Abstract
Increasing studies have revealed the importance of microRNAs (miRNAs) in tumorigenesis and tumor progression. miR-455-3p is a newly identified tumor suppressive RNA in various human cancers. However, the expression pattern and clinical significance of miR-455-3p in colorectal cancer (CRC) remains unclear. We found that expression of miR-455-3p was significantly reduced in CRC tissues and cell lines. In addition, we show that low miR-455-3p expression is associated with larger tumor size, advanced tumor stage, and poorer overall survival of CRC patients. Furthermore, in vitro experiments revealed that overexpression of miR-455-3p represses cell proliferation. Importantly, we show that the tumor protein translationally controlled 1 (TPT1) is a direct target of miR-455-3p. Moreover, expression of TPT1 was inversely correlated with the expression of miR-455-3p. Loss-of-function of TPT1 had a similar effect on CRC cell proliferation in vitro as gain-of-function of miR-455-3p. Taken together, these data suggest that miR-455-3p functions as tumor suppressive RNA by targeting TPT1 in CRC, and it might be a potential therapeutic target for CRC patients.
Keywords: miR-455-3p, TPT1, colorectal cancer, proliferation, cell cycle
Introduction
Colorectal cancer (CRC) ranks the third most common cancer with about 1.2 million new cases been diagnosed each year [1]. Over the past decade, we have witnessed the incidence and mortality rate steadily increased in Asian countries [2]. The improvement in the anti-cancer treatment methods has greatly the overall survival of CRC patients but it is still undesirable [3]. Therefore, understanding the mechanisms involved in CRC development will be necessary for the identification of new biomarkers for anti-cancer therapy strategies development.
The tumor protein translationally controlled 1 (TPT1) is a highly conserved protein and composed by three domains [4]. It was reported that the expression of TPT1 was upregulated when exposed to cellular stress including thermal shock or genotoxic reagent [5]. The significance of TPT1 in tumors was appreciated in recently years since it was identified as a target gene of the well-known tumor suppressor gene p53 [6]. In supporting this, numbers of studies have unraveled that TPT1 was highly expressed in many tumors including prostate cancer cell and colon cancer cell [7,8]. Notably, Ma et al. found the knockdown of TPT1 showed cell proliferation, migration, and invasion inhibition effect [8]. However, in CRC tissues, the expression status was not investigated yet.
miRNA was firstly identified in 1993 with a length of 18-25 nucleotides [9]. Since then, over 2,000 miRNAs have been identified and it is reported that about 65% of them were expressed in CRC [10,11]. The importance of miRNAs in regulating diverse cellular processes including cell proliferation, migration, and apoptosis has been recognized [12-14]. Results from previous studies have demonstrated the biological function of miRNAs was mainly rely on the binding with the 3’-untranlated region (3’-UTR) of target molecules [15]. By targeting these transcripts, miRNAs can either induce translation repression or mRNA degradation [16]. Therefore, uncovering the complex regulation network of miRNA will help us to understand how cancer was initiated or developed.
Recent studies have linked downregulation of miR-455-3p with progression of cancers including prostate cancer, non-small cell lung cancer, and esophageal squamous cell carcinoma, suggesting that miR-455-3p functions as a tumor suppressor [17-19]. There is also a study that reported down-regulation of miR-455-3p in a human colon cell line [20]. However, no published work has investigated the role and clinical significance of miR-455-3p in CRC. In this study, we demonstrate that miR-455-3p is downregulated in CRC tissues compared with in normal tissues. Moreover, we found thatmmiR-455 could inhibit cell proliferation and induce cell cycle arrested through directly targeting the expression of TPT1, a well-known oncogene.
Materials and methods
Tissue samples
Human CRC tissues and the matched noncancer tissues were collected from forty-eight patients who were diagnosed and received treatment at Tianjin Medical University Cancer Institute and Hospital between January 2010 and March 2012. This study was approved and monitored by the Ethic Committee of Tianjin Medical University Cancer Institute and Hospital. All the enrolled patients have provided the written informed consent. These tissues were frozen in liquid nitrogen and stored at -80°C for further usage.
