Abstract
The electrospray-MS analysis of oligonucleotides is hampered by non-volatile metal cations, which may produce adducts responsible for signal suppression and loss of resolution. Alternative to replacing metal cations with MS-friendly ammonium, we explored the utilization of nanospray emitters with submicrometer-diameter tips, which was shown to benefit the analysis of protein samples containing elevated salt concentrations. We demonstrated that such benefits are not limited to proteins, but extend also to oligonucleotide samples analyzed in the negative ion mode. At elevated Na+/Mg2+ concentrations, submicrometer tips produced significantly greater signal-to-noise ratios, as well as greatly reduced adducts and salt clusters, than observed when utilizing micrometer tips. These effects were marginally affected by emitter composition (i.e., borosilicate versus quartz), but varied according to salt concentration and number of oligonucleotide phosphates. The results confirmed that adduct formation is driven by the concentrating effects of the desolvation process, which leads to greatly increased solute concentrations as the volume of the droplet decreases. The process promotes cation-phosphate interactions that may not have necessarily existed in the initial sample, but nevertheless shape the observed adduct series. Therefore, such series may not accurately reflect the distribution of counter-ions surrounding the analyte in solution. No adverse effects were noted on specific metal interactions, such as those present in a model drug-DNA assembly. These observations indicate that the utilization of submicrometer tips represents an excellent alternative to traditional ammonium-replacement approaches, which enables the analysis of oligonucleotides in the presence of Na+/Mg2+ concentrations capable of preserving their structure and functional properties.
Introduction
Alkaline and alkaline-earth cations play essential roles in defining the structure and function of nucleic acids.1,2 The emergence of new classes of nucleic acids that do not owe their function to the encoded genetic information, but rather to their 3D structure and binding properties,3 has placed a premium on the ability to analyze these types of biopolymers under non-denaturing conditions, in sample solutions approaching physiological pH and ionic strength. Metal cations have general effects on solvation, which can be at least partially described by the relationship between chemical activity and ionic strength, as well as specific effects on both intra- and inter- molecular interactions that define the fold of nucleic acids.2 Metal cations are capable of neutralizing the negative charges on the highly acidic phosphate groups of the biopolymer backbone, which minimizes electrostatic repulsion between opposing strands to enable stable folding. Further, divalent cations such as Mg2+ can form salt bridges between distal phosphates to establish long-range contacts responsible for shaping overall structure topology.4 Finally, the mechanism of action of ribozymes and deoxyribozymes is proof that the roles of divalent cations are not only structural, but cover also the catalytic activities enacted by these classes of nucleic acids.5 For these reasons, the investigation of the structure-function relationships in these biopolymers cannot be accomplished in the absence of alkaline and alkaline-earth cations.
The mass spectrometric (MS) analysis of nucleic acids can be severely hampered by the presence of metal cations, which can lead to unwanted formation of stable adducts, loss of resolution, and extensive signal suppression [6,7 and references therein]. The discovery that electrospray ionization (ESI) can desorb intact oligonucleotide duplexes without strand dissociation8,9 has revealed this technique as an invaluable platform to investigate the binding of nucleic acids with other nucleic acids, proteins, and a variety of other ligands [10,11 and references therein]. Further, the fact that hydrogen bonding12 and stacking interactions13 can be preserved in the gas phase substantiates the potential of this approach to provide valid structural information. For this reason, beyond popular sequencing applications, ESI-MS has been successfully employed to assess structure,14 stability,15 and binding characteristics [10,11 and references therein] of target nucleic acids. These types of determinations must ensure the integrity of folding and binding interactions established in solution, while minimizing the deleterious effects of adduct formation on the quality of the analysis. These seemingly discording requirements are typically met by controlling pH and ionic strength through the utilization of MS-friendly electrolytes. The realization that ammonium adducts could readily release ammonia during desolvation, while leaving protons as counter-ions,16 has led to the preferred utilization of ammonium-based salts/buffers in place of physiological equivalents containing metal cations. Over the years, a variety of strategies have been developed to accomplish ammonium replacement, including ion exchange,17 ethanol precipitation,18 reversed- phase high-performance liquid chromatography,19 metal chelation,18 and ultrafiltration/microdialysis,20 which have met different levels of effectiveness.
Although ammonium replacement does not significantly alter the effects of ionic strength on the binding affinity of biomolecular complexes,21 the process may have dire consequences on specific interactions mediated by Mg2+, which cannot be properly replicated by monovalent ions. For this reason, we have been exploring alternative strategies for achieving controlled analyte desalting in the gas phase, rather than complete ion replacement in solution, which would help preserve essential metal interactions without jeopardizing the quality of MS determinations. In earlier studies, we employed a dual-spray setup that segregated analyte and chelator solutions in separate emitters, but allowed for the respective electrospray plumes to overlap in the same physical space.22 This modus operandi enabled the establishment of ion/ion reactions in the high-pressure region of the source, which could be finely tuned to eliminate non-specific adducts and preserve specific metal binding.22 In this report, we have tested the ability of submicrometer nanospray emitters to produce viable data from nucleic acid samples that had not been submitted to ammonium replacement and contained near physiological concentrations of metal ions. This investigation stemmed from the observation that the size of nanospray emitters significantly affected the incidence of metal adducts of proteins analyzed in solutions that contained common buffers and salts mimicking both intracellular and extracellular environments.23–25 Further, it has been recently shown that submicrometer emitters can facilitate the analysis of membrane proteins in the presence of both ionic and non-ionic detergents, thus promoting the application of native MS to traditionally challenging samples.26 Small diameter tips have been reported also to enable desalting of ions obtained from denaturing solutions that contained acid and/or organic solvents with a low concentration of nonvolatile salts.27,28 We have now evaluated the performance of such emitters in oligonucleotide analysis and compared the outcome with that afforded by regular size emitters. The results provided valuable new insights into the mechanism of adduct formation, which are discussed here in the context of the current knowledge of nucleic acid analysis.
