Abstract
Lung cancer is the leading cause of cancer-related mortality and non-small cell lung carcinoma (NSCLC) is the most common type. MicroRNAs (miRNAs) are small non-coding RNAs that negatively regulate gene expression at the post-transcriptional level. Aberrant expression of miRNAs has been demonstrated to be a prominent feature in NSCLC. The aim of this study was to determine the potential clinical value of serum miR-342-3p in NSCLC. We first evaluated the miR-342-3p levels in NSCLC cell lines, culture media of NSCLC cell lines, and serum samples from NSCLC patients as well as in their respective controls. The associations between serum miR-342-3p levels and clinicopathological parameters as well as clinical outcome were then determined. miR-342-3p expression was significantly downregulated in NSCLC cell lines, culture media of NSCLC cell lines, and the serum samples from NSCLC patients compared to their controls. Serum miR-342-3p discriminated NSCLC patients from healthy individuals. Low expression of serum miR-342-3p was significantly associated with advanced TNM stage and positive lymph node metastasis. In addition, NSCLC patients in the low serum miR-342-3p expression group had remarkably shorter overall survival than those in the high serum miR-342-3p expression group. Serum miR-342-3p was shown to be an independent prognosis factor. In conclusion, serum miR-342-3p might be a promising biomarker for NSCLC that can be used to improve diagnosis and prognosis.
Keywords: Non-small cell lung carcinoma, serum miR-342-3p, biomarker, prognosis, diagnosis
Introduction
Lung cancer is the most frequently diagnosed cancer and the leading cause of cancer mortality worldwide [1,2]. Non-small cell lung carcinoma (NSCLC) comprises approximately 80% of all lung cancer cases [3]. Lung adenocarcinoma and squamous cell lung carcinoma are the major subtypes of NSCLC, Despite the advances in surgical therapy, chemotherapy and radiotherapy as well as the advent of targeted therapies during recent decades, the mortality rate of NSCLC remains high mainly due to late diagnosis and the lack of effective treatment methodology [4]. Therefore, identifying novel biomarkers able to accurately detect NSCLC at the very early stage and characterize disease prognosis is important for NSCLC management.
MicroRNAs (miRNAs) are single-stranded, non-coding small RNA with 19 to 25 nucleotides in length. The mature miRNA forms an imperfect base pairing with its complementary sequence of the miRNA responsive element within the 3’ untranslated regions (3’UTRs) of the mRNA, leading to the downregulation of target genes [5]. An increasing number of miRNAs have been proven to actively involving in many crucial biological processes including, but not limited to, cell proliferation, differentiation, adhesion, migration, invasion, and apoptosis [6]. Alterations in miRNA expression have been reported in various diseased conditions including cancer of various types, indicating miRNA might be useful for the diagnosis and treatment of cancer [7]. In addition, the detectable miRNAs in tissue, blood, and other body fluids with high stability makes them attractive candidates for use in predictive and prognostic marker [8].
miRNAs are important for initiation and progression of NSCLC. For instance, the expression level of miR-338-3p was significantly reduced in NSCLC tissues and cell lines. Downregulated miR-338-3p was correlated with unfavorable clinical outcome of patients with NSCLC. In addition, miR-338-3p overexpression significantly suppressed the oncogenic behaviors of NSCLC cells, and vice versa, suggesting that miR-338-3p functions as a tumor suppressor in NSCLC [9]. Upregulation of miR-761 promotes progression and metastasis of NSCLC through regulating ING4 and TIMP2, indicating miR-761 plays an oncogenic role in the development of NSCLC [10]. Also, circulating miRNAs have been reported to be highly accurate for differentiating patients with NSCLC from healthy controls [11].
miR-342-3p regulate the carcinogenesis of various types of cancers such as liver cancer, cervical cancer, and NSCLC [12-14]. However, whether serum miR-342-3p levels are deregulated in patients with NSCLC and its potential clinical significance remain unknown. The purpose of this study was to determine the predictive and prognostic value of serum miR-342-3p for NSCLC.
