Abstract
Background
Bisphenol A is an important organic chemical as an intermediate, final and inert ingredient in manufacturing of many important products like polycarbonate plastics, epoxy resins, flame retardants, food–drink packaging coating, and other. BPA is an endocrine disruptor compound that mimics the function of estrogen causing damage to reproductive organs. Bacterial degradation has been consider as a cost effective and eco-friendly method for BPA degradation compared with physical and chemical methods. This study aimed to isolate and identify bacterial strain capable to degrade and tolerate high concentrations of this pollutant, studying the factors affecting the degradation process and study the degradation mechanism of this strain.
Results
YC-AE1 is a Gram negative bacterial strain isolated from soil and identified as Pseudomonas putida by 16S rRNA gene sequence and BIOLOG identification system. This strain found to have a high capacity to degrade the endocrine disruptor Bisphenol A (BPA). Response surface methodology using central composite design was used to statistically optimize the environmental factors during BPA degradation and the results obtained by significant model were 7.2, 30 °C and 2.5% for optimum initial pH, temperature and inoculum size, respectively. Prolonged incubation period with low NaCl concentration improve the biodegradation of BPA. Analysis of variance (ANOVA) showed high coefficient of determination, R2 and Adj-R2 which were 0.9979 and 0.9935, respectively. Substrate analysis found that, strain YC-AE1 could degrade a wide variety of bisphenol A-related pollutants such as bisphenol B, bisphenol F, bisphenol S, Dibutyl phthalate, Diethylhexyl phthalate and Diethyl phthalate in varying proportion. Pseudomonas putida YC-AE1 showed high ability to degrade a wide range of BPA concentrations (0.5–1000 mg l− 1) with completely degradation for 500 mg l− 1 within 72 h. Metabolic intermediates detected in this study by HPLC-MS were identified as 4,4-dihydroxy-alpha-methylstilbene, p-hydroxybenzaldeyde, p-hydroxyacetophenone, 4-hydroxyphenylacetate, 4-hydroxyphenacyl alcohol, 2,2-bis(4-hydroxyphenyl)-1-propanol, 1,2-bis(4-hydroxyphenyl)-2-propanol and 2,2-bis(4-hydroxyphenyl) propanoate.
Conclusions
This study reports Pseudomonas putida YC-AE1 as BPA biodegrader with high performance in degradation and tolerance to high BPA concentration. It exhibited strong degradation capacity and prominent adaptability towards a wide range of environmental conditions. Moreover, it degrades BPA in a short time via two different degradation pathways.
Keywords: Bisphenol A, Biodegradation, Pseudomonas putida, Endocrine disruptor, Degradation pathway, Response surface methodology
Background
Xenobiotic endocrine disruptor compounds are environmental pollutants that have health and environmental harms, raising important concerns [1, 2]. Bisphenol A (BPA), 2,2-bis(4-hydroxyphenyl) propane is a well-known endocrine disruptor that is showing a highly estrogenic activity and acute toxicity [3]. It can bind to receptors for other hormonally mediated processes and cause endocrine disruptions. EPA and the U.S. Food and Drug Administration established a safe reference dose (RfD) for humans at 50 μg/kg/day [4]. Since hormones levels in the human body are present in biologically active concentrations, exposure to these exogenous chemicals even in low doses can disrupt the proper functioning of the body’s endocrine system. In addition, dose scaling is valid for agents that follow linear dose-response relationships, many endocrine-disruptors, like their hormonal counterparts, demonstrate a non-monotonic dose-response curve and in this case, lower doses are as relevant as higher doses [5–7]. Chemically, BPA is an organic compound consisting of two phenolic rings connected by a carbon bridge with two methyl group attached to this bridge [8].
The production of BPA has increased in the last few decades because of its widespread application in many industries [9] to reach the high global demand which was approximately 6.5 million tons in 2012 [10, 11]. BPA is used as a major component (as plasticizer) for various consumer products including detergents, phenol resins, epoxy resins, polycarbonates, polyesters, polyacrylates, plastic packing materials and lacquer coatings on food cans [11]. There are large number of studies that reported many damaging effects of BPA for both human and animal organs [12]. Detectable concentrations of BPA were reported in amniotic fluid, fetal circulation and placental tissue of pregnant women [13]. Many studies reported that, exposure of aquatic animals to BPA cause reproductive toxicity like changing in sex hormone level which affected the development of ovary in rare minnow Gobiocypris rarus [14], decreased sperm count, female biased sex ratio, and increased malformation and mortality in zebra fish [15]. BPA was detected in a wide range of environments including landfill leachates [16], industrial wastewater [17], source water and drinking water [18], rivers, seas, and soils [19–22]. Therefore, special attention has been given to BPA toxicity in these environments [23]. The most common methods to remove pollutants from the environment included photodegradation, photoelectrocatalytic oxidation, oxidation [17] and biodegradation [24]. Among all these methods, bacterial biodegradation of BPA was the most important one [25] because of low costs and less disruption of the contaminated environment when compared to other cleanup methods. Many researchers reported several BPA-degrading bacteria and their degrading performance Achromobacter xylosoxidans strain B-16 [25], Pseudomonas sp. and Pseudomonas putida strain [26], Bacillus megaterium Strain ISO-2 [27], Sphingomonas bisphenolicum strain AO1 [28] and Pseudomonas sp. and Sphingomonas sp. [10].
Microorganisms and their products are very important in industrial processes such as in synthesis of a number of products and fermentation processes. Their use is influenced by environmental factors such as a temperature, pH, nutrients and as must be prior optimized before application. The optimization by one factor at time (OFAT) involves changing in one factor while other factors or variables at certain levels are fixed [29]. This method of optimization is time consuming especially with large number of variables in addition to, ignoring the interactions between different variables. These disadvantages of single-factor experiments can be overcome by using statistical optimization which allows all interactions of all variables and levels [30, 31].
In this study, a highly BPA tolerant strain with high BPA biodegradation capacity was isolated from soil sample beside the electronic wastes collection area in Guangdong province, China and identified as Pseudomonas putida YC-AE1, the environmental conditions affecting BPA degradation were studied using statistical methods and discussed. The ability of strain YC-AE1 to degrade other pollutants related to BPA was also studied. The metabolic intermediate compounds detected during the biodegradation process were identified and biodegradation pathways were proposed.
Results
Isolation and identification of BPA degrading strain
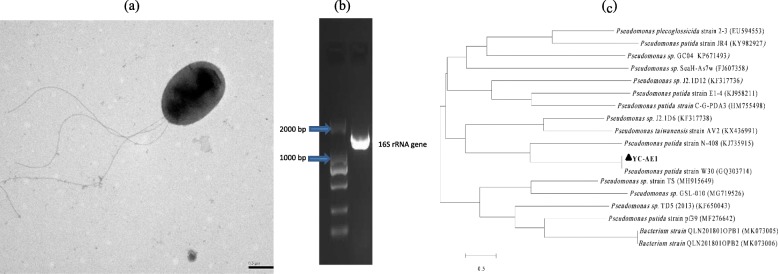
YC-AE1 strain isolated from the soil sample of Guangdong province showed high degradation and tolerance to BPA. It is a rod-shaped, flagellated, Gram-negative bacterium. The amplified 16S rRNA gene sequence (1439 bp) was deposited in Gen Bank (accession number, MK318658) and related strains were obtained by BLAST. The similarity between YC-AE1 and others was detected by constructing a phylogenetic tree (Fig.1) and the most closely related one was Pseudomonas putida strain W30 with 99% similarity. The BIOLOG tests was performed and showed consistent results with the 16S rRNA gene (PROB = 97.2%, SIM = 0.849, and DIST = 1.818 with Pseudomonas putida). According to morphology, BIOLOG tests and 16S rRNA gene sequencing, our strain YC-AE1 identified as Pseudomonas putida.
Fig. 1.
Identification of YC-AE1, (a) TEM investigation showing cell shape and lophotrichous flaggelation (b) amplified 16S rRNA gene (about 1500 bp) comparing with 2000 bp DNA ladder, (c) phylogenetic tree of Pseudomonas putida YC-AE1 with closely related strains
Statistical optimization of BPA degradation using RSM
The combined five factors with their levels and obtained experimental results are presented in Table 1, the highest BPA degradation (95%) was observed in the result obtained by the design (run No. 21). The design was analyzed by regression analysis resulted in a significant model with the following regression equation:
Table 1.
