Abstract
Epicardial adipose tissue (EAT) inflammation is implicated in the development and progression of coronary atherosclerosis. Dietary saturated and polyunsaturated fatty acids (SFAs and PUFA) can influence adipose tissue inflammation. We investigated the influence of dietary patterns, with emphasis on dietary fat type, and statin therapy, on EAT fatty acid (FA) composition and inflammatory gene expression. Thirty-two Ossabaw pigs were fed isocaloric amounts of a Heart Healthy (high in unsaturated fat) or Western (high in saturated fat) diets +/− atorvastatin for 6 months. EAT FA composition reflected dietary fat composition. There was no significant effect of atorvastatin on EAT FA composition. Total and long-chain SFAs were positively associated with inflammatory signaling (TLR2) and a gene involved in lipid mediator biosynthesis (PTGS2) (P<.0003). Medium-chain SFAs capric and lauric acids were negatively associated with IL-6 (all P<.0003). N-6 and n-3 PUFAs were positively associated with anti-inflammatory signaling genes (PPARG, FFAR4 and ADIPOQ) and long-chain n-3 PUFAs were positively associated with a gene involved in lipid mediator biosynthesis (ALOX5) (all P<.0003). These data indicate that dietary patterns, differing in fat type, influence EAT FA composition. Associations between EAT SFAs, PUFAs, and expression of genes related to inflammation provide a link between dietary quality and EAT inflammation.
Keywords: Epicardial adipose tissue, Fatty acids, Dietary patterns, Dietary fat, Ossabaw, Atorvastatin
1. Introduction
Chronic low-grade inflammation is thought to be a key mediator in the association between obesity and the development of cardiometabolic diseases [1]. Adipose tissue dysfunction, characterized by hypoxia, immune cell infiltration, increased free fatty acid release, and increased production of pro-inflammatory adipocytokines, is a key source of inflammation [1]. Epicardial adipose tissue (EAT) is an adipose tissue depot adjacent to the coronary arteries and in direct contact with the myocardium [2]. It is hypothesized that EAT with this inflammatory phenotype acts locally on the underlying coronary arteries, potentiating the development of coronary artery disease (CAD) [2]. Evidence supporting this hypothesis in humans includes positive associations of EAT volume with coronary artery calcification and plaque severity, as well as a higher expression of pro-inflammatory cytokines and macrophage infiltration in EAT from individuals with CAD, than those without CAD [3–5]. Thus, modulation of EAT inflammation is considered a potential therapeutic target for the management of CAD [6].
Fatty acids can alter adipose tissue inflammation via direct/indirect changes in gene expression and through their metabolism to lipid mediators [7,8]. Saturated fatty acids (SFAs) bind to surface toll-like receptors (TLRs) and trigger an innate inflammatory response and the production of pro-inflammatory adipocytokines [9]. SFAs have also been found to activate the NOD-like receptor pyrin domain containing 3 (NLRP3) inflammasome [10,11]. In contrast, long-chain n-3 polyunsaturated fatty acids (PUFAs) reduce adipose tissue inflammation. N-3 PUFAs, modulate inflammatory gene expression by signal transduction leading to the inhibition of pro-inflammatory transcription factors and activation of anti-inflammatory transcription factors [12,13]. N-3 PUFAs may also decrease the production of pro-inflammatory n-6 PUFA derived lipid mediators and increase the production of anti-inflammatory mediators [12]. Both de novo synthesized and dietary fatty acids may affect inflammatory status. The former offers a potential approach to alter inflammatory status.
In EAT, fatty acid composition and gene expression related to intracellular fatty acid trafficking appear to differ by metabolic state [14,15]. This suggests a relation between EAT fatty acid metabolism and metabolic dysfunction. A recent clinical trial has demonstrated that the reduced risk of cardiovascular death by high-dose EPA supplementation (icosapent ethyl) is not completely explained by reductions in triglyceride concentrations or systemic inflammation [16]. This raises the integrating possibility that the intervention may in part have provided cardio-protective effects by influencing local inflammation. It has previously been found in humans that adipose tissue fatty acid composition relates to local adipose inflammation [17]. Hence, dietary interventions that alter EAT fatty acid composition could influence EAT inflammation and the development of CAD. The collection of EAT in humans typically occurs during elective cardiac procedures, a situation not conducive to prior dietary intervention. To our knowledge, no study has examined the effect of dietary patterns, differing in fat quality, on EAT fatty acid composition or its association with EAT inflammatory gene expression.
The Ossabaw miniature pig develops an EAT depot similar to humans [18]. We previously evaluated the Ossabaw miniature pig as a translational model to study dietary patterns and atherosclerosis [19]. Using this experimental model our objectives were to 1) to determine the influence of Western-type diet (WD; high in saturated fat) and Heart Healthy diet (HHD; high in unsaturated fat), with and without atorvastatin, on EAT fatty acid composition; and 2) to determine if proportions of select EAT SFAs and PUFAs relate to EAT inflammatory gene expression.
2. Materials and methods
2.1. Study design, animals, and diets
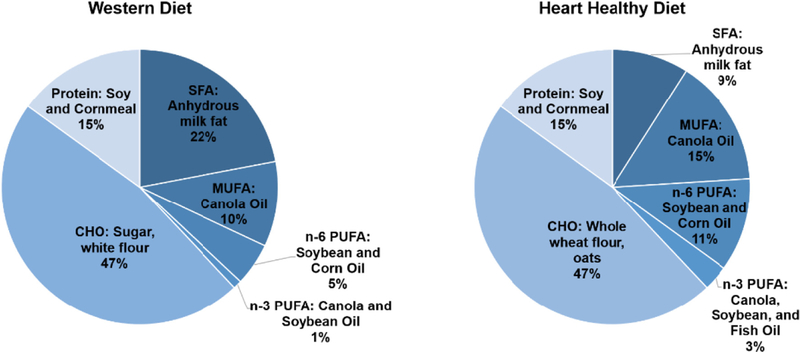
This study used samples generated from a previous investigation designed to determine the effect of dietary patterns and statin therapy on atherosclerosis development in the Ossabaw miniature pig [19]. Hence, the sample size was based on the original outcome of atherosclerosis. Research protocols and procedures have previously been reported [19] and were approved by the Beltsville Institutional Animal Care and Use Committee (IACUC). Additional approval for biological sample/tissue storage and analysis was obtained from the Tufts Medical Center/Tufts University IACUC. Thirty-two Ossabaw miniature pigs (16 boars, and 16 gilts, 5–8 weeks of age) were randomized into four groups using a 2×2 factorial design: WD, WD+statin (WD+S), and HHD+statin (HHD+S). Diets were designed to be similar to human dietary patterns and were fed in isocaloric amounts for 6 months following a 2-month acclimatization period. Both diets provided 38% energy from fat, 47% energy from carbohydrate and 15% energy from protein, and 2.5% wet weight (%w/w) of a vitamin and mineral mix [19]. Diets differed in the type and sources of dietary fat and carbohydrate (Fig. 1). The primary source of dietary fat in the WD was anhydrous milk fat (high in SFAs), while the primary sources in the HHD were canola, soybean and corn oils (high in MUFAs and PUFAs). The primary source of carbohydrate in the WD was refined, while the primary source in the HHD was whole grain. HHD-fed pigs were additionally fed a daily freeze-dried fruit and vegetable mix and supplemented with fish oil capsules (Epanova 1 g, 550 mg eicosapentaenoic acid [EPA, 20:5n-3]+200 mg docosahexaenoic acid [DHA, 22:6n-3], AstraZeneca, Wilmington, DE) 3 times per week. Diets differed in the amounts of fiber (HHD: 13 g/100 g, WD 7 g/100 g) and cholesterol (HHD: 0.1%w/w, WD: 1.5%w/w) [19]. The statin treated pigs received Atorvastatin (Lipitor, Pifzer, New York, NY) at 20 mg/day during months 1–3, and 40 mg/day during months 4–6 of the intervention period. Two pigs died during the course of the study; one in the WD group during the acclimatization period and one pig in the HHD group during the baseline blood draw, resulting in a final sample size of 30.