Materials and methods
Cell culture
Human NSCLC cell lines, A549, H1650, HCC827, and H1299 and the normal control cell line BEAS-2B were obtained from American Type Culture Collection (ATCC, Manassas, VA, USA). Cells were cultured in RPMI-1640 (Thermo Fisher Scientific, Waltham, MA, USA) with 10% fetal bovine serum (Thermo Fisher Scientific) and 100 U/ml penicillin-streptomycin (Thermo Fisher Scientific). All cells were cultured at 37°C in a 5% CO2 humidified environment.
Patients and samples
A total of 146 NSCLC patients including 106 males and 40 females were enrolled in the current study. The mean age at diagnosis of NSCLC was 62.50±15.30 years. Forty age and sex matched healthy people receiving physical examinations were randomly chosen as the controls. The clinicopathological information of all NSCLC patients are listed in Table 1. Informed consent was obtained from all subjects, and the study was approved by the Ethical Committee of Peking Union Medical College Hospital. Serum samples were collected from patients before receiving any kind of therapy. Blood samples were collected and centrifuged at 3,000 rpm for 10 min to separate serum. The serum samples were stored at -80°C for later use.
Table 1.
Correlation between serum miR-342-3p and clinicopathological parameters in patients with NSCLC
| Parameters | N | Serum miR-342-3p | p | |
|---|---|---|---|---|
|
| ||||
| Low (n = 72) | High (n = 74) | |||
| Age, years | ||||
| < 60 | 82 (56.2%) | 43 (29.5%) | 39 (26.7%) | 0.3928 |
| ≥ 60 | 64 (43.8%) | 29 (19.9%) | 35 (24.0%) | |
| Gender | ||||
| Male | 106 (72.6%) | 52 (35.6%) | 54 (37.0%) | 0.9190 |
| Female | 40 (27.4%) | 20 (13.7%) | 20 (13.7%) | |
| Smoking status | ||||
| No | 109 (74.7%) | 51 (34.9%) | 58 (39.7%) | 0.2947 |
| Yes | 37 (25.3%) | 21 (14.4%) | 16 (11.0%) | |
| Histological subtypes | ||||
| SSC | 78 (53.4%) | 37 (25.3%) | 41 (28.1%) | 0.6267 |
| AD | 68 (46.6%) | 35 (24.0%) | 33 (22.6%) | |
| Lymph node metastasis | ||||
| Negative | 91 (62.3%) | 38 (26.0%) | 53 (36.3%) | 0.0188 |
| Positive | 55 (37.7%) | 34 (23.3%) | 21 (14.4%) | |
| Tumor size (cm) | ||||
| < 5 | 97 (66.4%) | 45 (30.8%) | 52 (35.6%) | 0.3202 |
| ≥ 5 | 49 (33.6%) | 27 (18.5%) | 22 (15.1%) | |
| TNM stage | ||||
| I/II | 87 (59.6%) | 30 (20.5%) | 57 (39.0%) | < 0.001 |
| III/IV | 59 (40.4%) | 42 (28.8%) | 17 (11.6%) | |
Quantitative real-time PCR
Total miRNA from cell lines, culture median and serum sample were extracted using the mirVana miRNA Isolation Kit (Thermo Fisher Scientific, Waltham, MA, USA). The isolated RNA was first reverse transcribed using the TaqMan miRNA reverse transcription kit (Applied Biosystems, Foster City, CA, USA) according to the manufacturer’s protocol. Complementary DNA was amplified and quantified on a 7500 Real-Time PCR System (Applied Biosystems) using SYBR Green Master Mix (Thermo Fisher Scientific). The expression level of miR-342-3p was calculated by comparative threshold cycle (2-ΔΔCt) method with U6 as the endogenous controls. All reactions were performed in triplicate. The primers of miR-342-3p and U6 are as follows: miR-342-3p sense: 5’-TCCTCGCTCTCACACAGAAATC-3’, miR-342-3p antisense: 5’-TATGGTTGTTCACGACTCCTTCAC-3’, U6 sense: 5’-ATTGGAACGATACAGAGAAGATT-3’, U6 antisense: 5’-GGAACGCTTCACGAATTTG-3’.