Experimental
The samples examined in the study consisted of a 6mer oligo-deoxyribonucleotide, and 19mer and 38mer oligoribonucleotides with respective sequences: dCdGdCdGdCdG, CAC GAC CAG UCC GAC GAG C, and CAC GAC CAG UAA AAC GCG CCA CGA CCA CUC CGA CGA UC. All samples were purchased from IDT (Coralville, IA), ethanol precipitated, and reconstituted in trace analysis grade Fluka water from Honeywell (Morris Plains, NJ) (see Supporting Information for additional details). Cytochrome c from horse heart and chromomycin A3 from S. griseus were purchased from Sigma (St. Louis, MO) and used without further purification. All nanospray-MS analyses were carried out in either negative or positive ion mode on a Bruker Daltonics (Billerica, MA) solariX Fourier transform ion cyclotron resonance (FTICR) mass spectrometer equipped with a 12T superconducting magnet. Borosilicate and quartz emitters were produced in house by using either a Model P-87 or a Model P-2000 (Sutter Instruments, Novato, CA) pipette puller. The programs were adjusted to obtain tip diameters of either 1.90 ± 0.1 μm and 0.66 ± 0.02 μm for borosilicate, or 1.20 ± 0.16 μm and 0.61 ± 0.07 μm for quartz emitters, as determined by using a Hitachi (Schaumburg, IL) TM-1000 scanning electron microscope. A more detailed description of all experimental procedures is provided in Supporting Information.
Results and Discussion
Influence of tip diameter and emitter composition on salt adduction
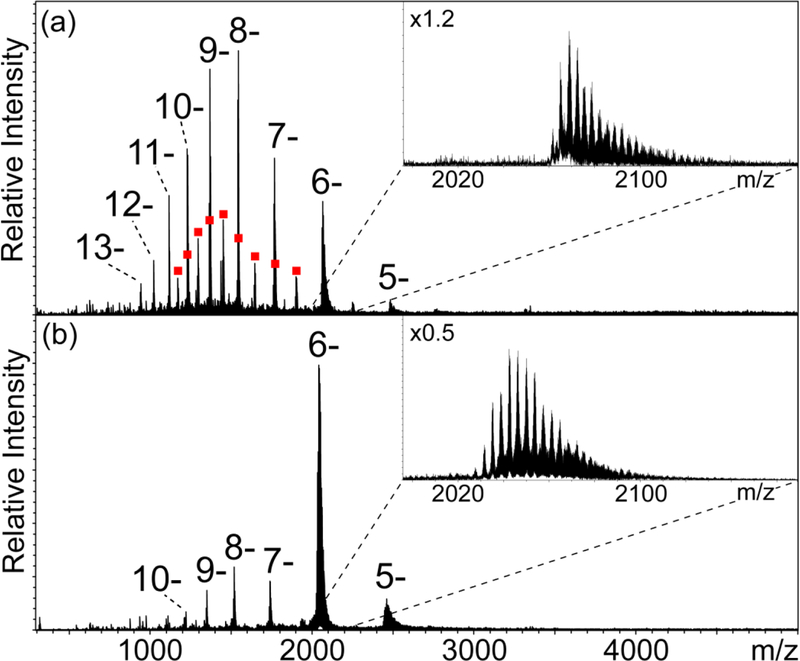
A 19mer RNA with mixed base composition to better mimic natural samples was selected as a model system to investigate the effects of tip diameter on salt adduction. The design of the construct was guided by structure-prediction algorithms to minimize the possibility that the selected sequence may form stable secondary structure with unpredictable effects on adduct formation (see Supporting Information for details). Fig. 1a depicts a representative mass spectrum obtained from a 3-μM sample in 1 mM NaCl aqueous solution with pH 7.5, which was analyzed by using a borosilicate emitter with micrometer-size tip (see Experimental and Supporting Information). The signal-to-noise ratio (S/N) of the RNA ions was significantly lower than typical data obtained from desalted samples of similar concentrations. Analyte signals corresponding to a charge state distribution spanning from 3- to 6- were immediately recognizable above 1,000 m/z, whereas numerous signals corresponding to salt clusters were observed in the lower range. Additionally, weaker signals were detected for a dimeric form, consistent with artifacts that are frequently present in ESI spectra obtained from oligonucleotide samples with relatively high concentrations (typically above ~1 μM).29 All analyte signals displayed broad distributions of sodium adducts, as exemplified by the representative 4- charge state in the inset of Fig. 1a. A curve fitting approach was employed to identify the centroid of the adduct distribution of each charge state in the spectrum (red curve in inset), which was then used to calculate the average mass of adducted analyte (red dashed line) and average number of adducts (see Supporting Information). The results reported in Table 1 indicated that the lower charge states afforded the greater incidence of adducts, consistent with the fact that metal cations neutralize the negative charges present on this type of analyte. However, it has been observed that lower charge states of protein ions also adduct more sodium ions than do higher charge states in positive ion mode,23–25 thus suggesting that other factors may also play a role.
Figure 1.
