Abstract
Porcine circovirus 2 (PCV2) has a major impact on the swine industry. Eight PCV2 genotypes (a-h) have been identified using capsid sequence analysis. PCV2d has been designated as the emerging genotype. The cryo-electron microscopy molecular envelope of PCV2d virus-like particles identifies differences between PCV2a, b and d genotypes that accompany the emergence of PCV2b from PCV2a, and PCV2d from PCV2b. These differences indicate that sequence analysis of genotypes is insufficient, and that it is important to determine the PCV2 capsid structure as the virus evolves. Structure-based sequence comparison demonstrate that each genotype possesses a unique combination of amino acids located on the surface of the capsid that undergo substitution. We also demonstrate that the capsid N-terminus moves in response to increasing amount of nucleic acid packaged into the capsid. Furthermore, we model a tetranucleotide between the 5- and 2-fold axes of symmetry that appears to be responsible for capsid stability.
Keywords: Circovirus, virus-like particle (VLP), cryo-electron microscopy, virus evolution, capsid structure
Introduction
Circoviruses are small nonenveloped icosahedral viruses that contain a circular, covalently closed, single stranded DNA (ssDNA) genome. Circoviruses have been identified in a variety of species and are known to cause infections in avian, aquatic, and terrestrial animals (Ellis, 2014; Rosario et al., 2017). The variation of the capsid morphology and genome organization has recently led to the classification of the Circoviridae family into two separate genera, Cyclovirus and Circovirus (Breitbart et al., 2017). Genome sequences associated with the Cyclovirus genus has been identified from several vertebrate and invertebrate species, although the recognition of definitive host for this group is still unclear (Rosario et al., 2017). The genus circovirus comprises porcine circoviruses types 1 (PCV1), 2 (PCV2), and 3 (PCV3) (Rosario et al., 2017). PCV2 infections are responsible for significant mortality among swine as the causative agent of porcine circovirus associated disease (PCVAD). Symptoms of PCVAD include weight loss, jaundice, stressed appearance, enlarged/depleted lymph nodes, pneumonia, enteritis, diarrhea, increased mortality and cull rates, abortions, stillbirths, and fetus mummification (Ellis, 2014; Palinski et al., 2017).
The PCV2 particle is approximately 19 nm in diameter, and the genome of PCV ranges from 1.7 kb to 2.3 kb in size. The circular nature of the genome has led to the viral family name circovirus. The evolutionary history has been explored and has allowed detailed phylogenetic trees and variations in the capsid surface structure to be described (Franzo et al., 2015; Wang et al., 2016; Wei et al., 2013; Xiao et al., 2015). The initial cryo-EM molecular envelope of several native circoviruses demonstrated that the capsid has T=1 icosahedral symmetry (Crowther et al., 2003; Khayat et al., 2011; Liu et al., 2016). The PCV genome encodes for one structural capsid protein (CP). The PCV2b CP expressed and purified from E. coli was shown to self-assemble into virus-like particle (VLP) during crystal formation for X-ray crystallography. The 2.3 Å structure of the VLP is similar to the morphology of the infectious virus determined to 25 Å resolution using cryo-EM (Crowther et al., 2003; Khayat et al., 2011). The CP fold can be described as a β-barrel, or a β-sandwich composed of two β-sheets. Each β-sheet is composed of four β-strands (BIDG and CHEF). The fold is similar to the canonical viral jelly roll first observed in tomato bushy stunt virus (Harrison et al., 1978). The loops connecting the β-strands form the features on the viral surface. The PCV2b CP was also expressed and purified as VLP from Trichoplasia ni insect cells (Khayat et al., 2011). The cryo-EM molecular envelope of this VLP demonstrated that the N-terminus is located inside the capsid, and the authors concluded that the antigenic property associated with the N-terminus is likely a result of the N-terminus transiently externalizing from the capsid via a process referred to as viral “breathing” (Guo et al., 2011; Khayat et al., 2011; Mahe et al., 2000). The externalization of the N-terminus may play an important role in the life cycle of the virus. While production of CP from E. coli and baculovirus expression systems have resulted in assembly of PCV2 VLPs that appear to resemble the structures of native PCV2 virus, the systems are quite different from a mammalian system which is a natural host for PCV2 virus. A mammalian expression system may provide better conditions for the examination of capsid assembly and structure; therefore, we have chosen HEK293 mammalian cells to evaluate the formation and structural characterization of the produced PCV2 VLPs.
The PCV2 CP entries in GenBank were recently categorized into eight different genotypes (PCV2a-h) (Franzo and Segales, 2018). The chronological deposition of PCV2 sequences into GenBank suggests that PCV2a was the dominant global genotype until early-2000 when a genotype shift to PCV2b was observed (Beach and Meng, 2012; Patterson and Opriessnig, 2010). The PCV2c genotype has only been reported from European countries and may have become extinct as there are only four depositions in GenBank. In 2013 Wei et al. reported and deposited a large quantity of PCV2d sequences into GenBank (Wei et al., 2013). Since then the number of PCV2d depositions in GenBank has significantly outpaced the deposition of PCV2b sequences (Franzo and Segales, 2018; Xiao et al., 2015). The increase in the deposition of PCV2d sequences may be a result of PCV2d becoming an emerging and predominant genotype in Asia, Europe, North and South America. While the cause(s) for the increase in depositions remains unclear, the presence of PCV2d in vaccinated herds suggests that either the vaccine is not appropriately administered or that PCV2d represents a genotype resistant to vaccination (Matzinger et al., 2016; Opriessnig et al., 2013). Despite the significant number of phylogenetic studies of the PCV2 genotypes and the emerging importance of PCV2d on the global swine industry there are no reports describing the structure of the PCV2d capsid. To determine if a structural difference between PCV2b and PCV2d may explain the shift in genotypes, we generated large quantities of the PVC2d VLPs for structure determination. Our HEK293 expression system is particularly advantageous for producing PCV2 VLP because it is rapid, robust, and simple. Study of the cytoplasmic and nuclear fractions of the mammalian cells indicates that PCV2 assembles and remains in the nucleus within 72 hours of transfection. The cryo-EM molecular envelope of the VLPs determined to a resolution of 3.3 Å allows us to confidently model the atomic coordinates for amino acids 36–231. Comparison of the PCV2d atomic coordinates to that of PCV2a and PCV2b provide a structural explanation for the PCV2a to PCV2b shift in genotypes. We model the N-terminus to be near the icosahedral 3-fold axes of symmetry. The position of the N-terminus agrees with uninterpreted molecular envelopes present in the deposited cryo-EM image reconstructions of PCV2a (EMDB-6555) and PCV2b (EMDB-8939). Surprisingly, this analysis shows the N-terminus to shift its position with the packaging of material into the capsid. We model a Pu-Pu-Py-Py tetranucleotide between the icosahedral 5- and 2-fold axes of symmetry. This is the first instance of nucleic acid modeled into a PCV capsid. We substitute the arginine amino acids that interact with the tetranucleotide and demonstrate that VLP formation is abrogated. We then compare the surface amino acid composition of PCV2a, b, d and determine that the amino acids that experience sequence diversity (amino acid percent identity) and variability (entropy-based conservation analysis). Analysis of 1,278 unique CP entries indicates that except for three amino acids (Met1, Pro15 and Arg147) every amino acid position has experienced a substitution. However, two groups of amino acids exhibit limited sequence variation and form distinct patches on the surface of the capsid. These patches may play important biological roles in the PCV2 life cycle.
Results
Recombinant PCV2 VLPs assemble and remain in the nucleus of expression system.
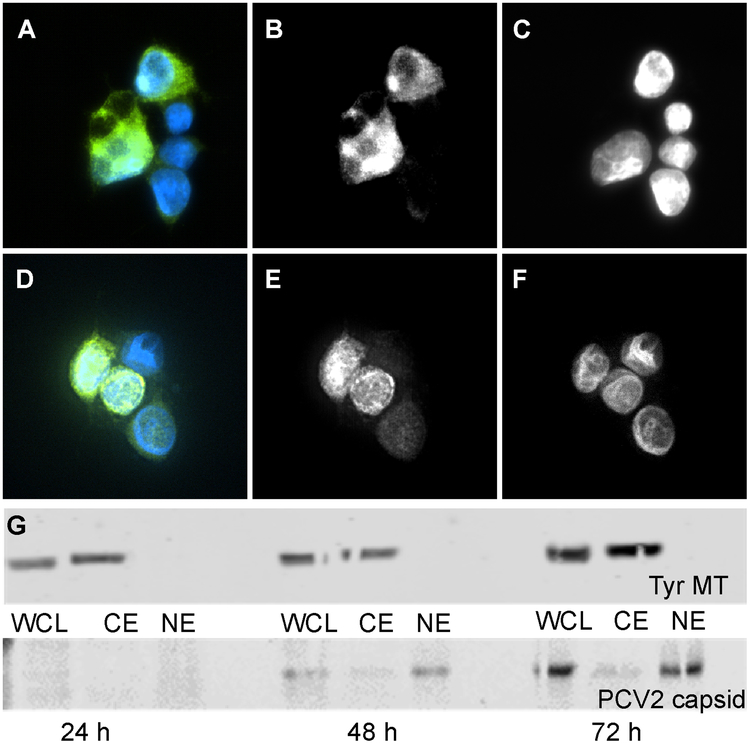
We sub-cloned the PCV2d capsid protein (CP) gene of C/2013/3 isolated in Taiwan (GenBank: AWD32058.1) into the mammalian expression plasmid pcDNA 3.4 in order to produce recombinant PCV2d VLP in mammalian cells (Fig. 1A). Transfection of mammalian cells HEK-293 suspension culture with the plasmid results in the production of the CP and assembly of the VLP. To determine the location of VLP assembly (nucleus versus cytoplasm), we used immunofluorescence and Western blot analysis of transfected cells. The analysis was performed at three time points post-transfection: 24, 48, and 72 hours. The mouse anti-PCV2 capsid monoclonal antibody was used as a probe directed to amino acids 49–125 of the CP (exact epitope location is proprietary information of GeneTex). The transfected cells were subsequently lysed at each of the time points and three cellular fractions isolated, whole cell lysate (WCL), cytoplasm (CE) and nuclear (NE) extracts. At 48 hours immunofluorescence expressed CP becomes evident. At this time point CP was found in both the nucleus and the cytoplasm; by 72 hours the nuclear compartment appeared enriched (Fig. 2). These results were confirmed by Western Blot analysis. These results demonstrate that while translation is, as expected, in the cell cytoplasm the protein is subsequently transported into the nucleus where capsid assembly occurs and the VLP remains. To date PCV2 VLP has been produced using insect cells, yeast, or E. coli (Duan et al., 2019; Khayat et al., 2011), thus expression of VLP using mammalian cells provides an analogous system to that utilized by natural PCV2 infection.
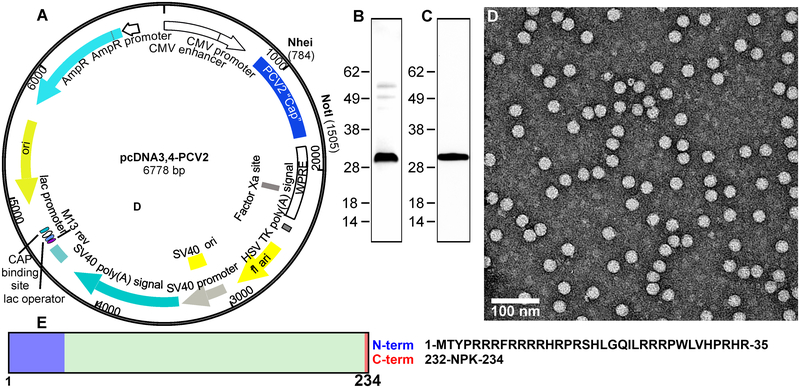
Figure 1. Expression of PCV2 virus-like particles in mammalian cells.