Statistical analysis
All statistical analyses were performed using the GraphPad Prism 7.0 (GraphPad Software Inc., San Diego, CA, USA) and MedCalc software (MedCalc, Mariakerke, Belgium). ANOVA was used to compare the miR-342-3p levels among different cell lines and their culture medium. Kruskal-Wallis test was carried out to analyze the serum miR-342-3p levels between NSCLC patients and normal controls. The area under the receiver operating characteristic (ROC) curve (AUC) was used to evaluate the predictive power of serum miR-342-3p. The correlation between clinicopathological variables and serum miR-342-3p was assessed using Chi-square test. OS curves were analyzed with the Kaplan-Meier method, and the log-rank test was used to assess survival differences between the groups. A Cox proportional hazards regression model was used to assess predictors related to overall survival. Differences were considered significant at a threshold of p < 0.05.
Results
miR-342-3p level was reduced in NSCLC cell lines and their culture media
We first used qRT-PCR to evaluate miR-342-3p expression in NSCLC cell lines and their culture media. Our results demonstrated that the expression level of miR-342-3p was significantly reduced in NSCLC cell lines (A549, H1650, HCC827, and H1299) in comparison with the normal control cell line BEAS-2B (**p < 0.01) (Figure 1A). Interestingly, miR-342-3p levels were also remarkably decreased in the culture medium of NSCLC cell lines (**p < 0.01) (Figure 1B).
Figure 1.
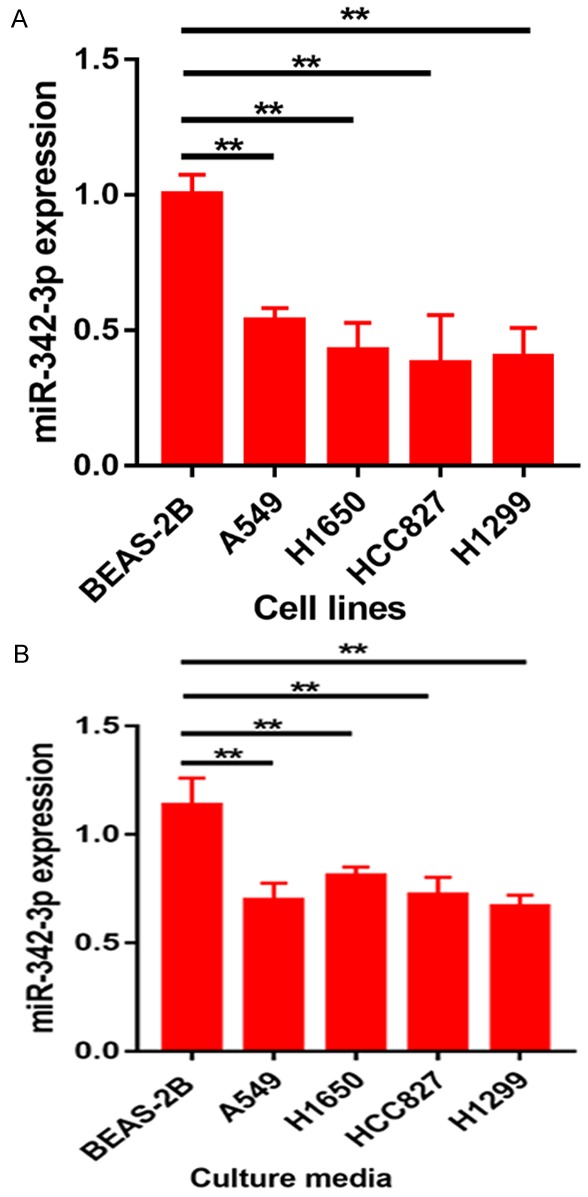
Expression level of miR-342-3p in NSCLC cell lines and their culture media.
Serum miR-342-3p discriminated NSCLC patients from healthy controls
Then we compared the expression level of serum miR-342-3p between NSCLC patients and healthy controls. The serum miR-342-3p levels were significantly reduced in NSCLC patients compared to healthy controls (**p < 0.01) (Figure 2A). ROC curve analysis was employed to evaluate the diagnostic value of serum miR-342-3p in discriminating NSCLC from healthy controls. The AUC value of serum miR-342-3p was 0.891 (specificity = 60.0%, sensitivity = 93.6%). The NSCLC patients were divided into two subgroups (patients with squamous cell carcinoma and patients with adenocarcinoma) (Figure 2B). Serum miR-342-3p could distinguish patients with squamous cell carcinoma (SCC) from healthy controls (AUC = 0.870, specificity = 82.5%, sensitivity = 75.6%) as well as patients with adenocarcinoma (AD) from healthy controls (AUC = 0.904, specificity = 82.5%, sensitivity = 82.1%) (Figure 2C, 2D).
