Abstract
Testicular cancer seminoma is one of the most common types of cancer among men of reproductive age. Patients with this condition usually present reduced semen quality, even before initiating cancer therapy. However, the underlying mechanisms by which testicular cancer seminoma affects male fertility are largely unknown. The aim of this study was to investigate alterations in the sperm proteome of men with seminoma undergoing sperm banking before starting cancer therapy, in comparison to healthy proven fertile men (control group). A routine semen analysis was conducted before cryopreservation of the samples (n = 15 per group). Men with seminoma showed a decrease in sperm motility (P = 0.019), total motile count (P = 0.001), concentration (P = 0.003), and total sperm count (P = 0.001). Quantitative proteomic analysis identified 393 differentially expressed proteins between the study groups. Ten proteins involved in spermatogenesis, sperm function, binding of sperm to the oocyte, and fertilization were selected for validation by western blot. We confirmed the underexpression of heat shock-related 70 kDa protein 2 (P = 0.041), ubiquinol-cytochrome C reductase core protein 2 (P = 0.026), and testis-specific sodium/potassium-transporting ATPase subunit alpha-4 (P = 0.016), as well as the overexpression of angiotensin I converting enzyme (P = 0.005) in the seminoma group. The altered expression levels of these proteins are associated with spermatogenesis dysfunction, reduced sperm kinematics and motility, failure in capacitation and fertilization. The findings of this study may explain the decrease in the fertilizing ability of men with seminoma before starting cancer therapy.
Keywords: male fertility, proteomics, seminoma, sperm proteins, sperm quality, testicular cancer
INTRODUCTION
Germ cell tumors (GCTs) represent the most common type of testicular cancer, accounting for about 90%–95% of all cases. The principal types of GCTs are nonseminomas and seminomas; the latter usually grows and spreads more slowly. In the last decades, there is a growing trend in the proportion of seminomas.1 The survival rate of men with seminoma is very high (over 95%); thus, it is generally not seen as a threat to public health. However, its impact on male fertility represents a major concern for reproductive medicine as it frequently affects men in reproductive age (20–44 years).2
Men with seminoma present impaired fertilizing ability, even before diagnosis.3 Testicular cancer seminoma affects the hypothalamic-pituitary-gonadal (HPG) axis and consequently disturbs spermatogenesis.4 These deleterious effects are dependent on the stage and type of seminoma, resulting in poor semen quality or even azoospermia.5 The treatment for this type of cancer, usually performed by surgery, chemotherapy, or radiotherapy, further affects semen quality5 and hormonal function,6 thus highly impairing male fertility. In fact, after cancer therapy, patients may become temporarily or permanently infertile.7 For that reason, it is strongly recommended that men diagnosed with seminoma undergo sperm banking to increase the probability to father a child in the future.8 The chances to establish a pregnancy by natural conception are 30% lower after the cancer therapy and the recovery of fertilizing ability usually takes several years.9 Therefore, in many surviving patients with seminoma, assisted reproductive technology (ART) with cryopreserved samples is the only option for having children.10 Still, sperm banking is not possible for many patients due to the high cost or lack of facilities, urgency to initiate the treatment, impaired spermatogenesis, and/or poor semen quality at the time of specimen collection.11
Proteomics studies have been recently used as a valuable tool to explore how certain health conditions affect male reproductive potential, especially by evaluating spermatozoa and seminal plasma proteome.12,13 Although spermatozoa are transcriptionally and translationally silent after being produced in the testis, the acquisition of sperm function occurs during maturation in the epididymis and transit through the female reproductive tract.14 Therefore, the sperm proteome is highly susceptible to alterations according to the health status of the individual, and this impacts the quality of sperm parameters. The deleterious effects of seminoma treatment represent a challenge to understand the mechanisms behind the impairment of male fertility caused by the disease. In this study, we used semen samples from men with testicular cancer seminoma that were cryopreserved before starting cancer therapy, to investigate alterations in the sperm proteome in comparison with healthy proven fertile men.
PARTICIPANTS AND METHODS
Semen analysis and cryopreservation
This study was conducted after approval by the Institutional Review Board (IRB) of Cleveland Clinic, Cleveland, OH, USA. Semen samples were obtained from healthy volunteers with proven fertility (control, n = 15) and patients with seminoma (n = 15). All the participants signed informed written consent to allow the use of their samples in this study. The inclusion criteria were as follows: (1) control group, healthy fertile men who had fathered a child in the last 2 years; (2) seminoma group, patients diagnosed with seminoma and undergoing sperm banking before starting cancer therapy. Following 2–3 days of abstinence, semen samples were collected at the Andrology Center, Cleveland Clinic. Samples were liquefied for 20–30 min in an incubator (Panasonic, Newark, NJ, USA) at 37°C, and a routine semen analysis was conducted according to the World Health Organization (WHO) 2010 guidelines.15 Semen volume, sperm motility, and sperm concentration were recorded. Total sperm count and total motile count were also calculated and the results were expressed as mean ± standard error of the mean (s.e.m.). Whole ejaculate samples were immediately cryopreserved in TEST-yolk buffer (TYB; Irvine Scientific, Santa Ana, CA, USA) in a ratio of 1:1 as previously described16 and finally labeled and stored in liquid nitrogen at −196°C.
Protein extraction and estimation
Samples were thawed on ice and centrifuged at 4000g for 10 min (Eppendorf, Hauppauge, NY, USA). To remove the freezing medium (TYB) as much as possible, the sperm pellet was washed four times in phosphate-buffered saline (PBS; Sigma-Aldrich, St. Louis, MO, USA) and centrifuged at 4000g for 10 min at 4°C. Total sperm protein was extracted overnight at 4°C with radioimmunoprecipitation assay (RIPA) buffer (Sigma-Aldrich). Subsequently, samples were centrifuged at 10 000g for 30 min at 4°C, to recover the protein fraction (supernatant). Pierce BCA Protein Assay kit (Thermo Fisher Scientific, Waltham, MA, USA) was used to estimate the protein concentration, according to the manufacturer's instructions.