A) Plasmid generated for expression of PCV2d CP. The codon optimized PCV2d capsid gene was synthesized (Blue Heron Technologies, Bothell, WA) and cloned into expression vector pcDNA3.4 (Fisher Scientific). B) Protein expression was conducted in transiently transfected suspension cultures of Expi293 cells (Life Technologies). SDS-PAGE analysis of purified PCV2d VLPs (1 ug protein) and stained with Coomassie blue. C) SDS-PAGE analysis of purified PCV2d VLPs (0.5 ug protein) transferred to a nitrocellulose membrane and probed for a Western Blot with primary rabbit anti PCV2 capsid polyclonal antibody (Cab 183908, Abcam, UK). D) Negative stained electron microscopy micrograph of purified VLP stained with uranyl acetate. Particle sizes are approximately19 nm diameter. E) Schematic for C (234 amino acids). The amino acids that could not be modeled into the molecular envelopes are shown in blue and red.
Analysis of purified PCV2d VLP and its protein composition was confirmed by SDS-PAGE and Western blot analysis (Fig. 1B and C). Further examination by negative stained electron microscopy showed homogeneous spherical particles with smooth edges, slightly rough surface with a diameter of ~19 nm (Fig. 1D). The CP described in this study possesses the amino acid sequence identified from a number of recently isolated and reported PCV2d virus genome entries in GenBank, such as W233–12 isolated in Japan (BBE28610.1), CN-FJC011 isolated in China (AVZ66019.1), and England/15-P0222-09-14 isolated in England (ATN97185.1). The schematic for CP is shown in Fig. 1E.
Cryo-electron microscopy and image reconstruction of PCV2d VLP.
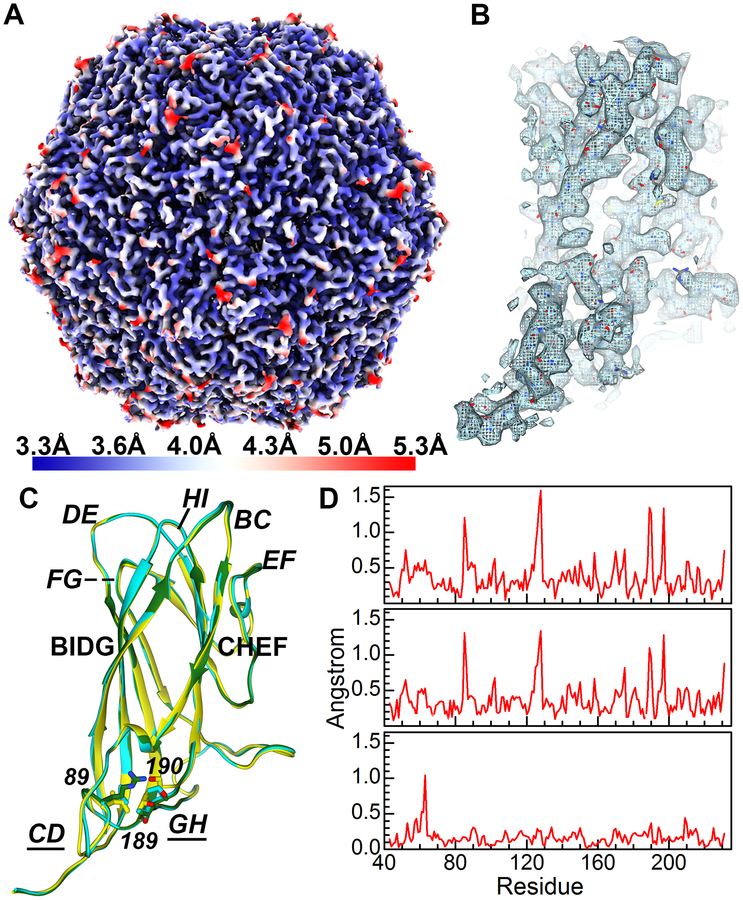
We determined an icosahedral cryo-EM molecular envelope of purified PCV2d VLP to a resolution of 3.3 Å (Fig. 3A). The molecular envelope of the side chains allows us to confidently model the atomic coordinates of PCV2d (Fig. 3B). The coordinates from the PCV2b crystal structure were manually fitted into the molecular envelope and amino acid substitutions were performed to reflect the PCV2d amino acid sequence. Additional amino acids were built for the N-terminus and the fitted model was refined through several iterations of automatic refinement with Phenix suite and manual modeling using the program Coot (Fig. 3B). The final refinement statistics are shown in Table 1. We were able to model atomic coordinates from residue 36–231. Uninterpretable molecular envelopes preceding amino acid 36 and following amino acid 231 could be observed, but no models were built due to interpretation concerns. The PCV2 CP fold is the canonical viral jelly roll first visualized for the tomato bushy stunt virus (Harrison et al., 1978; Khayat et al., 2011). The jelly roll can also be described as a β-sandwich that is composed of two β-sheets. Each sheet consists of four strands (BIDG and CHEF) (Fig. 3C) and has a slightly right-handed twist. The 60 CP pack together such that the β-sheets are normal to the T=1 icosahedral particle. The loops connecting the strands BC, DE, FG and HI are 4 to 9 amino acids, while the loops connecting strands CD, EF and GH are 21 to 36 amino acids. The shorter loops define the surface of the capsid, while the longer loops are predominantly involved in CP-CP interaction. Given the high sequence identity shared between the PCV2a, b and d sequences, it would be anticipated that no significant differences exist between their atomic coordinates. However, superimposing the subunits from these genotypes and generating distance plots of equivalent Cα atoms proves otherwise (Fig. 3C, 3D) (Dhindwal et al., 2019b; Liu et al., 2016). The regions that exhibit significant diversity correspond to two of the surface-exposed loops consisting of amino acids 85–91 (section of loop CD), and 188–194 (section of loop GH) (Fig. 3C, 3D). The movement in loop CD and GH are coordinated, and a result of a single amino acid substitution at position 89 (loop CD) that is situated under loop GH. The larger amino acids of PCV2d (Leu89) and PCV2b (Arg89), as compared to PCV2a (Val89), push loop GH of PCV2d and PCV2b further away from loop CD. The Cα atom of PCV2a Thr189-Ser190 and PCV2b Thr189-Ala190 (loop GH) are ~1.4 and 1.3 Å apart, respectively. The Cα atoms for amino acids 189–190 in PCV2a and PCV2d are closer to one another, because of the smaller size differential between PCV2d Leu89 and PCV2a Val89.
Figure 3. Structural study of the PCV2d VLP.
A) Icosahedral cryo-EM molecular envelope of the purified PCV2d VLP colored according to the local resolution. The gradient color map on the left-hand side indicates the resolution for the colors. B) Extracted molecular envelope for a subunit demonstrates the quality of the molecular envelope, where amino acid side chains can clearly be seen. The atomic coordinates have been modeled into the molecular envelope. C) Structural overlay of the PCV2a (yellow), PCV2b (green), and PCV2d (cyan). Amino acids 89, 189 and 190 are shown as stick models. The β-strands (bold) loops (bold-italic) are labeled. Loop CD (amino acids 75–94) and loop GH (163–194) where movement is observed. Figures generated using UCSF Chimera and ChimeraX (Goddard et al., 2018; Pettersen et al., 2004). D) Distances between equivalent amino acids (Cα atoms) are plotted after superposition of two PCV2 structures PCV2a-PCV2b (top), PCV2a-PCV2d (middle), and PCV2b-PCV2d (bottom).
Table 1.
Statistics for the cryo-EM image reconstruction of PCV2d and refinement of its atomic coordinates.
| PCV2d | |
|---|---|
| Cryo-EM image reconstruction | |
| Number of Micrographs | 799 |
| Defocus range | −0.68 to −2.81 μm |
| Total dose | 64 e- Å−2 |
| Dose rate | 6.4 e− Å−2 sec−1 |
| Number of Frames | 50 |
| Particles extracted | 23,358 |
| Particles used for reconstruction | 4,442 |
| FSC resolution * | 3.3 Å |
| B-factor sharpening† | −125.9 Å2 |
| Coordinate refinement | |
| CC | 84.6% |
| B-factor average | 64.6 Å2 |
| Bond angles (rmsd degrees)‡ | 0.013 |
| Bond lengths (rmsd Å)§ | 1.38 |
| Molprobity¶ | 2.18 |
| EMRinger | 5.1 |
| Clashscore | 1.7 |
| Ramachandran favored (%) | 91.8% |
| Ramachandran allowed (%) | 8.2% |
| EMDB | 20113 |
| PDB | 6OLA |
Fourier shell correlation reported by Relion 3.0 using the gold standard method at a CC of 0.143.
The B-factor sharpening reported by Relion 3.0 during the post-refinement process
The root-mean standard deviation for bonds reported by phenix.real_space_refine
The root-mean standard deviation for angles reported by phenix.real_space_refine
Molprobity Overall score
The capsid packages cellular nucleic acid and recognizes a Pu-Pu-Py-Py nucleotide sequence.
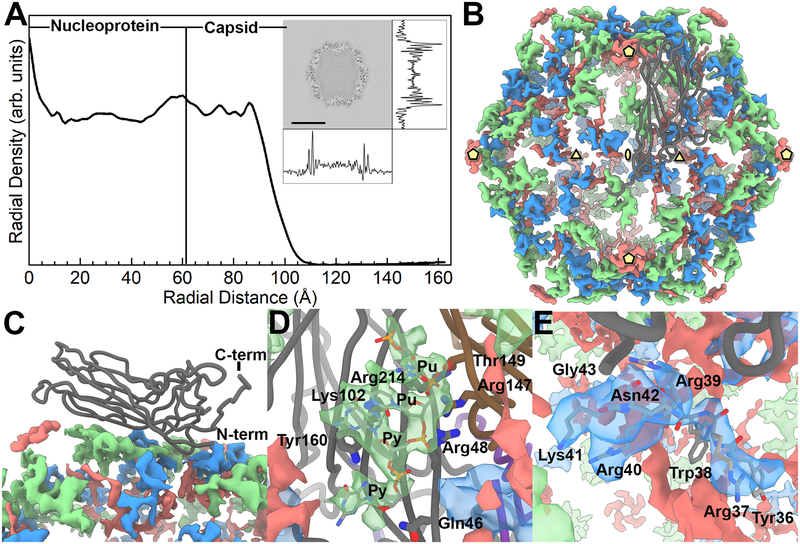
A near-central reconstruction slice was extracted from the cryo-EM molecular envelope and the density trace of pixel values was calculated in the horizontal and vertical directions (Fig. 4A). Based upon the central slice of the reconstruction, the PCV2 VLP outer diameter is approximately 18.5 nm assuming a roughly spherical shape for calculation. The outer volume is therefore estimated to be 3.3×103 nm3. Using the same spherical shape estimation, the inner diameter is approximately 13 nm. Therefore, the inner region volume is approximately 1.2×103 nm3. The radial profile of the molecular envelope indicates that a substantial amount of material is located within the capsid (i.e. compare the radial density of the nucleoprotein to the capsid) (Fig. 4A). We computationally measured the amount of material located inside the capsid by comparing the voxel count within the capsid shell to that inside the capsid -see methods and materials. A similar comparison performed for the asymmetric molecular envelope of the MS2 bacteriophage (EMD-8397) allowed us to calibrate the amount of material in our capsid to a sample with known RNA content. EMD-8397 is of sufficient quality to model the majority of the ssRNA genome (Dai et al., 2017). Our analysis indicates that in addition to the 60-copies of amino acids 1–42, which are believed to be in the interior of the capsid shell, there is scattering from an additional ~380 kDa of material. This is equivalent to ~1,200 nt of ribonucleic acid.