Fig. 1.
Macronutrient composition (% energy) and food sources of the Western and Heart Healthy diets. Macronutrient composition was confirmed by chemical analysis (Cumberland Valley Analytical Services, Hagerstown, MD, USA, and Eurofins Scientific, Des Moines, IA, USA). CHO: carbohydrate, SFA: saturated fatty acids, MUFA: monounsaturated fatty acids, PUFA: polyunsaturated fatty acids.
2.2. Sample collection
All samples were collected at the time of necropsy following euthanasia [19]. EAT adjacent to the proximal left anterior descending artery was collected, flash frozen in liquid nitrogen, and stored at −80°C until processing.
2.3. Measurement of EAT fatty acids
Lipids were extracted using a modified Folch procedure [20]. Individual fatty acids were separated by gas chromatography, identified using an authentic fatty acid internal standard (Nu-Chek Prep, Elysian, MN, USA), and expressed as molar percent (mol%) as described previously [21]. Thirty-six fatty acids were resolved. Fatty acid product-to-precursor ratios were calculated to estimate desaturase enzyme activities (stearoyl-CoA-desaturase 1 [SCD1; 161n-7/16:0], stearoyl-CoA-desaturase 2 [SCD2; 18:1n-9/18:0], delta-5-desaturase [D5D; 20:4n-6/20:3n-6]), and delta-6-desaturase [D6D; 20:3n-6/18:2n-6]) [22].
2.4. EAT gene expression
Gene expression of EAT homogenates was determined by RNA sequencing as described previously [23,24]. EAT RNA was isolated using RNeasy Universal Midi kit (Qiagen, Valencia, CA) per the manufacturer’s instructions. RNA was treated with Turbo DNase (Ambion, Waltham, MA) and RNA quality was assessed using an Experion RNA analysis electrophoresis kit (Bio-Rad, Hercules, CA). Only samples with an RNA Quality Indicator (RQI) greater than 7 were sequenced. Samples were prepared for sequencing using the Illumina TruSeq RNA Sample Preparation Kit v2 (Illumina, San Diego, CA) and AMPure XP beads (Beckman Coulter, Brea, CA) per the manufacturer’s instructions. DNA fragment size was determined using Experion DNA 1 K chips (Bio-Rad, Hercules, CA) and libraries were quantified using the KAPA Library Quantification kit (KAPA Biosystems, Wilmington, MA). Samples were sequenced using an Illumina NextSeq 500 sequencer (Illumina, San Diego, CA) with 100 base pair single end reads. Raw FASTQ data were processed for quality using CLC Bio Genomic Workbench (Qiagen, Valencia, CA). The EAT transcriptome was assembled using the annotated Sus scrofa 11.1-build as a reference genome [25]. Additional analysis was also performed using the Porcine Translational Research Database [26,27], as described below.
2.5. Statistical analysis
Data was analyzed using R (version 3.3.2) run on RStudio (version 1.0.153, RStudio: Integrated Development for R. RStudio, Boston, MA) and SAS for Windows (version 9.4; SAS Institute, Cary, NC). We removed data from one pig from the analysis due to sample oxidation that prevented the complete resolution of fatty acids. A two-way ANOVA (PROC GLM) with a Tukey–Kramer post hoc test was used to determine diet, statin, and diet × statin effects on EAT fatty acid composition. Residual plots were examined to determine normality and a Kruskal-Wallis test (PROC NPAR1WAY) was used for fatty acids not exhibiting a normal distribution. In all cases, the Kruskal-Wallis test and two-way ANOVA analyses were equivalent. Since results of the two-way ANOVA proved to be robust to departures from normality they are reported. A P≤.05 was considered statistically significant.
Spearman’s correlation coefficients were calculated between the EAT fatty acids and EAT gene expression using pigs pooled from all groups (n=29). Gene expression data from prior RNA sequencing to determine the effect of the respective dietary patterns and statin therapy on the EAT transcriptome was used for a targeted analysis of EAT gene expression and fatty acid composition [23]. In the present study we selected genes of interest on an a priori basis for their potential role in fatty acid mediated adipose tissue inflammation [28]. Genes were related to n-3 PUFA signaling (adiponectin [ADIPOQ], free fatty acid receptor 4 [FFAR4], and peroxisome proliferator activated receptor gamma [PPARG]), production of lipid-mediators (arachidonate–lipoxygenase [ALOX5 and ALOX15] and prostaglandin-endoperoxide synthase 2 [PTGS2]), and inflammation (interleukin 1 beta [IL-1β], toll like receptor 2 [TLR2], toll like receptor 4 [TLR4], interleukin 6 [IL-6], and monocyte chemotactic protein [MCP1]). We have previously published the independent effects of diet and statin therapy on EAT gene expression [23]. For the present study gene expression levels are summarized in Table A1. Gene expression of IL-1β was only available from analysis using the Porcine Translational Database. The following fatty acids were selected based on their role in diet induced inflammation: SFAs (total SFAs, capric acid [10:00], lauric, acid [12:0)] palmitic acid [16:0], stearic acid [18:0]), n-6 PUFAs (total n-6 PUFAs, linoleic acid [18:2n-6], arachidonic acid [20:4n-6], and n-3 PUFAs (total n-3 PUFAs, α-linolenic acid, EPA, docosapentaenoic acid [DPA, 22:5n-3], and DHA). Bonferroni adjusted p-values were used to correct for multiple testing (13 fatty acids and 11 genes) and associations with an r = +/−0.2 and P≤.0003 were considered statistically significant. Additionally, Spearman’s correlation coefficients were calculated to determine associations between estimated desaturase enzyme activities (SCD1, SCD2, D5D, and D6D) and expression of the aforementioned genes in EAT. Bonferroni adjusted p-values were used to correct for multiple testing (4 estimated desaturase activities and 11 genes) and we considered associations with an r = +/−0.2 and P≤.001 statistically significant. For all correlation analyses, values beyond 2 standard deviations of the mean were replaced with median proportion of the fatty acid for pigs in the respective diet +/− atorvastatin group.