Cell culture
Human CRC cell lines (HCT116, SW480) and immortalized human epithelial cell line (HIEC) obtained from ATCC (Manassas, VA, USA) and cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum (Gibco, Grand Island, NY, USA). These cells were cultured at 37°C in a humidified atmosphere of 5% CO2.
Cell transfection
The cells were seeded into 6-well plate and incubated overnight. Following, the chemical synthesized miR-455-3p mimic (5’-GCAGUCCAUGGGCAUAUACAC-3’), miR-455-3p inhibitor (5’-GUGUAUAUGCCCAUGGACUGC-3’), and control miRNA (miR-con) purchased from GenePharm (Shanghai, China) were transfected to the cultured cells using Lipo6000 (Beyotime, Jiangsu, China) per the manufacturer’s instructions to manipulate the expression of miR-455-3p. Then, the small-interfering RNA targeting TPT1 (si-TPT1, 5’-AAGGUACCGAAAGCACAGU-3’) and control siRNA (si-con) purchased from GenePharm (Shanghai, China) and TPT1 expression vector purchased from GenScript (Nanjing, China) were also transfected to the cultured cells using Lipo6000 (Beyotime, Jiangsu, China) to manipulate the expression of TPT1.
Quantitative real-time PCR assay
Tissue samples and cells were treated with Trizol reagent (Beyotime) to extract the total RNA according to the manufacturer’s instructions. Reverse transcription was conducted using Prime Script RT Master Mix (Takara, Dalian, China). qRT-PCR analysis was carried out with SYBR Green Master Mix (Applied Biosystems, Carlsbad, CA, USA) using ABI PRISM 7500 Real-Time PCR System (Applied Biosystems). The primers for miR-455-3p are: 5’-ACACTCCAGCTGGGGCAGTCCACGGGCATATACAC-3’(F), 5’-GTGCAGGGTCCGAGGT-3(R); for U6 snRNA are: 5’-GCGCGTCGTGAAGCGTTC-3’(F), 5’-GTGCAGGGTCCGAGGT-3’(R). The relative gene expression levels were calculated using the 2-ΔΔCt method with U6 snRNA as internal control.
Western blot assay
Tissue samples and cells were treated with RIPA lysis buffer (Beyotime) to extract the total protein according to the manufacturer’s instructions. Protein samples were separated on 10% SDS-PAGE gel and transferred to PVDF membrane and then the membtane was incubated with fat-free milk, followed by primary antibodies targeting TPT1 (ab133568, Abcam, Cambridge, MA, USA), and GAPDH (ab181602, Abcam) for overnight at 4°C. Next, the membrane was washed with TBST and then incubated with HRP-conjugated secondary antibodies (ab205718, Abcam) for 2 h at room temperature. The bands were detected using an enhanced chemiluminescence system (Beyotime). GAPDH was used as internal control.
Cell proliferation assay
The effect of miR-455-3p or TPT1 on cell proliferation was assessed by cell counting kit-8 (CCK-8) assay. The cells to be tested were seeded in 96-well plates at a density of about 2000 cells per well. Then, 10 μl CCK-8 (Beyotime) was added to each well in indicated time points and further incubated for at 37°C. The optical density of each well was measured at a wavelength of 450 nm.
Cell cycle assay
The effect of miR-455-3p or TPT1 on cell cycle was measured by flow cytometry. The cells to be tested were harvested, washed, and then fixed with 75% cold ethanol. Then, the cells were treated with 50 μg/ml propidium iodide (Sigma-Aldrich, St. Louis, MO, USA) and RNase A for 10 min. After filtration, the cells were subjected to cell cycle analysis using FCAScalibur flow cytometer (BD Biosciences, San Diego, CA, USA).