Figure 2.
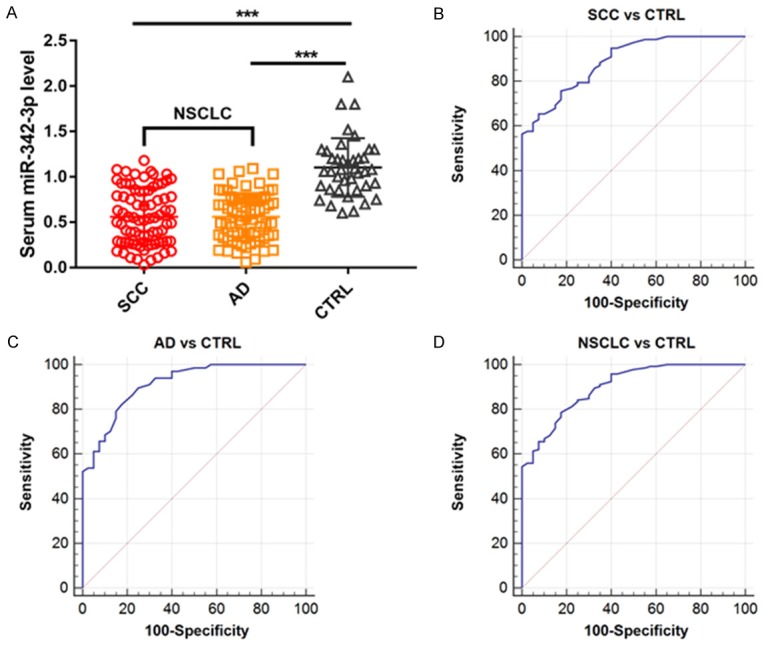
A. Expression level of serum miR-342-3p in NSCLC patients and healthy controls; B. Predictive value of serum miR-342-3p for discriminating SCC patients from healthy controls; C. AD patients from healthy controls and D. NSCLC patients from healthy controls.
Decreased serum miR-342-3p was associated with poor prognosis of NSCLC
All the NSCLC patients were divided into high serum miR-342-3p expression group and low serum miR-342-3p expression group. The median value of serum miR-342-3p in all NSCLC patients was used as the cut-off point. The association between serum miR-342-3p level and clinicopathological parameters was then analyzed. The results showed that reduced serum miR-342-3p was strongly associated with advanced TNM stage (p < 0.0001) and lymph node metastasis (p = 0.0188). However, no association was observed between serum miR-342-3p level and other parameters (Table 1).
OS in NSCLC patients with low serum miR-342-3p expression was shorter in those with high serum miR-342-3p expression (p = 0.0001) (Figure 3A). Then we analyzed the association between NSCLC and OS stratified by histological subtypes. The SCC patients in the low serum miR-342-3p expression group had clearly shorter 5-year OS compared with those in the high serum miR-342-3p expression group (p = 0.0219) (Figure 3B). Similarly, AD patients in the low serum miR-342-3p expression group had significantly shorter 5-year OS (p = 0.0015) (Figure 3C).
Figure 3.