Quantitative proteomic analysis
Three samples from the control or seminoma group were randomly selected for the proteomic analysis by liquid chromatography-tandem mass spectrometry (LC-MS/MS). Samples were pooled (n = 3) using the same amount of protein from each sample. Each pool was then evaluated as an individual sample in the proteomic analysis. The system used was a Finnigan LTQ-Orbitrap Elite hybrid mass spectrometer (Thermo Fisher Scientific) using the previously described conditions and software.17 Scaffold (version 4.0.6.1, Proteome Software Inc., Portland, OR, USA) was used for the identification of differentially expressed proteins (DEPs) between the control and seminoma groups. The spectral counts were used to determine the abundance of each protein (very low, low, medium, or high). The identified DEPs were categorized as underexpressed, overexpressed, or unique to one of the groups, based on the normalized spectral abundance factor (NSAF) ratio according to previously reported criteria.17
Bioinformatic analysis
Bioinformatic analysis of DEPs identified by LC-MS/MS was carried out using the Ingenuity Pathway Analysis software (IPA; Qiagen, Hilden, Germany). IPA was used to evaluate the canonical pathways, top diseases and bio-functions, and upstream regulators related to the identified DEPs. Proteins were selected for validation by western blot considering the following criteria: (1) proteins involved in reproductive system development and function; (2) proteins involved in the top canonical pathways; (3) proteins with a higher difference of abundance between the experimental groups; and (4) proteins with a well-described function in the literature. Only proteins that met all the above-mentioned criteria were subjected to western blot.
Western blot
Western blot was performed using individual samples from the control and seminoma groups (n = 6 per group). A total of 25 μg protein per sample was mixed with 4 × Laemmli sample buffer (Bio-Rad, Hercules, CA, USA) in a ratio of 1:3 and made up to 25 μl with PBS. Samples were boiled at 95°C for 10 min and immediately loaded into a 4%–15% (w/v) polyacrylamide gel (Bio-Rad). Electrophoresis was performed with constant voltage (90 V) for 2 h. Precision Plus Protein™ Dual Xtra Standard (Thermo Fisher Scientific) was used as the molecular weight marker. The resolved proteins were transferred (20 V for 30 min) to methanol-activated polyvinylidene difluoride (PVDF) membranes (GE Healthcare, Marlborough, MA, USA) and blocked for 90 min at room temperature, with a 5% (w/v) nonfat milk (Bio-Rad) solution prepared in tris-buffered saline with tween-20 (TBST; Sigma-Aldrich). Membranes were incubated overnight (4°C) with specific primary antibodies followed by the respective secondary antibodies at room temperature, for 90 min (Supplementary Table 1). Membranes were incubated with enhanced chemiluminescence (ECL) reagent (GE Healthcare) for 5 min, and the chemiluminescence signals were read in the ChemiDoc™ MP Imaging System (Bio-Rad). Densities from each band were quantified with Image Lab™ Software (version 6.0.1, Bio-Rad) and divided by the corresponding total protein lane density. Total protein density was obtained by incubation of the membranes with total colloidal gold protein stain (BioRad). The results were expressed as fold variation relative to the control group.
Supplementary Table 1.
List of the primary and secondary antibodies used in this study
| Antibody | Source | KDa | Dilution | Vendor | Catalog Number |
|---|---|---|---|---|---|
| ACE | Rabbit | 200 | 1:1000 | Abcam | ab85955 |
| ACR | Rabbit | 46 | 1:1000 | Abcam | ab203289 |
| ATP1A4 | Rabbit | 100 | 1:10000 | Abcam | ab76020 |
| ATP5A | Mouse | 54 | 1:1000 | Abcam | ab110411 |
| CCT3 | Rabbit | 61 | 1:2000 | Abcam | ab225878 |
| HSPA2 | Mouse | 70 | 1:500 | Abcam | ab89130 |
| NDUFS1 | Rabbit | 79 | 1:10000 | Abcam | ab157221 |
| PSME4 | Rabbit | 211 | 1:500 | Abcam | ab181203 |
| SPA17 | Rabbit | 17 | 1:1000 | Abcam | ab172626 |
| UQCRC2 | Mouse | 48 | 1:1000 | Abcam | ab110411 |
| Mouse* | Rabbit | - | 1:10000 | Abcam | ab6728 |
| Rabbit* | Goat | - | 1:10000 | Abcam | ab97051 |
*Secondary antibody. ACE: angiotensin-converting enzyme; ACR: acrosin precursor; CCT3: T-complex protein 1 subunit gamma; SPA17: sperm surface protein Sp17; ATP1A4: sodium/potassium-transporting ATPase subunit alpha-4; HSPA2: heat shock-related 70 kDa protein 2; PSME4: proteasome activator complex subunit 4; NDUFS1: NADH-ubiquinone oxidoreductase 75 kDa subunit; UQCRC2: cytochrome b-c1 complex subunit 2; ATP5A: ATP synthase subunit alpha
Statistical analyses
After testing normal distribution by the Kolmogorov–Smirnov test, semen parameters and western blot results were analyzed by Mann–Whitney U test for independent samples, using the MedCalc Software (version 17.8; MedCalc Software, Ostend, Belgium). All data are presented as mean ± s.e.m., and differences with P < 0.05 were considered statistically significant.
RESULTS
Semen quality in patients with testicular cancer seminoma
The average volume of the ejaculates was very similar between the control and seminoma groups (Table 1). However, there was a decrease in sperm motility (P = 0.019), sperm concentration (P = 0.003), total sperm count (P = 0.001), and total motile count (P = 0.001) in patients with seminoma relative to control (Table 1). Nevertheless, all the samples were considered normozoospermic according to the WHO 2010 criteria.15
Table 1.