Luciferase activity assay
By using the online prediction algorithm TargetScan, we identified the putative binding sequence in the 3’-UTR of TPT1 for miR-455-3p. Then, the predicted wild-type (wt) and mutate (mut) sequence in the predicted target sites were synthesized and cloned into pRL-TK (Promega, Madison, WI, USA). The cells were co-transfected with miR-455-3p mimic or miR-con and wt or mut 3’-UTR TPT1 constructs using Lipo6000 (Beyotime). After transfection, the relative luciferase activity was measured using Dual-luciferase activity assay system (Promega).
Statistical analysis
Data are presented mean ± standard deviation (SD) values. Data analysis was conducted using GraphPad Prism 5.0 software. Comparisons of two groups were conducted using Student’s t-tests; more than 2 independent groups were compared using one-way analysis of variance and Tukey test. The association between miR-455-3p and TPT1 in CRC tissues was calculated using the Spearman’s correlation test. Kaplan-Meier method was used to calculate overall survival of CRC patients. The difference of the survival curve was analyzed by log-rank test. P < 0.05 was considered statistically significant.
Results
miR-455-3p expression is downregulated in CRC cell lines and tissues
To investigate the potential role of miR-455-3p in CRC, we examined miR-455-3p levels in three CRC cell lines and the immortalized human epithelial cell line (HIEC). We found expression levels of miR-455-3p were reduced in CRC cell lines compared with in HIEC (Figure 1A). Additionally, expression of miR-455-3p in 48 tumor tissues and matched non-tumor tissues was determined. The results manifested that miR-455-3p was significantly lower in tumor tissues than in matched nontumor tissues (Figure 1B).
Figure 1.
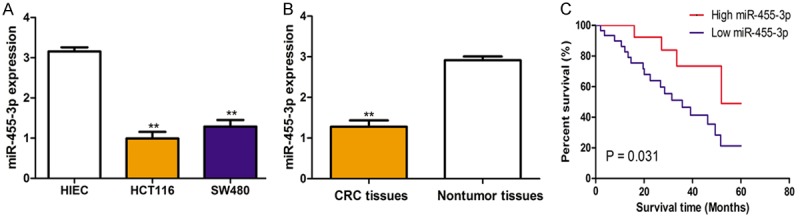
miR-455-3p expression in colorectal cancer tissues and cell lines and its relevance to overall survival. A. Expression of miR-455-3p in colorectal cancer cell lines (HCT116 and SW480) and immortalized human epithelial cell line (HIEC). B. Expression of miR-455-3p in colorectal cancer tissues and matched nontumor tissues. C. Kaplan-Meier curves for overall survival analysis by miR-455-3p expression in patients with colorectal cancer. (**P < 0.01) miR-455-3p: microRNA-455-3p.
miR-455-3p down-regulation is associated with poorer overall survival
In light of the finding that miR-455-3p was downregulated in tumor tissues, we further investigated the effect of miR-455-3p on overall survival. The Kaplan-Meier curve and log-rank test revealed that lower expression of miR-455-3p resulted in significantly poorer overall survival rate compared with those who with higher miR-455-3p expression (Figure 1C).
miR-455-3p upregulation inhibits cell proliferation and arrests cell cycle in vitro
In order to investigate the biological role of miR-455-3p in vitro, the CRC cell lines were transfected with miR-455-3p mimic, inhibitor, or miR-con, after which the cell proliferation rate and cell cycle distribution proportion were recorded. As expected, the results demonstrated that miR-455-3p mimic enhanced the expression of miR-455-3p in these CRC cell lines, while miR-455-3p inhibitor reduced the expression of miR-455-3p (Figure 2A). Further, the cell proliferation assay revealed that upregulation the expression of miR-455-3p reduced cell proliferation of the investigated cell lines (Figure 2B). Cell cycle analysis by flow cytometry showed that miR-455-3p upregulation could result the cells at G0/G1 phase increased and those at S phase decreased (Figure 2C). Additionally, the introduction increased the cells at S phase but decreased those at G0/G1 phase (Figure 2C).
Figure 2.