Nanospray mass spectra of 3-μM samples of 19mer RNA in water containing 1 mM NaCl at pH 7.5, which were acquired by using either (a) micrometer- or (b) submicrometer-size borosilicate emitters (see Supporting Information). The overall charge state is used to label each adduct series regardless of the actual balance between protonation and cation adduction. The representative 4- charge state displayed in the insets illustrates the utilization of curve fitting to calculate the average number of adducts manifested by each charge state (see Supporting Information). The results for the entire spectrum are summarized in Table 1.
Table 1.
Average sodium adduction and signal-to-noise ratios (S/N) observed in mass spectra obtained from 3-μM samples of 19mer RNA in water containing 1 mM NaCl at pH 7.5. The data were acquired by using either borosilicate or quartz emitters with large and small tip diameters (see Supporting Information).
| 19mer RNA | Average Na+ adduction and S/N observed in 1 mM NaCl | |||||||
|---|---|---|---|---|---|---|---|---|
| Micrometer borosilicate | Submicron borosilicate | Micrometer quartz | Submicron quartz | |||||
| z (−) | Adducts | S/N | Adducts | S/N | Adducts | S/N | Adducts | S/N |
| 3 | 11.6 | 3 | 3.8 | 8 | 12.2 | 6 | 4.2 | 5 |
| 4 | 9.9 | 23 | 3.3 | 212 | 6.8 | 74 | 3.7 | 450 |
| 5 | 8.1 | 20 | 1.8 | 196 | 3.7 | 14 | 1.6 | 350 |
| 6 | 6.3 | 8 | 1.5 | 116 | -- | -- | 1.3 | 169 |
| 7 | -- | -- | 1.1 | 108 | -- | -- | 0.9 | 100 |
| 8 | -- | -- | 0.6 | 84 | -- | -- | 0.8 | 48 |
| Ave. | 9.0 | 13.5 | 2.0 | 120.7 | 7.6 | 31.3 | 2.1 | 187.0 |
When the same sample was analyzed back-to-back on a submicrometer-size borosilicate emitter, the observed differences were striking (Fig. 1b). The charge distribution spanned from 3- to 8-, while the overall S/N was significantly greater and the abundance of salt clusters in the low m/z range was noticeably reduced. The S/N of the most abundant RNA charge state (4-) was nearly 10-fold higher with the submicrometer emitter (Table 1). The incidence of adduct formation was also noticeably reduced, as shown by the average number of sodium adducts to the representative 4- charge state, which decreased from 9.9 to 3.3 (Table 1 and Fig. 1b inset). The adduction decrease across the board enabled the detection of additional, higher charge states, in agreement with the presence of fewer charge-neutralizing cations. In this direction, the highest charge in the distribution (8-) manifested only one low-abundance adduct. Finally, the absence of dimeric artifacts may provide additional support to the mechanism proposed for the effects of submicrometer tips.23–25
These analyses were repeated by using quartz emitters to evaluate the possible impact of emitter composition on salt adduction. Representative data obtained from the same 19mer sample (i.e., 3 μM RNA in water containing 1 mM NaCl at pH 7.5) by using either a micrometer or submicrometer quartz emitter are compared in Fig. S-3 and S-4 of Supporting Information. Consistent with the results obtained from borosilicate emitters, tip size proved to significantly affect the outcome of the measurement. In particular, the larger tips produced a charge state distribution spanning from 3- to 5-, in addition to abundant salt clusters in the lower m/z range (Fig. S-3a). In contrast, the smaller tips produced a charge state distribution extending up to 8-, greatly improved S/N across the board and much reduced salt clusters (Fig. S-3b). Further, the incidence of salt adducts was significantly reduced, with analyte signals manifesting less than half the adduction displayed by corresponding species detected with the larger emitter and a 6- fold gain in the S/N of the most abundant charge state (4-) (Table 1). Analyzed by using the larger and smaller emitter, the representative 4-charge state provided 6.8 and 3.7 adducts, respectively. Also in this case, the highest charge state obtained with the submicrometer emitter (8-) displayed only a single low-abundance adduct (Fig. S-3b). The lowest charge state (3-) produced by the micrometer emitter also displayed distinct adduct series for either Na+ or NaCl (Fig. S-4b).
A close comparison of the data obtained from borosilicate and quartz emitters confirmed that the latter provided a definite advantage over the former, especially for the larger size ones (Table 1). The larger emitters provided an overall 16% reduction in average adduction (i.e., from 9.0 to 7.6 average adducts across all detected charge states) between borosilicate and quartz, but an extraordinary 132% increase of the average S/N (i.e., from 13.5 to 31.3 average S/N). In contrast, the submicrometer emitters afforded a comparable average adduction (i.e., 2.0 to 2.1 average adducts) between borosilicate and quartz, and a more limited 55% increase of S/N (i.e., from 120.7 to 187.0 average S/N). These results could be explained by the elemental composition of the borosilicate glass used in our study (i.e., Corning 7740), which consisted of SiO2 (80.6%) and B2O3 (13.0%), with significant amounts of Na2O (4.0%) and Al2O3 (2.3%). In contrast, the quartz glass (i.e., Heraeus HSQ300) consisted exclusively of SiO2, with only trace amounts of Li (0.5 ppm), Na (0.2 ppm), K (0.3 ppm), Ca (0.5 ppm), Fe (0.1 ppm), Ti (1.1 ppm), Zr (1.0 ppm), and Al (15 ppm). The much larger amount of Na present in the former suggests the possibility that leaching in solution may lead to significant increases of overall salt loads Regardless of composition, however, the smaller size emitters performed consistently better in terms of both average adduction and S/N (Table 1), thus corroborating the results described earlier for protein analysis in positive ion mode.23–28,30
Influence of analyte size and salt concentration on tip performance
Oligonucleotides can be observed in positive ion mode by virtue of so-called “wrong-way-round” ionization effects,31 but their analysis is more typically performed in negative ion mode to capitalize on the highly acidic nature of phosphate groups. As a first approximation, the greater number of phosphates afforded by the larger oligonucleotides tends to result in greater charging to support detection. At the same time, however, the greater number of negative charges may lead to greater incidence of adduction. In this context, adduct formation could be rationalized by a putative process in which the negative phosphates on the construct are partially neutralized by the formal addition of positive counter-ions present in solution. Such counter-ions may consist of metal cations, as well as protons provided by the aqueous environment or volatile ammonium ions, which by definition are not counted as adducts. If this holds true, increasing the number of phosphates should increase the incidence of adducts.