In a secondary analysis we examined associations between EAT fatty acids and gene expression (as described above) using gene expression values based on alignment to the Porcine Translational Research Database in place of the Sus scrofa 11.1-build reference genome. The Porcine Translational Research Database is a manually curated database with annotation on over 10,000 genes with full length RNA transcripts [26]. Analysis with this manually curated database helps to ensure results are robust to potential limitations in annotation in the Ensembl genome that relies on machine-based annotation [27].
3. Results
3.1. EAT fatty acid composition
Overall, there was a significant effect of the diet on proportions of EAT SFAs, PUFAs, and trans fatty acids, but no significant effect of atorvastatin or significant diet × statin interaction on EAT fatty acid composition (Table 1). Relative to the HHD, pigs fed the WD had significantly higher proportions of total SFAs and trans fatty acids; including palmitic acid, stearic acid, vaccenic acid (18:1n-7 t), and conjugated linoleic acid (18:2 CLA) (all P<.01). There were no significant differences in proportions of total MUFAs. However, pigs fed the HHD had higher proportions of oleic acid (18:1n-9), than WD fed pigs (P<.01). Pigs fed the HHD had higher proportions of total n-6 PUFAs and n-3 PUFAs; including linoleic acid, α-linolenic acid, EPA, and DPA (all P<.01) but not DHA. Estimated desaturase enzyme activity for SCD2 was higher in pigs fed the HHD, whereas estimated SCD1 and D6D activities was higher in pigs fed the WD (all P<.01). Estimated D5D activity was similar between diets.
Table 1.
EAT fatty acid composition (mean ± SD) of Ossabaw miniature pigs fed a HHD or WD +/− atorvastatin at the conclusion of the study1
| Fatty acid (mol%) |
P value | ||||||
|---|---|---|---|---|---|---|---|
| WD | WD+S | HHD | HHD+S | Diet | Statin | Diet × Statin | |
| SFAs | 47.95±1.31a | 48.69±1.32a | 40.27±2.51b | 38.87±2.13b | <.01 | .68 | .13 |
| 10:0 | 0.16±0.04a | 0.17±0.05a | 0.10±0.01b | 0.10±0.02b | <.01 | .58 | .88 |
| 12:0 | 0.27±0.05a | 0.30±0.06a | 0.14±0.02b | 0.15±0.01b | <.01 | .31 | .49 |
| 14:0 | 4.00±0.47a | 4.12±0.54a | 2.91±0.40b | 2.86±0.42b | <.01 | .85 | .65 |
| 15:0 | 0.46±0.07a | 0.45±0.08a | 0.32±0.05b | 0.32±0.05b | <.01 | .84 | .90 |
| 16:0 | 24.32±0.72a | 24.45±0.73a | 20.85±1.52b | 19.49±1.19b | <.01 | .16 | .07 |
| 18:0 | 18.44±1.09a | 18.89±1.21a | 15.63±1.06b | 15.52±1.3b | <.01 | .69 | .53 |
| 20:0 | 0.25±0.02 | 0.25±0.06 | 0.28±0.04 | 0.27±0.04 | .14 | .71 | .93 |
| 22:0 | 0.03±0.00 | 0.05±0.06 | 0.03±0.00 | 0.15±0.33 | .40 | .31 | .49 |
| 24:0 | 0.01±0.00 | 0.01±0.00 | 0.01±0.01 | 0.01±0.01 | .04 | .80 | 1.00 |
| MUFAs | 43.34±2.22 | 42.52±1.76 | 44.37±1.84 | 43.60±1.61 | .15 | .26 | .97 |
| 14:1n-5 | 0.10±0.02a | 0.11±0.03a | 0.03±0.01b | 0.03±0.01b | <.01 | .37 | .56 |
| 16:1n-9 | 0.39±0.05 | 0.39±0.05 | 0.33±0.07 | 0.38±0.05 | .14 | .16 | .19 |
| 16:1n-7 | 2.48±0.17a | 2.45±0.21a | 1.45±0.12b | 1.39±0.18b | <.01 | .49 | .78 |
| 18:1n-9 | 36.83±2.01 b | 36.27±1.81b | 38.97±1.893 | 38.24±1.34a | <.01 | .34 | .90 |
| 18:1n-7 | 2.70±0.20a | 2.55±0.11a | 2.36±0.09b | 2.42±0.18b | <.01 | .39 | .09 |
| 20:1n-9 | 0.77±0.14b | 0.67±0.09b | 0.96±0.17a | 0.87±0.14a | <.01 | .08 | .87 |
