma cells and the normal human skin HACAT cells were determined by qRT-PCR analysis. C: Survival curve was made of Kaplan-Meier analysis between the expression level of PANDAR and overall survival time in patients with melanoma. Data represent the mean ± SD from three independent experiments (*P<0.05, **P<0.001).
To further understand the significance of PANDAR in melanoma, we analyzed the correlation between PANDAR expression and the clinicopathological status of 62 melanoma patients. As indicated in Table 1, increased PANDAR expression level in melanoma tissues was closely correlated with tumor thickness (P=0.013), Clark level (P=0.002), histologic type (P=0.023) and TNM classification (P=0.029). Kaplan-Meier analysis revealed that high PANDAR expression was significantly associated with shorter overall survival (P=0.0) (Figure 1C).
Table 1.
Relationship between PANDAR expression and clinicopathological characteristics of melanoma patients (n=62)
| Characteristics | Total number | PANDAR expression | P value | |
|---|---|---|---|---|
|
| ||||
| Low (n=26) | High (n=36) | |||
| Age (years) | 0.951 | |||
| Mean | 67.12 | 67.68 | ||
| Median | 69.00 | 69.00 | ||
| Range | 47-82 | 46-83 | ||
| Gender | 0.303 | |||
| Male | 31 | 15 | 16 | |
| Female | 31 | 11 | 20 | |
| Clark level | 0.002 | |||
| II | 15 | 11 | 4 | |
| III | 25 | 12 | 13 | |
| IV-V | 21 | 3 | 18 | |
| Missing | 1 | 0 | 1 | |
| Tumor thickness (mm) | 0.013 | |||
| <1.0 | 39 | 21 | 18 | |
| ≥1.0 | 23 | 5 | 18 | |
| Location | 0.883 | |||
| Extremities | 22 | 9 | 13 | |
| Trunk | 29 | 13 | 16 | |
| Head and neck | 11 | 4 | 7 | |
| Histologic type | 0.023 | |||
| Superficial spreading | 38 | 21 | 17 | |
| Nodular | 16 | 4 | 12 | |
| Acral lentiginous | 8 | 1 | 7 | |
| Lymphocytic infiltrate | 0.462 | |||
| 0-1 | 25 | 12 | 13 | |
| 2-3 | 37 | 14 | 23 | |
| Ulceration | 0.017 | |||
| No | 45 | 23 | 22 | |
| Yes | 17 | 3 | 14 | |
| TNM classification | 0.029 | |||
| I | 49 | 24 | 25 | |
| II-IV | 13 | 2 | 11 | |
PANDAR regulates the proliferation, migration, and invasion of melanoma cells
Since our clinical data showed that PANDAR expression levels negatively correlated with tumor growth and metastasis, we further explored the function of PANDAR in proliferation, migration, and invasion in melanoma cell lines. Cells were transiently transfected with sh-PANDAR, which efficiently silenced endogenous expression of PANDAR (Figure 2A). Also, ectopic expression of PANDAR in cells by transfecting with pcDNA3.1-PANDAR markedly increased the PANDAR level than that in pcDNA3.1-NC-transfected cells. CCK-8 and colony formation assays were performed to assess the role of PANDAR in melanoma cell proliferation. The melanoma cells transfected with pcDNA3.1-PANDAR clearly grew faster compared to control cells (Figure 2B). The number and the mean size of the colonies formed by melanoma cells were significantly decreased following transfection with sh-PANDAR (Figure 2C), indicating that PANDAR exerts a promoting function in melanoma cell proliferation.
Figure 2.