Figure 4. The inner content of the PCV2 capsid.
A) A radial profile of the PCV2d cryo-EM molecular envelope. The capsid interior and shell are identified by assessing the cryo-EM molecular envelope. The inset is a central slice extracted from the cryo-EM molecular envelope, with the density trace of pixel values calculated in the horizontal and vertical directions. The radial profile demonstrates that number of voxels within the capsid is comparable to the capsid shell; thus, a substantial amount of material is located within the capsid interior. B) Strong difference peaks identified in the inner capsid. The icosahedral 5-, 3- and 2-fold axes of symmetry are identified by yellow pentagons, triangles and ellipses, respectively. A CP subunit is shown as a dark grey tube. We interpret the green colored difference peak to be a Pu-Pu-Py-Py tetranucleotide, the blue colored difference peak to be amino acids 36–41 of the PCV2 N-terminus, and the red colored difference peak to be “unidentified”. C) Side view showing the CP subunit and the difference peaks. D) Close up of the tetranucleotide that has been modeled into the difference peak (green) located near the 3-fold axes of symmetry, and the CP amino acids in proximity. Gln46 (strand B), Arg48 (strand B), Lys102 (strand D), and Arg214 (strand I) of one subunit, and Thr149 (strand F) and Arg147 (strand F) from a neighboring subunit form hydrogen bonds and electrostatic interaction with the phosphate backbones of the tetranucleotide. Tyr160 (strand I) forms π-bond overlap with the first Py in the tetranucleotide. E) Close up of the PCV2 N-terminus modeled into the difference peak (blue) located near the 3-fold axes of symmetry. Amino acids 36–42 are labeled.
We then used absorption spectroscopy to measure the content of the purified PCV2d VLP following the protocol described by Porterfield and Zlotnick (Porterfield and Zlotnick, 2010). The measurements allow one to subtract scattering (absorbances at 360 nm and 340 nm) from the sample and calculate the amount of RNA (absorbance at 260 nm) present as a ratio of the amount of protein (absorbance at 280 nm) present. Five measurements were made to yield an average of 23.8 nucleotides per CP. This is equivalent to 1,428 nucleotides per capsid, and in agreement with our estimations using the cryo-EM molecular envelope of PCV2d.
To interpret the inner composition of the PCV2d VLP molecular envelope, we generated a molecular envelope (difference map) representing the inner composition of the capsid by subtracting a 3.3 Å molecular envelope calculated from our coordinates (amino acids 43–231) from our 3.3 Å cryo-EM molecular envelope (Fig. 4B, C). Strong difference peaks are observed near the icosahedral 3-fold and between the 5- and 2-fold axes of symmetry. We model a short oligonucleotide Pu-Pu-Py-Py (Pu, purine; Py, pyrimidine) into the difference peak between the 5- and 2-fold axes of symmetry. This is the first instance where capsid-nucleic acid interaction has been reported for a PCV structure. The backbone ribose and phosphates of the oligonucleotide form hydrogen bonds and electrostatic interactions with Gln46, Arg48, Lys102 and Arg214 of one subunit, and Arg147 and Thr149 of a neighboring subunit. The side chains from Arg48, Arg147 and Arg214 stack like a zipper to form a positive patch on the inner surface of the capsid. The charge on the patch is neutralized by the two phosphates in the RNA backbone. The second Pu of the oligonucleotide forms π-interactions with Tyr160 of the PCV2d CP (Fig. 4D). Two observations support our model: the modeled coordinates demonstrate the nucleotide bases to stack and form π-bond overlap, and the coordinates of these nucleotides overlay with the coordinates of nucleotides that were modeled into the beak and feather disease virus (BFDV) crystal structure when the PCV2 and BFDV CP are superposed -BFDV is a member of the Circoviridae family and its CP shares 32% sequence identity with the PCV2 CP (Sarker et al., 2016).
Substitution of amino acids that interact with the Pu-Pu-Py-Py tetranucleotide sequence abrogates VLP formation.
Arg147 is flanked by Arg48 and Arg214 to form a zipper like structure on the inner surface of the capsid. Together, these amino acids form a positive patch where the phosphates of the tetranucleotide bind (Fig. 4D). To assess the significance of the interaction between Arg48, Arg147 and Arg214 with the Pu-Pu-Py-Py tetranucleotide sequence, we made a number of arginine to methionine substitutions. Methionine was selected for substitution due to its large size mimicking the side chain of arginine that lacks the guanidinium group. We made single substitutions Arg48Met, Arg147Met, Arg214Met, and the double substitution Arg147Met/Arg214Met. The mutant capsids were expressed and purified via the procedure developed for the parent capsid. Examination of gradient purified material from lysates by Western blot showed that the CP for each mutant was expressed to a level comparable to the parent construct. However, evaluation of VLP assembly via negative stain electron microscopy showed that VLPs were detected only in the fractions obtained from the Arg48Met substitution. This suggests that the positive charged patch and its interaction with nucleotide may play a role in capsid formation; however, further studies additional substitutions such as Arg48Met/Arg147Met or a triple mutant Arg48Met/Arg147Met/Arg214Met should allow us decipher the role of the inner zipper like structure in capsid assembly and possibly in genome encapsidation.
The N-terminus of the capsid protein is in the interior of the capsid and located near the icosahedral 3-fold axes of symmetry.
The N-termini of Circovirus CPs carry a large number of Arg and Lys and predicted to be an intrinsically disordered region -a region with no secondary structure (Rosario et al., 2015). We recently provided experimental support for this prediction by measuring the circular dichroism spectrum of the PCV2b N-terminus peptide (amino acids 1–42) (Dhindwal et al., 2019c). The crystal structure and cryo-EM molecular envelope of the PCV2b VLP and the cryo-EM molecular envelope of the PCV2a VLP both strongly suggested that the N-terminus is located inside the capsid (Dhindwal et al., 2019a; Khayat et al., 2011; Liu et al., 2016). Half of the amino acids in the N-terminus are Arg or Lys amino acids; thus, its high positive charge and potential location suggests that it may be capable of interacting with nucleic acid packaged in the capsid. The difference peak near the 3-fold axes of symmetry are adjacent to the modeled N-terminus of PCV2 (amino acid 43), and we have modeled six amino acids into this difference peak to extend the N-terminus to amino acid 36 (Fig. 4E). The N-terminus begins as a single turn helix that descends into the inner portion of the capsid. Additional molecular envelop for the N-terminus can be observed; however, we refrain from modeling this section as we cannot confidently interpret the direction of the polypeptide chain and identify the amino acids by the molecular envelope of their side chains. The reduced quality of the molecular envelope in this region is most likely due to the N-terminus not adopting the icosahedral symmetry imposed during data processing. Unfortunately, symmetry expansion, signal subtraction, and focused classification of this region was unable to improve the quality of the molecular envelope. Consequently, we do not know the location of amino acids 1–35 of the CP. A recent manuscript by Mo et al. models the N-terminus of PCV2 near the icosahedral 5-fold axes of symmetry (Mo et al., 2019). We will discuss the discrepancy between the two models in the discussion section.
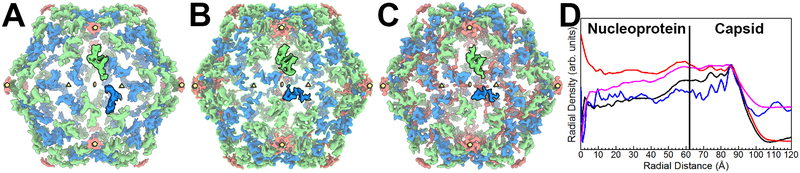
Difference maps were also calculated from the PCV2a and PCV2b cryo-EM molecular envelopes (Fig. 5A–C). The maps display a tetranucleotide molecular envelope located between the icosahedral 2- and 5-fold axes of symmetry. The location of these molecular envelopes are conserved between PCV2a, b, and d (Fig. 5A–C). Surprisingly, the molecular envelope for the PCV2a N-terminus is rotated ~90° with respect to its equivalents in the PCV2b and PCV2d molecular envelopes (Fig. 5A–C). Radial plots of the molecular envelopes suggest that the inner contents of the PCV2a molecular envelope consist of the least amount of packaged material (Fig. 5D). Perhaps the distinct positioning of the N-terminus in the PCV2a molecular envelope is a result of the lesser amount of packaged material. To assess if the different orientation of the Mo et al. expressed VLP N-terminus can be contributed to the inner contents of the capsid, we also generated a radial plot of this molecular envelope (Fig. 5D). The radial plot demonstrates that the molecular envelope for this VLP possesses the second highest content.
Figure 5. Inner capsid difference peaks of PCV2a, PCV2b and PCV2d.
Comparing the difference peaks in the inner capsid of PCV2a (A), PCV2b (B) and PCV2d (C) reveals a conserved location for the tetranucleotide molecular envelope (green) and differences for the N-termini molecular envelope (blue). The orientation of the PCV2b and PCV2d N-termini are conserved; however, the N-termini of the PCV2a are rotated ~90° clockwise. The icosahedral symmetry elements are shown using the same convention as Fig. 4B. The molecular envelopes for a single subunit (one of sixty) are painted with bold colors. D) Radial plots of PCV2a (blue, EMDB 6555), PCV2b (black, EMDB 8939), PCV2d (red, EMDB 20113), and PCV2 expressed in E. coli (light blue, EMDB 6746). The plots are normalized and scaled to one another to simplify the comparison. The PCV2d molecular envelope possesses the greatest amount of content within its capsid, while the PCV2a molecular envelope possesses the least amount of content within its capsid.
PCV2a, b and d genotypes demonstrate different diversity and variability.