| 22:1n-9 | 0.07±0.02b | 0.07±0.01b | 0.26±0.03a | 0.25±0.04a | <.01 | .80 | .82 |
| 24:1n-9 | 0.00±0.00 | 0.01±0.02 | 0.01±0.00 | 0.01±0.00 | .45 | .49 | .29 |
| n-6 PUFAs | 6.86±1.58b | 6.88±1.15b | 12.63±2.55a | 14.82±2.86a | <.01 | .20 | .19 |
| 18:2n-6 | 6.05±1.51b | 6.09±1.13b | 11.55±2.43a | 13.69±2.76a | <.01 | .18 | .18 |
| 18:3n-6 | 0.03±0.01 | 0.03±0.01 | 0.04±0.01 | 0.04±0.02 | .02 | .57 | .56 |
| 20:2n-6 | 0.28±0.06b | 0.26±0.04b | 0.56±0.09a | 0.58±0.09a | <.01 | .93 | .44 |
| 20:3n-6 | 0.12±0.01 | 0.13±0.01 | 0.13±0.02 | 0.12±0.02 | .86 | .91 | .68 |
| 20:4n-6 | 0.27±0.03 | 0.27±0.05 | 0.26±0.06 | 0.28±0.05 | .81 | .47 | .70 |
| 22:4n-6 | 0.09±0.01 | 0.09±0.01 | 0.09±0.01 | 0.09±0.01 | .19 | .53 | .95 |
| 22:5n-6 | 0.02±0.01a | 0.01±0.00a | 0.01±0.00b | 0.01±0.00b | .02 | .80 | .05 |
| n-3 PUFAs | 0.56±0.17b | 0.59±0.08b | 2.32±0.76a | 2.27±0.43a | <.01 | .94 | .82 |
| 18:3n-3 | 0.37±0.13b | 0.4±0.08b | 1.7±0.36a | 1.98±0.41a | <.01 | .18 | .24 |
| 18:4n-3 | 0.03±0.01a | 0.04±0.01a | 0.02±0.01b | 0.02±0.00b | <.01 | .63 | .58 |
| 20:5n-3 | 0.01±0.00b | 0.02±0.00b | 0.04±0.01a | 0.04±0.01a | <.01 | .22 | .65 |
| 22:5n-3 | 0.08±0.01b | 0.08±0.01b | 0.15±0.02a | 0.15±0.02a | <.01 | .89 | .96 |
| 22:6n-3 | 0.06±0.08 | 0.05±0.06 | 0.09±0.08 | 0.08±0.02 | .34 | .72 | .97 |
| Trans | 1.29±0.17a | 1.33±0.18a | 0.42±0.06b | 0.45±0.05b | <.01 | .51 | .92 |
| 16:1n-7t | 0.03±0.02a | 0.03±0.02a | 0.01±0.01b | 0.02±0.01b | <.01 | .62 | .96 |
| 16:1n-9t | 0.14±0.01a | 0.15±0.01a | 0.04±0.00b | 0.04±0.00b | <.01 | .42 | .58 |
| 18:1n-7 t | 0.22±0.06a | 0.23±0.05a | 0.06±0.01b | 0.07±0.02b | <.01 | .15 | .58 |
| 18:1n-9t | 0.20±0.04a | 0.22±0.04a | 0.07±0.02b | 0.08±0.01b | <.01 | .53 | .83 |
| 18:1n-10 −12 t | 0.27±0.02a | 0.28±0.03a | 0.12±0.01b | 0.12±0.01b | <.01 | .50 | .48 |
| 18:2 t | 0.14±0.01a | 0.15±0.02a | 0.04±0.01b | 0.04±0.01b | <.01 | .97 | .85 |
| 18:2CLA | 0.28±0.03a | 0.27±0.04a | 0.09±0.02b | 0.09±0.01b | <.01 | .81 | .71 |
| Ratios | |||||||
| SCD1 | 0.10±0.01a | 0.10±0.01a | 0.07±0.00b | 0.07±0.01b | <.01 | .90 | .56 |
| SCD2 | 2.01±0.20b | 1.93±0.16b | 2.50±0.24a | 2.48±0.17a | <.01 | .45 | .73 |
| D5D | 2.17±0.32 | 2.19±0.43 | 2.09±0.22 | 2.32±0.28 | .74 | .34 | .40 |
| D6D | 0.02±0.00a | 0.02±0.00a | 0.01±0.00b | 0.01±0.00b | <.01 | .29 | .46 |
Values presented as mean ± SD. Superscripts denote a significant diet effect determined by a two-way ANOVA in which means without a common superscript differ (p≤.05). EAT: epicardial adipose tissue, WD: Western diet, HHD: heart healthy diet, WD+S: Western diet+atorvastatin, HHD+S: heart healthy diet+atorvastatin, SFAs: saturated fatty acids, MUFAs: monounsaturated fatty acids, PUFAs: polyunsaturated fatty acids, Trans: trans fatty acids, CLA: conjugated linolenic acid, SCD1: stearoyl-CoA-desaturase 1 (161n-7/16:0), SCD2: stearoyl-CoA-desaturase (18:1n-9/18:0), D5D: delta-5-desaturase (20:4n-6/20:3n-6), D6D: delta-6-desaturase (20:3n-6/18:2n-6).
3.2. Associations between EAT fatty acids and gene expression
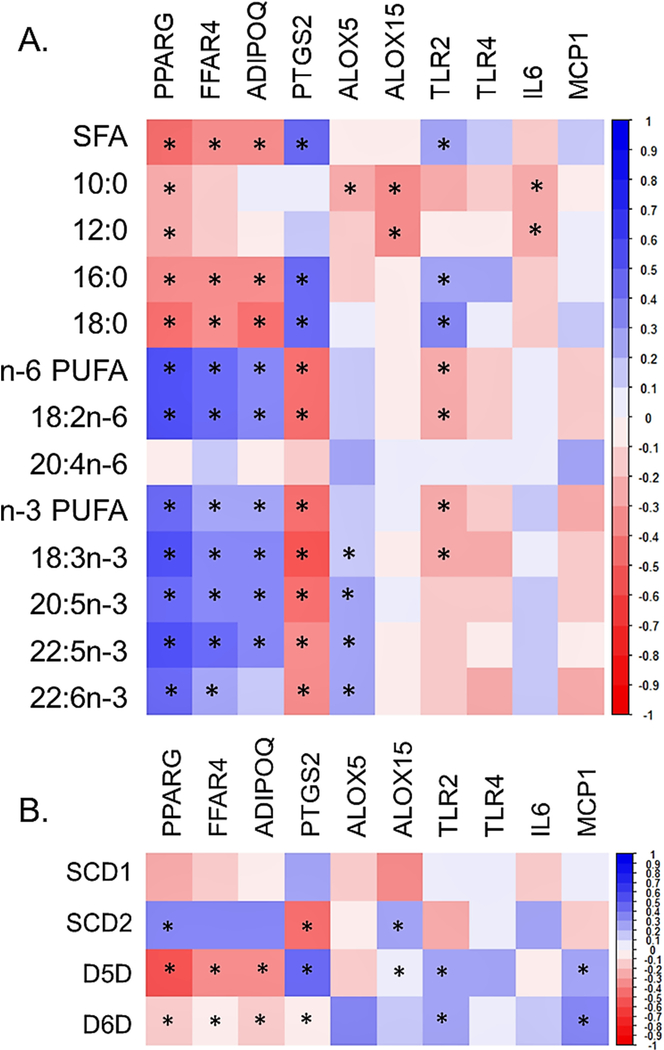
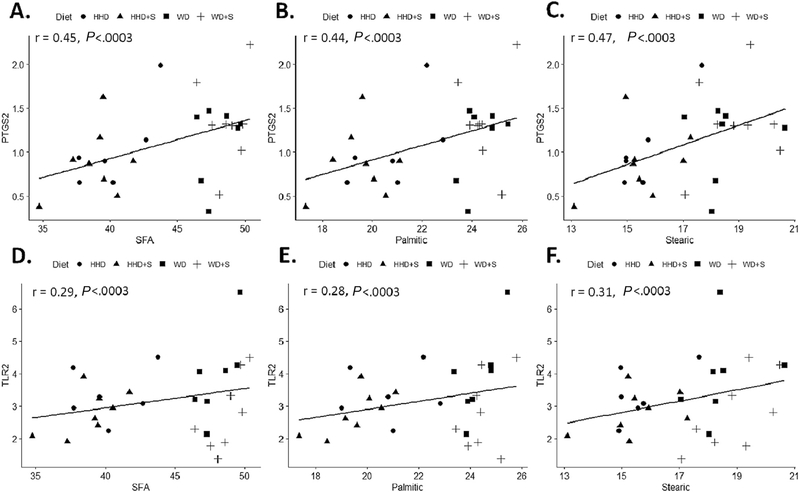
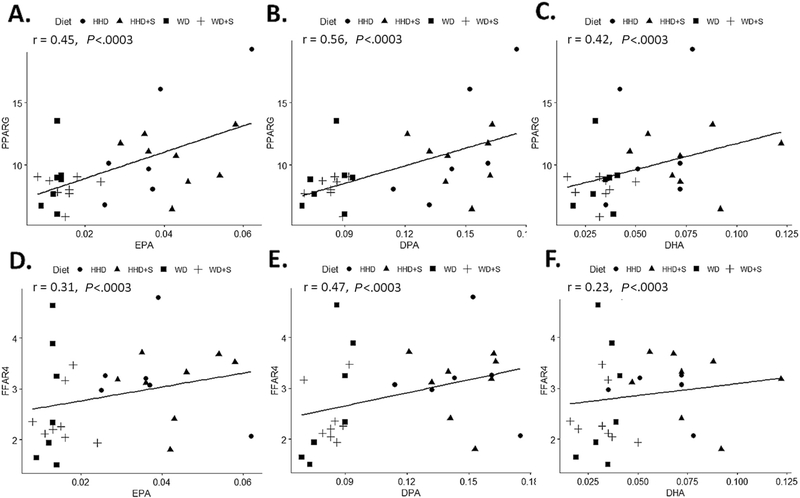