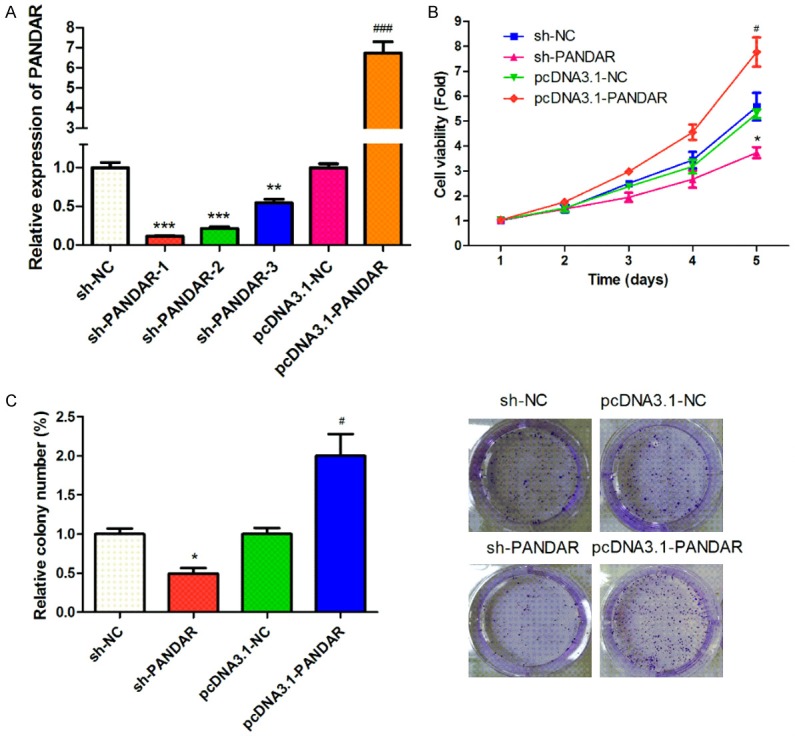
Effect of PANDAR expression on cell viability and clone formation abilities. A: Knockdown and overexpression efficiencies of PANDAR were evaluated by qRT-PCR. B: CCK-8 was used to test the effect of PANDAR expression on cell viability. C: Colony formation assays was used to evaluate the clone formation ability. Data represent the mean ± SD from three independent experiments (*, #P<0.05).
Furthermore, as shown in Figure 3, melanoma cells with reduced PANDAR expression showed increased ability of migration and invasion than that in control cells, whereas elevated PANDAR suppressed cell migration and invasion when compared to control cells. These data suggested that PANDAR inhibits melanoma cells migration and invasion.
Figure 3.
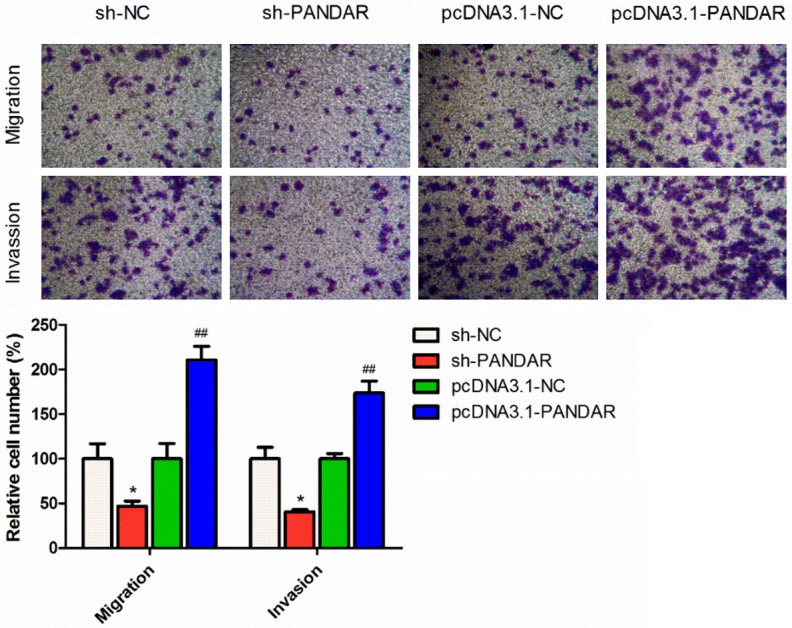
Effects of PANDAR expression on cell migration and invasion assessed by Transwell assay. Data represent the mean ± SD from three independent experiments (*P<0.05, ##P<0.001).