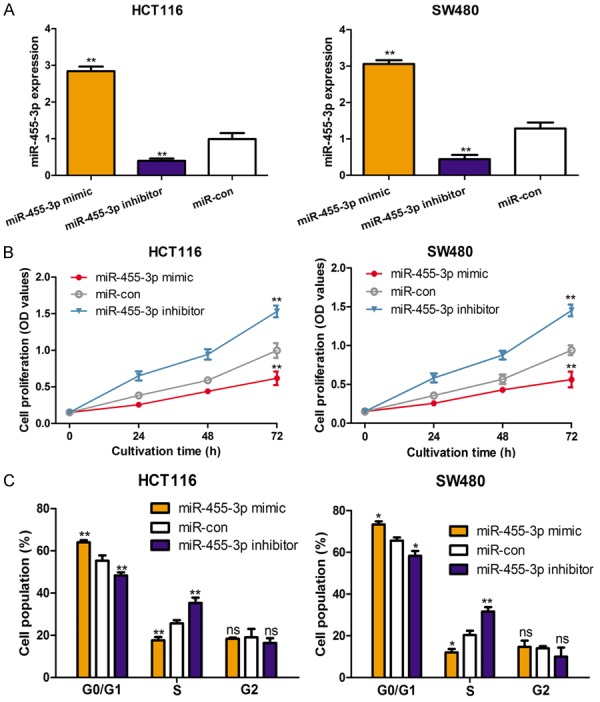
Overexpression of miR-455-3p inhibits cell proliferation and induces cell cycle arrest in colorectal cancer cell lines. A. Expression of miR-455-3p was measured by qRT-PCR; B. Cell proliferation was measured by CCK-8 assay; C. Cell cycle was measured by flow cytometry in HCT116 and SW480 cell lines with miR-455-3p mimic, inhibitor, or miR-con transfection. (**P < 0.01, *P < 0.05, ns not significant) miR-455-3p: microRNA-455-3p; qRT-PCR: quantitative real-time PCR; CCK-8: cell counting kit-8; miR-con: control miRNA.
TPT1 is a direct target of miR-455-3p in CRC
To identify the effector by which miR-455-3p exerts its suppressive role, we searched the possible target genes of miR-455-3p using TargetScan. As presented in Figure 3A, we show that TPT1 acts as a potential target of miR-455-3p. To validate this prediction, we analyzed the relative luciferase activity in CRC cell lines co-transfected with miR-455-3p mimic or miR-con and wt or mut 3’-UTR TPT1 constructs. The results show that miR-455-3p significantly reduced the luciferase activity of the wt 3’-UTR TPT1 construct but did not alter the activity of mut 3’-UTR TPT1 construct (Figure 3B). To further confirm TPT1 as a direct target of miR-455-3p, we analyzed the protein expression of TPT1 in the CRC cell lines transfected with miR-455-3p mimic or miR-con. Western blot analysis revealed that miR-455-3p significantly reduced TPT1 expression at the protein level (Figure 3C).
Figure 3.