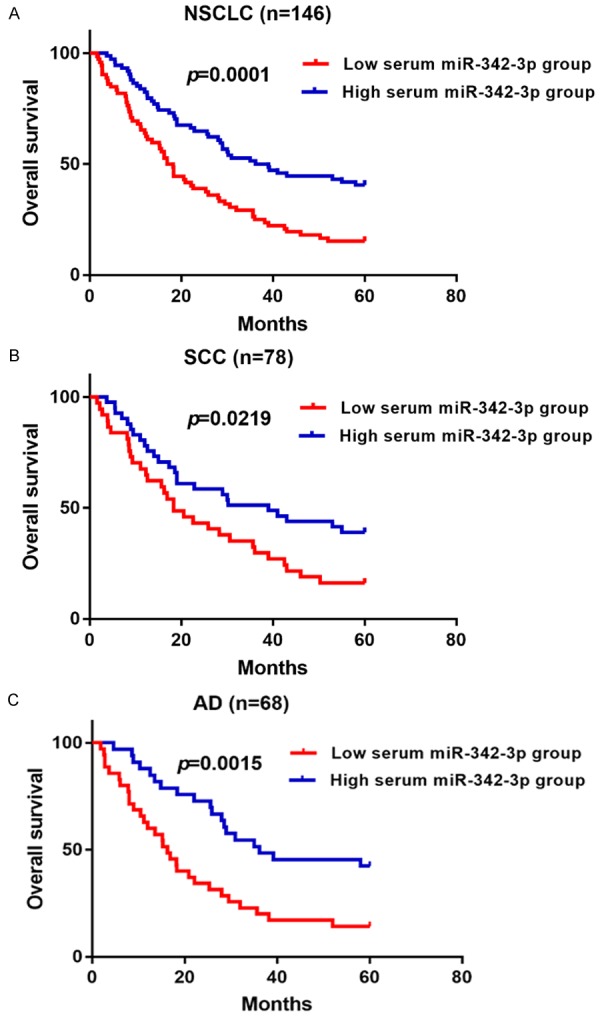
Association between serum miR-342-3p level and overall survival in NSCLC.
In multivariate analysis, serum miR-342-3p was an independent prognostic factor for OS (HR 1.903, 95% CI 1.052-2.714; p = 0.043), together with TNM stage (p = 0.015) (Table 2).
Table 2.
Multivariate analyses of independent prognostic factors on overall survival
| Parameters | Overall survival | ||
|---|---|---|---|
|
| |||
| Hazard ratio | 95% CI | p | |
| TNM stage (III/IV vs I/II) | 2.763 | 1.153-3.920 | 0.015 |
| Serum miR-342-3p (low vs high) | 1.903 | 1.052-2.714 | 0.043 |
Discussion
In this study, we have demonstrated that the miR-342-3p levels are significantly reduced in lung cancer cell lines and their culture medium as well as the serum samples from patients with NSCLC. In addition, serum miR-342-3p could accurately discriminate NSCLC patients from healthy controls. NSCLC patients with lower expression of serum miR-342-3p levels had worse prognosis, indicating that miR-342-3p might function as a tumor suppressor in tumorigenesis of NSCLC and serum miR-342-3p might serve as a predictive and prognostic biomarker for NSCLC. We propose that miR-342-3p is less synthesized in the lung cancer cells, thus its level is reduced in the serum samples derived from NSCLC patients.
In line with our results, the expression level of miR-342-3p was downregulated in NSCLC cell lines and tissues, and ectopic expression of miR-342-3p inhibited cell proliferation and invasion in vitro and tumor growth in vivo [14,15]. MYC is among the most frequently amplified and overexpressed oncogenes in lung cancer. miR-342-3p was demonstrated to indirectly regulating MYC activity via direct repression of E2F1 [16]. The expression level of miR-342-3p was especially decreased in NSCLC patients with ALK rearrangement compared to those with EGFR or KRAS mutation [17]. miR-342 was also shown to inhibit the growth of small cell lung cancer cells [18]. Due to the limited samples of our study, further studies with larger cohorts are needed to validate our findings. In addition, the molecular mechanisms accounting for the role of miR-342-3p in NSCLC needs further investigation.
miR-342-3p has also been reported as a tumor suppressor miRNA in multiple cancers.
miR-342 is encoded in an intron of the gene EVL and it is frequently suppressed in colorectal cancer. Overexpression of miR-342-3p induced apoptosis of colorectal cancer cells [19]. The expression level of miR-342-3p was significantly reduced in osteosarcoma tissues and cell lines. miR-342-3p overexpression suppressed the malignant behavior of osteosarcoma cells and miR-342-3p under-expression exhibited the opposite effects [20]. Dramatic downregulation of miR-342 was observed tamoxifen resistant breast cancer cell lines. Restoration of miR-342 could sensitize these cells to tamoxifen-induced apoptosis and cell cycle progression [21]. Ectopic expression of miR-342-3p suppressed the proliferation, migration, and invasion of cervical cell lines by directly regulating FOXM1 [22]. The expression level of serum miR-342 was reduced in patients with acute myeloblastic leukemia compared with the healthy controls [23].