PANDAR promotes melanoma tumorigenesis in vivo
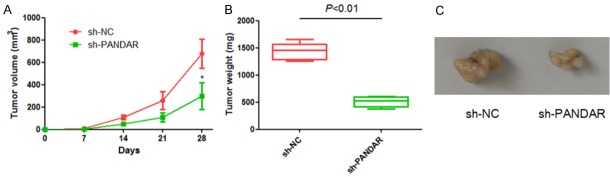
To confirm the above data in vivo, melanoma cells transfected with sh-PANDAR or sh-NC were injected subcutaneously into nude mice, respectively. Xenograft tumor volumes were measured each week after a palpable tumor formed, and mice were sacrificed five weeks after tumor implantation. As shown in Figure 4, the final tumor volume and weight of sh-PANDAR group were markedly smaller than that of sh-NC group. Therefore, silencing of PANDAR could significantly inhibit tumorigenesis of melanoma cells in vivo.
Figure 4.
Down-regulated expression of PANDAR inhibits melanoma tumorigenesis in vivo. A: The tumor volumes in sh-PANDAR and sh-NC groups. B: The tumor weights in sh-PANDAR and sh-NC groups. C: The tumor sizes in sh-PANDAR and sh-NC groups. Data represent the mean ± SD from six independent experiments (*P<0.05).
PANDAR facilitates EMT in CRC cells
EMT is an important factor in cell invasion. Thus, we next determined whether EMT markers were altered in melanoma cells with deregulation of PANDAR. The expression of E-cadherin, N-cadherin, vimentin, and fibronectin protein level was analyzed by Western blot. As shown in Figure 5, we found that expression of N-cadherin and vimentin was increased while E-cadherin expression was decreased, in melanoma cells with overexpressed PANDAR, whereas opposite results were obtained when PANDAR was knocked down in melanoma cells.
Figure 5.
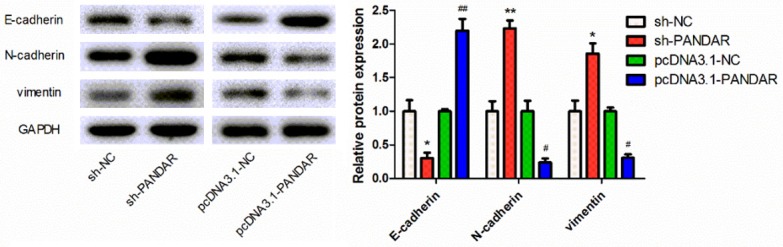
Effect of PANDAR on EMT-related gene expression in melanoma cells. The expression levels of E-cadherin, N-cadherin and vimentin in cells with different treatments were analyzed by Western blotting. Data represent the mean ± SD from three independent experiments (*, #P<0.05, **, ##P<0.01).
Discussion
Melanoma is one of the most aggressive malignant carcinomas and generally has a poor cure rate because of its metastasis and invasive behavior [29]. Many lncRNAs, such as lncRNAs CCAT1 [19], GAS5 [30], SLNCR1 [31], were discovered to be dysregulated in melanoma tissues and play a vital role in the metastasis or invasion of melanoma. However, the function of lncRNA PANDAR in melanoma has not been previously clarified.
In the present study, we show for the first time that PANDAR is upregulated in melanoma tissues more than in normal tissues based on qRT-PCR analysis. Consistently, similar results were obtained from melanoma cell lines. Also, we investigated the association of PANDAR expression with clinicopathological characteristics of patients with melanoma and the results showed higher expression level of PANDAR, higher Clark level, thicker tumor, and advancer TNM classification. Also, we determined the relationship between PANDAR expression level and prognosis of melanoma patients through evaluating the correlation between PANDAR expression and clinical outcomes. Kaplan-Meier analysis showed that patients with high levels of PANDAR expression had remarkably shorter survival time than those with low levels. All date suggested that PANDAR might involve in the tumorigenesis and the development of melanoma. Furthermore, in vitro, we performed CCK-8 and colony formation assays to investigate the biological function of PANDAR in melanoma cells. PANDAR knockdown showed low cell viability compared with the control group, while overexpressed PANDAR showed high cell viability. In addition, Transwell assay and Western blotting analysis showed PANDAR could increase cell migration and invasion abilities, as well as regulate EMT. In vivo, we carried out a melanoma-bearing model to determine the tumorigenesis of PANDAR and PANDAR knockdown showed low tumorigenesis compared with the control group.