Semen parameters of fertile men (control) and patients with testicular cancer seminoma
| Parameter | Control | Seminoma | P |
|---|---|---|---|
| Semen volume (ml) | 3.53±0.35 | 3.33±0.42 | 0.541 |
| Sperm motility (%) | 67±3 | 54±5 | 0.019 |
| Sperm concentration (106 ml−1) | 95.49±7.79 | 46.72±12.19 | 0.003 |
| Total sperm count (106) | 316.92±45.41 | 136.11±41.55 | 0.001 |
| Total motile count (106) | 211.88±30.09 | 75.63±22.44 | 0.001 |
Results are presented as mean±s.e.m. (n=15 per group). Statistical significance was considered for P < 0.05. s.e.m.: standard error of the mean
Differentially expressed proteins
Proteomic analysis identified 1149 proteins in the control group and 911 in the seminoma group. After comparative analysis between the experimental groups, a total of 1192 proteins were quantified and 393 were found to be differentially expressed (Supplementary Table 2). More than half (52.7%) of the DEPs were underexpressed, while 20.1% were overexpressed in spermatozoa of patients with seminoma. Furthermore, 4.1% of the DEPs were unique to the seminoma group and 23.1% unique to the control group (Figure 1).
Supplementary Table 2.
List of the differentially expressed proteins identified by the bioinformatic analysis when comparing the sperm proteome of fertile men (control) and patients with testicular cancer seminoma
| Protein | Accession | Average SC | Abundance | NSAF ratio | t-test | Expression | |||
|---|---|---|---|---|---|---|---|---|---|
| Control | Seminoma | Control | Seminoma | Seminoma/Control | P | ||||
| 1 | Transmembrane and coiled-coil domain-containing protein 2 | 56847610 | 23.3 | 0 | M | ni | 0.00 | 0.00000 | Unique to Control |
| 2 | Isocitrate dehydrogenase (NAD) subunit alpha, mitochondrial precursor | 5031777 | 48.0 | 0 | M | ni | 0.00 | 0.00000 | Unique to Control |
| 3 | Succinyl-CoA ligase (ADP-forming) subunit beta, mitochondrial precursor | 11321583 | 25.7 | 0 | M | ni | 0.00 | 0.00001 | Unique to Control |
| 4 | Short-chain specific acyl-CoA dehydrogenase, mitochondrial precursor | 4557233 | 50.0 | 0 | M | ni | 0.00 | 0.00001 | Unique to Control |
| 5 | Probable serine carboxypeptidase CPVL isoform X1 | 530384848 | 27.0 | 0 | M | ni | 0.00 | 0.00006 | Unique to Control |
| 6 | ATP synthase subunit O, mitochondrial precursor | 4502303 | 33.7 | 0 | M | ni | 0.00 | 0.00018 | Unique to Control |
| 7 | Doublecortin domain-containing protein 2C | 566006166 | 21.7 | 0 | M | ni | 0.00 | 0.00021 | Unique to Control |
| 8 | Bifunctional glutamate/proline-tRNA ligase | 62241042 | 21.0 | 0 | M | ni | 0.00 | 0.00088 | Unique to Control |
| 9 | Exportin-7 | 154448892 | 27.3 | 0 | M | ni | 0.00 | 0.00197 | Unique to Control |
| 10 | Uncharacterized protein KIAA1683 isoform X1 | 530415216 | 23.3 | 0 | M | ni | 0.00 | 0.00606 | Unique to Control |
| 11 | Leucine-rich repeat-containing protein 37A3 isoform X14 | 578840218 | 12.3 | 0 | L | ni | 0.00 | 0.00000 | Unique to Control |
| 12 | Heme oxygenase 2 isoform a | 555943918 | 11.3 | 0 | L | ni | 0.00 | 0.00001 | Unique to Control |
| 13 | Actin-related protein T3 | 221139714 | 17.7 | 0 | L | ni | 0.00 | 0.00001 | Unique to Control |
| 14 | Ubiquitin carboxyl-terminal hydrolase 7 isoform 1 | 150378533 | 18.3 | 0 | L | ni | 0.00 | 0.00001 | Unique to Control |
| 15 | Tetratricopeptide repeat protein 25 | 13899233 | 12.7 | 0 | L | ni | 0.00 | 0.00003 | Unique to Control |
| 16 | Actin-like protein 7A | 5729720 | 16.7 | 0 | L | ni | 0.00 | 0.00005 | Unique to Control |
| 17 | Dynein intermediate chain 2, axonemal isoform X4 | 530412670 | 15.7 | 0 | L | ni | 0.00 | 0.00008 | Unique to Control |
| 18 | Four and a half LIM domains protein 1 isoform 5 | 228480205 | 18.7 | 0 | L | ni | 0.00 | 0.00008 | Unique to Control |
| 19 | Putative lipoyltransferase 2, mitochondrial precursor | 221554520 | 9.0 | 0 | L | ni | 0.00 | 0.00011 | Unique to Control |
| 20 | Tubulin polymerization-promoting protein family member 2 | 226491350 | 16.3 | 0 | L | ni | 0.00 | 0.00012 | Unique to Control |