Experimental design (conditions and responses) for BPA degradation by Pseudomonas putida YC-AE1
| Run No. | A | B | C | D | E | BPA degradation (%)a |
|---|---|---|---|---|---|---|
| Initial pH (values) | NaCl Conc. (%) | Incubation period (hours) | Inoculum size (%) | Incubation temp. (°C) | ||
| 7 | 5.00 (−2) | 0.75 (0) | 20 (0) | 2.5 (0) | 31 (0) | 2.2 |
| 5 | 6.13 (−1) | 1.13 (+ 1) | 15 (− 1) | 3.25 (+ 1) | 37 (+ 1) | 2.7 |
| 8 | 6.13 (−1) | 1.13 (+ 1) | 15 (−1) | 1.75 (− 1) | 25 (− 1) | 7.8 |
| 10 | 6.13 (−1) | 0.38 (− 1) | 25 (+ 1) | 1.75 (− 1) | 25 (− 1) | 81.7 |
| 11 | 6.13(− 1) | 1.13 (+ 1) | 25 (+ 1) | 1.75 (− 1) | 37 (+ 1) | 0 |
| 12 | 6.13 (−1) | 0.38 (− 1) | 15 (− 1) | 1.75 (− 1) | 37 (+ 1) | 0 |
| 14 | 6.13 (− 1) | 1.13 (+ 1) | 25 (+ 1) | 3.25 (+ 1) | 25 (− 1) | 6.4 |
| 28 | 6.13 (−1) | 0.38 (− 1) | 25 (+ 1) | 3.25 (+ 1) | 37 (+ 1) | 13.38 |
| 29 | 6.13 (−1) | 0.38 (− 1) | 15 (− 1) | 3.25 (+ 1) | 25 (− 1) | 93.5 |
| 1 | 7.25 (0) | 0.75 (0) | 10 (−2) | 2.5 (0) | 31 (0) | 2 |
| 2 | 7.25 (0) | 1.5 (+ 2) | 20 (0) | 2.5 (0) | 31 (0) | 39.4 |
| 3 | 7.25 (0) | 0.75 (0) | 20 (0) | 2.5 (0) | 31 (0) | 77 |
| 4 | 7.25 (0) | 0.75 (0) | 20 (0) | 2.5 (0) | 19 (−2) | 17.8 |
| 15 | 7.25 (0) | 0.75 (0) | 20 (0) | 2.5 (0) | 31 (0) | 77.2 |
| 16 | 7.25 (0) | 0.75 (0) | 30 (+ 2) | 2.5 (0) | 31 (0) | 90 |
| 19 | 7.25 (0) | 0.75 (0) | 20 (0) | 4 (+ 2) | 31 (0) | 63.2 |
| 20 | 7.25 (0) | 0 (−2) | 20 (0) | 2.5 (0) | 31 (0) | 90 |
| 22 | 7.25 (0) | 0.75 (0) | 20 (0) | 2.5 (0) | 31 (0) | 77.3 |
| 23 | 7.25 (0) | 0.75 (0) | 20 (0) | 2.5 (0) | 43 (+ 2) | 0 |
| 24 | 7.25 (0) | 0.75 (0) | 20 (0) | 1 (−2) | 31 (0) | 17.5 |
| 6 | 8.38 (+ 1) | 0.38 (−1) | 25 (+ 1) | 1.75 (−1) | 37 (+ 1) | 1.7 |
| 9 | 8.38 (+ 1) | 1.13 (+ 1) | 15 (−1) | 3.25 (+ 1) | 25 (− 1) | 17.5 |
| 13 | 8.38(+ 1) | 1.13 (+ 1) | 25 (+ 1) | 1.75 (−1) | 25 (− 1) | 15 |
| 18 | 8.38(+ 1) | 0.38 (−1) | 15 (−1) | 3.25 (+ 1) | 37 (+ 1) | 7 |
| 21 | 8.38 (+ 1) | 0.38 (−1) | 25 (+ 1) | 3.25 (+ 1) | 25 (− 1) | 95 |
| 25 | 8.38 (+ 1) | 1.13 (+ 1) | 15 (−1) | 1.75 (− 1) | 37 (+ 1) | 0 |
| 26 | 8.38 (+ 1) | 1.13 (+ 1) | 25 (+ 1) | 3.25 (+ 1) | 37 (+ 1) | 0 |
| 27 | 8.38 (+ 1) | 0.38 (−1) | 15 (− 1) | 1.75 (− 1) | 25 (− 1) | 78.3 |
| 17 | 9.5 (+ 2) | 0.75 (0) | 20 (0) | 2.5 (0) | 31 (0) | 60 |
aAll response (BPA degradation percentage) values are shown as means for three independent experiments
where, A, B, C, D and E are the actual values of initial pH, NaCl concentration, incubation period, inoculum size and Incubation temperature, respectively. Regression analysis for the biodegradation shown in Table 2, indicate that the model used in our study was highly significant with very low P-value (0.0001) and high F-value (229.53). As shown in Table 2, all linear and quadratic variables were highly significant with p-value close to (0.00) indicating the model success. Two of variable interactions; pH and NaCl concentration (AB) and pH and incubation temperature (AE) were slightly non-significant with p-values 0.0944 and 0.0760 respectively, indicating the interactions were not favorable to the response while, the other two variables, NaCl concentration and inoculum size (BD) and NaCl concentration and incubation temperature (BE) were highly significant. The model fitness was expressed by the coefficient of determination, R2 and Adj-R2 which were 0.9979 and 0.9935, respectively.
Table 2.
Regression analysis for the biodegradation of BPA by Pseudomonas putida YC-AE1
| Source | Coefficient | Sum of Squares | Degrees of freedom | Mean Square | F Value |
P value Prob > F |
|---|---|---|---|---|---|---|
| Model | 38,370.26 | 19 | 2019.48 | 229.53 | < 0.0001 | |
| pH, A | 14.45 | 1670.42 | 1 | 1670.42 | 189.86 | < 0.0001 |
| NaCl Conc., B | −12.65 | 1280.18 | 1 | 1280.18 | 145.50 | < 0.0001 |
| Incubation period, C | 22 | 3872 | 1 | 3872 | 440.09 | < 0.0001 |
| Inoculum size, D | 11.425 | 1044.24 | 1 | 1044.24 | 118.68 | < 0.0001 |
| Incubation temp., E | −4.45 | 158.42 | 1 | 158.42 | 18.00 | 0.0022 |
| AB | 1.38625 | 30.74 | 1 | 30.74 | 3.49 | 0.0944 |
| AE | −1.48625 | 35.34 | 1 | 35.34 | 4.01 | 0.0760 |
| BD | −2.71125 | 117.61 | 1 | 117.61 | 13.36 | 0.0053 |
| BE | 17.65125 | 4985.06 | 1 | 4985.06 | 566.60 | < 0.0001 |
| A2 | −11.9997 | 3532.64 | 1 | 3532.64 | 401.52 | < 0.0001 |
| B2 | −3.59973 | 317.90 | 1 | 317.90 | 36.13 | 0.0002 |
| C2 | −8.27473 | 1679.82 | 1 | 1679.82 | 190.92 | < 0.0001 |
| D2 | −9.68723 | 2302.26 | 1 | 2302.26 | 261.67 | < 0.0001 |
| E2 | −17.5497 | 7556.09 | 1 | 7556.09 | 858.82 | < 0.0001 |
| A2B | −7.42375 | 293.93 | 1 | 293.93 | 33.40 | 0.0003 |
| A2C | −21.6013 | 2488.60 | 1 | 2488.60 | 282.85 | < 0.0001 |
| A2D | −8.23875 | 362.01 | 1 | 362.01 | 41.14 | 0.0001 |
| A2E | −18.7013 | 1865.26 | 1 | 1865.26 | 212.00 | < 0.0001 |
| AB2 | −13.8863 | 1028.41 | 1 | 1028.41 | 116.89 | < 0.0001 |
| Residual | 79.18 | 9 | 8.79 | |||
| Lack of Fit | 79.13 | 7 | 11.30 | 484.50 | 0.0021 | |
| Pure Error | 0.04 | 2 | 0.02 | |||
| Total | 38,449.44 | 28 | R2 = 0.9979 | Adj-R2 = 0.9935 | ||
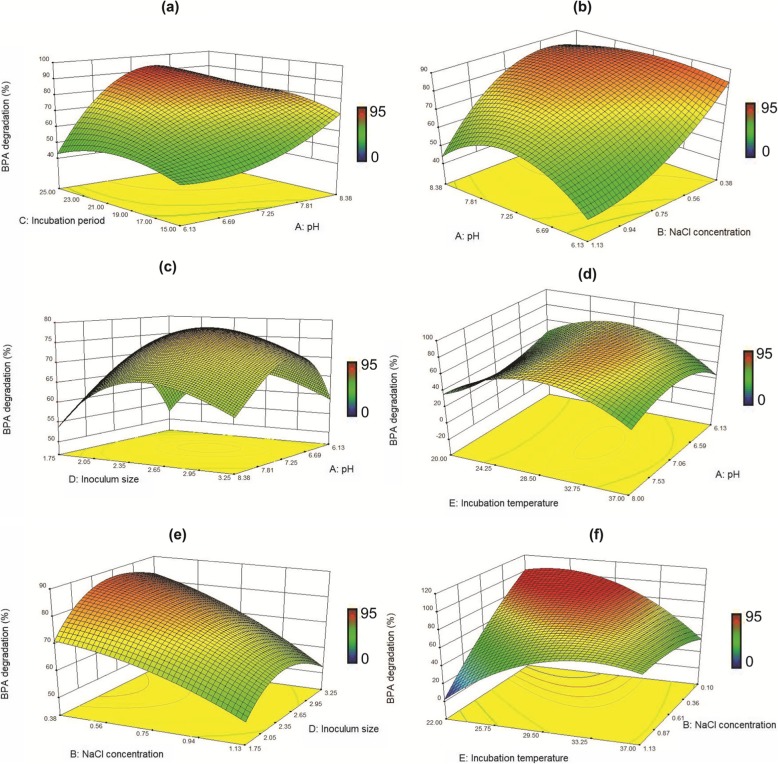
To determine the optimum values of BPA degradation conditions for obtaining the highest and fastest degradation rate, three-dimensional (3-D) response surface curves were plotted as shown in Fig. 2. These results showed that an optimum was observed near the central value of pH, temperature and inoculum size. Pseudomonas putida YC-AE1 could grow and degrade BPA over a wide range of pH values with optimum value 7.2, Fig. 2a-d. The biodegradation of BPA was highly affected by the incubation period, it can be easily observed this effect from the high positive coefficient value (+ 22) of the variable as shown in Table 2. From Fig. 2a, prolonged incubation period was shown to increase the degradation of BPA by our strain YC-AE1.