Cancer metastases represent a multistep biological process that is driven by the acquisition of genetic and/or epigenetic alterations within tumor cells. Many studies showed that cancer metastasis is related to the cancer cell EMT program [32-34]. EMT refers to the biological process of epithelial cells transformed into interstitial phenotype cells through specific procedures, which play an important role in embryonic development, chronic inflammation, tissues reconstruction, fibrosis disease, as well as cancer metastasis. Metastasis is characterized by reduce expression of cell adhesion molecules (such as E-cadherin) and increased expression of cells cytoskeleton protein (Vimentin) and N-cadherin. The epithelial cells will lose epithelial phenotype of connection with the basement membrane, and obtain the mesenchymal phenotype with higher migration and invasion, anti-apoptosis, and the ability of degradation of extracellular matrix. Meanwhile, lncRNAs were reported to have close relationship with EMT progression, serving as a promoter or an inhibitor [35-37]. For example, lncRNA-ROR played an important role in the development of gallbladder cancer and mediates the EMT in gallbladder cancer [38]. Long noncoding RNA LINC01133 inhibited EMT in colorectal cancer by interacting with SRSF6 [39]. BRAF-activated lncRNA contributed to colorectal cancer migration by inducing epithelial-mesenchymal transition [40]. Additionally, high expression of lncRNA PANDAR indicated a poor prognosis for colorectal cancer and promoted metastasis by EMT pathway [27]. However, the function of PANDAR on EMT in melanoma still remained unknown. Thus, to further seek the molecular mechanism through which PANDAR promoted the metastasis of melanoma, we determined the expression level of EMT-related markers following knockdown/overexpression of PANDAR. Our results show that knockdown of PANDAR significantly weaken the expression of N-cadherin, vimentin, and increases the expression level of E-cadherin, however overexpression of PANDAR increased expression of N-cadherin, vimentin, and decreased the expression level of E-cadherin. These results demonstrate that PANDAR might promote melanoma metastasis and invasion through regulating EMT.
In conclusion, we first discovered that the PANDAR expression was strikingly upregulated in melanoma tissues compared with paired-adjacent non-tumorous tissues and elevated PANDAR showed a positive correlation with short overall survival time. These results indicate that PANDAR might play a pivotal oncogenic role in the occurrence and development of melanoma. Also, we identified PANDAR might serve as an indicator of poor survival rate and a negative prognostic factor for patients with melanoma. We also illuminated that knockdown of PANDAR could inhibit cell viability, migration, invasion, tumorigenesis, and EMT, whereas overexpression of PANDAR presented the opposite results, by promoting cell viability, migration, invasion, tumorigenesis, and EMT. These new findings suggested that PANDAR might promote melanoma cell invasion through regulating EMT and it might be used as a potential diagnostic and therapeutic target of melanoma.
Acknowledgements
The research was supported by Anhui Provincial Natural Science Foundation (No. 3482192).
Disclosure of conflict of interest
None.