Although miR-342-3p is widely regarded as a tumor suppressive miRNA, it might be not the case under all circumstances. For instance, miR-342-3p was highly expressed in exosomes from oral cancer cells, and miR-342-3p overexpression increased cell motility and invasive ability, suggesting that miR-342-3p might play an oncogenic role in oral cancer [24]. The expression level of serum miR-342-3p was significantly upregulated in patients with triple-negative breast cancer (TNBC) compared with healthy controls and patients with NTNBC [25]. miR-342 plays no role in the development, growth rate, or pathogenicity of pancreatic acinar carcinoma [26]. Therefore, the role of miR-342-3p in the initiation and development of cancers is closely associated with cancer subtypes and tumor microenvironment.
In conclusion, our results demonstrate that serum miR-342-3p level is decreased in NSCLC patients and its downregulation is associated with poor prognosis. Therefore, serum miR-342-3p might be a promising biomarker for monitoring NSCLC.
Acknowledgements
This study was supported by Major Project of Scientific Research of Ministry of Education (NO. 311037).
Disclosure of conflict of interest
None.
References
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, Thun MJ. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 2.Patel JN, Ersek JL, Kim ES. Lung cancer biomarkers, targeted therapies and clinical assays. Transl Lung Cancer Res. 2015;4:503–514. doi: 10.3978/j.issn.2218-6751.2015.06.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Molina JR, Yang P, Cassivi SD, Schild SE, Adjei AA. Non-small cell lung cancer: epidemiology, risk factors, treatment, and survivorship. Mayo Clin Proc. 2008;83:584–594. doi: 10.4065/83.5.584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Henschke CI, Yankelevitz DF. CT screening for lung cancer: update 2007. Oncologist. 2008;13:65–78. doi: 10.1634/theoncologist.2007-0153. [DOI] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 6.Esquela-Kerscher A, Slack FJ. Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 7.Soifer HS, Rossi JJ, Saetrom P. MicroRNAs in disease and potential therapeutic applications. Mol Ther. 2007;15:2070–2079. doi: 10.1038/sj.mt.6300311. [DOI] [PubMed] [Google Scholar]
- 8.Zen K, Zhang CY. Circulating microRNAs: a novel class of biomarkers to diagnose and monitor human cancers. Med Res Rev. 2012;32:326–348. doi: 10.1002/med.20215. [DOI] [PubMed] [Google Scholar]
- 9.Zhang P, Shao G, Lin X, Liu Y, Yang Z. MiR-338-3p inhibits the growth and invasion of non-small cell lung cancer cells by targeting IRS2. Am J Cancer Res. 2017;7:53–63. [PMC free article] [PubMed] [Google Scholar]
- 10.Yan A, Yang C, Chen Z, Li C, Cai L. MiR-761 promotes progression and metastasis of non-small cell lung cancer by targeting ING4 and TIMP2. Cell Physiol Biochem. 2015;37:55–66. doi: 10.1159/000430333. [DOI] [PubMed] [Google Scholar]
- 11.Wang C, Ding M, Xia M, Chen S, Van Le A, Soto-Gil R, Shen Y, Wang N, Wang J, Gu W, Wang X, Zhang Y, Zen K, Chen X, Zhang C, Zhang CY. A five-miRNA panel identified from a multicentric case-control study serves as a novel diagnostic tool for ethnically diverse non-small-cell lung cancer patients. EBioMedicine. 2015;2:1377–1385. doi: 10.1016/j.ebiom.2015.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao Y, Zhang SG, Wang ZH, Liao JC. Down-regulation of miR-342-3p in hepatocellular carcinoma tissues and its prognostic significance. Eur Rev Med Pharmacol Sci. 2017;21:2098–2102. [PubMed] [Google Scholar]
- 13.Li XR, Chu HJ, Lv T, Wang L, Kong SF, Dai SZ. miR-342-3p suppresses proliferation, migration and invasion by targeting FOXM1 in human cervical cancer. FEBS Lett. 2014;588:3298–3307. doi: 10.1016/j.febslet.2014.07.020. [DOI] [PubMed] [Google Scholar]