Fig. 2.
Response surface plots showing the effect of incubation period and pH (a); pH and NaCl concentration (b); inoculum size and pH (c); incubation temperature and pH (d); NaCl concentration and inoculum size (e); incubation temperature and NaCl (f)
The effect of temperature on BPA degradation by YC-AE1 showed in Fig. 2d. There was a gradual increase in BPA degradation by YC-AE1 by increasing the temperature from 15 to 25 °C and the maximum degradation was observed between 25 and 30 °C. The degradation then decreased beyond increasing temperature degrees. As shown in Table 2 the addition of NaCl to the degradation medium (TEM) has a reverse effect on BPA degradation, as described by a negative coefficient value (− 12). Data illustrated in Fig. 2b, e, f showed a sever decrease in BPA degradation by YC-AE1 with increasing NaCl concentration. The optimum inoculum size for BPA degradation was 2.5% and this can be clearly observed in the data plotted in Fig. 2c, e.
Substrate utilization test
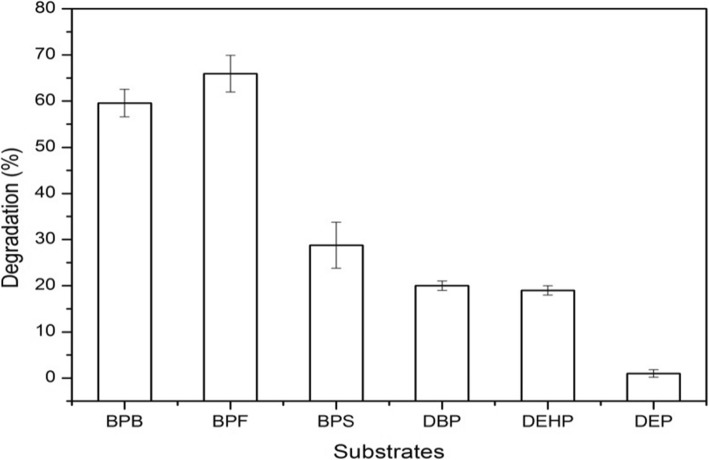
The capability of Pseudomonas putida strain YC-AE1 to degrade six different organic pollutants (BPB, BPF, BPS, DBP, DEP and DEHP) was examined. Data presented in Fig. 3 showed that, the YC-AE1 has an ability to degrade all examined pollutants in varying proportions. Pseudomonas putida YC-AE1 showed high ability to degrade about 60 and 67% of bisphenol B and F (100 mg l− 1), respectively, while lower degradation was observed in both bisphenol S, DBP and DEHP with about 30, 20 and 18% respectively.
Fig. 3.
Degradation efficiency for different pollutants (100 mg l−1) by Pseudomonas putida YC-AE1. Error bars indicate the standard deviation obtained in three independent experiments
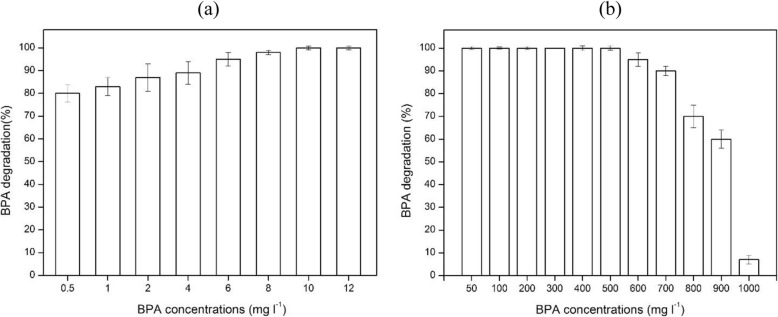
High and low BPA concentration for efficient biodegradation
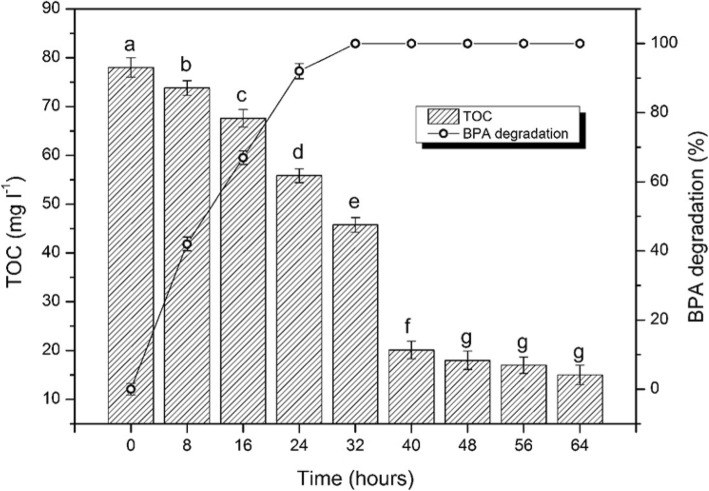
The degradation rates in low and high BPA concentration are demonstrated in Fig. 4a, b after 15 and 72 h cultivation, respectively. As shown in Fig. 4a, the strain was able to degrade more than 80% of 0.5 mg l− 1 BPA then, the degradation gradually increased with increasing the concentration to reach 100% with 10 and 12 mg l− in that short incubation time (15 h). On the other hand of high concentrations (Fig. 4b, the strain YC-AE1 was able to degrade 100% of BPA (50–500 mg l− 1). However, when the BPA concentration increased (600, 700, 800, 900 and 1000 mg l− 1) the degradation rate decreased to (95, 90, 70, 60 and 7%), respectively. Further incubation of strain YC-AE1 for another 2 days resulted in complete degradation of BPA (600–1000 mg l− 1). For detection of BPA mineralization, TOC experiment was performed, and the data obtained illustrated in Fig. 5. There was a dramatical significant decreasing in TOC starting from 78 to 20 mg l− 1 after 0 and 32 h incubation, respectively, (i.e., 75% depletion in TOC). With the degradation time further going on, there was non-significant decreasing in TOC to reach finally 82% at 64 h incubation.
Fig. 4.