References
- 1.Deeks ED. Nivolumab: a review of its use in patients with malignant melanoma. Drugs. 2014;74:1233–9. doi: 10.1007/s40265-014-0234-4. [DOI] [PubMed] [Google Scholar]
- 2.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 4.Robert C, Karaszewska B, Schachter J, Rutkowski P, Mackiewicz A, Stroiakovski D, Lichinitser M, Dummer R, Grange F, Mortier L, Chiarion-Sileni V, Drucis K, Krajsova I, Hauschild A, Lorigan P, Wolter P, Long GV, Flaherty K, Nathan P, Ribas A, Martin AM, Sun P, Crist W, Legos J, Rubin SD, Little SM, Schadendorf D. Improved overall survival in melanoma with combined dabrafenib and trametinib. N Engl J Med. 2015;372:30–9. doi: 10.1056/NEJMoa1412690. [DOI] [PubMed] [Google Scholar]
- 5.Weber JS, D’Angelo SP, Minor D, Hodi FS, Gutzmer R, Neyns B, Hoeller C, Khushalani NI, Miller WH Jr, Lao CD, Linette GP, Thomas L, Lorigan P, Grossmann KF, Hassel JC, Maio M, Sznol M, Ascierto PA, Mohr P, Chmielowski B, Bryce A, Svane IM, Grob JJ, Krackhardt AM, Horak C, Lambert A, Yang AS, Larkin J. Nivolumab versus chemotherapy in patients with advanced melanoma who progressed after anti-CTLA-4 treatment (CheckMate 037): a randomised, controlled, open-label, phase 3 trial. Lancet Oncol. 2015;16:375–84. doi: 10.1016/S1470-2045(15)70076-8. [DOI] [PubMed] [Google Scholar]
- 6.Robert C, Schachter J, Long GV, Arance A, Grob JJ, Mortier L, Daud A, Carlino MS, McNeil C, Lotem M, Larkin J, Lorigan P, Neyns B, Blank CU, Hamid O, Mateus C, Shapira-Frommer R, Kosh M, Zhou H, Ibrahim N, Ebbinghaus S, Ribas A KEYNOTE-006 investigators. Pembrolizumab versus ipilimumab in advanced melanoma. N Engl J Med. 2015;372:2521–32. doi: 10.1056/NEJMoa1503093. [DOI] [PubMed] [Google Scholar]
- 7.Dubey AK, Gupta U, Jain S. Breast cancer statistics and prediction methodology: a systematic review and analysis. Asian Pac J Cancer Prev. 2015;16:4237–45. doi: 10.7314/apjcp.2015.16.10.4237. [DOI] [PubMed] [Google Scholar]
- 8.Dinger ME, Pang KC, Mercer TR, Mattick JS. Differentiating protein-coding and noncoding RNA: challenges and ambiguities. PLoS Comput Biol. 2008;4:e1000176. doi: 10.1371/journal.pcbi.1000176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Singh DK, Prasanth KV. Functional insights into the role of nuclear-retained long noncoding RNAs in gene expression control in mammalian cells. Chromosome Res. 2013;21:695–711. doi: 10.1007/s10577-013-9391-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mizutani R, Wakamatsu A, Tanaka N, Yoshida H, Tochigi N, Suzuki Y, Oonishi T, Tani H, Tano K, Ijiri K, Isogai T, Akimitsu N. Identification and characterization of novel genotoxic stress-inducible nuclear long noncoding RNAs in mammalian cells. PLoS One. 2012;7:e34949. doi: 10.1371/journal.pone.0034949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Q, Wang Y, Liang J, Tian Y, Zhang Y, Tao K. Bioinformatics analysis to identify the critical genes, microRNAs and long noncoding RNAs in melanoma. Medicine (Baltimore) 2017;96:e7497. doi: 10.1097/MD.0000000000007497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Q, Gao S, Li H, Lv M, Lu C. Long noncoding RNAs (lncRNAs) in triple negative breast cancer. J Cell Physiol. 2017;232:3226–3233. doi: 10.1002/jcp.25830. [DOI] [PubMed] [Google Scholar]