| 21 | Isocitrate dehydrogenase (NAD) subunit beta, mitochondrial isoform a precursor | 28178821 | 19.3 | 0 | L | ni | 0.00 | 0.00019 | Unique to Control |
| 22 | Protein DPCD | 39930355 | 18.3 | 0 | L | ni | 0.00 | 0.00028 | Unique to Control |
| 23 | Long-chain-fatty-acid-CoA ligase 3 | 42794754 | 9.7 | 0 | L | ni | 0.00 | 0.00031 | Unique to Control |
| 24 | Sodium/potassium-transporting ATPase subunit alpha-3 isoform 1 | 22748667 | 17.3 | 0 | L | ni | 0.00 | 0.00047 | Unique to Control |
| 25 | 26S proteasome non-ATPase regulatory subunit 4 | 5292161 | 9.0 | 0 | L | ni | 0.00 | 0.00047 | Unique to Control |
| 26 | ATP synthase subunit g, mitochondrial | 51479156 | 9.0 | 0 | L | ni | 0.00 | 0.00049 | Unique to Control |
| 27 | Acyl-CoA dehydrogenase family member 9, mitochondrial | 21361497 | 18.0 | 0 | L | ni | 0.00 | 0.00061 | Unique to Control |
| 28 | Transcription factor A, mitochondrial isoform 1 precursor | 4507401 | 15.0 | 0 | L | ni | 0.00 | 0.00065 | Unique to Control |
| 29 | Elongation factor Tu, mitochondrial precursor | 34147630 | 10.7 | 0 | L | ni | 0.00 | 0.00066 | Unique to Control |
| 30 | Eukaryotic translation elongation factor 1 epsilon-1 isoform 2 | 208879470 | 8.0 | 0 | L | ni | 0.00 | 0.00068 | Unique to Control |
| 31 | Voltage-dependent calcium channel subunit alpha-2/delta-2 isoform X1 | 530373385 | 8.0 | 0 | L | ni | 0.00 | 0.00075 | Unique to Control |
| 32 | Armadillo repeat-containing protein 12 isoform X1 | 530381603 | 12.0 | 0 | L | ni | 0.00 | 0.00079 | Unique to Control |
| 33 | Deoxyuridine 5’- triphosphate nucleotidohydrolase, mitochondrial isoform 3 | 70906444 | 13.0 | 0 | L | ni | 0.00 | 0.00081 | Unique to Control |
| 34 | Probable inactive serine protease 37 isoform 1 precursor | 285394164 | 9.0 | 0 | L | ni | 0.00 | 0.00087 | Unique to Control |
| 35 | 26S proteasome non-ATPase regulatory subunit 14 | 5031981 | 9.0 | 0 | L | ni | 0.00 | 0.00088 | Unique to Control |
| 36 | Mitochondria-eating protein isoform X4 | 530376736 | 16.7 | 0 | L | ni | 0.00 | 0.00097 | Unique to Control |
| 37 | Mitochondrial fission 1 protein | 151108473 | 8.7 | 0 | L | ni | 0.00 | 0.00110 | Unique to Control |
| 38 | Alpha-soluble NSF attachment protein | 47933379 | 8.0 | 0 | L | ni | 0.00 | 0.00137 | Unique to Control |
| 39 | Maleylacetoacetate isomerase isoform 1 | 22202624 | 9.0 | 0 | L | ni | 0.00 | 0.00179 | Unique to Control |
| 40 | 40S ribosomal protein S15 | 4506687 | 10.0 | 0 | L | ni | 0.00 | 0.00184 | Unique to Control |
| 41 | Aladin isoform 2 | 291045307 | 8.3 | 0 | L | ni | 0.00 | 0.00186 | Unique to Control |
| 42 | Ubiquitin carboxyl-terminal hydrolase isozyme L1 | 21361091 | 14.3 | 0 | L | ni | 0.00 | 0.00195 | Unique to Control |
| 43 | Stomatin-like protein 2, mitochondrial isoform a | 7305503 | 12.3 | 0 | L | ni | 0.00 | 0.00200 | Unique to Control |
| 44 | Protein FAM209B isoform X2 | 578835992 | 8.0 | 0 | L | ni | 0.00 | 0.00205 | Unique to Control |
| 45 | Putative protein FAM71E2 | 223972704 | 12.3 | 0 | L | ni | 0.00 | 0.00229 | Unique to Control |
| 46 | Acyl-protein thioesterase 1 isoform 1 | 5453722 | 11.7 | 0 | L | ni | 0.00 | 0.00240 | Unique to Control |
| 47 | Histone H1t | 20544168 | 8.0 | 0 | L | ni | 0.00 | 0.00244 | Unique to Control |
| 48 | Armadillo repeat-containing protein 4 isoform X3 | 578818430 | 18.7 | 0 | L | ni | 0.00 | 0.00324 | Unique to Control |
| 49 | Dnaj homolog subfamily B member 1 isoform X1 | 578833210 | 13.3 | 0 | L | ni | 0.00 | 0.00375 | Unique to Control |
| 50 | Calcium-binding mitochondrial carrier protein Aralar2 isoform 1 | 237649019 | 14.0 | 0 | L | ni | 0.00 | 0.00450 | Unique to Control |
| 51 | Long-chain-fatty-acid-CoA ligase ACSBG2 isoform a | 574584557 | 17.7 | 0 | L | ni | 0.00 | 0.00479 | Unique to Control |
| 52 | Methionine-tRNA ligase, cytoplasmic | 14043022 | 10.0 | 0 | L | ni | 0.00 | 0.00512 | Unique to Control |
| 53 | 60S acidic ribosomal protein P0 | 4506667 | 13.3 | 0 | L | ni | 0.00 | 0.00722 | Unique to Control |
| 54 | Cytoplasmic dynein 1 heavy chain 1 | 33350932 | 11.3 | 0 | L | ni | 0.00 | 0.00728 | Unique to Control |
| 55 | ADP-ribosylation factor 6 | 4502211 | 9.0 | 0 | L | ni | 0.00 | 0.00763 | Unique to Control |