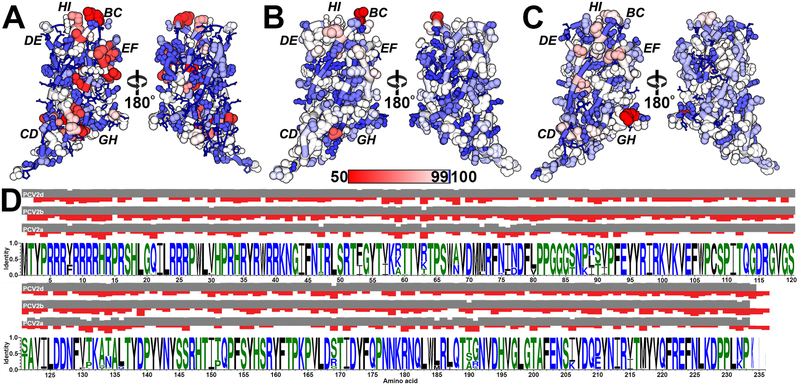
Several phylogenetic studies have compared the amino acid sequence of PCV2 CPs to differentiate PCV2 into genotypes (Franzo et al., 2015; Franzo and Segales, 2018; Xiao et al., 2015). Indeed, Franzo and Segales recently identified eight PCV2 genotypes (a-h) (Franzo and Segales, 2018). A report by Wang et al. mapped the sequence variation in 50 PCV2 CP sequences onto the crystal structure and identified regions of variability (Wang et al., 2016); however, since then a significant number of CP sequences have been deposited into GenBank that requires revisiting this analysis. We asked, do amino acids that exhibit the greatest diversity (lowest percent identity) or greatest variability (highest entropy-based measure) cluster on the 3D structure of the CP, and is the clustering conserved among PCV2a, b, and d genotypes (Pei and Grishin, 2001). Percent identity at an amino acid position in a set of aligned sequences is the frequency of the most occurring amino acid multiplied by 100. Entropy-based conservation analysis at an amino acid position in a set of aligned sequences describes the frequency of different amino acids, with the greatest entropy occurring when all amino acids exhibit identical frequencies. The alignment conservation server (AL2CO) calculates entropy-based measurement as follows (Pei and Grishin, 2001).
We chose to concentrate on these three genotypes because of the larger number of GenBank entries. We initiated our analysis by downloading the nucleotide sequences used by Franzo and Segales in their study (PCV2a: 675; PCV2b: 1,984; PCV2d: 1,491) from GenBank (Franzo and Segales, 2018). We note that sequences arising from recombination have been removed by Franzo and Segales. We then converted the nucleic acid sequences to amino acid sequences using the NCBI ORFfinder program (Sayers et al., 2011) and removed all redundant amino acid sequences from each genotype using the CD-HIT server (Niu et al., 2010). A total of 317 PCV2a, 501 PCV2b and 368 PCV2d unique sequences were aligned for each genotype and were mapped onto the PCV2a, PCV2b and PCV2d atomic coordinates (Fig. 6). The PCV2a genotype demonstrates the greatest sequence diversity with the lowest sequence variability -the two most diverse sequences share 86.3% identity (AF364094.1 vs. AIQ85146.1) (Fig. 6A and 6D). The most diverse positions are in loops BC, CD, EF, GH, and HI while the most variable positions are in loop CD. The high sequence diversity in the apposed loops BC and HI is suggestive of a conformational epitope (conf1). Similarly, the high sequence diversity in the apposed loops CD and GH is suggestive of a conformational epitope (conf2). Indeed, Shang et al. demonstrated that PCV2 neutralizing antibodies 2B1, 7F4, and 6H9 bound to PCV2 isolates NB0301, SX0201, HZ0301, HZ0201, TZ0601, JH0602, but not ISU-31 (Shang et al., 2009). The difference in sequence between ISU-31 and the remaining PCV2 isolates locates to loops CD, GH and HI. Consequently, antibodies 2B1, 7F4, and 6H9 may bind to either conf1 or conf2. The PCV2b genotype exhibits less sequence diversity, but greater sequence variability –the two most diverse sequences share 84.5% identity (ABL07437.1 vs. ABR14586.1) (Fig. 6B and 6D). The most diverse positions are in loop BC and the most variable positions are in loops BC, CD, DE, and HI. The PCV2d genotype exhibit distinct sequence diversity and variability when compared to the PCV2a and b genotypes -the two most diverse sequences share 83.3% identity (KP824717.1 vs. AVR51420.1). For example, the PCV2b genotype exhibits greater sequence diversity in loop BC and variability in loop CD, while PCV2d exhibits greater sequence diversity in loop GH and variability in loop EF (Fig. 6C and 6D). The diversity/variability observed in loop EF, and the significance of this loop in antibody neutralization, was first described by a series of antigenic subtyping experiments by Lefebvre et al. and Saha et al., where panels of neutralizing monoclonal antibodies were tested for their ability to bind to different strains of PCV2. These experiments demonstrated that amino acid substitutions in loop EF affected antibody binding (Lefebvre et al., 2008; Saha et al., 2012). Not shown in Fig. 4 are the last few C-terminal amino acids of the CP, as none of the structural studies have been successful in visualizing these amino acids. These amino acids are highly variable within each genotype, and between genotypes (Franzo and Segales, 2018; Rosario et al., 2017; Wei et al., 2013; Xiao et al., 2015). PCV2b and PCV2d also exhibit greater sequence variability in the inner surface of the capsid when compared to PCV2a (Fig. 6A–C).
Figure 6. Sequence diversity and variability in the PCV2a, b, d genotypes.
A) Amino acid sequence alignment information plotted on the PCV2a, b, and d coordinates. The red-to-blue color gradient represents diversity (sequence identity) -gradient shown in the middle of the image. The size of the atoms and tube represent variation (AL2CO: sequence entropy), with smaller atoms/tubes representing lower entropy/variation and larger atoms/tubes representing greater entropy/variation. Lower entropy indicates fewer different amino acids present in the alignment at a position, and larger entropy indicates more different amino acids present at a position. The plotting of variation allows one to appreciate the frequency of different amino acids at each position. Left, amino acids facing the capsid exterior. Right, amino acids facing the capsid interior. A) Attained from 317 unique CP entries plotted on the surface of the PCV2a atomic coordinates (PDB entry 3JCI). B) attained from 501 unique CP entries plotted on the surface of the PCV2b atomic coordinates (PDB entry 6DZU). C) attained from 368 unique CP entries plotted on the surface of the PCV2d atomic coordinates (PDB entry 6OLA). D) WebLogo image of the PCV2a, PCV2b, and PCV2d CP entries (Schneider and Stephens, 1990). The grey and red bars represent the sequence diversity (identity) and variability (entropy) for each amino acid position. Values for sequence diversity and variability were calculated using the ALCO server (Pei and Grishin, 2001).
Highly conserved amino acids form patches on the capsid surface.
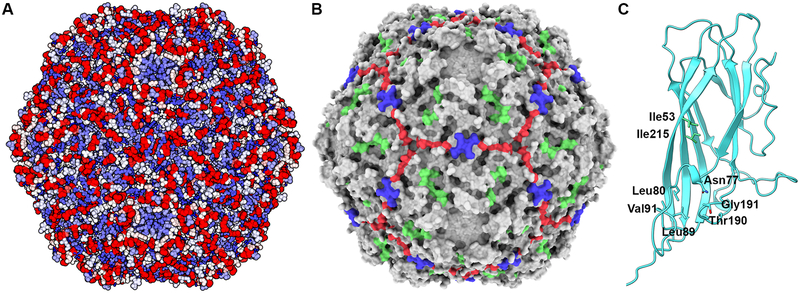
The sequences discussed above were expanded to include all PCV2 GenBank entries, then reduced to a unique set of amino acid sequences using the same strategy discussed above (1,278 entries). The sequences with the greatest diversity (GenBank entries: AVZ65995.1 and ALK04312.1) share 74.4% sequence identity. Closer inspection of the aligned sequences indicates that substitutions are observed within the hydrophobic core of the protein, at the intersubunit interface, and amino acids on the outer and inner surface of the capsid. Only Met1, Pro15 and Arg147 are absolutely conserved. The analysis suggests that PCV2 is capable of tolerating substitutions at nearly every position within the capsid while remaining an infectious virus. To visualize the sequence conservation at each amino acid position, we plotted the sequence alignment information onto the PCV2d atomic coordinates using the AL2CO procedure described above (Fig. 7A) (Pei and Grishin, 2001). We then concentrated on amino acids that are located on the surface of the capsid with their side chains exposed to solvent (Table 2). Of these amino acids, positions that demonstrate more than 99% sequence identity form three patches on the surface of the capsid (Table 2 and Fig. 7B).
Figure 7. Sequence comparison of 1,278 PCV2 capsid protein entries plotted on the PCV2d atomic coordinates.
A) The sequence diversity and variation plotted on the PCV2d atomic coordinates -same coloring scheme as in Fig. 6. Image made with UCSF Chimera (Pettersen et al., 2004). B) Surface representation of the PCV2d atomic coordinates with the three conserved patches colored in green (Tyr55, Thr56, Asp70, Met71, Arg73, Asp127), blue (Pro82, Gly83, Gly85) and red (Asp168, Thr170, Gln188, Thr189). Image made with UCSF ChimeraX and colored using flat lighting (Goddard et al., 2018). C) Ribbon cartoon of a PCV2d subunit. Amino acids in stick are evolutionary coupled together, as determined using the plmc. MATLAB 2019, and EVzoom programs. Figures generated using UCSF Chimera (Pettersen et al., 2004).
Table 2. Conserved amino acids on the surface of PCV2.
Sequence alignment of 1278 PCV2 CP amino acid sequences identifies positions that are conserved (more than 99% sequence identity) on the surface of the capsid with the side chains of the amino acid exposed to solvent.
| Surface exposed amino acids | 51, 53, 55–56, 58–64, 66, 70–71, 73, 75, 77–78, 82–83, 85, 88–89, 112–113, 115, 127–128, 131–138, 144, 166, 168–170, 188–191, 194, 204, 206–208, 210, 227, 229–234 |
| Patch 1 | Tyr55, Thr56, Asp70, Met71, Arg73, Asp127 |
| Patch 2 | Pro82, Gly83, Gly85: |
| Patch 3 | Asp168, Thr170, Gln188, Thr189 |
Evolutionary coupled substitutions differentiate the PCV2 genotypes.
The large number of CP sequences allowed us to ask if any of the amino acid positions are evolutionary constrained (Hopf et al., 2014). Evolutionary constrained amino acids require that substitution of one amino acid to be coupled to substitution of another amino acids. In the simplest of cases this may be because the amino acids pack against one another in the structure of the protein, such that substitution to a larger amino acid in one position requires substitution to a smaller amino acid in the second position for proper packing to occur. Such information can be used to predict the fold of a protein or identify functionally important sites (Hopf et al., 2017; Hopf et al., 2014; Shamsi et al., 2017). Evolutionary coupling (EC) measurements determined from the 1,278 unique sequences indicates that two independent locations in the structure demonstrate coupling. The first location consists of amino acids 53 and 215, and the second location consists of amino acids 77, 80, 89, 90, 190 and 191 (Fig. 7C). Sequence differences in the second location are responsible for the structural diversity observed between PCV2a and PCV2b/d in loops CD and GH (Fig. 3C) (Dhindwal et al., 2019b; Liu et al., 2016). The EC results may indicate that the coupled amino acids are functionally relevant. Solvent accessible surface area calculations with the GetArea server (http://curie.utmb.edu/getarea.html) indicates that amino acids 77, 80, 89, and 90 are more than 75% buried, while amino acids 190 and 191 are exposed to the solvent (Fraczkiewicz and Braun, 1998). Amino acids 190 and 191 help define a neutralizing epitope on the surface of the capsid -see below.