Ability of Pseudoonas putida YC-AE1 to degrade Low (a) and High (b) BPA concentrations. Error bars indicate the standard deviation obtained in three independent experiments
Fig. 5.
Total organic carbon depletion and BPA degradation by Pseudomonas putida YC-AE1. The different letters represent significant differences at P ≤ 0.05
Metabolic intermediates and metabolic pathway
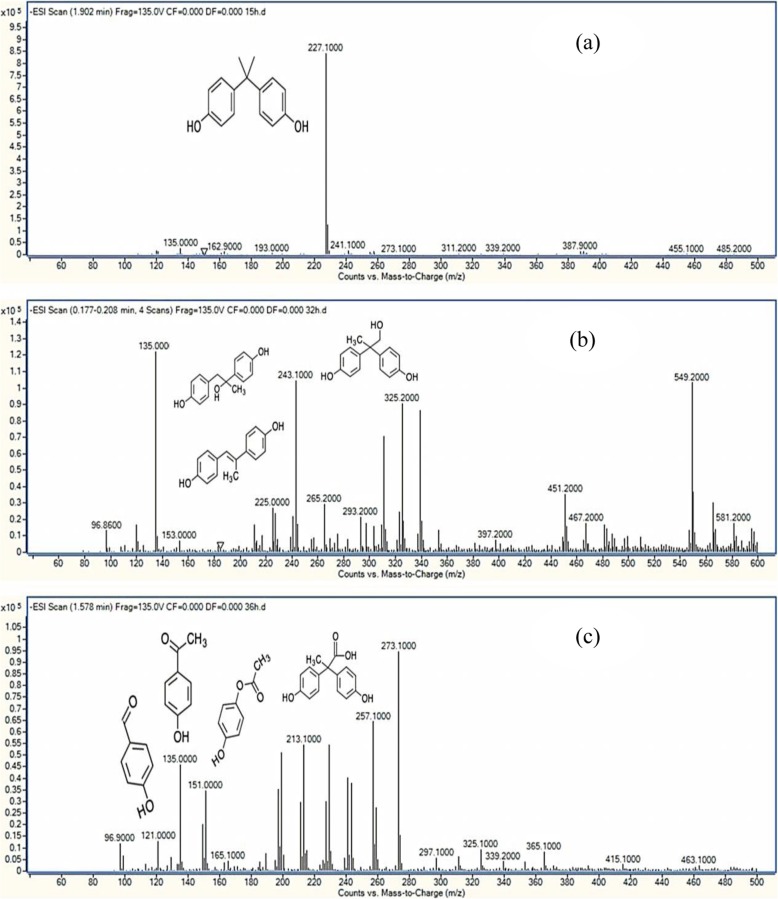
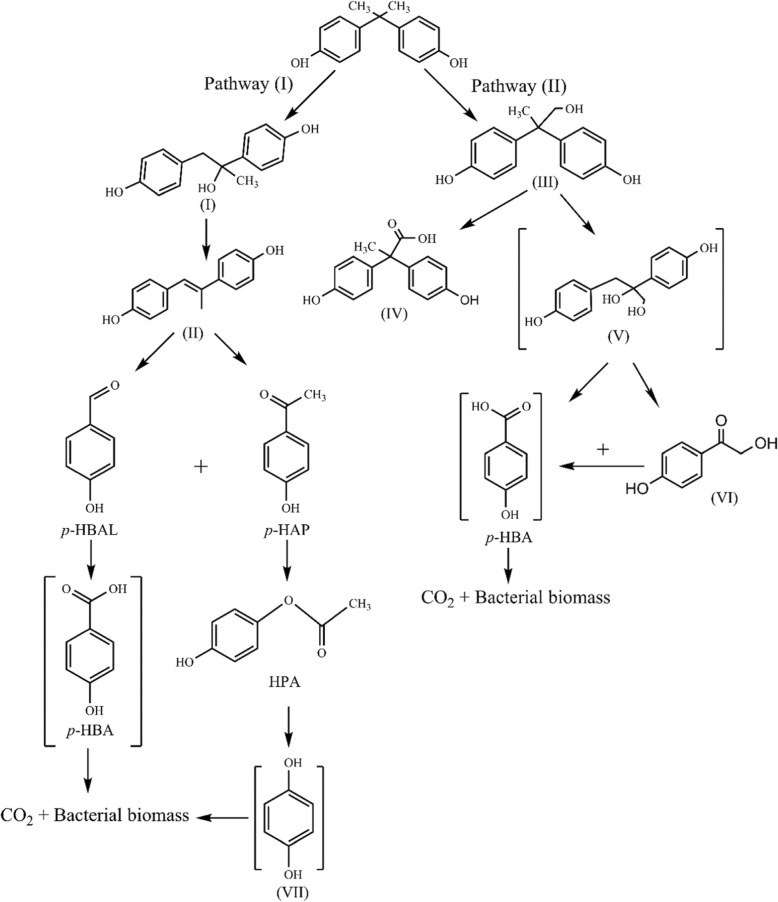
Detection of metabolic intermediates during BPA degradation with optimum conditions (200 mg l− 1 BPA, pH 7.2, 30 °C and 2.5% inoculum size) was performed. Based on the mass spectra (LC-MS) as shown in Fig. 6, eight compounds relating to BPA degradation were identified as following, BPA (m/z 227.1), 4,4-Dihydroxy-alpha-methylstilbene (m/z 225), p-hydroxybenzaldeyde (p-HBAL) (m/z 122), p-hydroxyacetophenone (p-HAP) (m/z 136), 4-hydroxyphenylacetate (HPA) and 4-Hydroxyphenacylalcohol (same m/z, 152), 2,2-bis(4-hydroxyphenyl)-1-propanol and 1,2-bis (4-hydroxyphenyl)-2-propanol (same m/z, 244) and 2,2-bis (4-hydroxyphenyl) propanoate (m/z 258). Based on the intermediates obtained during the biodegradation of BPA, the proposed pathway for BPA biodegradation by strain YC-AE1 is represented in Fig. 7. There were two different proposed pathways for BPA degradation by YC-AE1, both of them start by hydroxylation for BPA to form 1,2-Bis(4-hydroxyphenyl)-2-propanol and 2,2-Bis(4-hydroxyphenyl)-1-propanol in pathway (I) and (II), respectively. In pathway (I), 1,2-Bis(4-hydroxyphenyl)-2-propanol dehydrated to 4,4-Dihydroxy-alpha-methylstilbene [32]. The previously mentioned compound was further oxidized to form p- HBAL and p- HAP. p- HAP was metabolized to HPA and then to HQ. Both of hydroxybenzoic acid (HBA) and HQ were assumed mineralized to carbon dioxide (CO2) and bacterial biomass through benzoate degradation pathway. HBA and HQ were not detected and presumed in the pathway. In pathway (II), 2,2-Bis(4-hydroxyphenyl)-1-propanol is metabolized to form 2,2-bis (4-hydroxyphenyl) propanoate and 2,3-Bis(4-hydroxyphenyl)-1,2-propanediol. The intermediate (2, 3-Bis (4-hydroxyphenyl)-1,2-propanediol) was further metabolized through many steps (as shown in Fig. 7) to form finally HBA which mineralized to CO2 and bacterial biomass through benzoate degradation pathway.
Fig. 6.
Mass spectra from HPLC-MS during BPA degradation by Pseudomonas putida YC-AE1 and detected intermediate compounds, BPA (a), 4,4-Dihydroxy-alpha-methylstilbene, 1,2-bis (4-hydroxyphenyl)-2-propanol and 2,2-bis(4-hydroxyphenyl)-1-propanol (b), 2,2-bis (4-hydroxyphenyl) propanoate, p-HBAL, p-HAP and HPA (c)
Fig. 7.