| 56 | Glycine-tRNA ligase precursor | 116805340 | 15.0 | 0 | L | ni | 0.00 | 0.00770 | Unique to Control |
| 57 | BAG family molecular chaperone regulator 5 isoform b | 6631077 | 9.3 | 0 | L | ni | 0.00 | 0.00818 | Unique to Control |
| 58 | 60S ribosomal protein L7a | 4506661 | 5.3 | 0 | VL | ni | 0.00 | 0.00000 | Unique to Control |
| 59 | Isobutyryl-CoA dehydrogenase, mitochondrial | 7656849 | 7.0 | 0 | VL | ni | 0.00 | 0.00000 | Unique to Control |
| 60 | cAMP-dependent protein kinase catalytic subunit gamma | 15619015 | 7.0 | 0 | VL | ni | 0.00 | 0.00000 | Unique to Control |
| 61 | Vitamin K epoxide reductase complex subunit 1-like protein 1 isoform 1 | 46309463 | 3.7 | 0 | VL | ni | 0.00 | 0.00000 | Unique to Control |
| 62 | Translocation protein SEC63 homolog | 6005872 | 2.0 | 0 | VL | ni | 0.00 | 0.00001 | Unique to Control |
| 63 | UDP-N-acetylhexosamine pyrophosphorylase | 156627575 | 3.0 | 0 | VL | ni | 0.00 | 0.00001 | Unique to Control |
| 64 | Guanine nucleotide-binding protein subunit beta-2-like 1 | 5174447 | 2.0 | 0 | VL | ni | 0.00 | 0.00003 | Unique to Control |
| 65 | Dynein intermediate chain 1, axonemal isoform 2 | 526479830 | 7.0 | 0 | VL | ni | 0.00 | 0.00003 | Unique to Control |
| 66 | Fibronectin type III domain-containing protein 8 | 8922138 | 2.0 | 0 | VL | ni | 0.00 | 0.00009 | Unique to Control |
| 67 | 40S ribosomal protein S26-like | 530438702 | 3.0 | 0 | VL | ni | 0.00 | 0.00009 | Unique to Control |
| 68 | Mitochondrial import receptor subunit TOM22 homolog | 9910382 | 6.0 | 0 | VL | ni | 0.00 | 0.00009 | Unique to Control |
| 69 | Cation channel sperm-associated protein subunit beta precursor | 51339295 | 2.0 | 0 | VL | ni | 0.00 | 0.00009 | Unique to Control |
| 70 | Maestro heat-like repeat-containing protein family member 7 | 223278410 | 3.3 | 0 | VL | ni | 0.00 | 0.00010 | Unique to Control |
| 71 | ADP-ribosylation factor-like protein 2 isoform 1 | 148612885 | 2.7 | 0 | VL | ni | 0.00 | 0.00015 | Unique to Control |
| 72 | protein NDRG1 isoform 1 | 207028748 | 4.0 | 0 | VL | ni | 0.00 | 0.00016 | Unique to Control |
| 73 | Speriolin isoform 1 | 197276668 | 6.3 | 0 | VL | ni | 0.00 | 0.00017 | Unique to Control |
| 74 | Radial spoke head protein 6 homolog A | 13540559 | 3.3 | 0 | VL | ni | 0.00 | 0.00018 | Unique to Control |
| 75 | DCN1-like protein 1 | 36030883 | 4.7 | 0 | VL | ni | 0.00 | 0.00025 | Unique to Control |
| 76 | dnaJ homolog subfamily C member 3 precursor | 5453980 | 3.7 | 0 | VL | ni | 0.00 | 0.00025 | Unique to Control |
| 77 | Sialic acid synthase | 12056473 | 3.0 | 0 | VL | ni | 0.00 | 0.00028 | Unique to Control |
| 78 | Glutamine-tRNA ligase isoform b | 441478305 | 3.7 | 0 | VL | ni | 0.00 | 0.00028 | Unique to Control |
| 79 | Mimitin, mitochondrial | 29789409 | 4.3 | 0 | VL | ni | 0.00 | 0.00031 | Unique to Control |
| 80 | 60S ribosomal protein L22 proprotein | 4506613 | 5.0 | 0 | VL | ni | 0.00 | 0.00032 | Unique to Control |
| 81 | EF-hand calcium-binding domain-containing protein 14 | 7662160 | 6.7 | 0 | VL | ni | 0.00 | 0.00033 | Unique to Control |
| 82 | Iron-sulfur cluster assembly enzyme ISCU, mitochondrial isoform X1 | 530400013 | 4.7 | 0 | VL | ni | 0.00 | 0.00036 | Unique to Control |
| 83 | Growth hormone-inducible transmembrane protein | 118200356 | 4.7 | 0 | VL | ni | 0.00 | 0.00037 | Unique to Control |
| 84 | S-phase kinase-associated protein 1 isoform b | 25777713 | 4.0 | 0 | VL | ni | 0.00 | 0.00040 | Unique to Control |
| 85 | Calcium-binding mitochondrial carrier protein Aralar1 | 21361103 | 3.3 | 0 | VL | ni | 0.00 | 0.00050 | Unique to Control |
| 86 | diphosphomevalonate decarboxylase | 4505289 | 2.3 | 0 | VL | ni | 0.00 | 0.00051 | Unique to Control |
| 87 | V-type proton ATPase subunit E 2 isoform X1 | 530368260 | 4.0 | 0 | VL | ni | 0.00 | 0.00052 | Unique to Control |
| 88 | Nucleosome assembly protein 1-like 1 | 21327708 | 4.3 | 0 | VL | ni | 0.00 | 0.00056 | Unique to Control |
| 89 | 26S protease regulatory subunit 4 | 24430151 | 6.0 | 0 | VL | ni | 0.00 | 0.00056 | Unique to Control |