Discussion
Porcine circovirus 2 genome encodes for four known proteins: a replicase (ORF1) responsible for genome replication, a CP (ORF2) responsible for generating the capsid shell, and an ORF3 and ORF4 that are believed to regulate cellular apoptosis (Pan et al., 2018; Teras et al., 2018). PCV2 demonstrates the ability to rapidly mutate and evolve into novel genotypes -an ability that is governed by the accumulation of point mutations (Duffy et al., 2008; Rodriguez-Carino et al., 2011) and genome recombination (Cai et al., 2012). Phylogenetic analysis of the PCV2 CP sequence indicates that eight genotypes are distributed globally (PCV2a-h) (Franzo and Segales, 2018). Deposition of entries into Genbank suggests that until early-2000 PCV2a was the dominant genotype, when a shift to PCV2b was observed (Beach and Meng, 2012; Patterson and Opriessnig, 2010). The larger deposition of entries for PCV2b in GenBank suggests that it is the dominant genotype; however, the recent decrease in PCV2b and increase in PCV2d depositions suggests that there may be a shift from PCV2b to PCV2d (Wei et al., 2013). Given the history of genotype shifts, the high mutation rate and genome recombination of PCV2, it would not be too surprising if additional genotype shifts occurred. To understand why genotype shifts are occurring, it is necessary to establish an expression and purification protocol to rapidly acquire large quantities of PCV2 VLP for structural and biophysical studies. It is important to structurally characterize the PCV2 capsid as it is the entity under selective pressure by the environment (e.g. immune system and cellular interaction), and sequence information sometimes is not enough to distinguish differences between genotypes. Consequently, we have established a rapid and robust mammalian expression system for producing large quantity of PCV2 VLP. The study described in this paper is the first system where mammalian cells have been utilized to generate large quantity of VLP. The simple, robust, and rapid protocol of the mammalian expression system provides a tremendous advantage to the time-consuming and involved protocol necessary for baculovirus generation (Fig. 1). The mammalian expression system described here can generate VLP four days from when the expression plasmid is obtained, whereas the fastest baculovirus expression protocol can generate VLP in eight days (Scholz and Suppmann, 2017). The yield of purified VLP for this expression system is 1 mg per liter of growth. The decrease in time necessary to generate VLP is particularly advantageous when mutagenesis experiments are to be performed. Immunofluorescence and Western blot analysis of CP expression revealed that CP can be detected in the cytoplasm at 24 hours, but only detected in the nucleus at 72 hours. This suggests that the CP is unable to exit from the nucleus of mammalian cells within the first 72 hours of expression, and that capsid assembly likely occurs in the nucleus (Fig. 2). The assembly of capsids in the nucleus is supported by immunofluorescence studies that follow the kinetics of PCV2 infection showing the CP of PCV2 to enter the cytoplasm followed by the nucleus of infected PK-15 cells (Meerts et al., 2005). These studies demonstrate altered kinetics for different PCV2 strains. For example, the CP of strain Stoon-1010 could be observed in the nucleus for up to 72 hours post infection (hpi), whereas the CP of strain Stoon-1103 begins exiting the nucleus at 24 hpi. Comparable studies using thin-section electron microscopy visualize the morphogenesis of PCV2 during infection of L35 lymphoblastoid cell lines (Rodriguez-Carino et al., 2011). These studies identify PCV2 like particles entering the nucleus for genome replication, viral assembly and encapsidation.
Figure 2. Location of expressed PCV2d capsid protein in transfected HEK293 cell.
Transfected cells were fixed with acetone and labeled with mouse anti-PCV2 capsid monoclonal antibody to probe the capsid (green) and with DAPI (blue) to localize the nuclei. A) images from cells 48 hours post transfection with immunofluorescence from the capsid shown by the green channel and fluorescence from DAPI (nucleus labeling) by the blue channel. B) same image showing information from the green channel (capsid), C) same image showing information from the blue channel labeling (nucleus). D) images from cells 72 hours post transfection with immunofluorescence identical to A). E) same image showing information from the green channel (capsid), F) same image showing information from the blue channel (nucleus). G) Western blot analysis of whole cell lysate (WCL), cytoplasm (CE) and nuclear extracts (NE) collected 24, 48 and 72 hours post transfection. Top panel) The samples were probed for tyrosinated microtubules. Bottom panel) The samples were probed for PCV2 CP. The presence of tyrosinated microtubules in CE but not in NE provided the quality control of cell fractionation.
The 3.3 Å PCV2d cryo-EM molecular envelope allows us to confidently model amino acids 36 to 231, and a Pu-Pu-Py-Py tetranucleotide (Fig. 3). Comparison of the PCV2a, b and d atomic coordinates attained from cryo-EM studies identifies differences in the conformations of the surface-exposed loops CD (amino acids 86–91) and GH (amino acids 188–194) (Fig. 3C). The movement of these loops are coupled to one another. The difference between PCV2a and PCV2b in the positions of these loops provides a structural description for the genotype shift observed in 2003 from PCV2a to PCV2b (Beach and Meng, 2012; Patterson and Opriessnig, 2010). Such a difference could not have been identified by sequence analysis alone. Indeed, the structures indicate that substitution at a single position (amino acid 89) is responsible for the difference in loop positions. The Pu-Pu-Py-Py tetranucleotide coordinates modeled into our PCV2 molecular envelope is the first observation of ordered nucleic acid in a PCV2 capsid (Fig. 4B and 4D). The nucleic acid is likely to be RNA as VLP assembly occurs in the nucleus of the mammalian expression system. Hydrogen bonds and electrostatic interactions are observed between Gln46, Arg48, Lys102 and Arg214 of one subunit, and Arg147 and Thr149 of a neighboring subunit with the backbone phosphates of the tetranucleotide. The side chains of amino acids Arg48, Arg147 and Arg214 form a positive patch on the inner surface of the capsid that forms electrostatic interaction with the negative charges of the nucleic acid phosphates. The importance of this interaction is demonstrated by Arg147Met and Arg147Met/Arg214Met mutants that are unable to produce VLP. The base from the second Pu of the tetranucleotide forms π-interactions with the side chain of Tyr160. All four bases of the tetranucleotide make π-interactions. Interestingly, the placement of the tetranucleotide coordinates with respect to the PCV2 CP is nearly identical to the placement of a tetranucleotide observed in the beak feather disease virus (BFDV) -another member of the Circoviridae family (Sarker et al., 2016). The position for some of the amino acids that interact with the PCV2 nucleic acid overlaps with the position of amino acids that interact with the BFDV ssDNA, while others are unique to PCV2 or BFDV. PCV2 and BFDV share 32% sequence identity. Analysis of the PCV2d cryo-EM molecular envelope suggests that ~1,200 nt of RNA is packaged into the capsid. Absorption spectroscopy experiments support this by identifying 23.8 nt of RNA per CP for a total of 1,428 nt packaged into the capsid. This is slightly smaller than the 1,768 nt PCV2 genome.
Amino acids 36–42 modeled into the N-terminus of the PCV2d molecular envelope are located inside the PCV2 capsid near the icosahedral 3-fold axes of symmetry (Fig. 4B, and 4E). Amino acids 43–50 form the B β-strand of the viral jelly-roll remain in the interior of the capsid until amino acid 51, which is exposed on the surface of the capsid (Fig. 4C) (Dhindwal et al., 2019a; Khayat et al., 2011). Interestingly, there is a discrepancy between our model and the model recently described by Mo et al. (Mo et al., 2019). Mo et al. model amino acids 33–42 to interact with amino acids 17–27 near the icosahedral 5-fold axes of symmetry. Moreover, Mo et al. model amino acids 43–49 to form a loop that points to the exterior of the capsid near the 3-fold axes of symmetry. The different models may be a result of the different expression and assembly systems used by these studies; we use a mammalian expression system where the VLP assemble in the nucleus of the cell whereas Mo et al. use an E. coli. expression system with in vitro VLP assembly. We note that our model is also consistent with the cryo-EM molecular envelopes of PCV2a and PCV2b VLP attained from insect cell expression systems -where amino acids 43–50 are modeled in the interior rather than exterior of the capsid (Dhindwal et al., 2019a; Liu et al., 2016). Further studies are necessary to clarify the discrepancies between the two models.
Comparison of difference maps calculated from subtracting the cryo-EM molecular envelopes of PCV2a, b, and d from their respective atomic coordinates (amino acids 42–231) reveals similarities and differences between the interior of PCV2a and PCV2b/d (Fig. 5A–C). All difference maps identify similar molecular volumes describing the tetranucleotide we’ve modeled into the PCV2d molecular envelope. However, the positions of the molecular envelopes for the PCV2a N-termini are different than those of PCV2b/d. While the N-termini for the three genotypes locate near the icosahedral 3-fold axes of symmetry, the PCV2a molecular envelope is rotated 90° clockwise such that it points away from the 3-fold axes of symmetry (Fig. 5A–C). Possibly, the position of the PCV2a N-terminus is correlated to the lesser amount of material present inside the PCV2a capsid (Fig. 5D). If so, the N-terminus may act as a sensing switch for regulating capsid assembly. We are exploring this possibility with biochemical and structural studies. The amount of material packaged into the capsids may be a result of the different PCV2 sequences, the different expression systems, the purification protocol, or the fact that these are VLP. It remains to be determined if the amount of packaged material into PCV2 virion is consistent between particles.
Phylogenetic analysis of the PCV2 CP sequences has recently been used to identify eight genotypes (a-h) (Franzo and Segales, 2018). We mapped the diversity and variability present in the CP sequences of the PCV2a, b, and d genotypes onto their respective atomic coordinates (Fig. 6A–C) (Dhindwal et al., 2019a; Liu et al., 2016). The analysis, coupled with published work, suggest a conformational epitope composed of loops BC and HI, and another composed of loops CD and GH (Shang et al., 2009). The analysis also demonstrates that PCV2a exhibits the greatest sequence diversity with lowest sequence variability (Fig. 6A and 6D). Both PCV2b and d exhibit high sequence variability (Fig. 6B, 6C and 6D). The increased variability in the PCV2 genotypes may be a result of the selective pressure induced by the vaccination program in 2006 (Karuppannan and Opriessnig, 2017). The number of CP sequence entries in GenBank was dominated by PCV2a prior to the introduction of the program but was quickly shadowed by PCV2b entries after introduction of the vaccination program. The number of PCV2d entries is greater than PCV2b entries for 2016 (Franzo and Segales, 2018). We note that the sequence diversity and variability observed in these three genotypes are sufficiently high such that a single sequence is not truly representative of the genotype when one considers antibody escape substitutions. The diversity and variability in CP sequences suggests that PCV2 may indeed be regarded as viral quasi-species (Correa-Fiz et al., 2018).
Alignment of 1,278 unique PCV2 CP sequences indicates that only three amino acids (Met1, Pro15 and Arg147) are absolutely conserved, and that the remaining positions in the sequence have undergone substitutions (Fig. 7A). This demonstrates the remarkable plasticity of the capsid structure to undergo substitution while maintaining an infectious virus. The sequence analysis coupled with the structural information identify conserved patches on the capsid surface that may play important roles in the life cycle of the virus (Fig. 7B). Evolutionary coupling analysis of the sequence alignment identified two regions that underwent co-evolution. One of these regions correlates with the structural diversity observed between PCV2a and PCV2b/d in loops CD and GH (Fig. 7C).
The VLP expression system has additional applications. For example, the system can be used to generate capsids assembled as a multivalent mosaic that simultaneously displays neutralizing epitopes of several genotypes (i.e. transfect cells with multiple plasmids expressing different CP genotypes). Additionally, the 40 amino acids at the N-terminus can be replaced with specific tags to package biologics, expressed in the same cells, into the capsid. Such nanostructures could be utilized for diagnostics or delivery systems. There have been multiple reports demonstrating that VLP can assemble if the N-terminus of the CP is replaced with other sequences (Khayat et al., 2011; Zhang et al., 2014).