Proposed pathway for BPA degradation by Pseudomonas putida YC-AE1, 1,2-bis(4-hydroxyphenyl)-2-propanol (I), 4,4-dihydroxy-alpha-methylstilbene (II), 2,2-bis(4-hydroxyphenyl)-1-propanol (III), 2,2-bis (4-hydroxyphenyl) propanoate (IV), 2,3-bis(4-hydroxyphenyl)-1,2-propanediol (V), 4-hydroxyphenacyl alcohol (VI), hydroquinone (HQ) (VII). The compounds in parentheses were not detected in our study but assumed to be produced
Discussion
There currently is increasing interest in endocrine disruptors, and their biodegradation by bacteria has been extensively studied. BPA is a known endocrine disruptor that displays estrogenic activity and acute toxicity [3]. Finding new bacterial strains with strong degradation capacity and prominent adaptability towards a wide range of environmental conditions is of great importance. In our study, one bacterial strain showed excellent biodegradation performance was isolated and identified as Pseudomonas putida strain YC-AE1 according to identification methods. YC-AE1 could degrade 200 mgl− 1 within 20 h under optimum environmental factors. For optimizing the factors affecting the biodegradation process, RSM was performed using CCD. RSM is used to determine the combined effect of several variables and their interactions. A statistical optimization approach using CCD was used to study the linear, quadratic and interactive effects of various parameters on BPA degradation by Pseudomonas putida strain YC-AE1. The F-value and probability value (p-value) are tools for evaluating the significance of each of the parameters in the model equation. The pattern of interactions between the variables is indicated by these coefficients. The larger F- value and the smaller P-value are an indication of the high significance of the corresponding coefficient [30]. The high value of R2 (so close to 1) supports the accuracy of the model and demonstrates a good correlation between actual and predicted values of BPA degradation. The significance of the model and high R2, support the model to predict responses. For microbial effective process, it is necessary to monitor and control parameters that affect the process like, inoculum volume, salt concentration, pH, temperature, incubation period, etc. Maximum BPA degradation was observed at pH of 7.2 and our results was different from that obtained by [25, 33] which reported that maximum BPA degradation was shown at pH 7 by Achromobacter xylosoxidans strain B-16 and Arthrobacter sp. YC-RL1.
The inoculums to be used in degradation medium must be in a healthy and active state moreover being of optimum size. Such conditions possibly minimize the length of the log phase. Lower inoculum volumes decrease the degradation efficiency of YC-AE1 and, this is may be due to low number of bacterial cells that decrease the amount of enzymes for biodegradation. Also, inoculum size higher than the optimum value (2.5%) had a little increasing effect on BPA degradation by YC-AE1, and this may be because the reduction of dissolved oxygen and increased competition towards nutrients [34]. Zhang et al. [25] reported that, increasing the size of inoculum leads to increase the degradation of BPA by Achromobacter xylosoxidans strain B-16 isolated from compost leachate of municipal solid waste.
Temperature is one of the most important parameters that affect any microbial process. The growth rate of microorganisms becomes slow below or above the optimum growth temperature because of a reduced rate of cellular production [35]. Enzyme thermal stability and activity is correlated to an organism’s growth temperature and also, degradation is an enzyme-controlled activity hence as the temperature increases, the cellular growth and physiological functions increase to an optimum value [36]. Fouda [37] reported a relatively high optimum temperature (35–40 °C) for BPA biodegradation by Klebsiella pneumoniae J2 and Enterobacter asburiae L4. The biodegradation of BPA was greatly reduced by addition of NaCl to the medium component and this result may be due to the effect of NaCl on the growth curve of our strain by elongating the lag phase and delaying the degradation process [38, 39] and this explains why the strain regains its ability to degrade BPA in the presence of salt by further incubation for another 2 days resulted in degradation of 70% of BPA (Data not shown).
It is very important for bacterial strain intended to use in natural remediation to has ability to degrade many pollutants or at least many derivatives of one pollutant. For that purpose, our strain Pseudomonas putida YC-AE1 was examined to degrade and tolerate 6 different BPA-related organic pollutants. BPS was the hardest one in the examined bisphenols to degrade because it contains S=O double chemical bond, which gives the structure chemical durability. The steric effects between the substrates and the responsible enzymes declines from BPB to BPF, which may be the reason why BPF is easier to degrade. The lowest degradation ability was shown with DEP with degradation rate not more than 3%. Many Pollutants were reported to be degraded by Pseudomonas sp by researchers. For example, phenols [40], phenolics like pentachlorophenol [41] and Catechol [42]. Under normal environmental conditions, the concentration of environmental pollutants is always very low compared with that examined in vitro [43, 44] and these pose many problems for bioremediation process because, the concentration of pollutants is too low to sustain microbial growth and ensure the accessibility to microbe and even more than that to induce metabolic genes [33]. Therefore, determining the survival ability of Pseudomonas putida YC-AE1 against both high and low BPA concentrations is important. Strain YC-AE1 could survive in both conditions and remain active in all BPA concentrations. This performance and ability of strain YC-AE1 to degrade and tolerate these extremely low and high concentrations are important for application in BPA bioremediation especially with fast degradation rate (200 mg l− 1 in 20 h). This quality makes strain YC-AE1 a promising bacterium compared with other reported strains. Suyamud et al. [27] reported Bacillus megaterium strain ISO-2, which can degrade 5 mg l− 1 of BPA within 72 h on mineral salt medium supplemented with yeast extract as co-substrate. Sphingomonas bisphenolicum strain AO1 was reported to degrade 100 mg l− 1 BPA to undetectable level within 48 h in minimum medium with 1% glucose [28].
Biodegradation of BPA and its metabolic intermediates could not fully support the mineralization of BPA, TOC experiment could show the mineralization rate of BPA. Although, faster BPA mineralization rate has generally been reported for microbial communities than the culture of BPA-degrading lonely strains [45], interestingly, our strain Pseudomonas putida YC-AE1 lonely demonstrated high BPA mineralization compared with microbial communities. Although, the TOC depletion percentage in our experiment (82% after 64 h) by Pseudomonas putida YC-AE1 was closely similar to that reported by Yu et al. [45] that used co-culture of sphingomonas sp. (Sph-2) and Pseudomonas sp. (84% after 72 h), Surprisingly, our experiment was conducted with 100 mg l− 1 compared with 50 mg l− 1 BPA by Yu et al. [45], and these results reflect the high degradation and mineralization rate of our strain YC-AE1.
The intermediates detected in our study have a good consistency with those reported by Eio et al. [32]. Das et al. [46] reported many different intermediate compounds detected during BPA degradation by recombinant laccase from Bacillus sp. GZB. Three different pathways were proposed based on three intermediates, Hydroxybenzaldeyde, p-hydroxybenzoic acid and p-hydroquinone (HQ) detected during biodegradation of BPA by Achromobacter xylosoxidans strain B-16 isolated from compost leachate [25]. Based on the intermediates obtained in our study using LC-MS and in light of literature [25, 27, 32, 46, 47], two degradation pathway by Pseudomonas putida YC-AE1 was proposed. The intermediates were detected from both two degradation pathways at same experiment and this is an evidence that our strain use both of pathways in the degradation and this may interpret the high degradation rate of YC-AE1.
Conclusion
This study has reported the ability of promising pseudomonas putida strain YC-AE1 isolated from Guangdong province (China) to degrade BPA as the sole source for carbon with excellent adaptation to a wide range of environmental conditions. This strain can degrade both of extremely high and low concentrations of BPA and mineralize it in a short time compared with reported strains. Furthermore, it can degrade many BPA-related pollutants. Analysis of intermediates showed two degradation pathways for BPA which may interpret the fast degradation rate of our strain. These criteria recommend pseudomonas putida YC-AE1 as a promising strain for natural remediation of environmental pollutant BPA. Also, our study provides a good microbial source for further studies on bacterial BPA degradation metabolism and molecular mechanism responsible for that degradation.
Methods
Chemicals and medium
The Bisphenols (BPA, BPB, BPF and BPS), DBP, DEHP and DEP used in this study were purchased from Shanghai Macklin Biochemical Co., Ltd. China, (all purities above 99%). Stock solutions (20,000 mg l− 1) of all these substrates were prepared by dissolving each individual in methanol (99%). Other chemicals used were of analytical reagent grade, all solvents used were high performance liquid chromatography (HPLC) grade. Two types of media were used in this study: (1) Luria-Bertani (LB) medium for growth and enrichment. It consisted of 10.0 g l− 1 peptone, 5.0 g l− 1 yeast extract, and 10.0 g l− 1 NaCl. (2) Trace Elements Medium (TEM) as described by [48] were used for isolation of BPA biodegradable strains and statistical optimization experiments.