A heat map of summarizing correlation coefficients between selected SFAs and PUFAs (mol%), and gene expression (rpkm) is displayed in Fig. 2A, and the associated correlation coefficients in Table A2. Proportions of total and long-chain SFAs (palmitic and stearic acid) were positively associated with expression of PTGS2 and TLR2 (all P<.0003, Fig. 3). The medium-chain SFAs (capric and lauric acid) had inverse associations with expression of ALOX15 and IL-6 (all P<.0003). Proportions of all n-6 and n-3 PUFAs, except arachidonic acid, were positively associated with the expression of PPARG and FFAR4 (all P<.0003, Fig. 4). With the exception of arachidonic acid and DHA, all other PUFAs were positively associated with expression of ADIPOQ (all P<.0003). Proportions of long-chain n-3 PUFAs (EPA, DPA, and DHA) were positively associated with ALOX5 expression (all P<.0003). No statistically significant associations were identified between proportions of SFAs or PUFAs and expression of TLR4 and MCP1.
Fig. 2.
(A) Heat map representing Spearman’s correlation coefficients between proportions (mol%) of SFAs, capric acid, lauric acid, palmitic acid, stearic acid, PUFAs n-6, linoleic acid, arachidonic acid, n-3 PUFAs, α-linolenic acid, EPA, DPA, and DHA and EAT gene expression (rpkm). A Bonferroni correction was used to adjust for multiple comparisons (*P≤.0003). (B) Heat map representing Spearman’s correlation coefficients between EAT estimated desaturase enzyme activities and EAT gene expression (rpkm). A Bonferroni correction was used to adjust for multiple comparisons (*P≤.001).
Fig. 3.
Scatter plots displaying the correlations between (A) the proportion of total SFA (mol %) and PTGS2 gene expression (rpkm) in EAT. (B) the proportion of palmitic acid (mol %) and PTGS2 gene expression (rpkm) in EAT. (C) the proportion of stearic acid (mol %) and PTGS2 gene expression (rpkm) in EAT. (D) the proportion of total SFA (mol %) and TLR2 gene expression (rpkm) in EAT. (E) the proportion of palmitic acid (mol %) and TLR2 gene expression (rpkm) in EAT. (F) the proportion of stearic acid (mol %) and TLR2 gene expression (rpkm) in EAT. HHD: Heart Healthy diet, WD: Western diet, WD+S: Western diet+atorvastatin, HHD+S: heart healthy diet+atorvastatin, PTGS2: prostaglandin-endoperoxide synthase 2, SFA: saturated fatty acids, TLR2: toll like receptor 2, EAT: epicardial adipose tissue. All correlations were statistically significant (P≤.0003).
Fig. 4.
Scatter plots displaying the correlations between (A) the proportion of EPA (mol %) and PPARG gene expression (rpkm) in EAT. (B) the proportion of DPA (mol %) and PPARG gene expression (rpkm) in EAT. (C) the proportion of DHA (mol %) and PPARG gene expression (rpkm) in EAT (rpkm) in EAT. between (D) the proportion of EPA (mol %) and FFAR4 gene expression (rpkm) in EAT. (E) the proportion of DPA (mol %) and FFAR4 gene expression (rpkm) in EAT. (F) the proportion of DHA (mol %) and FFAR4 gene expression (rpkm) in EAT (rpkm) in EAT. HHD: Heart Healthy diet, WD: Western diet, WD+S: Western diet+atorvastatin, HHD+S: heart healthy diet+atorvastatin, EPA: eicosapentaenoic acid, DPA: docosapentaenoic acid, DHA: docosahexaenoic acid FFAR4: free fatty acid receptor 4, PPARG: peroxisome proliferator activated receptor, EAT: epicardial adipose tissue. All correlations were statistically significant (P≤.0003).