Conclusion
In summary, we describe a mammalian assembly system capable of producing large quantities of PCV2 VLP, perform cryo-EM image analysis of the PCV2d VLP, demonstrate structural differences between the PCV2a, b, and d genotypes, model a tetranucleotide and a section of the N-terminus into the cryo-EM molecular envelope, demonstrate that the capsid packages nucleic acid, visualize the diversity and variability of the deposited PCV2 CP sequences on the CP structure, and identify conserved patches on the surface of the capsid. We are uncertain what role, if any, these patches may play in the life cycle of PCV2.
Materials and methods
Cells, Capsid Gene, Plasmid and antibody.
Suspension cultures of Expi293F human cells (Life Technologies, CA) were grown in serum-free Expi293 expression medium (Life Technologies, CA) at 37°C in a 5% CO2 environment and agitated at 150 rpm in Erlenmeyer flasks. The porcine circovirus type 2 (PCV2) gene encoding the CP (GenBank: AWD32058.1) was chemically synthesized using a codon-optimized sequence by Blue Heron Technologies (Bothell, WA). The recognition site for NheI and the Kozak sequence were added right upstream from the start codon, and the recognition site for NotI was incorporated after the termination codon. The synthesized Cap gene was recovered from the transport plasmid by a double digestion with NheI and NotI restriction enzymes and sub-cloned after gel purification into the mammalian expression plasmid pcDNA3.4 cut with the same enzymes. The ligated plasmid was transformed into MAX Efficiency Stbl2 Cells (Life Technologies) and a correct clone was identified via restriction enzyme analysis and verified by sequencing.
Virus Like Particle (VLP) Production and Purification.
PCV2 VLPs were produced in a suspension culture of Expi293F mammalian cells following transient transfection with the plasmid pcDNA3.4-PCV2 (Fig. 1). Expi293F cells were seeded at the concentration of 2×106 cells/ml and cultured for 16h prior to transfection. Plasmid DNA (1 μg/ml) was diluted in a volume of Opti-MEM representing 5% of the total volume of the culture. Separately, polyethylenimine (PEI) was prepared in an equivalent volume of Opti-MEM (4 μg/ml). After 5 min of incubation at room temperature, the PEI solution was added dropwise to the tube containing the DNA and after 30 min of incubation at RT the mixture was added to the cell suspension in a dropwise manner. Twenty-four hours after transfection, Valproic acid sodium salt (VPA) was added to the cell culture to a final concentration of 3.75 mM to inhibit cell proliferation. Seventy-two hours post-transfection the cells were pelleted by centrifugation at 2,000g for 15 min, and then washed one time with phosphate buffered saline (PBS) and spun again at 2,000g for 15 min. The cell pellet was re-suspended in PBS and then subjected to three freeze (−80°C) and thaw (37°C) cycles. Subsequently, the cells were further fragmented by three cycles of sonication and clarified by two successive centrifugations, first at 2,000g for 15 min followed by 8,000g for 15 min. The PCV2 VLPs contained in the clarified supernatant were further purified by ultracentrifugation on a two-layer CsCl density gradient: lower layer, 5 ml of 1.4 g/ml CsCl and upper layer, 10ml of 1.25 g/ml of CsCl both prepared in 10mM Tris-HCl, (pH 7.9). Samples were loaded onto the gradient and spun at 15°C for 4h at 140,000g using a SW28 rotor (Beckman Coulter, CA). The VLPs appeared as an opaque band at the interface of the 1.25 and 1.4 g/ml CsCl layers and were collected by piercing the tube with an 18G needle and syringe. The collected solution was mixed with 37 % CsCl in 10 mM Tris-HCl (pH 7.9) to final volume of 12 ml and then spun at 15°C for 16hr at 155,000g using a SW 41Ti rotor (Beckman Coulter, CA). The VLPs were detected at the lower part of the tube and recovered as described above. Collected VLP material was dialyzed against 10mM Tris-HCl pH 7.9 and 150mM NaCl at 10°C overnight using a Slide-A-Lyzer Cassette. Purified PCV2 VLPs were concentrated and buffer exchanged to phosphate buffered saline (PBS) using Amicon Utra-4 centrifugal filter devices (Merck Millipore, MA). PCV2 VLP samples were stored in 50–100 μl aliquots at −80°C.
Western Blot and Coomassie Blue Stain.
Purified VLPs were mixed with loading buffer, heated at 100°C for 5 min and run on a 4 to 12% Bis-Tris SDS-polyacrylamide gel (Life Technologies, CA). Loading amounts of proteins were 1 μg for Coomassie staining and 0.5 μg – for Western Blot. After electrophoretic separation, the gel was stained with Coomassie blue or proteins electro-transferred onto a 0.45 μm nitrocellulose membrane (Life Technologies LC2001). The membrane was then blocked with 5 % non-fat milk in TBST (10 mM Tris-HCl, 130 mM NaCl, and 0.05% Tween-20, pH 7.4) for 1h at (20°C) followed by an overnight incubation at (20°C) in primary Rabbit anti-porcine circovirus antibody (Cab 183908, Abcam, UK) diluted with blocking buffer. Membranes were washed 3 times with TBST and then incubated for 2 h with secondary antibody (goat anti-rabbit IgG HRP conjugated, 1:1,000) diluted in blocking buffer. Finally, membranes were washed 3 times with TBST and developed with ECL Western blot system (Life Technologies, CA) according to manufacturer’s instructions. The stained gel and immune blot images were acquired with a FluorChem Imager instrument (Protein Simple, CA).
Immunofluorescence.
The HEK293 adherent culture cells were transfected with pcDNA3.4-PCV2d plasmid using Amaxa Nucleofector II instrument by Lonza (program A-23). The transfected cell were plated on 6-well plate, each well contained cover slip to performed immunofluorescence study on transfected cells. The cover slips were removed 24, 48 and 72 hours post transfection and fixed with ice-cold acetone for 10 min. Expressed PCV2d CP was detected by using Mouse anti-PCV2 CP monoclonal antibody (GeneTex, GTX634211). The Donkey anti Mouse IgG conjugated with Alexa488 dye (Jackson ImmunoResearch) used as a secondary antibody. DAPI staining allowed us to localize the nuclei.
Images were acquired with a Neo sCMOS camera (6.45μm pixels, 560MHz, Andor Technology) on a Nikon TiE inverted microscope (Nikon Inc., Mellville, NY) using 40X (NA 0.95) plan apochromat objectives. 14–16bit images were scaled linearly to highlight features of interest and converted to 8-bit copies for figure assembly. Devices were controlled by Elements software (Nikon Instruments).
Negative Staining and TEM Examination.
5 uL of pCV2–2 VLP samples was applied to CF200-CU carbon film 200 mesh copper grids (Electron Microscopy Science) for 1 min and the grids were then washed with 200 μl of 50 mM of Na Cacodylate buffer and then strained immediately with 50 μl of 0.5% uranyl acetate for 1 min. The grids were examined by a JOEL 2100 transmission electron microscope operating at 200 kV with an Orius 2048 × 2048 pixel CCD (Gatan Inc., Pleasanton, CA).
Cryo-EM Data Collection.
Frozen hydrated samples of PCV2d VLPs were prepared on Quantifoil R 2/2, 200 mesh copper grids (Electron Microscopy Science). A 4 μl sample of the VLP was applied to the grid blotted for 3 seconds and flash frozen in liquid ethane using a FEI Vitrobot instrument. The grids were stored in liquid nitrogen until data collection. Data was collected at cryogenic temperatures on a FEI Titan Krios, operating at 300 kV, with Gatan K2 camera post a GIF quantum energy filter with a width of 15 eV. Data collection was performed with the Leginon suite (Suloway et al., 2009).
Image Reconstruction.
The MotionCor2 package was used for correct for particle motion (Zheng et al., 2017). Default parameters and dose weighting were used for the correction, with the patch 5 option, and the first frame of each movie was discarded during the alignment. The particles were selected automatically using Gautomatch v0.53, and contrast transfer function (CTF) estimation was performed on the aligned micrographs using Gctf v0.50 (Zhang et al., 2016). Relion 3.0 was used to extract 23,358 300×300 pixel particles from the dose-weighted micrographs using the coordinates identified by Gautomatch. Reference free 2D classification was performed with Relion 3.0 (Scheres, 2012a). Non-default options for this step included a diameter of 250 Å and 128 classes were requested (Scheres, 2012b). An initial model was generated to 60 Å resolution using the PCV2 crystal structure (PDB entry 3R0R) with the molmap function of UCSF Chimera (Pettersen et al., 2004). 3D classification was carried out on 7196 particles using Relion 3.0 with a diameter of 250 Å, 3 classes, and C1 symmetry. A single class with 4,442 particles exhibited the highest resolution. These particles were used for a high-resolution image reconstruction with Relion 3.0. Again, a diameter of 250 Å and I1 symmetry were used with the remaining default parameters of Relion 3.0. A binary mask was created using the relion_mask_create program of Relion 3.0. The binary mask for postprocessing was generated as follows: 1) the high resolution molecular envelope was low pass filtered to 15 Å resolution using relion_image_handler (Relion 3.0), 2) the lowest threshold at which noise exterior to the PCV2 capsid was identified for this volume using UCSF Chimera, 3) relion_mask_create (Relion 3.0) was used to convert this volume into a binary mask with the identified threshold, mask dialation by 7 pixels and 2 soft edge pixels. The resulting mask was then inspected with UCSF Chimera to ensure that no internal cavities existed.
Local resolution was calculated with the program MonoRes. The same binary mask used during the postprocessing with Relion was used for the calculation. A resolution range of 3.3 Å to 5.3 Å was used (Vilas et al., 2018).
Structure refinement.
The atomic coordinates for the crystal structure of PCV2b crystal structure (PDB entry 3R0R) were modified using Coot (Emsley and Cowtan, 2004). The biological matrices necessary to generate a VLP are present in the PDB and were used by Coot to generate a VLP of PCV2d. UCSF Chimera was used to manually dock the VLP into the symmetrized molecular envelopes. The resulting coordinates were iteratively refined using phenix.real_space_refine from the Phenix software package with non-crystallographic symmetry (NCS) constraints applied, and manual fitting with Coot (Afonine et al., 2013).
The Pu-Pu-Py-Py sequence was manually fitted into the cryo-EM molecular envelope. The sequence was selected based on the shape of the molecular envelope and the correlation coefficient reported by phenix.real_space_refine after real space refinement.
Site directed mutagenesis.
Mutations in the CP gene were introduced by site-directed mutagenesis using specific primers containing codon substitution from Arg48Met, Arg147Met and Arg214Met for single position changes. The forward (F) and reverse (R) oligonucleotides used to generate the mutants are:
Arg48Met F/R (CTTCAACACCatgCTCTCCCGCAC / ATGCCATTTTTCCTTCTC),
Arg147Met F/R (CTACTCCTCCatgCATACCATAACCCAG / TTTACATAGGGGTCATAG);
Arg214Met F/R (CTACAATATCatgATAACCATGTATGTACAATTCAGAGAATTTAATC / TCCTGGTCGTAGATACTG).
The double mutant was generated by changing Arg214Met in the already mutated Arg147Met. The sequence of the mutated genes was confirmed using the services of Genewiz. Capsid production utilizing the mutated plasmids was carried out in the same manner as that used for the wild type capsid.