Isolation, screening and culture conditions for high tolerant BPA degrading strain
Soil and water samples were collected from five different provinces in China and these provinces were Heilongjiang, Hebei, Shandong, Guangdong and Anhui. Five grams or 5 ml of soil or water samples respectively, were separately added to 100 ml of TEM in 250 ml Erlenmeyer flask and supplied with 100 mg l− 1 of BPA. The flasks were incubated in a rotary shaker at 30 °C with 180 rpm. After 7 days, 5 ml of the culture (each flask) was transferred to fresh TEM with 200 mg l− 1 and incubated at the same condition mentioned above for 7 days. This process was repeated three more times with increasing BPA concentrations in each run to reach 500 mg l− 1 [49]. The final cultures were spread on solid TEM containing 100 mg l− 1 BPA as a sole source of carbon and incubated for 5 days at 30 °C, single colonies were picked and inoculated into TEM supplemented with 100 mg l− 1 BPA and incubated (30 °C and 180 rpm) to test the biodegradation. Uninoculated fresh culture of TEM with 100 mg l− 1 BPA was set as abiotic control. The residual concentrations of BPA were determined by using HPLC.
Identification of BPA degrading strain
The total genomic DNA of bacterial isolate was extracted by using Bacterial Genomic Extraction Kit (Takara, Japan). The 16S rRNA gene was amplified from genomic DNA by using two universal primers (27F: AGAGTTTGATCCTGGCTCAG, 1492R: AAGGAGGTGATCCAGCC). The PCR product was sequenced (Sangon Biotech Co., Ltd., Beijing, China) and the obtained sequence was deposited to GenBank, and aligned to get the similar sequences by BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi). The phylogenetic tree was constructed using MEGA 5 software [50] with a neighbor-joining algorithm [51]. The tree was drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the maximum composite likelihood method [52]. To confirm 16S rRNA gene identification and determining the biochemical tests of the bacterial isolate, Biolog tests (BIOLOG inc., USA), was accomplished and the results were analyzed by Microlog 3 (version 5.2) software.
Statistical optimization of BPA degradation by response surface methodology (RSM)
RSM was conducted using a factorial central composite design (CCD). CCD is one of the most common design used in RSM which has the same predictability in all directions from the center [53]. Five experimental factors selected to design the experiment included pH (A), NaCl concentration (B), incubation period (C), inoculum size (D) and incubation temperature (E) with 5 levels for each factor, generating 29 trials using Design-Expert software (version 8). The Reduced Cubic Model was fitted for biodegradation response analysis. The value of the dependent response was the mean of three replications. The detailed experimental designs of five parameters are shown in Table 1. In this experiment, TEM with BPA (200 mg l− 1) as sole source of carbon was used and inoculum was prepared by growing the bacterial strain on L. B medium to reach (O.D(600) = 0.8), the bacterial cells were collected by centrifugation at 5000 rpm for 5 min and washed twice by fresh TEM and resuspended using the same original volume with TEM.
Substrate utilization test
To determine the ability of selected strain to degrade different organic pollutants other than BPA, the bacterial strain was inoculated (1% inoculum size) into TEM (15 ml) supplemented with 100 mg l− 1 of each of the following substrates: bisphenol B (BPB), bisphenol F (BPF), bisphenol S (BPS), Dibutyl phthalate (DBP), Diethylhexyl phthalate (DEHP) and Diethyl phthalate (DEP). The cultures were incubated in a rotary shaker (30 °C and 180 rpm) for 72 h. Cultures without inoculation were set as abiotic control. The residual concentrations of bisphenols (B, F, and S) and (DBP, DEHP) were measured by HPLC and gas chromatography (GC) respectively.
Detection of intermediates
To determine the biodegradation intermediates, the selected bacterial strain was grown in TEM supplemented with BPA (200 mg l− 1) under optimal culture conditions. 100 ml of sample was taken from the culture at specific intervals as following (15, 20, 24, 28, 32, 36 h) and centrifuged at 10000 rpm for 10 min, the cell-free supernatant was acidified (pH: 2–3) using HCl. The samples were extracted twice with the same volume of ethyl acetate, organic layer separated and dried by Rotary evaporator (BUCHI, Switzerland) and re-suspended in 1 ml methanol (99%). The samples were subjected to high performance liquid chromatography-mass spectroscopy (HPLC-MS) to detect the intermediates.
BPA degradation at low and high concentrations and total organic carbon (TOC) determination
The concentration of BPA in natural polluted environments is very low compared with that always examined in vitro. Therefore, the ability of bacterial strain to degrade and utilize BPA at low and high concentrations is a very important character especially in strains intended to use in environmental bioremediation. The high and low concentrations used were as follows (50, 100, 200, 300, 400, 500, 600, 700, 800, 900, and 1000 mg l− 1) and (0.5, 1, 2, 4, 6, 8, 10 and 12 mg l− 1) respectively. The residual BPA concentration and degradation rate were determined after 15 and 72 h for low and high concentrations by HPLC, respectively. In low concentration experiment, the relatively short incubation time was to detect the differences in degradation rates compared with BPA concentrations. To detect the BPA mineralization rate, TOC depletion was determined during the degradation time. The selected bacterial strain was grown in TEM supplemented with 100 mg l− 1 of BPA at 30 °C. Samples (20 ml) were taken in triplicate each 8 h intervals as following (8, 16, 24, 32, 40, 48, 56 and 64 h) and centrifugated at 15000 rpm for 30 min to eliminate the bacterial cells. The cell-free supernatant was kept at − 20 °C. TOC was determined for all preserved samples by Walkley-Black method [54].
Analysis methods
Cell growth was measured as optical density (OD) at 600 nm using a UV-Visible spectrophotometer (Thermo-Scientific, USA). For detection of bisphenols, an Agilent 1200 system HPLC (Agilent, USA) with visible ultraviolet detector was used equipped with Eclipse XDB C18 column (4.6 × 150 mm × 5 μm). The mobile phase was acetonitrile 85% and (water, 0.1% acetic acid) 15%, with a flow rate 1 ml/minute and detecting wavelength 220 nm. The retention time was 1.55 min and 2 μl for sample injection, the detection limits were 0.3, 0.4, .4, 0.5 μgml− 1 for BPA, BPB, BPF and BPS, respectively. For detection of DBP, DEHP and DEP, a GC system (GC-2010 SHIMADZU, Kyoto, Japan) with a flame ionizing detector (FID) was used. Samples were extracted twice by an equal volume of n-hexane and the hexan phase was separated and analyzed by GC [55], the detection limits were 0.1 μgml− 1. The intermediate metabolites of BPA degradation were analyzed by (HPLC–MS, 1290, USA). The mobile phase was a mixture of (50:50%) of water and acetonitrile (with 0.1% formic acid for each one), running at a flow rate of 1 mL/min. The sample was injected (2 μL) as methanolic extract and scan mode with 3 kV capillary voltage was used to analyze negative ionization. The dry gas flow was 5 L/min and the dry nitrogen was heated to 325 °C. Mass hunter (version A.02.00, Agilent, USA) was used to analyze and collect the data.
Acknowledgments
We are grateful to Mr. Xianjun Li for his assistance in the collection of samples and Khaled S. Nassar for his helping in designing the experiment with Design Expert software. Also, we thank Ebrahim Shehata for his technical assistance in the TOC experiment.
Abbreviations
- BPA
Bisphenol A
- BPB
Bisphenol B
- BPF
Bisphenol F
- BPS
Bisphenol S
- CCD
Central composite design
- DBP
Dibutyl phthalate
- DEHP
Diethylhexyl phthalate
- DEP
Diethyl phthalate
- GC
Gas chromatography
- HPA
Hydroxyphenylacetate
- HPLC
High performance liquid chromatography
- HQ
Hydroquinone
- p-HAP
p-hydroxyacetophenone
- p-HBAL
p-hydroxybenzaldeyde
- RSM
Response Surface Methodology
- TEM
Trace elements medium
- TOC
Total organic acrbon
Authors’ contributions
AE conducted the practical work for all experiments; YJ designed the statistical optimization experiments; JW analyzed the HPLC-MS results. YJ and JW contributed reagents/materials/analysis tools; AE and MAAK wrote the manuscript. NR, IK and YY revised the manuscript. All authors have read and approved the final manuscript.
Funding
This study is financially supported by National Natural Science Foundation of China [Nos. 31540067, 21876201] and the Basic Research Fund of Chinese Academy of Agricultural Sciences [Nos. 1610042017001, 1610042018005 and 1610042018006]. The funding body has no role in the design of the study and collection, analysis, interpretation of data and in writing the manuscript.
Availability of data and materials
All data generated or analyzed during this study are included in this published article except the sequence of 16S rRNA gene was deposited to Gen Bank (NCBI) with accession number, MK318658.