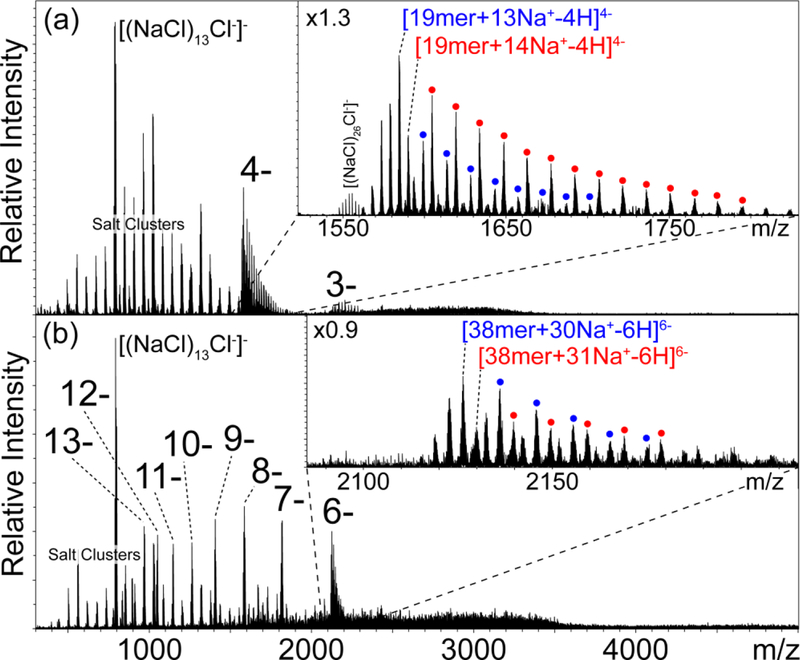
To test this hypothesis, we designed a 38mer RNA oligonucleotide that approximately doubled the number of phosphates present in the above analyte. Assuming that these functional groups are the only source of negative charges and considering that the synthetic oligonucleotides lacked terminal phosphates, the 38mer and 19mer RNA afforded respectively 37 and 18 putative negative sites. Also in this case, the model possessed a mixed base composition and a sequence that prevented the folding of obvious secondary structure. Representative data obtained from a 3-μM sample in aqueous 1 mM NaCl at pH 7.5 are provided in Fig. S-5 of Supporting Information, which were recorded by using emitters with either micrometer- or submicrometer-diameter tips (panel a and b, respectively). At first sight, the former displayed significantly lower S/N and greater incidence of adducts than the latter, as corroborated by the respective quantitative data in Table S-1 (see Supporting Information). The assessment revealed that the average S/N increased from 28.4 to 75.1 (corresponding to a 168% increase) between the larger- and smaller-tip data, whereas average adduction dropped from 21.6 to 2.2 (a 90% decrease). The adducts formed by the representative 6- charge state dropped from 26.0 to 5.0 (an 81% decrease), as evident also from the sizeable mass shift observed in the insets of Fig. S-5. The S/N of the most abundant charge state with the submicrometer tip (6-) was more than 5-fold higher than the most abundant charge state with the micrometer tip (6-), indicating that the detection limit was improved by this same factor. Consistent with the 19mer data, the highest charge state (12-) produced by the submicrometer emitter displayed only a single adduct with relatively low abundance, and the extensive salt clusters in the low range were also significantly reduced. Comparing the results obtained from the two analytes revealed that the incidence of adduct formation increased with oligonucleotide size only when the analysis was performed with the larger tips. Indeed, the values of average adduction were 9.0 and 21.6 for the 19mer and 38mer, respectively, when larger tips were used, but only 2.0 and 2.2 with the smaller ones (compare Table 1 and S-1). These data clearly show that the average adduction produced by the larger tips almost exactly doubled when doubling the analyte’s size, and thus the number of potentially negative sites.
We next determined the effects of increasing salt concentration to test whether the formation of higher order adducts might be promoted by mass action. Representative data obtained from samples containing 20 mM NaCl at pH 7.5 are respectively provided in Fig. S-6 and S-7 of Supporting Information, and the corresponding values are summarized in Table S-2. As expected, these conditions produced noticeable deterioration of S/N across the board and increased salt clustering in the low/intermediate range, which were only partially remediated by the submicrometer tips. Close data comparison revealed that the values of average adduction afforded by the larger tips varied very little at higher salt concentrations, whereas those afforded by the submicrometer ones displayed significant differences. For instance, the values observed for the 19mer were 7.6 and 7.7 for the 1 and 20 mM NaCl samples analyzed by using the micrometer tips, but 2.1 and 7.8 when the submicrometer ones were used. In similar fashion, the 38mer provided 21.6 and 21.0 with the larger tips, and 2.2 and 21.2 with the smaller ones. For both analytes, the benefits of the submicrometer tips were minimized by the higher salt concentration in solution. Further, average adduction seemed to converge toward a common value for each construct (i.e., ~7.8 and ~21.1 for 19mer and 38mer, respectively), which was already met at the lower salt concentration when the larger tips were used. These observations suggested the existence of a possible cap of average adduction above which the submicrometer tips lost their effectiveness.