3.3. Associations between EAT estimated desaturase activities and gene expression
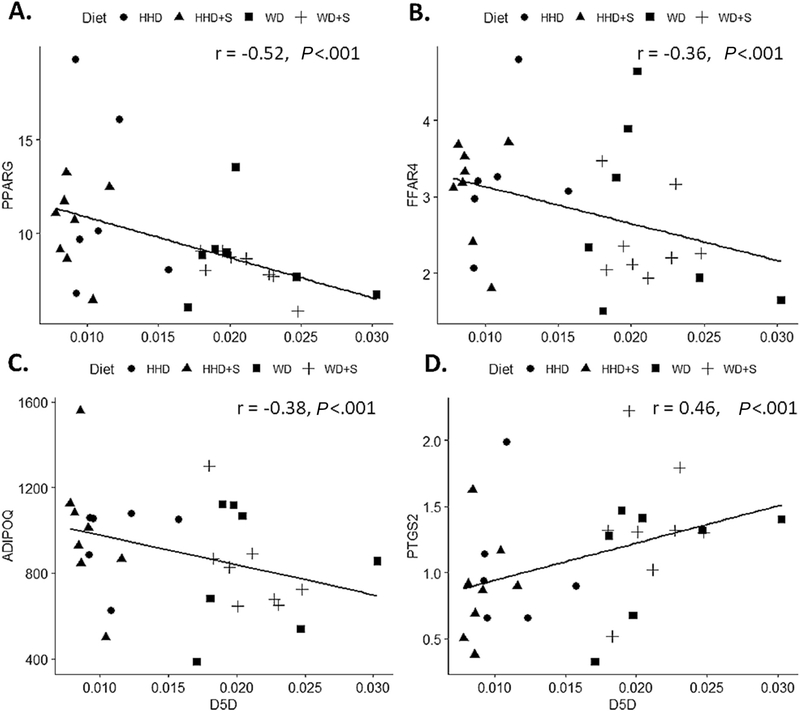
Associations between EAT estimated desaturase enzyme activities and EAT gene expression are represented in Fig. 2B, all correlation coefficient values are included in Table A2. Estimated SCD2 and D5D activities were more commonly associated with gene expression. Estimated SCD2 activity was positively associated with the expression of genes involved in n-3 PUFA signaling (PPARG, FFAR4, and ADIPOQ), and ALOX15 but were inversely associated with PTGS2 and TLR2 (all P<.001). In contrast, estimated D5D activity was inversely associated with the expression of genes involved in n-3 PUFA signaling (PPARG, FFAR4, and ADIPOQ) and, positively associated with PTGS2, TLR2, and MCP1 (all P<.001, Fig. 5). There were no significant associations between estimated desaturase activities and ALOX5, IL-1β, and TLR4 expression.
Fig. 5.
Scatterplots displaying the correlations between (A) estimated D5D activity and PPARG gene expression (rpkm) in EAT. (B) estimated D5D activity and FFAR4 gene expression (rpkm) in EAT (C) estimated D5D activity and ADIPOQ gene expression (rpkm) in EAT (D) estimated D5D activity and PTGS2 gene expression (rpkm) in EAT. HHD: Heart Healthy diet, WD: Western diet, WD+S: Western diet+atorvastatin, HHD+S: heart healthy diet+atorvastatin, D5D: delta-5-desaturase (20:4n-6/20:3n-6), PPARG: peroxisome proliferator activated receptor gamma, FFAR4: free fatty acid receptor 4, ADIPOQ: adiponectin, PTGS2: prostaglandin-endoperoxide synthase 2, EAT: epicardial adipose tissue. All correlations were statistically significant (P≤.001).
3.4. Secondary analysis
Correlations between proportions of SFAs, PUFAs, and estimated desaturase activities with gene expression were similar using the Porcine Translational Research Database (Table A3). In this analysis, we additionally examined associated between SFAs, PUFAs and expression of IL-1β and found no statistically significant associations (Table S3).
4. Discussion
Adipose tissue fatty acid composition may be related to adipose inflammation. However, little is known about the influence of dietary patterns, differing in fat type, and statin therapy on EAT fatty acid composition. We investigated the effect of dietary patterns differing in fatty acid profile fat on EAT fatty acid composition and the relationship between proportions of EAT fatty acids and inflammatory gene expression in the Ossabaw pig model. Our results indicate that dietary patterns, differing in dietary fat quality, are a significant determinant of EAT fatty acid composition and that proportions of SFAs and PUFAs associated with the expression of genes related to inflammation.
While EAT fatty acid composition was reflective of the respective dietary fat sources, we cannot rule out the possible contribution of endogenous synthesis and preferential storage of SFAs which accounted for a large proportion (38–48%) of total fatty acids across all groups of pigs [29]. Similarly, as MUFAs are endogenously synthesized and preferentially stored in adipose tissue [29], we observed no differences in the proportion of total MUFAs between diet groups. We did however note a significant diet effect on proportions of oleic acid, which may in part be due to the significant increase of estimated SCD2 activity, in pigs fed the HHD.
In general, proportions of EAT PUFAs were positively associated with the expression of genes involved in n-3 PUFA signaling (FFAR4, PPARG and ADIPOQ). N-3 PUFAs serve as ligands for both PPARG and FFAR4 [30]. Following induction of PPARG downstream signaling may lead to increased expression of ADIPOQ and consequently the production of adiponectin; which is associated with reductions in adipose tissue inflammation and improvements insulin sensitivity [28]. Induction of FFAR4 (also known as GPR120) by n-3 PUFAs inhibits inflammatory signaling [30,31]. Together these associations suggest dietary PUFAs may induce anti-inflammatory signaling in EAT.
PTGS2 and ALOX (ALOX5 and ALOX15) genes are involved in the synthesis of lipid mediators derived from n-6 and n-3 PUFAs [32,33]. The n-6 PUFA arachidonic acid is considered the major substrate for these enzymes. When available, n-3 PUFAs can displace arachidonic acid in the cell membrane shifting the balance from arachidonic acid derived pro-inflammatory lipid mediators to n-3 PUFA derived less-inflammatory or anti-inflammatory mediators [32,33]. Positive associations between long-chain n-3 PUFAs (EPA, DPA, and DHA) and ALOX5 expression support their role as both substrates and inducers of n-3 derived lipid mediators [34]. Notably, we did not observe associations between proportions of arachidonic acid and expression of any genes involved in lipid mediator synthesis.
Total and long-chain SFAs (palmitic and stearic) were positively correlated with PTGS2. This observation is consistent with prior work suggesting that SFAs induce PTGS2 expression [35,36]. Total and long-chain SFAs were also associated with the expression of TLR2. SFA induced activation of TLR signaling evokes inflammatory signaling that contributes to adipose tissue inflammation [37]. We observed non-statistically significant trends towards a positive association between total SFAs and palmitic acid with TLR4. Unexpectedly, no significant associations were observed between any of the SFAs and expression of inflammatory cytokines IL-6, IL-1β, and MCP1. This lack of significant associations between specific inflammatory genes and fatty acids may be due to greater than expected weight gain during the diet intervention period [19], independently resulting in induction of EAT inflammatory genes. Additionally, we measured gene expression in EAT homogenates which is reflective of multiple cells types. Hence, gene expression from cells responsible for production of inflammatory cytokines may have been diluted.
The estimated enzyme activity of D5D was more strongly associated with gene expression than other estimated enzyme activities. D5D facilitates the conversion of dihomo-y-linolenic acid (20:3n-6) to arachidonic acid and eicosatetraenoic acid (20:4n-3) to EPA [38]. Hence, the positive association between D5D and PTGS2 could be reflective of higher substrate availability. Dietary PUFAs can inhibit desaturase (delta-5 and delta-6 desaturase) gene expression [39] which may explain inverse associations between estimated desaturase activity and genes involved in n-3 PUFA signaling (FFAR4, PPARG and ADIPOQ).