Ethics approval and consent to participate
Sampling was conducted on private lands after taking the permissions from the owners of these lands.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Adel Eltoukhy, Email: adelaly@azhar.edu.eg.
Yang Jia, Email: 845576584@qq.com.
Ruth Nahurira, Email: ruth.nahurira@yahoo.com.
M. A. Abo-Kadoum, Email: Abokadoum@qq.com
Ibatsam Khokhar, Email: ibatsamk@yahoo.com.
Junhuan Wang, Email: wangjunhuan_1993@163.com.
Yanchun Yan, Email: yanyanchun@caas.cn.
References
- 1.Atacag EH. Biodegradation and detoxification of BPA: involving laccase and a mediator. CLEAN Soil Air Water. 2015;43:932–939. doi: 10.1002/clen.201400628. [DOI] [Google Scholar]
- 2.Wang Q, Chen M, Shan G, Chen P, Cui S, Yi S, et al. Bioaccumulation and biomagnification of emerging bisphenol analogues in aquatic organisms from Taihu Lake, China. Sci Total Environ. 2017;598:814–820. doi: 10.1016/j.scitotenv.2017.04.167. [DOI] [PubMed] [Google Scholar]
- 3.Alexander HC, Dill DC, Smith LW, Guiney PD, Dorn P. Bisphenol A: acute aquatic toxicity. Environ Toxicol Chem. 1988;7:19–26. doi: 10.1002/etc.5620070104. [DOI] [Google Scholar]
- 4.Seachrist DD, Bonk KW, Ho SM, Prins GS, Soto AM, Keri RA. A review of the carcinogenic potential of bisphenol a. Reprod Toxicol. 2016;59:167–182. doi: 10.1016/j.reprotox.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Welshons WV, Thayer KA, Judy BM, Taylor JA, Curran EM, Vom Saal FS. Large effects from small exposures. I. Mechanisms for endocrine-disrupting chemicals with estrogenic activity. Environ Health Perspect. 2003;111:994–1006. doi: 10.1289/ehp.5494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vom Saal FS, Hughes C. An extensive new literature concerning low-dose effects of bisphenol a shows the need for a new risk assessment. Environ Health Perspect. 2005;113:926–933. doi: 10.1289/ehp.7713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vandenberg LN, Colborn T, Hayes TB, Heindel JJ, Jacobs DR, Jr, Lee D-H, et al. Hormones and endocrine-disrupting chemicals: low-dose effects and nonmonotonic dose responses. Endocr Rev. 2012;33:378–455. doi: 10.1210/er.2011-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kang JH, Kondo F. Bisphenol a in the surface water and freshwater snail collected from rivers around a secure landfill. Bull Environ Contam Toxicol. 2006;76:113–118. doi: 10.1007/s00128-005-0896-4. [DOI] [PubMed] [Google Scholar]
- 9.Ye X, Pierik FH, Angerer J, Meltzer HM, Jaddoe VWV, Tiemeier H, et al. Levels of metabolites of organophosphate pesticides, phthalates, and bisphenol a in pooled urine specimens from pregnant women participating in the Norwegian mother and child cohort study (MoBa) Int J Hyg Environ Health. 2009;212:481–491. doi: 10.1016/j.ijheh.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Larsson K, Lindh CH, Jönsson BAG, Giovanoulis G, Bibi M, Bottai M, et al. Phthalates, non-phthalate plasticizers and bisphenols in Swedish preschool dust in relation to children’s exposure. Environ Int. 2017;102:114–124. doi: 10.1016/j.envint.2017.02.006. [DOI] [PubMed] [Google Scholar]
- 11.Zhou NA, Kjeldal H, Gough HL, Nielsen JL. Identification of putative genes involved in bisphenol a degradation using differential protein abundance analysis of Sphingobium sp. BiD32. Environ Sci Technol. 2015;49:12232–12241. doi: 10.1021/acs.est.5b02987. [DOI] [PubMed] [Google Scholar]
- 12.Chouhan S, Yadav SK, Prakash J, Singh SP. Effect of Bisphenol a on human health and its degradation by microorganisms: a review. Ann Microbiol. 2014;64:13–21. doi: 10.1007/s13213-013-0649-2. [DOI] [Google Scholar]
- 13.Yamada H, Furuta I, Kato EH, Kataoka S, Usuki Y, Kobashi G, et al. Maternal serum and amniotic fluid bisphenol a concentrations in the early second trimester. Reprod Toxicol. 2002;16:735–739. doi: 10.1016/S0890-6238(02)00051-5. [DOI] [PubMed] [Google Scholar]
- 14.Zhang Y, Tao S, Yuan C, Liu Y, Wang Z. Non-monotonic dose–response effect of bisphenol a on rare minnow Gobiocypris rarus ovarian development. Chemosphere. 2016;144:304–311. doi: 10.1016/j.chemosphere.2015.08.079. [DOI] [PubMed] [Google Scholar]
- 15.Chen J, Xiao Y, Gai Z, Li R, Zhu Z, Bai C, et al. Reproductive toxicity of low level bisphenol a exposures in a two-generation zebrafish assay: evidence of male-specific effects. Aquat Toxicol. 2015;169:204–214. doi: 10.1016/j.aquatox.2015.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamamoto T, Yasuhara A, Shiraishi H, Nakasugi O. Bisphenol a in hazardous waste landfill leachates. Chemosphere. 2001;42:415–418. doi: 10.1016/S0045-6535(00)00079-5. [DOI] [PubMed] [Google Scholar]
- 17.Lee H-B, Peart TE. Bisphenol a contamination in Canadian municipal and industrial wastewater and sludge samples. Water Qual Res J. 2000;35:283–298. doi: 10.2166/wqrj.2000.018. [DOI] [Google Scholar]
- 18.Kleywegt S, Pileggi V, Yang P, Hao C, Zhao X, Rocks C, et al. Pharmaceuticals, hormones and bisphenol a in untreated source and finished drinking water in Ontario, Canada—occurrence and treatment efficiency. Sci Total Environ. 2011;409:1481–1488. doi: 10.1016/j.scitotenv.2011.01.010. [DOI] [PubMed] [Google Scholar]
- 19.Bolz U, Hagenmaier H, Körner W. Phenolic xenoestrogens in surface water, sediments, and sewage sludge from Baden-Württemberg, south-West Germany. Environ Pollut. 2001;115:291–301. doi: 10.1016/S0269-7491(01)00100-2. [DOI] [PubMed] [Google Scholar]
- 20.Kolpin DW, Furlong ET, Meyer MT, Thurman EM, Zaugg SD, Barber LB, et al. Pharmaceuticals, hormones, and other organic wastewater contaminants in US streams, 1999–2000: A national reconnaissance. Environ Sci Technol. 2002;36:1202–1211. doi: 10.1021/es011055j. [DOI] [PubMed] [Google Scholar]
- 21.Staples CA, Dome PB, Klecka GM, Oblock ST, Harris LR. A review of the environmental fate, effects, and exposures of bisphenol a. Chemosphere. 1998;36:2149–2173. doi: 10.1016/S0045-6535(97)10133-3. [DOI] [PubMed] [Google Scholar]
- 22.Yamauchi K, Ishihara A, Fukazawa H, Terao Y. Competitive interactions of chlorinated phenol compounds with 3, 3’, 5-triiodothyronine binding to transthyretin: detection of possible thyroid-disrupting chemicals in environmental waste water. Toxicol Appl Pharmacol. 2003;187:110–117. doi: 10.1016/S0041-008X(02)00045-5. [DOI] [PubMed] [Google Scholar]
- 23.Wang R, Diao P, Chen Q, Wu H, Xu N, Duan S. Identification of novel pathways for biodegradation of bisphenol a by the green alga Desmodesmus sp. WR1, combined with mechanistic analysis at the transcriptome level. Chem Eng J. 2017;321:424–431. doi: 10.1016/j.cej.2017.03.121. [DOI] [Google Scholar]
- 24.Kang J-H, Katayama Y, Kondo F. Biodegradation or metabolism of bisphenol a: from microorganisms to mammals. Toxicology. 2006;217:81–90. doi: 10.1016/j.tox.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 25.Zhang C, Zeng G, Yuan L, Yu J, Li J, Huang G, et al. Aerobic degradation of bisphenol a by Achromobacter xylosoxidans strain B-16 isolated from compost leachate of municipal solid waste. Chemosphere. 2007;68:181–190. doi: 10.1016/j.chemosphere.2006.12.012. [DOI] [PubMed] [Google Scholar]