- 14.Xie X, Liu H, Wang M, Ding F, Xiao H, Hu F, Hu R, Mei J. miR-342-3p targets RAP2B to suppress proliferation and invasion of non-small cell lung cancer cells. Tumour Biol. 2015;36:5031–5038. doi: 10.1007/s13277-015-3154-3. [DOI] [PubMed] [Google Scholar]
- 15.Xue X, Fei X, Hou W, Zhang Y, Liu L, Hu R. miR-342-3p suppresses cell proliferation and migration by targeting AGR2 in non-small cell lung cancer. Cancer Lett. 2018;412:170–178. doi: 10.1016/j.canlet.2017.10.024. [DOI] [PubMed] [Google Scholar]
- 16.Tai MC, Kajino T, Nakatochi M, Arima C, Shimada Y, Suzuki M, Miyoshi H, Yatabe Y, Yanagisawa K, Takahashi T. miR-342-3p regulates MYC transcriptional activity via direct repression of E2F1 in human lung cancer. Carcinogenesis. 2015;36:1464–1473. doi: 10.1093/carcin/bgv152. [DOI] [PubMed] [Google Scholar]
- 17.Kim H, Yang JM, Jin Y, Jheon S, Kim K, Lee CT, Chung JH, Paik JH. MicroRNA expression profiles and clinicopathological implications in lung adenocarcinoma according to EGFR, KRAS, and ALK status. Oncotarget. 2017;8:8484–8498. doi: 10.18632/oncotarget.14298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu H, Cai T, Carmona GN, Abuhatzira L, Notkins AL. Small cell lung cancer growth is inhibited by miR-342 through its effect of the target gene IA-2. J Transl Med. 2016;14:278. doi: 10.1186/s12967-016-1036-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grady WM, Parkin RK, Mitchell PS, Lee JH, Kim YH, Tsuchiya KD, Washington MK, Paraskeva C, Willson JK, Kaz AM, Kroh EM, Allen A, Fritz BR, Markowitz SD, Tewari M. Epigenetic silencing of the intronic microRNA hsa-miR-342 and its host gene EVL in colorectal cancer. Oncogene. 2008;27:3880–3888. doi: 10.1038/onc.2008.10. [DOI] [PubMed] [Google Scholar]
- 20.Zhang S, Liu L, Lv Z, Li Q, Gong W, Wu H. MicroRNA-342-3p inhibits the proliferation, migration, and invasion of osteosarcoma cells by targeting astrocyte-elevated gene-1 (AEG-1) Oncol Res. 2017;25:1505–1515. doi: 10.3727/096504017X14886485417426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cittelly DM, Das PM, Spoelstra NS, Edgerton SM, Richer JK, Thor AD, Jones FE. Downregulation of miR-342 is associated with tamoxifen resistant breast tumors. Mol Cancer. 2010;9:317. doi: 10.1186/1476-4598-9-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fayyad-Kazan H, Bitar N, Najar M, Lewalle P, Fayyad-Kazan M, Badran R, Hamade E, Daher A, Hussein N, ElDirani R, Berri F, Vanhamme L, Burny A, Martiat P, Rouas R, Badran B. Circulating miR-150 and miR-342 in plasma are novel potential biomarkers for acute myeloid leukemia. J Transl Med. 2013;11:31. doi: 10.1186/1479-5876-11-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao L, Zhang Y. miR-342-3p affects hepatocellular carcinoma cell proliferation via regulating NF-κB pathway. Biochem Biophys Res Commun. 2015;457:370–377. doi: 10.1016/j.bbrc.2014.12.119. [DOI] [PubMed] [Google Scholar]
- 24.Sakha S, Muramatsu T, Ueda K, Inazawa J. Exosomal microRNA miR-1246 induces cell motility and invasion through the regulation of DENND2D in oral squamous cell carcinoma. Sci Rep. 2016;6:38750. doi: 10.1038/srep38750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shin VY, Siu JM, Cheuk I, Ng EK, Kwong A. Circulating cell-free miRNAs as biomarker for triple-negative breast cancer. Br J Cancer. 2015;112:1751–1759. doi: 10.1038/bjc.2015.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dooley J, Lagou V, Pasciuto E, Linterman MA, Prosser HM, Himmelreich U, Liston A. No functional role for microRNA-342 in a mouse model of pancreatic acinar carcinoma. Front Oncol. 2017;7:101. doi: 10.3389/fonc.2017.00101. [DOI] [PMC free article] [PubMed] [Google Scholar]