- 13.Beaver LM, Kuintzle R, Buchanan A, Wiley MW, Glasser ST, Wong CP, Johnson GS, Chang JH, Löhr CV, Williams DE, Dashwood RH, Hendrix DA, Ho E. Long noncoding RNAs and sulforaphane: a target for chemoprevention and suppression of prostate cancer. J Nutr Biochem. 2017;42:72–83. doi: 10.1016/j.jnutbio.2017.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lanzos A, Carlevaro-Fita J, Mularoni L, Reverter F, Palumbo E, Guigó R, Johnson R. Discovery of cancer driver long noncoding RNAs across 1112 tumour genomes: new candidates and distinguishing features. Sci Rep. 2017;7:41544. doi: 10.1038/srep41544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Serviss JT, Johnsson P, Grander D. An emerging role for long non-coding RNAs in cancer metastasis. Front Genet. 2014;5:234. doi: 10.3389/fgene.2014.00234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu Q, Zhao N, Zha G, Wang H, Tong Q, Xin S. LncRNA HOXA11-AS exerts oncogenic functions by repressing p21 and miR-124 in uveal melanoma. DNA Cell Biol. 2017;36:837–844. doi: 10.1089/dna.2017.3808. [DOI] [PubMed] [Google Scholar]
- 17.Li J, Chen Y, Chen Z, He A, Xie H, Zhang Q, Cai Z, Liu Y, Huang W. SPRY4-IT1: A novel oncogenic long non-coding RNA in human cancers. Tumour Biol. 2017;39:1010428317711406. doi: 10.1177/1010428317711406. [DOI] [PubMed] [Google Scholar]
- 18.Zhao H, Xing G, Wang Y, Luo Z, Liu G, Meng H. Long noncoding RNA HEIH promotes melanoma cell proliferation, migration and invasion via inhibition of miR-200b/a/429. Biosci Rep. 2017;37 doi: 10.1042/BSR20170682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lv L, Jia JQ, Chen J. LncRNA CCAT1 upregulates proliferation and invasion in melanoma cells via suppressing miR-33a. Oncol Res. 2018;26:201–208. doi: 10.3727/096504017X14920318811749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang BJ, Ding HW, Ma GA. Long Noncoding RNA PVT1 promotes melanoma progression via endogenous sponging MiR-26b. Oncol Res. 2017 doi: 10.3727/096504017X14920318811730. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bian D, Shi W, Shao Y, Li P, Song G. Long non-coding RNA GAS5 inhibits tumorigenesis via miR-137 in melanoma. Am J Transl Res. 2017;9:1509–1520. [PMC free article] [PubMed] [Google Scholar]
- 22.Bian D, Gao C, Bao K, Song G. The long non-coding RNA NKILA inhibits the invasion-metastasis cascade of malignant melanoma via the regulation of NF-kB. Am J Cancer Res. 2017;7:28–40. [PMC free article] [PubMed] [Google Scholar]
- 23.Hung T, Wang Y, Lin MF, Koegel AK, Kotake Y, Grant GD, Horlings HM, Shah N, Umbricht C, Wang P, Wang Y, Kong B, Langerød A, Børresen-Dale AL, Kim SK, van de Vijver M, Sukumar S, Whitfield ML, Kellis M, Xiong Y, Wong DJ, Chang HY. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat Genet. 2011;43:621–9. doi: 10.1038/ng.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhan Y, Lin J, Liu Y, Chen M, Chen X, Zhuang C, Liu L, Xu W, Chen Z, He A, Zhang Q, Sun X, Zhao G, Huang W. Up-regulation of long non-coding RNA PANDAR is associated with poor prognosis and promotes tumorigenesis in bladder cancer. J Exp Clin Cancer Res. 2016;35:83. doi: 10.1186/s13046-016-0354-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma P, Xu T, Huang M, Shu Y. Increased expression of LncRNA PANDAR predicts a poor prognosis in gastric cancer. Biomed Pharmacother. 2016;78:172–6. doi: 10.1016/j.biopha.2016.01.025. [DOI] [PubMed] [Google Scholar]
- 26.Peng W, Fan H. Long non-coding RNA PANDAR correlates with poor prognosis and promotes tumorigenesis in hepatocellular carcinoma. Biomed Pharmacother. 2015;72:113–8. doi: 10.1016/j.biopha.2015.04.014. [DOI] [PubMed] [Google Scholar]