Analogous determinations were also performed on samples that contained increasing MgCl2 concentrations to ascertain the possible effects of the charge borne by the cation. The outcomes observed with divalent adducts were qualitatively identical to those afforded by monovalent ones. For example, Fig. 2 and S-8 of Supporting Information provide representative data obtained from 38mer RNA in either 0.5 or 5 mM MgCl2 solutions at pH 6.7. In this case, micrometer tips produced average adduction of 11.2 and 13.8 at lower and higher Mg2+, respectively, whereas submicrometer ones provided corresponding values of 6.0 and 15.2 (Table S-3 of Supporting Information). Consistent with the NaCl experiments, the small-diameter tips significantly improved the data obtained at lower salt concentrations, but similar results between the two sizes were obtained at higher concentrations. The putative adduction cap for Mg2+ fell around ~14.5, as compared instead to ~21.1 for Na+. The fact that one was not exactly twice the other suggested that the charge of the cation may not represent the only determinant factor in adduct formation. This is consistent with the observation that Mg2+ and Na+ interactions have different effects on the conformation of nucleic acids in solution, which may result in widely different binding stoichiometries. This fact is reflected also by the increased observation of dimeric artifacts, which may be promoted by the ability of Mg2+ to stabilize nucleic acid binding interactions (see for example Fig. 2a).
Figure 2.
Nanospray mass spectra of 3-μM samples of 38mer RNA in water containing 0.5 mM MgCl2 at pH 6.7, which were acquired by using either (a) micrometer- or (b) submicrometer-size emitters (see Supporting Information). The overall charge state is used to label each adduct series regardless of the actual balance between protonation and cation adduction. The distributions of signals corresponding to dimer formation are indicated by red squares ( ). The insets enlarge the regions of the 6- charge state to better appreciate the adduct series. The information provided by the spectrum is summarized in Table S-3 of Supporting Information.
). The insets enlarge the regions of the 6- charge state to better appreciate the adduct series. The information provided by the spectrum is summarized in Table S-3 of Supporting Information.
Effects of adduction levels on analyte detection
The observation of a possible cap on adduct formation prompted a closer examination of the maximum number of adducts observed for each analyte, which may reveal possible correlations with their respective maximum number of negative sites. The data summarized in Table S-4 of Supporting Information were obtained from 19mer and 38mer samples that contained greater concentrations of NaCl and MgCl2 to further promote adduct formation (see Fig. 3 and S-9 of Supporting Information). The maximum number of Na+ matched very closely the balance between the number of phosphates on the constructs (i.e., 18 and 37 for 19mer and 38mer, respectively) and the actual experimental charge. A similar outcome was obtained for the maximum number of Mg2+ adducts when its doubly charged character was accounted for. Aside for minor match imprecisions for the weaker charge states, which may be ascribed to the inability to detect the next higher-order adduct above noise level, the maximum number was consistently lower than the total number of phosphates, in agreement with the neutralization mechanism. This outcome corroborates the fact that actual signal detection depends on the existence of subpopulations of ions that are not fully neutralized by counter-ions under the selected conditions. The charge-balancing act was also reflected in the observation that the lower charge states were consistently the more heavily adducted species in each spectrum. Taken a step further, full saturation of available negative charges by adduct counter-ions could lead to complete analyte neutralization, which could explain the undesirable signal suppression plaguing the analysis of oligonucleotides at high salt concentrations.
Figure 3.
Nanospray mass spectra of 3-μM samples of (a) 19mer RNA in 30 mM NaCl at pH 7.5, and (b) 38mer RNA in 40 mM NaCl at pH 7.5, which were acquired by using submicrometer-size emitters (see Supporting Information). The overall charge state is used to label each adduct series regardless of the actual balance between protonation and cation adduction. Signals indicated with colored dots correspond to subsequent NaCl adduction to the ion labeled with the same color. The information provided by the spectrum is summarized in Table S-4 of Supporting Information.
While the neutralization process can properly describe adduct formation in negative ion mode, it cannot fully explain the experimental behavior observed in the opposite polarity. For example, the representative data in Fig. S-10 of Supporting Information were obtained from a 38mer sample in 25 mM NaCl at pH 7.5, which was analyzed in either negative or positive ion mode. Micrometer emitters were employed to maximize the incidence of adduction observable at this salt concentration. The general quality of the positive ion data was significantly inferior, as evidenced by abundant salt clusters across the low/intermediate range and detection of only one charge state (5+) with rather limited S/N. Abundant adduction was observed in either polarity, as reflected by the values of average and maximum adductions reported in Table S-5. However, the principle of charge balance was only followed by the negatively charged adducts, which never exceeded the 37 phosphate groups present on this construct. In contrast, the lone charge state observed in positive ion mode displayed values of average and maximum adduction of 39.6 and 40, which can be achieved only upon neutralization of all phosphates and further addition of five Na+ ions to reach the observed charge state. If binding of additional Na+ ions cannot be attributed to full-fledged ionic bonds with negative phosphates, then it must involve different types of interactions with other electron-rich functional groups on the construct. The possible implication of alterative interactions would also explain the abundant formation of NaCl adducts observed at high salt concentrations (see for example Fig. S-4).