Sequence alignment, entropy calculation and evolution coupling.
The nucleotide sequences used by Franzo and Segales were downloaded from GenBank (Franzo and Segales, 2018). The NCBI ORFfinder suite was used to identify ORFs in each sequence. The ORFs identified from each entry was filtered to identify the CP ORF. The resulting ORFs were submitted to CD-HIT to identify a unique set of sequence for each genotype identified by Franzo and Segales (Niu et al., 2010). The sequence diversity and variability (entropy) for each genotype was calculated using the AL2CO routine built into the UCSF Chimera suite (Pettersen et al., 2004). The atomic coordinates of PCV2a, b, and d were used for the three genotypes studied (PDB entries 3JCI, 6DZU and 6OLA).
The PCV2 CP sequences were expanded by performing a protein Blast search with the sequence of PCV2d and the organism common name Porcine circovirus 2 (taxid:85708) filter (Johnson et al., 2008). A total of 1,966 PCV2 sequences were identified. Partial sequences, sequences with names containing “putative”, “P3”, “unknown”, and “P27.9” were manually removed. Sequence alignment was performed on the remaining sequences using MUSCLE with default parameters (Edgar, 2004). Sequences that generated gaps, possessed more than ten amino acids with a distinct sequence, or had no similar sequences were manually removed. This was done in order to remove spurious errors/artifacts that may have occurred during the sequencing process (i.e. artificial recombination during PCR with Taq polymerase (Martin et al., 2010)). Several rounds of alignment and deletion were performed to remove such sequences. The final set of sequences was combined with the pool of sequences identified by Franzo and Segales, and submitted to the CD-HIT server to attain a unique set of 1,278 sequences. The final round of alignment was performed with Clustal Omega with default parameters (Sievers et al., 2011). Evolutionary coupling calculations could not be successfully performed with the EVcouplings server (evfold.org) because of the limited range in the Expect (E) value present the sequence (i.e. the sequences are too similar). Consequently, plmc was used to generate coevolution and covariation within the sequences. The L2 lambda for fields and couplings used were 0.01 and 16.0, respectively, and a maximum number of 100 iterations were performed. The results were converted for visualization with EVzoom using MATLAB 2019 and the scripts provided by the plmc program (Hopf et al., 2017). The resulting matrix was visualized with EVzoom (Hopf et al., 2017), and structural covariance was visually confirmed with UCSF Chimera (Pettersen et al., 2004).
Measuring the interior content of the capsid.
The radial profile of the molecular envelope was calculated using the bradial program from the Bsoft package (Heymann, 2001). The radial profile in the interior of the capsid is quite strong and likely to arise from the PCV2 N-terminus (amino acids 1–36) and additional material. We assumed that cellular RNA was present in the capsid to stabilize the positive charge of the capsid interior (Tarasova et al., 2017). To calculate the amount of RNA present in the PCV2 capsid, we used the asymmetric molecular envelope of MS2 (EMD-8397) as a standard -as the ssRNA genome of MS2 is resolved in this molecular envelope (Dai et al., 2017). The intent is to adjust the threshold of the MS2 molecular envelope to account for 100% mass content in the capsid shell, then identify the threshold at which 100% mass content of the genome is accounted for. This threshold value is then used to measure the RNA content of the PCV2 molecular envelope after the PCV2 molecular envelope has been adjusted such that 100% mass content in the capsid shell is accounted for.
First, a 15 Å molecular envelope was generated for the MS2 capsid shell (PDB entry: 5TC1) with the UCSF Chimera vop command (Pettersen et al., 2004). The envelope was converted to the binary mask with the Relion 3.0 relion_mask_create program (Scheres, 2012a). The mask was applied to the molecular envelope using the Bsoft bmask program to extract the voxels pertaining to the capsid shell (Heymann, 2001). The EMAN volume program was used to rescale the map densities to a threshold of 1.0 for a mass of 2.54 MDa -the total mass of 180 CPs (T=3) and one maturation protein (Dai et al., 2017). The map densities of the unmasked molecular envelope was adjusted, via visual inspection with UCSF Chimera and scaling with Bsoft bimg program, such that its shell region was identical to the masked molecular envelope at threshold of 1.0 (2.54 MDa) (Heymann, 2001; Pettersen et al., 2004). The molecular envelope of the genome was isolated by applying an inverted mask of the capsid shell to remove the capsid shell, and a spherical mask to remove the solvent. The masks were applied with the Bsoft bmask program (Heymann, 2001). The mass content of the remaining volume was calculated using the EMAN volume program (Ludtke et al., 1999). The Oligo Calc: Oligonucleotide Properties Calculator was used to determine the molecular weight of the MS2 genome (GenBank entry: NC_001417.2) to be 1.10 MDa (Kibbe, 2007). From this value we determined that at a threshold of 0.781 the MS2 genome is fully accounted for in the EMD-8397 molecular envelope whose map distribution has been adjusted to account for 100% mass content of the capsid shell.
Following a similar procedure for the PCV2 molecular envelope (amino acids 43–231), we determined that a threshold of 0.781 corresponds to a mass of 730 kDa in the capsid interior. From this mass we subtracted the mass of the 60 N-termini (amino acids 1–42, 5.8 kDa) that are believed to be inside the capsid and contribute to the envelope. If we assume that the remaining mass of 384 kDa is from RNA, this equates to approximately 1,200 nt. Approximate MW of ssRNA = (#nucleotides × 320.5) + 159.
Measurement of nucleic acid content in purified VLP.
The ratio of nucleic acid to protein (260nm:280nm) was determined after correction for scattering at 340nm and 360nm according to the protocol established by Porterfield and Zlotnick (Porterfield and Zlotnick, 2010). Briefly, an adsorption scan of three sample were collected two times each from 640nm to 220nm using a VWR UV-1600 PC Spectrophotometer. The mass of the CP was calculated to be 28,057.04 Da using the amino acid sequence and the Expasy Protparam server (Gasteiger et al., 2005). The extinction coefficient of the CP was calculated using the same server (Pace et al., 1995), which yielded a value of 48,360 M−1 cm−1. A 260 nm / 280 nm ratio for pure protein (0.6) was used to calculate the extinction coefficient for RNA 29,016 M−1 cm−1.
Highlights.
PCV2 is the causative agent of a variety of swine illnesses, and economic impact.
PCV2 rapidly shifts genotypes, and structures from three genotypes are different.
We report an expression system capable of rapidly producing virus-like particles.
The capsid protein N-terminus senses and responds to the packaged nucleic acid.
A tetranucleotide within the capsid appears to stabilize the capsid.
Acknowledgement
Funds responsible for supporting these studies were provided by NIH National Institute of General medical Sciences and National institute of Allergy and Infectious Diseases (5SC1AI114843) to RK, by Grant Number 5G12MD007603-30 from the National Institute on Minority Health and Health Disparities to RK and PG, and by TechnoVax, Inc. and Huvepharma to JMG, KW, and BG. Data collection for the PCV2d VLP was performed at the Simons Electron Microscopy Center and National Resource for Automated Molecular Microscopy located at the New York Structural Biology Center, supported by grants from the Simons Foundation (349247), NYSTAR, and the NIH National Institute of General Medical Sciences (GM103310).
Refereces
- Afonine PA, Headd JJ, Terwilliger TC, Adams PC, 2013. Computational Crystallography Newsletter 4, 43–44. [Google Scholar]
- Beach NM, Meng XJ, 2012. Efficacy and future prospects of commercially available and experimental vaccines against porcine circovirus type 2 (PCV2). Virus Res 164, 33–42. [DOI] [PubMed] [Google Scholar]
- Breitbart M, Delwart E, Rosario K, Segales J, Varsani A, Ictv Report C, 2017. ICTV Virus Taxonomy Profile: Circoviridae. J Gen Virol 98, 1997–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai L, Ni J, Xia Y, Zi Z, Ning K, Qiu P, Li X, Wang B, Liu Q, Hu D, Yu X, Zhou Z, Zhai X, Han X, Tian K, 2012. Identification of an emerging recombinant cluster in porcine circovirus type 2. Virus Res 165, 95–102. [DOI] [PubMed] [Google Scholar]
- Correa-Fiz F, Franzo G, Llorens A, Segales J, Kekarainen T, 2018. Porcine circovirus 2 (PCV-2) genetic variability under natural infection scenario reveals a complex network of viral quasispecies. Sci Rep 8, 15469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowther RA, Berriman JA, Curran WL, Allan GM, Todd D, 2003. Comparison of the structures of three circoviruses: chicken anemia virus, porcine circovirus type 2, and beak and feather disease virus. J Virol 77, 13036–13041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X, Li Z, Lai M, Shu S, Du Y, Zhou ZH, Sun R, 2017. In situ structures of the genome and genome-delivery apparatus in a single-stranded RNA virus. Nature 541, 112–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhindwal S, Avila B, Feng S, Khayat R, 2019a. Porcine Circovirus 2 Uses a Multitude of Weak Binding Sites To Interact with Heparan Sulfate, and the Interactions Do Not Follow the Symmetry of the Capsid. J Virol 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhindwal S, Avila B, Feng S, Khayat R, 2019b. Porcine circovirus 2 uses a multitude of weak binding sites to interact with heparan sulfate, and the interactions do not follow the symmetry of the capsid. Journal of Virology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhindwal S, Feng S, Khayat R, 2019c. The Arginines in the N-terminus of the Porcine circovirus 2 virus like particles are responsible for disrupting the membranes at neutral and acidic pH Journal of Molecular Biology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan J, Yang D, Chen L, Yu Y, Zhou J, Lu H, 2019. Efficient production of porcine circovirus virus-like particles using the nonconventional yeast Kluyveromyces marxianus. Appl Microbiol Biotechnol 103, 833–842. [DOI] [PubMed] [Google Scholar]
- Duffy S, Shackelton LA, Holmes EC, 2008. Rates of evolutionary change in viruses: patterns and determinants. Nat Rev Genet 9, 267–276. [DOI] [PubMed] [Google Scholar]
- Edgar RC, 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32, 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis J, 2014. Porcine circovirus: a historical perspective. Vet Pathol 51, 315–327. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K, 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60, 2126–2132. [DOI] [PubMed] [Google Scholar]
- Fraczkiewicz R, Braun W, 1998. Exact and efficient analytical calculation of the accessible surface areas and their gradients for macromolecules. Journal of Computational Chemistry 19, 319–333. [Google Scholar]
- Franzo G, Cortey M, Olvera A, Novosel D, Castro AM, Biagini P, Segales J, Drigo M, 2015. Revisiting the taxonomical classification of Porcine Circovirus type 2 (PCV2): still a real challenge. Virol J 12, 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzo G, Segales J, 2018. Porcine circovirus 2 (PCV-2) genotype update and proposal of a new genotyping methodology. PLoS One 13, e0208585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A, 2005. Protein Identification and Analysis Tools on the ExPASy Server. Humana Press. [DOI] [PubMed] [Google Scholar]