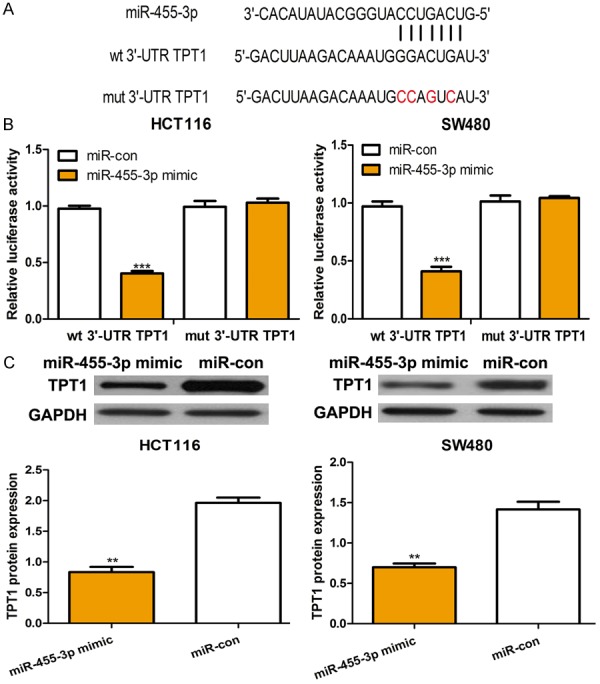
TPT1 was a direct target of miR-455-3p. A. The putative binding sequences of miR-455-3p in the 3’-UTR of TPT1. B. Luciferase reporter assay of HCT116 and SW480 cell lines with wt 3’-UTR TPT1 or mut 3’-UTR TPT1 and miR-455-3p mimic or miR-con transfection. C. Expression of TPT1 in HCT116 and SW480 cell lines with miR-455-3p mimic or miR-con transfection. (***P < 0.001, **P < 0.01) miR-455-3p: microRNA-455-3p; UTR: untranslated region; TPT1: The tumor protein translationally controlled 1; wt: wild-type; mut: mutant; miR-con: control miRNA.
TPT1 expression is negatively correlated with miR-455-3p expression in CRC
We then measured the expression of TPT1 in CRC tissues. Figure 4A revealed that the expression of TPT1 was upregulated in CRC tissues compared with in matched noncancer tissues. Subsequently, we analyzed the expression of TPT1 and miR-455-3p in CRC tissues. We found TPT1 expression was negatively correlated with miR-455-3p expression in CRC (Figure 4B).
Figure 4.
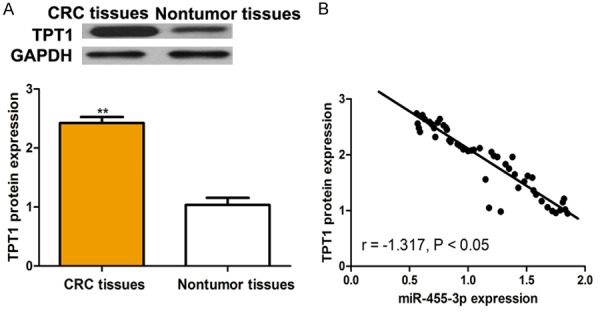
Expression of TPT1 was inversely correlated with the expression of miR-455-3p in colorectal cancer. A. Expression of miR-455-3p in colorectal cancer tissues and matched nontumor tissues. B. An inverse relationship between miR-455-3p and TPT1 expression levels. (**P < 0.01) miR-455-3p: microRNA-455-3p; TPT1: The tumor protein translationally controlled 1.
TPT1 down-regulation suppresses cell proliferation and arrests cell cycle in vitro
To determine the effect of TPT1 on CRC cell lines, the si-TPT1 and si-con were transfected into investigated CRC cell lines. As shown in Figure 5A, si-TPT1 decreased the protein expression of TPT1 in CRC cell lines. In addition, we found that down-regulation of TPT1 repressed cell proliferation and partially reverse the stimulation effect of miR-455-3p inhibitor (Figure 5B). Furthermore, cell cycle analysis demonstrated that si-TPT1 induce cell cycle arrest in G0/G1 phase, which is similar with the effect of miR-455-3p mimic (Figure 5C). Furthermore, we found miR-455-3p inhibitor relieve the inhibitory effect of si-TPT1 on cell cycle (Figure 5C).
Figure 5.