The possibility that adduct formation may not be supported exclusively by ionic interactions leading to charge neutralization was further tested by examining protein analytes that, by virtue of their more diverse functional groups, may offer greater opportunities for alternative binding modes. We selected cytochrome c as a possible model based on the limited number of acidic residues (i.e., 13 total D, E, and C-terminus) capable of providing discrete negative charges and a molecular mass comparable to that of the 38mer RNA (i.e., 12,360 vs. 12,106 Da, respectively). Fig. S-11 provides representative data obtained in either negative or positive ion mode from a 10-μM sample in water containing 25 mM sodium acetate at pH 6.5. Emitters with micrometer-size tips were employed to favor adduct formation. Values of average and maximum adductions are included in Table S-5. Consistent with the high salt content, both spectra displayed extensive clusters, low S/N across the board, and detection of only one charge state with numerous Na+ adducts. The fact that the maximum adduction in either polarity (i.e., 13 and 18 in negative and positive ion modes, respectively) matched or exceeded the number of acidic residues (i.e., 13) implicates other types of interactions in establishing Na+ adducts.
This consideration is particularly significant in the context of the mechanism proposed to explain the properties of submicrometer tips,23–26 which is based on the concentrating effects of solvent evaporation on electrospray droplets. During the desolvation process, rapid solvent evaporation leads to a decrease of the volume of a droplet with a corresponding increase of the concentration of any solute therein. The larger droplets produced by micrometer tips contains more solute ions than the smaller ones produced by submicrometer ones and, thus, the concentration reached during desolvation can be much higher. In the context of adduct formation, higher Na+ concentrations could force greater binding by mere mass action, with the stronger electrostatic interactions favored over weaker alternative types. Consistent with this possibility, the data produced by submicrometer tips for both 38mer RNA and cytochrome c displayed maximum adducts in either polarity, which did not exceed the number of negative sites on the respective analyte (see Fig. S-12 and S-13, and Table S-5 in Supporting Information). In positive mode, for instance, the 40 adducts displayed by the 38mer (5+) did not exceed the number necessary to neutralize 37 negative sites and achieve a final 5+ charge state. Likewise, the 3 adducts displayed by cytochrome c (6+) did not exceed the number necessary to neutralize 13 negative sites and impart a 6+ overall charge. In both samples, the Na+ concentration reached during desolvation was not sufficient to saturate all possible sites of electrostatic interactions and, thus, weaker alternative interactions were not operational.
Effects of tip size on specific Mg2+ binding
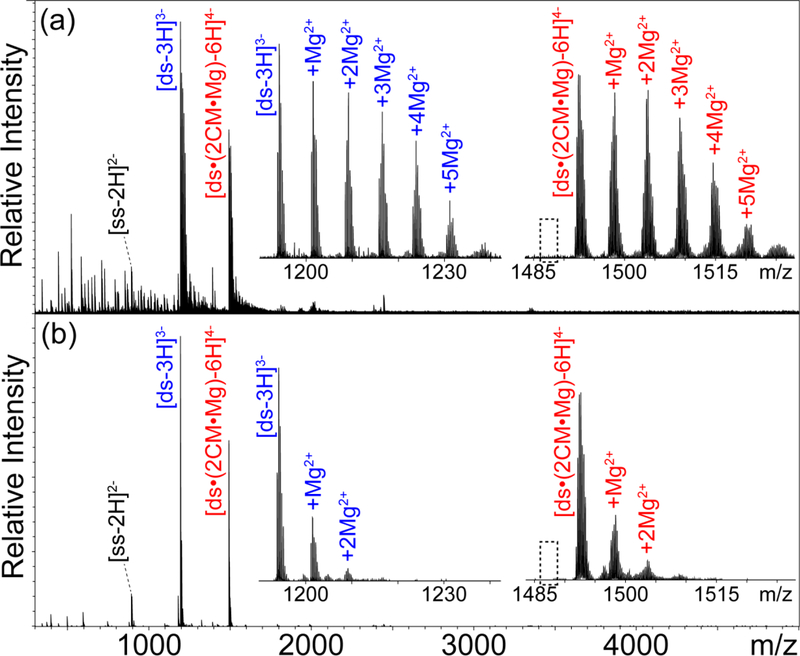
We finally explored the effects of tip size on the detection of specific interactions between metal cations and nucleic acid structures, which may be affected by the concentrating effects of desolvation. We revisited a non-covalent complex consisting of a duplex DNA construct (ds), two equivalents of chromomycin A3 (CM), and one of Mg(II)22 (see Fig. S-14 of Supporting Information), which was here analyzed by using either micrometer or submicrometer tips. In this system, the presence of Mg2+ is required to stabilize an initial CM dimer that can subsequently bind the helical DNA structure. In agreement with our earlier work,22 the spectra displayed full-fledged ds•(2CM•Mg) assembly in equilibrium with unbound ds substrate (Fig. 4). The spectra contained also a signal corresponding to the initial self-complementary 6mer (labeled ss) involved in the duplex-formation equilibrium. No signals were detected for partially assembled complexes lacking Mg2+ or one equivalent of CM ligand, consistent with the excellent stability exhibited by the full-fledged assembly.
Figure 4.
Nanospray MS spectra of 5 μM double-stranded DNA substrate (ds), 20 μM chromomycin A3 (CM), 0.5 mM magnesium acetate, and 150 mM ammonium acetate at pH 7.0, which were acquired by using either (a) micrometer- or (b) submicrometer-size emitters (see Supporting Information). The overall charge state is used to label each adduct distribution regardless of the actual balance between protonation and cation adduction. The insets enlarge the m/z regions containing signals that correspond to both unbound ds substrate (labeled in blue) and full-fledged ds•(2CM•Mg) assembly (labeled in red) to highlight matching adduction patterns. The dashed box indicates the position that would be occupied by a hypothetical assembly lacking the necessary coordinated Mg2+.