| 90 | Mitochondrial ornithine transporter 1 | 7657585 | 5.3 | 0 | VL | ni | 0.00 | 0.00057 | Unique to Control |
| 91 | 60S ribosomal protein L5 | 14591909 | 3.7 | 0 | VL | ni | 0.00 | 0.00097 | Unique to Control |
| 92 | Dynein heavy chain 17, axonemal | 256542310 | 88.0 | 1.0 | H | VL | 0.01 | 0.00001 | UE in Seminoma |
| 93 | L-amino-acid oxidase isoform 2 precursor | 384381475 | 76.0 | 0.3 | M | VL | 0.01 | 0.00000 | UE in Seminoma |
| 94 | Sperm-associated antigen 6 isoform X1 | 530392552 | 58.0 | 0.3 | M | VL | 0.01 | 0.00000 | UE in Seminoma |
| 95 | Nuclear pore complex protein Nup93 isoform X1 | 530424559 | 37.7 | 0.3 | M | VL | 0.01 | 0.00129 | UE in Seminoma |
| 96 | Valine-tRNA ligase | 5454158 | 87.7 | 1.7 | H | VL | 0.01 | 0.00004 | UE in Seminoma |
| 97 | Sperm surface protein Sp17 | 8394343 | 31.3 | 0.3 | M | VL | 0.02 | 0.00010 | UE in Seminoma |
| 98 | Exportin-2 isoform 1 | 29029559 | 16.7 | 0.3 | L | VL | 0.02 | 0.00026 | UE in Seminoma |
| 99 | 26S proteasome non-ATPase regulatory subunit 13 isoform 1 | 157502193 | 19.7 | 0.3 | L | VL | 0.02 | 0.00142 | UE in Seminoma |
| 100 | Cathepsin F precursor | 6042196 | 21.0 | 0.3 | M | VL | 0.03 | 0.00007 | UE in Seminoma |
| 101 | 26S proteasome non-ATPase regulatory subunit 7 | 25777615 | 13.7 | 0.3 | L | VL | 0.03 | 0.00230 | UE in Seminoma |
| 102 | Uncharacterized protein C7orf61 | 51972226 | 14.3 | 0.3 | L | VL | 0.03 | 0.00109 | UE in Seminoma |
| 103 | Vacuolar protein sorting-associated protein 13A isoform C | 66346672 | 19.7 | 0.3 | L | VL | 0.03 | 0.00122 | UE in Seminoma |
| 104 | Mitochondrial pyruvate carrier 1-like protein | 306922396 | 18.0 | 0.3 | L | VL | 0.03 | 0.00012 | UE in Seminoma |
| 105 | Plasma membrane calcium-transporting ATPase 4 isoform 4b | 48255957 | 52.3 | 2.7 | M | VL | 0.03 | 0.00001 | UE in Seminoma |
| 106 | Presequence protease, mitochondrial isoform 2 precursor | 41352061 | 50.3 | 1.0 | M | VL | 0.03 | 0.00003 | UE in Seminoma |
| 107 | Exportin-1 isoform X1 | 530368070 | 8.3 | 0.3 | L | VL | 0.03 | 0.00004 | UE in Seminoma |
| 108 | Ras-related protein Rab-11B | 190358517 | 15.7 | 0.3 | L | VL | 0.03 | 0.00012 | UE in Seminoma |
| 109 | Phosphatidylethanolamine-binding protein 4 precursor | 116812622 | 15.0 | 0.3 | L | VL | 0.04 | 0.00029 | UE in Seminoma |
| 110 | Protein FAM71A | 282721094 | 12.3 | 0.3 | L | VL | 0.04 | 0.00258 | UE in Seminoma |
| 111 | Puromycin-sensitive aminopeptidase | 158937236 | 45.0 | 1.3 | M | VL | 0.04 | 0.00124 | UE in Seminoma |
| 112 | Epimerase family protein SDR39U1 isoform 1 | 116812630 | 13.3 | 0.3 | L | VL | 0.04 | 0.00151 | UE in Seminoma |
| 113 | V-type proton ATPase catalytic subunit A | 19913424 | 15.7 | 0.3 | L | VL | 0.04 | 0.00313 | UE in Seminoma |
| 114 | Cullin-associated NEDD8-dissociated protein 1 | 21361794 | 143.3 | 7.0 | H | VL | 0.05 | 0.00000 | UE in Seminoma |
| 115 | Low molecular weight phosphotyrosine protein phosphatase isoform c | 4757714 | 8.7 | 0.3 | L | VL | 0.05 | 0.00005 | UE in Seminoma |
| 116 | Dynein heavy chain 8, axonemal isoform X1 | 578811443 | 132.3 | 6.3 | H | VL | 0.05 | 0.00003 | UE in Seminoma |
| 117 | Heat shock protein 75, mitochondrial isoform 1 precursor | 155722983 | 8.3 | 0.3 | L | VL | 0.05 | 0.00081 | UE in Seminoma |
| 118 | Cullin-3 isoform 3 | 380714665 | 58.3 | 2.3 | M | VL | 0.05 | 0.00012 | UE in Seminoma |
| 119 | Lysosomal alpha-glucosidase isoform X1 | 530411863 | 5.0 | 0.3 | VL | VL | 0.05 | 0.00074 | UE in Seminoma |
| 120 | Isoleucine-tRNA ligase, mitochondrial precursor | 46852147 | 40.7 | 1.7 | M | VL | 0.05 | 0.00001 | UE in Seminoma |
| 121 | Protein FAM71B | 222418633 | 46.7 | 1.3 | M | VL | 0.05 | 0.00050 | UE in Seminoma |
| 122 | Actin-related protein T2 | 29893808 | 45.7 | 1.7 | M | VL | 0.06 | 0.00004 | UE in Seminoma |
| 123 | Thioredoxin domain-containing protein 3 | 148839372 | 18.3 | 1.0 | L | VL | 0.06 | 0.00016 | UE in Seminoma |
| 124 | Carnitine O-palmitoyltransferase 1, muscle isoform isoform a | 4758050 | 11.3 | 1.0 | L | VL | 0.06 | 0.00107 | UE in Seminoma |