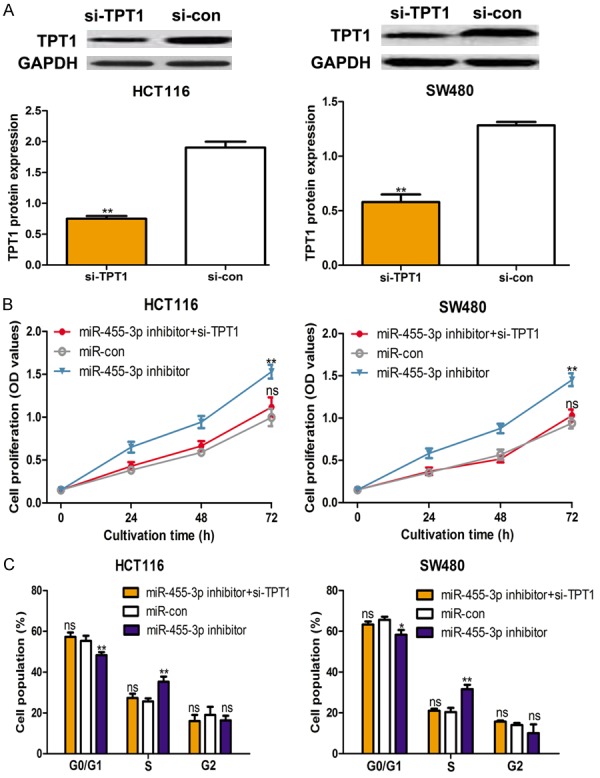
TPT1 down-regulation suppresses cell proliferation and induces cell cycle arrest in colorectal cancer cell lines. A. Expression of TPT1 in HCT116 and SW480 cell lines with si-TPT1 and si-con transfection. B. Cell proliferation was measured by CCK-8 assay and C. Cell cycle was measured by flow cytometry in HCT116 and SW480 cell lines with control or miR-455-3p inhibitor or miR-455-3p inhibitor and si-TPT1 transfection. (**P < 0.01, *P < 0.05, ns not significant) miR-455-3p: microRNA-455-3p; TPT1: The tumor protein translationally controlled 1; CCK-8: cell counting kit-8; si-TPT1: siRNA targeting TPT1; si-con: control siRNA.
Discussion
In 2003, it was first reported that miRNAs are associated with CRC [21]. From then on, a number of miRNAs were identified to be associated with the initiation or progression of CRC [22,23]. In light of these findings, the potential of miRNAs as the therapy targets of CRC patients has been investigated and showed a relatively bright future for targeted therapy [24]. In this study, we found that miR-455-3p was remarkedly reduced in CRC cell lines compared with the immortalized human epithelial cell line. Moreover, we also found reduced expression of miR-455-3p in CRC tissues compared to the matched noncancer tissues. Importantly, we found lower expression of miR-455-3p was associated with poorer overall survival of CRC patients. Additionally, functional assay revealed that cell proliferation was inhibited and cell cycle was arrested by ectopic expression of miR-455-3p. Moreover, expression of TPT1 was found to be inversely correlated with expression of miR-455-3p. Therefore, the results of this study indicate that miR-455-3p might be a novel biomarker for CRC treatment.
Emerging evidence has demonstrated that miRNAs play an important role in cancer initiation and development through regulating cell behavior [15]. Several miRNAs were found dysregulated in CRC and could regulate cell proliferation, migration, and apoptosis recently [25-27]. For example, it was reported that miR-598 expression was upregulated in CRC [25]. Additionally, cell proliferation and cell cycle progression were promoted by miR-598 upregulation via regulation of expression of INPP5E [25]. However, for the role of miR-455-3p in CRC, it has been rarely studied. In our study, we show that miR-455-3p expression was reduced in CRC, and thus we deduce that miR-455-3p might be a tumor suppressor in CRC.
In vitro functional assay has revealed that cell proliferation of CRC could be inhibited by miR-455-3p. We investigated whether cell proliferation inhibition was due to the arrest of cell cycle by measuring the cell cycle distribution using flow cytometry. We found that cell cycle was arrested in G0/G1 phase by miR-455-3p overexpression. To further study the mechanisms via which miR-455-3p inhibits cell proliferation and cell cycle progression in CRC, TPT1 was validated as a target of miR-455-3p by bioinformatics and expression correlation analyses. Our findings also revealed that inhibition of TPT1 partially reverse the stimulation effect of miR-455-3p inhibitor on cell proliferation and cell cycle progression.