- 26.Kang JH, Kondo F. Bisphenol a degradation by bacteria isolated from river water. Arch Environ Contam Toxicol. 2002;43:265–269. doi: 10.1007/s00244-002-1209-0. [DOI] [PubMed] [Google Scholar]
- 27.Suyamud B, Inthorn D, Panyapinyopol B. Biodegradation of Bisphenol a by a newly isolated Bacillus megaterium strain ISO-2 from a polycarbonate industrial wastewater. 2018. [Google Scholar]
- 28.Oshiman KI, Tsutsumi Y, Nishida T, Matsumura Y. Isolation and characterization of a novel bacterium, Sphingomonas bisphenolicum strain AO1, that degrades bisphenol a. Biodegradation. 2007;18:247–255. doi: 10.1007/s10532-006-9059-5. [DOI] [PubMed] [Google Scholar]
- 29.Xu C-P, Kim S-W, Hwang H-J, Choi J-W, Yun J-W. Optimization of submerged culture conditions for mycelial growth and exo-biopolymer production by Paecilomyces tenuipes C240. Process Biochem. 2003;38:1025–1030. doi: 10.1016/S0032-9592(02)00224-8. [DOI] [Google Scholar]
- 30.Khambhaty Y, Mody K, Jha B, Gohel V. Statistical optimization of medium components for κ-carrageenase production by Pseudomonas elongata. Enzym Microb Technol. 2007;40:813–822. doi: 10.1016/j.enzmictec.2006.06.015. [DOI] [Google Scholar]
- 31.Kumar P, Satyanarayana T. Optimization of culture variables for improving glucoamylase production by alginate-entrapped Thermomucor indicae-seudaticae using statistical methods. Bioresour Technol. 2007;98:1252–1259. doi: 10.1016/j.biortech.2006.05.019. [DOI] [PubMed] [Google Scholar]
- 32.Eio EJ, Kawai M, Tsuchiya K, Yamamoto S, Toda T. Biodegradation of bisphenol a by bacterial consortia. Int Biodeterior Biodegrad. 2014;96:166–173. doi: 10.1016/j.ibiod.2014.09.011. [DOI] [Google Scholar]
- 33.Ren L, Jia Y, Ruth N, Shi Y, Wang J, Qiao C, et al. Biotransformations of bisphenols mediated by a novel Arthrobacter sp. strain YC-RL1. Appl Microbiol Biotechnol. 2016;100:1967–1976. doi: 10.1007/s00253-015-7076-1. [DOI] [PubMed] [Google Scholar]
- 34.Smita GS, Ray P, Mohapatra S. Quantification and optimization of bacterial isolates for production of alkaline protease. Asian J Exp Biol Sci. 2012;3:180–186. [Google Scholar]
- 35.Karthikeyan RS, Rakshit SK, Baradarajan A. Optimization of batch fermentation conditions for dextran production. Bioprocess Eng. 1996;15:247–251. doi: 10.1007/BF02391585. [DOI] [Google Scholar]
- 36.Engqvist MKM. Correlating enzyme annotations with a large set of microbial growth temperatures reveals metabolic adaptations to growth at diverse temperatures. BMC Microbiol. 2018;18:1–14. doi: 10.1186/s12866-018-1320-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fouda A. Biodegradation of Bisphenol a by some bacterial species and significance role of plasmids. Int J Adv Res Biol Sci. 2015;2:93–108. [Google Scholar]
- 38.Ulrich AC, Guigard SE, Foght JM, Semple KM, Pooley K, Armstrong JE, et al. Effect of salt on aerobic biodegradation of petroleum hydrocarbons in contaminated groundwater. Biodegradation. 2009;20:27–38. doi: 10.1007/s10532-008-9196-0. [DOI] [PubMed] [Google Scholar]
- 39.de Carvalho CC, da Fonseca MM. Degradation of hydrocarbons and alcohols at different temperatures and salinities by Rhodococcus erythropolis DCL14. FEMS Microbiol Ecol. 2005;51:389–399. doi: 10.1016/j.femsec.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 40.Basha KM, Rajendran A, Thangavelu V. Recent advances in the biodegradation of phenol: a review. Asian J Exp Biol Sci. 2010;1:219–234. [Google Scholar]
- 41.Lee SG, Yoon BD, Park YH, Oh HM. Isolation of a novel pentachlorophenol-degrading bacterium, Pseudomonas sp. Bu34. J Appl Microbiol. 1998;85:1–8. doi: 10.1046/j.1365-2672.1998.00456.x. [DOI] [PubMed] [Google Scholar]
- 42.Kumar A, Kumar S, Kumar S. Biodegradation kinetics of phenol and catechol using Pseudomonas putida MTCC 1194. Biochem Eng J. 2005;22:151–159. doi: 10.1016/j.bej.2004.09.006. [DOI] [Google Scholar]
- 43.Peng X, Qu X, Luo W, Jia X. Co-metabolic degradation of tetrabromobisphenol a by novel strains of Pseudomonas sp. and Streptococcus sp. Bioresour Technol. 2014;169:271–276. doi: 10.1016/j.biortech.2014.07.002. [DOI] [PubMed] [Google Scholar]
- 44.Yamada T, Takahama Y, Yamada Y. Biodegradation of 2, 4, 6-tribromophenol by Ochrobactrum sp. strain TB01. Biosci Biotechnol Biochem. 2008;72:1264–1271. doi: 10.1271/bbb.70755. [DOI] [PubMed] [Google Scholar]
- 45.Yu K, Yi S, Li B, Guo F, Peng X, Wang Z, et al. An integrated meta-omics approach reveals substrates involved in synergistic interactions in a bisphenol a (BPA)-degrading microbial community. Microbiome. 2019;7:1–13. doi: 10.1186/s40168-019-0634-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Das R, Li G, Mai B, An T. Spore cells from BPA degrading bacteria Bacillus sp. GZB displaying high laccase activity and stability for BPA degradation. Sci Total Environ. 2018;640–641:798–806. doi: 10.1016/j.scitotenv.2018.05.379. [DOI] [PubMed] [Google Scholar]
- 47.Wang J, Ren L, Jia Y, Ruth N, Shi Y, Qiao C, et al. Degradation characteristics and metabolic pathway of 4-nitrophenol by a halotolerant bacterium Arthrobacter sp. CN2. Toxicol Environ Chem. 2016;98:226–240. doi: 10.1080/02772248.2015.1115507. [DOI] [Google Scholar]
- 48.Nahurira R, Ren L, Song J, Jia Y, Wang J, Fan S, et al. Degradation of Di(2-Ethylhexyl) phthalate by a novel Gordonia alkanivorans strain YC-RL2. Curr Microbiol. 2017;74:309–319. doi: 10.1007/s00284-016-1159-9. [DOI] [PubMed] [Google Scholar]
- 49.Ren L, Jia Y, Ruth N, Qiao C, Wang J, Zhao B, et al. Biodegradation of phthalic acid esters by a newly isolated Mycobacterium sp. YC-RL4 and the bioprocess with environmental samples. Environ Sci Pollut Res. 2016;23:16609–16619. doi: 10.1007/s11356-016-6829-4. [DOI] [PubMed] [Google Scholar]
- 50.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 52.Tamura K, Nei M, Kumar S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci. 2004;101:11030–11035. doi: 10.1073/pnas.0404206101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gao Y-L, Ju X-R, Jiang H-H. Use of response surface methodology to investigate the effect of food constituents on Staphylococcus aureus inactivation by high pressure and mild heat. Process Biochem. 2006;41:362–369. doi: 10.1016/j.procbio.2005.06.023. [DOI] [Google Scholar]
- 54.Walkley A, Black IA. An examination of the Degtjareff method for determining soil organic matter, and a proposed modification of the chromic acid titration method. Soil Sci. 1934;37:29–38. doi: 10.1097/00010694-193401000-00003. [DOI] [Google Scholar]
- 55.Zhang Y, Chen H, Liu J, Geng G, Liu D, Geng H, et al. Genome sequencing and biodegradation characteristics of the n-butyl benzyl phthalate degrading rdthetghteyt/‘lm ;n HS-D2 rdthetghteyt. Int Biodeterior Biodegradation. 2016;128:56–62. doi: 10.1016/j.ibiod.2016.08.024. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article except the sequence of 16S rRNA gene was deposited to Gen Bank (NCBI) with accession number, MK318658.