- 27.Lu M, Liu Z, Li B, Wang G, Li D, Zhu Y. The high expression of long non-coding RNA PANDAR indicates a poor prognosis for colorectal cancer and promotes metastasis by EMT pathway. J Cancer Res Clin Oncol. 2017;143:71–81. doi: 10.1007/s00432-016-2252-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu Y, Jiang X, Cui Y. Upregulated long noncoding RNA PANDAR predicts an unfavorable prognosis and promotes tumorigenesis in cholangiocarcinoma. Onco Targets Ther. 2017;10:2873–2883. doi: 10.2147/OTT.S137044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yan H, Ren MY, Wang ZX, Feng SJ, Li S, Cheng Y, Hu CX, Gao SQ, Zhang GQ. Zerumbone inhibits melanoma cell proliferation and migration by altering mitochondrial functions. Oncol Lett. 2017;13:2397–2402. doi: 10.3892/ol.2017.5742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen L, Yang H, Xiao Y, Tang X, Li Y, Han Q, Fu J, Yang Y, Zhu Y. LncRNA GAS5 is a critical regulator of metastasis phenotype of melanoma cells and inhibits tumor growth in vivo. Onco Targets Ther. 2016;9:4075–87. doi: 10.2147/OTT.S98203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmidt K, Joyce CE, Buquicchio F, Brown A, Ritz J, Distel RJ, Yoon CH, Novina CD. The lncRNA SLNCR1 mediates melanoma invasion through a conserved SRA1-like region. Cell Rep. 2016;15:2025–37. doi: 10.1016/j.celrep.2016.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xin B, He X, Wang J, Cai J, Wei W, Zhang T, Shen X. Nerve growth factor regulates CD133 function to promote tumor cell migration and invasion via activating ERK1/2 signaling in pancreatic cancer. Pancreatology. 2016;16:1005–1014. doi: 10.1016/j.pan.2016.09.005. [DOI] [PubMed] [Google Scholar]
- 33.Huang M, Wu S, Hu Q, Wu H, Wei S, Xie H, Sun K, Li X, Fang L. Agkihpin, a novel SVAE may inhibit the migration and invasion of liver cancer cells associated with the inversion of EMT induced by Wnt/beta-catenin signaling inhibition. Biochem Biophys Res Commun. 2016;479:283–289. doi: 10.1016/j.bbrc.2016.09.060. [DOI] [PubMed] [Google Scholar]
- 34.He J, Shen S, Lu W, Zhou Y, Hou Y, Zhang Y, Jiang Y, Liu H, Shao Y. HDAC1 promoted migration and invasion binding with TCF12 by promoting EMT progress in gallbladder cancer. Oncotarget. 2016;7:32754–64. doi: 10.18632/oncotarget.8740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xiao C, Wu CH, Hu HZ. LncRNA UCA1 promotes epithelial-mesenchymal transition (EMT) of breast cancer cells via enhancing Wnt/beta-catenin signaling pathway. Eur Rev Med Pharmacol Sci. 2016;20:2819–24. [PubMed] [Google Scholar]
- 36.Liu S, Song L, Yao H, Zhang L, Xu D, Gao F, Li Q. MiR-375 is epigenetically downregulated by HPV-16 E6 mediated DNMT1 upregulation and modulates EMT of cervical cancer cells by suppressing lncRNA MALAT1. PLoS One. 2016;11:e0163460. doi: 10.1371/journal.pone.0163460. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 37.Li Y, Huang S, Li Y, Zhang W, He K, Zhao M, Lin H, Li D, Zhang H, Zheng Z, Huang C. Decreased expression of LncRNA SLC25A25-AS1 promotes proliferation, chemoresistance, and EMT in colorectal cancer cells. Tumour Biol. 2016;37:14205–14215. doi: 10.1007/s13277-016-5254-0. [DOI] [PubMed] [Google Scholar]
- 38.Wang SH, Zhang MD, Wu XC, Weng MZ, Zhou D, Quan ZW. Overexpression of LncRNA-ROR predicts a poor outcome in gallbladder cancer patients and promotes the tumor cells proliferation, migration, and invasion. Tumour Biol. 2016;37:12867–12875. doi: 10.1007/s13277-016-5210-z. [DOI] [PubMed] [Google Scholar]
- 39.Kong J, Sun W, Li C, Wan L, Wang S, Wu Y, Xu E, Zhang H, Lai M. Long non-coding RNA LINC01133 inhibits epithelial-mesenchymal transition and metastasis in colorectal cancer by interacting with SRSF6. Cancer Lett. 2016;380:476–84. doi: 10.1016/j.canlet.2016.07.015. [DOI] [PubMed] [Google Scholar]
- 40.Guo Q, Zhao Y, Chen J, Hu J, Wang S, Zhang D, Sun Y. BRAF-activated long non-coding RNA contributes to colorectal cancer migration by inducing epithelial-mesenchymal transition. Oncol Lett. 2014;8:869–875. doi: 10.3892/ol.2014.2154. [DOI] [PMC free article] [PubMed] [Google Scholar]