The outcomes provided by the different tips matched those obtained from the other models in the study. Spectra produced by micrometer tips displayed the usual salt clusters, limited S/N, and extensive analyte adduction, which were all greatly improved by using submicrometer tips. At this Mg2+ concentration (i.e., 0.5 mM), up to six adducts were detected in the data produced by larger tips, whereas up to two were observed with smaller ones. The same adduction patterns were observed for both full-fledged ds•(2CM•Mg) assembly and its unbound ds counterpart (compare insets of Fig. 4), consistent with the fact that free and bound analytes underwent adduct formation together, in similar size-shrinking droplets. This observation confirmed that the incorporation of coordinated Mg2+ in the assembly was independent from the adduction process and indeed preceded it in the initial sample solution. At the same time, this observation suggested that the rapid changes of solute concentration occurring during desolvation might not significantly affect pre-exiting interactions with specific Mg2+. The fact that non-specific adduction of additional Mg2+ produced by larger emitters can be largely eliminated by the submicron ones suggests that the latter could be useful for unambiguously determining the absolute number of specific metal ions (vs. the minimum number) necessary for the formation of a specific complex in solution.
Conclusions
The benefits of submicrometer tips in the analysis of anionic oligonucleotides matched those observed for proteins in positive ion mode.23–26 Regardless of the analysis’ polarity, the smaller droplets produced by such tips contain fewer solute ions than the larger ones obtained from the same sample by using larger tips. As droplets evaporate during the electrospray process, the shrinking volumes lead to rapid increases of metal ion concentration that may affect binding interactions through mass action. The final outcome is determined by the nature of the interaction, the type of analyte and counter-ion, and the actual concentration reached before fully desolvated ions are generated. Phosphate groups confer oligonucleotides their unique anionic character, which is defined by the interplay between the dissociation equilibria of the acidic protons and the establishment of putative ionic interactions with metal cations. For this reason, the observed adducts reflect the balance between different types of counter-ions, which is influenced by pH and salt concentration. Complete neutralization of all available phosphates results in signal suppression, whereas further binding of additional cations –supported by non-ionic interactions with other types of functional groups– may enable the detection of more highly adducted ions in positive ion mode. The presence of weakly basic sites on the nucleobases could account for further protonation, whereas that of electron-rich functional groups could explain the formation of additional metal adducts.
Although it is very difficult to unambiguously discriminate which metal interactions may pre-exist in solution, or arise from the significant environmental changes induced by the desolvation process, our data provide some interesting observations. In pH-neutral solutions, a dissociation constant (pKa) approaching 0 ensures that phosphate groups in phosphodiester linkages are completely dissociated, which implies the absence of virtually any pre-existing H+-phosphate interactions before electrospray. The detection of species that included both H+ and Na+/Mg2+ to produce the final charging suggests a possible competition for the negative phosphates, which is triggered by the desolvation-induced concentrating effects. Such competition would imply that the detected adduct series may not provide an accurate representation of the distribution of metal cations in direct contact with the oligonucleotide in solution. This observation is consistent with the rather loose character of the interactions between phosphate and counter-ions, which establish what has been defined as a non-specific “atmosphere” surrounding nucleic acid species.2 In contrast, metals involved in specific coordination interactions, such as the constitutive Mg2+ in the chromomycin-duplex complex, are immune from such competition by virtue of their high kinetic stability, which makes them less sensitive to the rapid changes of concentration occurring during desolvation.
Our data indicate that the favorable properties of submicrometer tips are marginally affected by emitter composition (i.e., borosilicate versus quartz), but are dependent on initial salt concentration and number of phosphates on the analyte. If the initial salt concentration exceeds a certain value, the droplets may be already saturated with potential adducting ions regardless of their initial size/volume. Such value is greater when the number of phosphates available for cation interactions is greater. Within the boundaries determined by these factors, however, the benefits of using submicron emitters include greater S/N and reduced adduct formation. Important practical consequences are expected to include a much-improved analytical sensitivity and the ability to unambiguously identify the signal of un-adducted species, which is key to achieving accurate mass determinations, especially for unknown samples. These findings are particularly significant for the analysis of oligonucleotides in the presence of Mg2+ ions, which is essential to stabilize the structure and binding interactions of functional nucleic acids. Our experiments show that submicrometer tips facilitate the analysis of samples reaching up to 10 mM Mg2+ concentrations. Although such figure falls somewhat short of physiological values, which have been determined to be in the 17–20 mM range in the majority of mammalian cells,32 this concentration is sufficient to support the establishment of specific Mg2+ interactions involved in catalysis or tertiary contacts, which manifest typical dissociation constants lower than micromolar.33 Therefore, the utilization of submicrometer tips represents an excellent alternative to traditional ammonium-replacement approaches. Future work will test the performance of submicrometer tips in the analysis of progressively larger protein-nucleic acid complexes, which will be expected to enable the utilization of the high Mg2+ concentrations necessary to ensure their stability during lengthy sample preparation steps.
Supplementary Material
Acknowledgements
Funding for this work was provided by the University at Albany (to D.F.); NIH-NIAID [R21 AI133617-01 to D.F.]; NIH-NIGMS [R01 GM121844-01 to D.F.]; and NIH-NIDA [R01DA046113-01 to D.F.]. This material is also based upon work supported by the National Science Foundation Division of Chemistry under grant number CHE-1609866 (E.R.W.).
Footnotes
Supporting Information
Experimental details, Figures S-1 – S-14 and Tables S-1 – S-5.