| 125 | Phosphoglycolate phosphatase | 108796653 | 15.3 | 0.3 | L | VL | 0.06 | 0.00014 | UE in Seminoma |
| 126 | Ecto-ADP-ribosyltransferase 3 isoform X8 | 530377706 | 38.3 | 1.7 | M | VL | 0.06 | 0.00004 | UE in Seminoma |
| 127 | EF-hand calcium-binding domain-containing protein 1 isoform a | 13375787 | 11.3 | 0.3 | L | VL | 0.07 | 0.00341 | UE in Seminoma |
| 128 | Izumo sperm-egg fusion protein 2 isoform X1 | 578833932 | 9.0 | 0.3 | L | VL | 0.07 | 0.00273 | UE in Seminoma |
| 129 | Sodium/potassium-transporting ATPase subunit alpha-4 isoform 1 | 153946397 | 59.7 | 5.3 | M | VL | 0.07 | 0.00006 | UE in Seminoma |
| 130 | Enoyl-CoA delta isomerase 2, mitochondrial isoform 2 | 260274832 | 25.3 | 1.0 | M | VL | 0.07 | 0.00081 | UE in Seminoma |
| 131 | Casein kinase II subunit beta isoform 1 | 23503295 | 9.3 | 0.3 | L | VL | 0.07 | 0.00181 | UE in Seminoma |
| 132 | Small membrane A-kinase anchor protein | 110349742 | 9.3 | 0.3 | L | VL | 0.07 | 0.00193 | UE in Seminoma |
| 133 | 60S ribosomal protein L12 | 4506597 | 14.3 | 0.7 | L | VL | 0.07 | 0.00044 | UE in Seminoma |
| 134 | Leucine-rich repeat-containing protein 37A3 precursor | 75677612 | 20.3 | 1.3 | M | VL | 0.07 | 0.00021 | UE in Seminoma |
| 135 | NADH dehydrogenase (ubiquinone) iron-sulfur protein 8, mitochondrial isoform X1 | 530396818 | 8.0 | 0.3 | L | VL | 0.07 | 0.00305 | UE in Seminoma |
| 136 | Heat shock 70 protein 4L | 31541941 | 93.3 | 2.3 | H | VL | 0.08 | 0.00012 | UE in Seminoma |
| 137 | Sperm equatorial segment protein 1 precursor | 21717832 | 100.7 | 5.0 | H | VL | 0.08 | 0.00000 | UE in Seminoma |
| 138 | Pyruvate dehydrogenase E1 component subunit beta, mitochondrial isoform 1 precursor | 156564403 | 67.3 | 3.3 | M | VL | 0.08 | 0.00002 | UE in Seminoma |
| 139 | Choline transporter-like protein 5 isoform B | 194239633 | 8.0 | 1.3 | L | VL | 0.08 | 0.00340 | UE in Seminoma |
| 140 | 6-Phosphofructokinase type C isoform 1 | 11321601 | 131.3 | 10.7 | H | L | 0.09 | 0.00000 | UE in Seminoma |
| 141 | 26S proteasome non-ATPase regulatory subunit 12 isoform 1 | 4506221 | 10.7 | 0.7 | L | VL | 0.09 | 0.00427 | UE in Seminoma |
| 142 | ruvB-like 2 | 5730023 | 137.3 | 7.7 | H | VL | 0.09 | 0.00007 | UE in Seminoma |
| 143 | T-complex protein 1 subunit gamma isoform a | 63162572 | 128.7 | 7.7 | H | VL | 0.09 | 0.00000 | UE in Seminoma |
| 144 | ATP synthase subunit beta, mitochondrial precursor | 32189394 | 354.3 | 21.7 | H | M | 0.09 | 0.00000 | UE in Seminoma |
| 145 | Phosphatidylglycerophosphatase and protein-tyrosine phosphatase 1 isoform 1 | 148224884 | 11.7 | 0.7 | L | VL | 0.09 | 0.00097 | UE in Seminoma |
| 146 | 26S proteasome non-ATPase regulatory subunit 3 | 25777612 | 35.3 | 3.0 | M | VL | 0.09 | 0.00001 | UE in Seminoma |
| 147 | Importin-5 isoform X2 | 530423350 | 24.7 | 1.7 | M | VL | 0.10 | 0.00181 | UE in Seminoma |
| 148 | Mitochondrial dicarboxylate carrier isoform 2 | 20149598 | 56.0 | 3.7 | M | VL | 0.10 | 0.00026 | UE in Seminoma |
| 149 | TMEM189-UBE2V1 fusion protein | 40806190 | 8.3 | 0.7 | L | VL | 0.10 | 0.00394 | UE in Seminoma |
| 150 | Dynein heavy chain 7, axonemal | 151301127 | 18.0 | 1.0 | L | VL | 0.11 | 0.00033 | UE in Seminoma |
| 151 | Lysozyme-like protein 1 | 73390143 | 9.7 | 0.7 | L | VL | 0.11 | 0.00451 | UE in Seminoma |
| 152 | Importin subunit alpha-1 | 4504897 | 54.7 | 3.3 | M | VL | 0.11 | 0.00004 | UE in Seminoma |
| 153 | Nuclear pore complex protein Nup155 isoform 1 | 24430149 | 86.0 | 8.3 | H | L | 0.12 | 0.00002 | UE in Seminoma |
| 154 | Mitochondrial 2-oxoglutarate/malate carrier protein isoform 1 | 21361114 | 39.0 | 2.7 | M | VL | 0.12 | 0.00399 | UE in Seminoma |
| 155 | Hyaluronidase PH-20 isoform 2 | 23510418 | 35.3 | 2.3 | M | VL | 0.12 | 0.00063 | UE in Seminoma |
| 156 | 40S ribosomal protein S16 | 4506691 | 10.7 | 0.7 | L | VL | 0.12 | 0.00186 | UE in Seminoma |
| 157 | 26S proteasome non-ATPase regulatory subunit 11 | 28872725 | 13.0 | 1.0 | L | VL | 0.12 | 0.00032 | UE in Seminoma |
| 158 | 26S proteasome non-ATPase regulatory subunit 6 isoform 2 | 7661914 | 18.7 | 1.7 | L | VL | 0.13 | 0.00236 | UE in Seminoma |