- Goddard TD, Huang CC, Meng EC, Pettersen EF, Couch GS, Morris JH, Ferrin TE, 2018. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci 27, 14–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Lu Y, Huang L, Wei Y, Liu C, 2011. Identification of a new antigen epitope in the nuclear localization signal region of porcine circovirus type 2 capsid protein. Intervirology 54, 156–163. [DOI] [PubMed] [Google Scholar]
- Harrison SC, Olson AJ, Schutt CE, Winkler FK, Bricgone G, 1978. Tomato bushy stunt virus at 2.9Å resolution. Nature 279, 368–373. [DOI] [PubMed] [Google Scholar]
- Heymann JB, 2001. Bsoft: image and molecular processing in electron microscopy. J Struct Biol 133, 156–169. [DOI] [PubMed] [Google Scholar]
- Hopf TA, Ingraham JB, Poelwijk FJ, Schärfe CPI, Springer M, Sander C, Marks DS, 2017. Mutation effects predicted from sequence co-variation. Nature Biotechnology 35, 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopf TA, Scharfe CP, Rodrigues JP, Green AG, Kohlbacher O, Sander C, Bonvin AM, Marks DS, 2014. Sequence co-evolution gives 3D contacts and structures of protein complexes. Elife 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson M, Zaretskaya I, Raytselis Y, Merezhuk Y, McGinnis S, Madden TL, 2008. NCBI BLAST: a better web interface. Nucleic Acids Res 36, W5–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karuppannan AK, Opriessnig T, 2017. Porcine Circovirus Type 2 (PCV2) Vaccines in the Context of Current Molecular Epidemiology. Viruses 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khayat R, Brunn N, Speir JA, Hardham JM, Ankenbauer RG, Schneemann A, Johnson JE, 2011. The 2.3-angstrom structure of porcine circovirus 2. J Virol 85, 7856–7862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kibbe WA, 2007. OligoCalc: an online oligonucleotide properties calculator. Nucleic Acids Res 35, W43–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre DJ, Costers S, Van Doorsselaere J, Misinzo G, Delputte PL, Nauwynck HJ, 2008. Antigenic differences among porcine circovirus type 2 strains, as demonstrated by the use of monoclonal antibodies. J Gen Virol 89, 177–187. [DOI] [PubMed] [Google Scholar]
- Liu Z, Guo F, Wang F, Li TC, Jiang W, 2016. 2.9 A Resolution Cryo-EM 3D Reconstruction of Close-Packed Virus Particles. Structure 24, 319–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludtke SJ, Baldwin PR, Chiu W, 1999. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol 128, 82–97. [DOI] [PubMed] [Google Scholar]
- Mahe D, Blanchard P, Truong C, Arnauld C, Le Cann P, Cariolet R, Madec F, Albina E, Jestin A, 2000. Differential recognition of ORF2 protein from type 1 and type 2 porcine circoviruses and identification of immunorelevant epitopes. J Gen Virol 81, 1815–1824. [DOI] [PubMed] [Google Scholar]
- Martin DP, Lemey P, Lott M, Moulton V, Posada D, Lefeuvre P, 2010. RDP3: a flexible and fast computer program for analyzing recombination. Bioinformatics 26, 2462–2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzinger SR, Opriessnig T, Xiao CT, Catanzaro N, Beach NM, Slade DE, Nitzel GP, Meng XJ, 2016. A chimeric virus created by DNA shuffling of the capsid genes of different subtypes of porcine circovirus type 2 (PCV2) in the backbone of the non-pathogenic PCV1 induces protective immunity against the predominant PCV2b and the emerging PCV2d in pigs. Virology 498, 82–93. [DOI] [PubMed] [Google Scholar]
- Meerts P, Misinzo G, McNeilly F, Nauwynck HJ, 2005. Replication kinetics of different porcine circovirus 2 strains in PK-15 cells, fetal cardiomyocytes and macrophages. Arch Virol 150, 427–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo X, Li X, Yin B, Deng J, Tian K, Yuan A, 2019. Structural roles of PCV2 capsid protein N-terminus in PCV2 particle assembly and identification of PCV2 type-specific neutralizing epitope. PLoS Pathog 15, e1007562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu B, Fu L, Li W, Gao Y, Huang Y, 2010. CD-HIT Suite: a web server for clustering and comparing biological sequences. Bioinformatics 26, 680–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opriessnig T, Xiao CT, Gerber PF, Halbur PG, 2013. Emergence of a novel mutant PCV2b variant associated with clinical PCVAD in two vaccinated pig farms in the U.S. concurrently infected with PPV2. Vet Microbiol 163, 177–183. [DOI] [PubMed] [Google Scholar]
- Pace CN, Vajdos F, Fee L, Grimsley G, Gray T, 1995. How to measure and predict the molar absorption coefficient of a protein. Protein Sci 4, 2411–2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palinski R, Pineyro P, Shang P, Yuan F, Guo R, Fang Y, Byers E, Hause BM, 2017. A Novel Porcine Circovirus Distantly Related to Known Circoviruses Is Associated with Porcine Dermatitis and Nephropathy Syndrome and Reproductive Failure. J Virol 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y, Li P, Jia R, Wang M, Yin Z, Cheng A, 2018. Regulation of Apoptosis During Porcine Circovirus Type 2 Infection. Front Microbiol 9, 2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson AR, Opriessnig T, 2010. Epidemiology and horizontal transmission of porcine circovirus type 2 (PCV2). Anim Health Res Rev 11, 217–234. [DOI] [PubMed] [Google Scholar]
- Pei J, Grishin NV, 2001. AL2CO: calculation ofpositional conservation in a protein sequence alignment. Bioinformatics 17, 700–712. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE, 2004. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- Porterfield JZ, Zlotnick A, 2010. A simple and general method for determining the protein and nucleic acid content of viruses by UV absorbance. Virology 407, 281–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Carino C, Duffy C, Sanchez-Chardi A, McNeilly F, Allan GM, Segales J, 2011. Porcine circovirus type 2 morphogenesis in a clone derived from the l35 lymphoblastoid cell line. J Comp Pathol 144, 91–102. [DOI] [PubMed] [Google Scholar]
- Rosario K, Breitbart M, Harrach B, Segales J, Delwart E, Biagini P, Varsani A, 2017. Revisiting the taxonomy of the family Circoviridae: establishment of the genus Cyclovirus and removal of the genus Gyrovirus. Arch Virol 162, 1447–1463. [DOI] [PubMed] [Google Scholar]
- Rosario K, Schenck RO, Harbeitner RC, Lawler SN, Breitbart M, 2015. Novel circular single-stranded DNA viruses identified in marine invertebrates reveal high sequence diversity and consistent predicted intrinsic disorder patterns within putative structural proteins. Front Microbiol 6, 696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha D, Huang L, Bussalleu E, Lefebvre DJ, Fort M, Van Doorsselaere J, Nauwynck HJ, 2012. Antigenic subtyping and epitopes’ competition analysis of porcine circovirus type 2 using monoclonal antibodies. Vet Microbiol 157, 13–22. [DOI] [PubMed] [Google Scholar]
- Sarker S, Terron MC, Khandokar Y, Aragao D, Hardy JM, Radjainia M, Jimenez-Zaragoza M, de Pablo PJ, Coulibaly F, Luque D, Raidal SR, Forwood JK, 2016. Structural insights into the assembly and regulation of distinct viral capsid complexes. Nat Commun 7, 13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayers EW, Barrett T, Benson DA, Bolton E, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Federhen S, Feolo M, Fingerman IM, Geer LY, Helmberg W, Kapustin Y, Landsman D, Lipman DJ, Lu Z, Madden TL, Madej T, Maglott DR, Marchler-Bauer A, Miller V, Mizrachi I, Ostell J, Panchenko A, Phan L, Pruitt KD, Schuler GD, Sequeira E, Sherry ST, Shumway M, Sirotkin K, Slotta D, Souvorov A, Starchenko G, Tatusova TA, Wagner L, Wang Y, Wilbur WJ, Yaschenko E, Ye J, 2011. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 39, D38–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres SH, 2012a. A Bayesian view on cryo-EM structure determination. J Mol Biol 415, 406–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres SH, 2012b. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol 180, 519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider TD, Stephens RM, 1990. Sequence logos: a new way to display consensus sequences. Nucleic Acids Research 18, 6097–6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholz J, Suppmann S, 2017. A new single-step protocol for rapid baculovirus-driven protein production in insect cells. BMC Biotechnol 17, 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamsi Z, Moffett AS, Shukla D, 2017. Enhanced unbiased sampling of protein dynamics using evolutionary coupling information. Sci Rep 7, 12700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang SB, Jin YL, Jiang XT, Zhou JY, Zhang X, Xing G, He JL, Yan Y, 2009. Fine mapping of antigenic epitopes on capsid proteins of porcine circovirus, and antigenic phenotype of porcine circovirus type 2. Mol Immunol 46, 327–334. [DOI] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, Thompson JD, Higgins DG, 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7, 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suloway C, Shi J, Cheng A, Pulokas J, Carragher B, Potter CS, Zheng SQ, Agard DA, Jensen GJ, 2009. Fully automated, sequential tilt-series acquisition with Leginon. J Struct Biol 167, 11–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarasova E, Farafonov V, Khayat R, Okimoto N, Komatsu TS, Taiji M, Nerukh D, 2017. All-Atom Molecular Dynamics Simulations of Entire Virus Capsid Reveal the Role of Ion Distribution in Capsid’s Stability. J Phys Chem Lett 8, 779–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teras M, Viisileht E, Pahtma-Hall M, Rump A, Paalme V, Pata P, Pata I, Langevin C, Ruutel Boudinot S, 2018. Porcine circovirus type 2 ORF3 protein induces apoptosis in melanoma cells. BMC Cancer 18, 1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilas JL, Gomez-Blanco J, Conesa P, Melero R, Miguel de la Rosa-Trevin J, Oton J, Cuenca J, Marabini R, Carazo JM, Vargas J, Sorzano COS, 2018. MonoRes: Automatic and Accurate Estimation of Local Resolution for Electron Microscopy Maps. Structure 26, 337–344 e334. [DOI] [PubMed] [Google Scholar]
- Wang N, Zhan Y, Wang A, Zhang L, Khayat R, Yang Y, 2016. In silico analysis of surface structure variation of PCV2 capsid resulting from loop mutations of its capsid protein (Cap). J Gen Virol 97, 3331–3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei C, Zhang M, Chen Y, Xie J, Huang Z, Zhu W, Xu T, Cao Z, Zhou P, Su S, Zhang G, 2013. Genetic evolution and phylogenetic analysis of porcine circovirus type 2 infections in southern China from 2011 to 2012. Infect Genet Evol 17, 87–92. [DOI] [PubMed] [Google Scholar]
- Xiao CT, Halbur PG, Opriessnig T, 2015. Global molecular genetic analysis of porcine circovirus type 2 (PCV2) sequences confirms the presence of four main PCV2 genotypes and reveals a rapid increase of PCV2d. J Gen Virol 96, 1830–1841. [DOI] [PubMed] [Google Scholar]
- Zhang H, Qian P, Liu L, Qian S, Chen H, Li X, 2014. Virus-like particles of chimeric recombinant porcine circovirus type 2 as antigen vehicle carrying foreign epitopes. Viruses 6, 4839–4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Gu Y, Fan H, 2016. Effect of impurities and post-experimental purification in SAD phasing with serial femtosecond crystallography data. Acta Crystallogr D Struct Biol 72, 789–794. [DOI] [PubMed] [Google Scholar]
- Zheng SQ, Palovcak E, Armache JP, Verba KA, Cheng Y, Agard DA, 2017. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods 14, 331–332. [DOI] [PMC free article] [PubMed] [Google Scholar]