Conflict of interest statement. None declared
References
- (1).Record MT; Lohman TM; de Haseth PJ Mol. Biol 1976, 107 (2), 145–158. [DOI] [PubMed] [Google Scholar]
- (2).Draper DE; Grilley D; Soto AM Ann. Rev. Biophys. Biomol. Struct 2005, 34, 221–243. [DOI] [PubMed] [Google Scholar]
- (3).Cech TR; Steitz JA Cell 2014, 157 (1), 77–94. [DOI] [PubMed] [Google Scholar]
- (4).Auffinger P; Grover N; Westhof E Met. Ions Life Sci 2011, 9, 1–35. [PubMed] [Google Scholar]
- (5).Piccirilli JA; Vyle JS; Caruthers MH; Cech TR Nature 1993, 361 (6407), 85–88. [DOI] [PubMed] [Google Scholar]
- (6).Nordhoff E; Kirpekar F; Roepstorff P Mass Spectrom. Rev 1996, 15, 67–138. [DOI] [PubMed] [Google Scholar]
- (7).Fabris D; Turner KB; Hagan NA in Mass Spectrometry of Nucleosides and Nucleic Acids; Banoub JH and Limbach PA, Ed.s, CRC Press, Boca Raton: 2009; pp 303–327. [Google Scholar]
- (8).Ganem B; Li Y-T; Henion JD Tetrahedron Lett. 1993, 34, 1445–1448. [Google Scholar]
- (9).Light-Wahl KJ; Springer DL; Winger BE; Edmonds CG; Camp DG; Thrall BD; Smith RD J. Am. Chem. Soc 1993, 115, 803–804. [Google Scholar]
- (10).Hofstadler SA; Griffey RH Chem. Rev 2001, 101 (2), 377–390. [DOI] [PubMed] [Google Scholar]
- (11).Fabris D Anal. Chem 2011, 83 (15), 5810–5816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Schnier PD; Klassen JS; Strittmatter EF; Williams ER J. Am. Chem. Soc 1998,120 (37), 9605–9613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Stephenson W; Asare-Okai PN; Chen AA; Keller S; Santiago R; Tenenbaum SA; Garcia AE; Fabris D; Li PTX J. Am. Chem. Soc 2013, 135 (15), 5602–5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Gidden J; Baker ES; Ferzoco A; Bowers MT Int. J. Mass Spectrom 2005, 240 (3), 183–193. [Google Scholar]
- (15).Gabelica V; De Pauw E J. Mass Spectrom 2001, 36 (4), 397–402. [DOI] [PubMed] [Google Scholar]
- (16).Amad MH; Cech NB; Jackson GS; Enke CG J. Mass Spectrom 2000, 35 (7), 784–789. [DOI] [PubMed] [Google Scholar]
- (17).Nordhoff E; Ingendoh A; Cramer R; Overberg A; Stahl B; Karas M; Hillenkamp F; Crain PF Rapid Commun. Mass Spectrom 1992, 6 (12), 771–776. [DOI] [PubMed] [Google Scholar]
- (18).Stults JT; Marsters JC; Carr SA Rapid Commun. Mass Spectrom 1991, 5 (8), 359–363. [Google Scholar]
- (19).Little DP; Thannhauser TW; McLafferty FW Proc. Natl. Acad. Sci. U.S.A 1995, 92 (6), 2318–2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Liu C; Wu Q; Harms AC; Smith RD Anal. Chem 1996, 68 (18), 3295–3299. [DOI] [PubMed] [Google Scholar]
- (21).Gavriilidou AFM; Gülbakan B; Zenobi R Anal. Chem 2015, 87 (20), 10378–10384. [DOI] [PubMed] [Google Scholar]
- (22).Turner KB; Monti S; Fabris D J. Am. Chem. Soc 2008, 30, 13353–13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Susa AC; Xia Z; Williams ER Anal. Chem 2017, 89 (5), 3116–3122. [DOI] [PubMed] [Google Scholar]
- (24).Susa AC; Xia Z; Williams ER Angew. Chem. Int. Ed. Engl 2017, 56 (27), 7912–7915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Xia Z; Williams ER J. Am. Soc. Mass Spectrom 2018, 29 (1), 194–202. [DOI] [PubMed] [Google Scholar]
- (26).Susa AC; Lippens JL; Xia Z; Loo JA; Campuzano IDG; Williams ER J. Am. Soc. Mass Spectrom 2018, 29 (1), 203–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Schmidt A; Karas M; Dülcks T J. Am. Soc. Mass Spectrom 2003, 14 (5), 492–500. [DOI] [PubMed] [Google Scholar]
- (28).Hu J; Guan Q-Y; Wang J; Jiang X-X; Wu Z-Q; Xia X-H; Xu J-J; Chen H-Y Anal. Chem 2017, 89 (3), 1838–1845. [DOI] [PubMed] [Google Scholar]
- (29).Ding J; Anderegg RJ J. Am. Soc. Mass Spectrom 1995, 6 (3), 159–164. [DOI] [PubMed] [Google Scholar]
- (30).Yuill EM; Sa N; Ray SJ; Hieftje GM; Baker LA Anal. Chem 2013, 85 (18), 8498–8502. [DOI] [PubMed] [Google Scholar]
- (31).Zhou S; Cook KD J. Am. Soc. Mass Spectrom 2000, 11, 961–966. [DOI] [PubMed] [Google Scholar]
- (32).Romani AM Arch. Biochem. Biophys 2011, 512 (1), 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Grilley D; Soto AM; Draper DE Proc. Natl. Acad. Sci. U.S.A 2006, 103 (38), 14003–14008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.