In summary, our results indicate that miR-455-3p is a tumor suppressor in CRC, at least in part, by directly regulating TPT1. To the best of our knowledge, this is the first paper to demonstrate the cell proliferation and cell cycle can be regulated by miR-455-3p/TPT1 axis. Our study provides new evidence to understand how CRC develops and might lead to new therapeutic targets for CRC treatment in the near future.
Acknowledgements
This project was supported by Key Project of Tianjin Science and Technology Support Program (15ZCZDSY00890).
Disclosure of conflict of interest
None.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Ng SC, Wong SH. Colorectal cancer screening in Asia. Br Med Bull. 2013;105:29–42. doi: 10.1093/bmb/lds040. [DOI] [PubMed] [Google Scholar]
- 3.Kumar R, Price TJ, Beeke C, Jain K, Patel G, Padbury R, Young GP, Roder D, Townsend A, Bishnoi S, Karapetic CS. Colorectal cancer survival: an analysis of patients with metastatic disease synchronous and metachronous with the primary tumor. Clin Colorectal Cancer. 2014;13:87–93. doi: 10.1016/j.clcc.2013.11.008. [DOI] [PubMed] [Google Scholar]
- 4.Brioudes F, Thierry AM, Chambrier P, Mollereau B, Bendahmane M. Translationally controlled tumor protein is a conserved mitotic growth integrator in animals and plants. Proc Natl Acad Sci U S A. 2010;107:16384–9. doi: 10.1073/pnas.1007926107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gnanasekar M, Dakshinamoorthy G, Ramaswamy K. Translationally controlled tumor protein is a novel heat shock protein with chaperone-like activity. Biochem Biophys Res Commun. 2009;386:333–7. doi: 10.1016/j.bbrc.2009.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen WM, Wang HH, Tao SS, Zheng Y, Wu W, Lian FR, Jaramilo M, Fang DY, Zhang DD. Tumor protein translationally controlled 1 is a p53 target gene that promotes cell survival. Cell Cycle. 2013;12:2321–8. doi: 10.4161/cc.25404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.nanasekar M, Thirugnanam S, Zheng G, Chen A, Ramaswamy K. Gene silencing of translationally controlled tumor protein (TCTP) by siRNA inhibits cell growth and induces apoptosis of human prostate cancer cells. Int J Oncol. 2009;34:1241–6. [PubMed] [Google Scholar]
- 8.Ma Q, Geng Y, Xu W, Wu Y, He F, Shu W, Huang M, Du H, Li M. The role of translationally controlled tumor protein in tumor growth and metastasis of colon adenocarcinoma cells. J Proteome Res. 2010;9:40–9. doi: 10.1021/pr9001367. [DOI] [PubMed] [Google Scholar]
- 9.Carrington JC, Ambros V. Role of microRNAs in plant and animal development. Science. 2003;301:336–8. doi: 10.1126/science.1085242. [DOI] [PubMed] [Google Scholar]
- 10.Xi Y, Formentini A, Chien M, Weir DB, Russo JJ, Ju J, Kornmann M, Ju J. Prognostic values of microRNAs in colorectal cancer. Biomark Insights. 2006;2:113–21. [PMC free article] [PubMed] [Google Scholar]
- 11.Slattery ML, Pellatt AJ, Lee FY, Herrick JS, Samowitz WS, Stevens JR, Wolff RK, Mullany LE. Infrequently expressed miRNAs in influence survival after diagnosis with colorectal cancer. Oncotarget. 2017;8:83845–59. doi: 10.18632/oncotarget.19863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li N, Zhao X, Wang L, Zhang S, Cui M, He J. miR-494 suppresses tumor growth of epithelial ovarian carcinoma by targeting IGF1R. Tumour Biol. 2016;37:7767–76. doi: 10.1007/s13277-015-4603-8. [DOI] [PubMed] [Google Scholar]