The original investigation was designed to compare the effects of two dietary patterns, a HHD and WD, on the development of atherosclerosis in Ossabaw miniature pigs, rather than dietary fat type, per se [19]. This approach increases the translational value of our study. However, we cannot rule out the possibility that other dietary components influenced EAT fatty acid composition and gene expression. While differences in human and porcine fatty acid metabolism exist our results are in agreement with prior work suggesting the preferential storage of oleic and palmitic acid in human EAT [40]. One key mechanism by which PUFAs may modulate inflammation is via the production of lipid mediators. Though our results suggest a shift in balance from arachidonic acid to n-3 PUFAs derived lipid mediators with the HHD, we cannot confirm this without a direct measurement. Additionally, the only adipose tissue depot assessed was EAT. Future work comparing additional adipose tissue depots will help determine unique influences of diet on EAT. Difficulties associated with porcine research make it challenging to achieve a large sample size. However, these models allow for comprehensive assessment of the interplay between diet and tissues that are challenging to investigate in humans.
We conclude that HHD and WD, differing in dietary fat quality, have differential influences EAT fatty acid composition and subsequent gene expression. Associations between EAT SFAs with genes involved in inflammatory signaling, and EAT PUFAs with genes involved in anti-inflammatory signaling, provide a link between diet and EAT inflammation. Targeted changes in dietary quality may be a viable nutritional strategy to reduce EAT inflammation and in turn effect the development of CAD.
Supplementary Material
Acknowledgments
☆☆ Funding source: This work was supported by the NHLBI T32 Nutrition and Cardiometabolic Disorders Pre-doctoral Research Training Grant (NIH T32HL069772-15), the NIH Multidisciplinary Training Program in Cardiovascular Epidemiology (5T32HL125232), USDA agreement 588-1950-9-001, and USDA-ARS project 8040-51530-056-00-D. Any opinions, findings, conclusions or recommendations expressed in this publication are those of the author(s) and do not necessarily reflect the view of the NIH or USDA.
Footnotes
☆ Declaration of interest: None.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jnutbio.2019.04.013.
References
- [1].Saltiel AR, Olefsky JM. Inflammatory mechanisms linking obesity and metabolic disease. J Clin Invest 2017;127:1–4. 10.1172/JCI92035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Iacobellis G. Local and systemic effects of the multifaceted epicardial adipose tissue depot. Nat Rev Endocrinol 2015;11:363–71. 10.1038/nrendo.2015.58. [DOI] [PubMed] [Google Scholar]
- [3].Nerlekar N, Brown AJ, Muthalaly RG, Talman A, Hettige T, Cameron JD, et al. Association of Epicardial Adipose Tissue and High-Risk Plaque Characteristics: a systematic review and meta-analysis. J Am Heart Assoc 2017;6 10.1161/JAHA.117.006379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Goeller M, Achenbach S, Marwan M, Doris MK, Cadet S, Commandeur F, et al. Epicardial adipose tissue density and volume are related to subclinical atherosclerosis, inflammation and major adverse cardiac events in asymptomatic subjects. J Cardiovasc Comput Tomogr 2017. 10.1016/j.jcct.2017.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Baker AR, Silva da NF, Quinn DW, Harte AL, Pagano D, Bonser RS, et al. Human epicardial adipose tissue expresses a pathogenic profile of adipocytokines in patients with cardiovascular disease. Cardiovasc Diabetol 2006;5:1 10.1186/1475-2840-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Akoumianakis I, Tarun A, Antoniades C. Perivascular adipose tissue as a regulator of vascular disease pathogenesis: identifying novel therapeutic targets. Br J Pharmacol 2017;174:3411–24. 10.1111/bph.13666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Rocha DM, Bressan J, Hermsdorff HH. The role of dietary fatty acid intake in inflammatory gene expression: a critical review. Sao Paulo Med J Rev Paul Med 2017;135:157–68. 10.1590/1516-3180.2016.008607072016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Teng K-T, Chang C-Y, Chang LF, Nesaretnam K. Modulation of obesity-induced inflammation by dietary fats: mechanisms and clinical evidence. Nutr J 2014; 13:12 10.1186/1475-2891-13-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Rocha DM, Caldas AP, Oliveira LL, Bressan J, Hermsdorff HH. Saturated fatty acids trigger TLR4-mediated inflammatory response. Atherosclerosis 2016;244:211–5. 10.1016/j.atherosclerosis.2015.11.015. [DOI] [PubMed] [Google Scholar]
- [10].Wen H, Gris D, Lei Y, Jha S, Zhang L, MT H Huang, et al. Fatty acid-induced NLRP3-ASC inflammasome activation interferes with insulin signaling. Nat Immunol 2011;12:408–15. 10.1038/ni.2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Robblee MM, Kim CC, Porter Abate J, Valdearcos M, Sandlund KLM, Shenoy MK, et al. Saturated fatty acids engage an IRE1α-dependent pathway to activate the NLRP3 Inflammasome in myeloid cells. Cell Rep 2016;14:2611–23. 10.1016/j.celrep.2016.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Calder PC. Omega-3 fatty acids and inflammatory processes: from molecules to man. Biochem Soc Trans 2017;45:1105–15. 10.1042/BST20160474. [DOI] [PubMed] [Google Scholar]
- [13].Rodríguez-Cruz M, Serna DS. Nutrigenomics of ω−3 fatty acids: regulators of the master transcription factors. Nutrition 2017;41:90–6. 10.1016/j.nut.2017.04.012. [DOI] [PubMed] [Google Scholar]
- [14].Pezeshkian M, Mahtabipour M- R. Epicardial and subcutaneous adipose tissue fatty acids profiles in diabetic and non-diabetic patients candidate for coronary artery bypass graft. BioImpacts BI 2013;3:83–9. 10.5681/bi.2013.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Vural B, Atalar F, Ciftci C, Demirkan A, Susleyici-Duman B, Gunay D, et al. Presence of fatty-acid-binding protein 4 expression in human epicardial adipose tissue in metabolic syndrome. Cardiovasc Pathol Off J Soc Cardiovasc Pathol 2008;17: 392–8. 10.1016/j.carpath.2008.02.006. [DOI] [PubMed] [Google Scholar]