- 13.Tian J, Hu L, Li X, Geng J, Dai M, Bai X. MicroRNA-130b promotes lung cancer progression via PPAR γ/VEGF-A/BCL-2-mediated suppression of apoptosis. J Exp Clin Cancer Res. 2016;35:105. doi: 10.1186/s13046-016-0382-3. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14.Sheng X, Chen H, Wang H, Ding Z, Xu G, Zhang J, Lu W, Wu T, Zhao L. MicroRNA-130b promotes cell migration and invasion by targeting peroxisome proliferator-activated receptor gamma in human glioma. Biomed Pharmacother. 2015;76:121–6. doi: 10.1016/j.biopha.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 15.Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 16.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 17.Zhao Y, Yan M, Yun Y, Zhang J, Zhang R, Li Y, Wu X, Liu Q, Miao W, Jiang H. MicroRNA-455-3p functions as a tumor suppressor by targeting eIF4E in prostate cancer. Oncol Rep. 2017;37:2449–58. doi: 10.3892/or.2017.5502. [DOI] [PubMed] [Google Scholar]
- 18.Gao X, Zhao H, Diao C, Wang X, Xie Y, Liu Y, Han J, Zhang M. miR-455-3p serves as prognostic factor and regulates the proliferation and migration of non-small cell lung cancer through targeting HOXB5. Biochem Biophys Res Commun. 2018;495:1074–80. doi: 10.1016/j.bbrc.2017.11.123. [DOI] [PubMed] [Google Scholar]
- 19.Yang H, Wei YN, Zhou J, Hao TT, Liu XL. MiR-455-3p acts as a prognostic marker and inhibits the proliferation and invasion of esophageal squamous cell carcinoma by targeting FAM83F. Eur Rev Med Pharmacol Sci. 2017;21:3200–6. [PubMed] [Google Scholar]
- 20.Zheng J, Lin Z, Zhang L, Chen H. MicroRNA-455-3p inhibits tumor cell proliferation and induces apoptosis in HCT116 human colon cancer cells. Med Sci Monit. 2016;22:4431–7. doi: 10.12659/MSM.898452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Michael MZ, O’Connor SM, van Holst Pellekaan NG, Young GP, James RJ. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol Cancer Res. 2003;1:882–91. [PubMed] [Google Scholar]
- 22.Shirafkan N, Mansoori B, Mohammadi A, Shomali N, Ghasbi M, Baradaran B. MicroRNAs as novel biomarkers for colorectal cancer: New outlooks. Biomed Pharmacother. 2018;97:1319–30. doi: 10.1016/j.biopha.2017.11.046. [DOI] [PubMed] [Google Scholar]
- 23.Dong Y, Yu J, Ng SS. MicroRNA dysregulation as a prognostic biomarker in colorectal cancer. Cancer Manag Res. 2014;6:405–22. doi: 10.2147/CMAR.S35164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li X, Nie J, Mei Q, Han WD. MicroRNAs: Novel immunotherapeutic targets in colorectal carcinoma. World J Gastroenterol. 2016;22:5317–31. doi: 10.3748/wjg.v22.i23.5317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li KP, Fang YP, Liao JQ, Duan JD, Feng LG, Luo XZ, Liang ZJ. Upregulation of miR-598 promotes cell proliferation and cell cycle progression in human colorectal carcinoma by suppressing INPP5E expression. Mol Med Rep. 2018;17:2991–7. doi: 10.3892/mmr.2017.8207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang C, Luo H. miR-19-5p enhances tumorigenesis in human colorectal cancer cells by targeting TSPYL5. DNA Cell Biol. 2018;37:23–30. doi: 10.1089/dna.2017.3804. [DOI] [PubMed] [Google Scholar]
- 27.Liu M, Yin K, Guo X, Feng H, Yuan M, Liu Y, Zhang J, Guo B, Wang C, Zhou G, Zhou Z, Zhang CY, Chen X. Diphthamide biosynthesis 1 is a novel oncogene in colorectal cancer cells and is regulated by MiR-218-5p. Cell Physiol Biochem. 2017;44:505–14. doi: 10.1159/000485087. [DOI] [PubMed] [Google Scholar]