- [16].Bhatt DL, Steg PG, Miller M, Brinton EA, Jacobson TA, Ketchum SB, et al. Cardiovascular risk reduction with Icosapent ethyl for hypertriglyceridemia. N Engl J Med 2019;380:11–22. 10.1056/NEJMoa1812792. [DOI] [PubMed] [Google Scholar]
- [17].Vaittinen M, Männistö V, Käkelä P, Ågren J, Tiainen M, Schwab U, et al. Interorgan cross talk between fatty acid metabolism, tissue inflammation, and FADS2 genotype in humans with obesity. Obes Silver Spring 2017;25:545–52. 10.1002/oby.21753Md. [DOI] [PubMed] [Google Scholar]
- [18].McKenney ML, Schultz KA, Boyd JH, Byrd JP, Alloosh M, Teague SD, et al. Epicardial adipose excision slows the progression of porcine coronary atherosclerosis. J Cardiothorac Surg 2014;9:2. 10.1186/1749-8090-9-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Matthan NR, Solano-Aguilar G, Meng H, Lamon-Fava S, Goldbaum A, Walker ME, et al. The Ossabaw pig is a suitable translational model to evaluate dietary patterns and coronary artery disease risk. J Nutr 2018;148:542–51. 10.1093/jn/nxy002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Folch J, Lees M, Stanley GHS. A simple method for the isolation and purification of total LIPIDES from ANIMAL tissues. J Biol Chem 1957;226:497–509. [PubMed] [Google Scholar]
- [21].Lichtenstein AH, Matthan NR, Jalbert SM, Resteghini NA, Schaefer EJ, Ausman LM. Novel soybean oils with different fatty acid profiles alter cardiovascular disease risk factors in moderately hyperlipidemic subjects. Am J Clin Nutr 2006;84: 497–504. [DOI] [PubMed] [Google Scholar]
- [22].Matthan NR, Ooi EM, Horn LV, Neuhouser ML, Woodman R, Lichtenstein AH. Plasma phospholipid fatty acid biomarkers of dietary fat quality and endogenous metabolism predict coronary heart disease risk: a nested case-control study within the Women’s Health Initiative observational study. J Am Heart Assoc 2014; 3:e000764. 10.1161/JAHA.113.000764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Walker ME, Matthan NR, Solano-Aguilar G, Jang S, Lakshman S, Molokin A, et al. A Western dietary pattern and atorvastatin induce Epicardial adipose tissue interferon signaling in the Ossabaw pig. J Nutr Biochem 2019. 10.1016/j.jnutbio.2019.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Solano-Aguilar G, Molokin A, Botelho C, Fiorino A- M, Vinyard B, Li R, et al. Transcriptomic profile of whole blood cells from elderly subjects fed probiotic Bacteria lactobacillus rhamnosus GG ATCC 53103 (LGG) in a phase I open label study. PLoS One 2016;11:e0147426. 10.1371/journal.pone.0147426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Sus scrofa - Ensembl genome browser 91 n.d. https://useast.ensembl.org/Sus_scrofa/Info/Index, Accessed date: 5 February 2018.
- [26].DGIL Porcine Translational Research Database. USDA ARS n.d. https://www.ars.usda.gov/northeast-area/beltsville-md-bhnrc/beltsville-human-nutrition-research-center/diet-genomics-and-immunology-laboratory/docs/dgil-porcine-translational-research-database/. [accessed January 31, 2019].
- [27].Dawson HD, Chen C, Gaynor B, Shao J, Urban JF. The porcine translational research database: a manually curated, genomics and proteomics-based research resource. BMC Genomics 2017;18:643 10.1186/s12864-017-4009-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Kalupahana NS, Claycombe KJ, Moustaid-Moussa N. (n-3) fatty acids alleviate adipose tissue inflammation and insulin resistance: mechanistic insights. Adv Nutr 2011;2:304–16. 10.3945/an.111.000505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Hodson L, Skeaff CM, Fielding BA. Fatty acid composition of adipose tissue and blood in humans and its use as a biomarker of dietary intake. Prog Lipid Res 2008; 47:348–80. 10.1016/j.plipres.2008.03.003. [DOI] [PubMed] [Google Scholar]
- [30].Calder PC. Mechanisms of action of (n-3) fatty acids. J Nutr 2012;142:592S–9S. 10.3945/jn.111.155259. [DOI] [PubMed] [Google Scholar]
- [31].Oh DY, Talukdar S, Bae EJ, Imamura T, Morinaga H, Fan W, et al. GPR120 is an Omega-3 fatty acid receptor mediating potent anti-inflammatory and insulin-sensitizing effects. Cell 2010;142:687–98. 10.1016/j.cell.2010.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Calder PC. N–3 Polyunsaturated fatty acids, inflammation, and inflammatory diseases n.d. http://ajcn.nutrition.org, Accessed date: 10 January 2018. [DOI] [PubMed]
- [33].Calder PC. Marine omega-3 fatty acids and inflammatory processes: effects, mechanisms and clinical relevance. Biochim Biophys Acta 2015;1851:469–84. 10.1016/j.bbalip.2014.08.010. [DOI] [PubMed] [Google Scholar]
- [34].Calder PC. Omega-3 fatty acids and inflammatory processes. Nutrients 2010;2: 355–74. 10.3390/nu2030355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Lee JY, Sohn KH, Rhee SH, Hwang D. Saturated fatty acids, but not unsaturated fatty acids, induce the expression of cyclooxygenase-2 mediated through toll-like receptor 4. J Biol Chem 2001;276:16683–9. 10.1074/jbc.M011695200. [DOI] [PubMed] [Google Scholar]
- [36].Hellmann J, Zhang MJ, Tang Y, Rane M, Bhatnagar A, Spite M. Increased saturated fatty acids in obesity alter resolution of inflammation in part by stimulating prostaglandin production. J Immunol Baltim 1950;2013(191):1383–92. 10.4049/jimmunol.1203369Susscrofa. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Fessler MB, Rudel LL, Brown JM. Toll-like receptor signaling links dietary fatty acids to the metabolic syndrome. Curr Opin Lipidol 2009;20:379–85. 10.1097/MOL.0b013e32832fa5c4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Russo GL. Dietary n–6 and n–3 polyunsaturated fatty acids: from biochemistry to clinical implications in cardiovascular prevention. Biochem Pharmacol 2009; 77:937–46. 10.1016/j.bcp.2008.10.020. [DOI] [PubMed] [Google Scholar]
- [39].Nakamura MT, Nara TY. Structure, function, and dietary regulation of Δ6, Δ5, and Δ9 desaturases. Annu Rev Nutr 2004;24:345–76. 10.1146/annurev.nutr.24.121803.063211. [DOI] [PubMed] [Google Scholar]
- [40].Pezeshkian M, Noori M, Najjarpour-Jabbari H, Abolfathi A, Darabi M, Darabi M, et al. Fatty acid composition of epicardial and subcutaneous human adipose tissue. Metab Syndr Relat Disord 2009;7:125–31. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.