| 159 | T-complex protein 1 subunit zeta-2 isoform X1 | 578830267 | 36.7 | 3.3 | M | VL | 0.13 | 0.00001 | UE in Seminoma |
| 160 | Bifunctional ATP-dependent dihydroxyacetone kinase/FAD-AMP lyase (cyclizing) isoform X1 | 530396576 | 29.0 | 3.0 | M | VL | 0.13 | 0.00104 | UE in Seminoma |
| 161 | Ropporin-1B | 59891409 | 92.7 | 7.3 | H | VL | 0.13 | 0.00003 | UE in Seminoma |
| 162 | Dynactin subunit 2 isoform 3 | 387527974 | 15.7 | 1.7 | L | VL | 0.13 | 0.00423 | UE in Seminoma |
| 163 | ras-related protein Rab-14 | 19923483 | 19.0 | 1.3 | L | VL | 0.13 | 0.00925 | UE in Seminoma |
| 164 | Proteasome activator complex subunit 4 | 163644283 | 52.7 | 5.7 | M | VL | 0.13 | 0.00058 | UE in Seminoma |
| 165 | T-complex protein 1 subunit alpha isoform a | 57863257 | 132.3 | 13.0 | H | L | 0.13 | 0.00002 | UE in Seminoma |
| 166 | Pyruvate dehydrogenase E1 component subunit alpha, testis-specific form, mitochondrial precursor | 4885543 | 50.0 | 3.7 | M | VL | 0.13 | 0.00018 | UE in Seminoma |
| 167 | Dynein light chain roadblock-type 2 | 18702323 | 8.7 | 0.7 | L | VL | 0.13 | 0.00338 | UE in Seminoma |
| 168 | Nuclear transport factor 2 | 5031985 | 9.7 | 0.7 | L | VL | 0.13 | 0.00959 | UE in Seminoma |
| 169 | Metalloreductase STEAP4 isoform 1 | 100815815 | 13.3 | 1.7 | L | VL | 0.13 | 0.00942 | UE in Seminoma |
| 170 | Prenylated Rab acceptor protein 1 | 222144309 | 8.3 | 1.0 | L | VL | 0.13 | 0.00233 | UE in Seminoma |
| 171 | Heat shock protein 105 isoform 1 | 42544159 | 8.7 | 0.7 | L | VL | 0.14 | 0.00595 | UE in Seminoma |
| 172 | ATP synthase subunit gamma, mitochondrial isoform L (liver) precursor | 50345988 | 48.7 | 3.3 | M | VL | 0.14 | 0.00014 | UE in Seminoma |
| 173 | 3-Hydroxyisobutyryl-CoA hydrolase, mitochondrial isoform 1 precursor | 37594471 | 11.7 | 1.0 | L | VL | 0.14 | 0.00055 | UE in Seminoma |
| 174 | Transmembrane protein 89 precursor | 56847630 | 12.7 | 1.0 | L | VL | 0.14 | 0.00529 | UE in Seminoma |
| 175 | T-complex protein 1 subunit beta isoform 1 | 5453603 | 120.7 | 12.0 | H | L | 0.14 | 0.00005 | UE in Seminoma |
| 176 | T-complex protein 1 subunit zeta isoform a | 4502643 | 71.7 | 7.3 | M | VL | 0.15 | 0.00013 | UE in Seminoma |
| 177 | Inactive serine protease 54 precursor | 122937420 | 19.0 | 1.7 | L | VL | 0.15 | 0.00060 | UE in Seminoma |
| 178 | Nucleoporin p54 isoform 1 | 26051237 | 21.7 | 2.3 | M | VL | 0.16 | 0.01278 | UE in Seminoma |
| 179 | T-complex protein 1 subunit theta isoform 1 | 48762932 | 77.7 | 8.3 | M | L | 0.16 | 0.00032 | UE in Seminoma |
| 180 | Sperm protein associated with the nucleus on the X chromosome B/F | 190570192 | 22.0 | 2.7 | M | VL | 0.16 | 0.00428 | UE in Seminoma |
| 181 | Histone H2A-Bbd type 2/3 | 63029935 | 21.7 | 2.0 | M | VL | 0.16 | 0.00688 | UE in Seminoma |
| 182 | Transcription elongation factor B polypeptide 2 isoform a | 6005890 | 11.0 | 1.3 | L | VL | 0.16 | 0.00017 | UE in Seminoma |
| 183 | Protein MENT isoform X1 | 578801150 | 97.7 | 11.0 | H | L | 0.17 | 0.00005 | UE in Seminoma |
| 184 | ATP synthase subunit d, mitochondrial isoform a | 5453559 | 33.0 | 3.0 | M | VL | 0.17 | 0.00091 | UE in Seminoma |
| 185 | Ropporin-1A isoform X1 | 530374814 | 55.7 | 5.7 | M | VL | 0.17 | 0.00001 | UE in Seminoma |
| 186 | ATP synthase F (0) complex subunit B1, mitochondrial precursor | 21361565 | 35.3 | 3.7 | M | VL | 0.17 | 0.00076 | UE in Seminoma |
| 187 | NADH dehydrogenase (ubiquinone) flavoprotein 1, mitochondrial isoform 1 precursor | 20149568 | 14.0 | 1.7 | L | VL | 0.17 | 0.00400 | UE in Seminoma |
| 188 | Apolipoprotein O isoform X1 | 578837961 | 40.3 | 4.3 | M | VL | 0.17 | 0.00011 | UE in Seminoma |
| 189 | 26S proteasome non-ATPase regulatory subunit 1 isoform 1 | 25777600 | 49.0 | 8.3 | M | L | 0.17 | 0.00016 | UE in Seminoma |
| 190 | Elongation factor 1-delta isoform 1 | 304555581 | 32.3 | 3.7 | M | VL | 0.18 | 0.00006 | UE in Seminoma |
| 191 | 26S proteasome non-ATPase regulatory subunit 8 | 156631005 | 25.7 | 2.3 | M | VL | 0.18 | 0.00001 | UE in Seminoma |
| 192 | ATP synthase subunit alpha, mitochondrial isoform a precursor | 50345984 | 265.3 | 33.0 | H | M | 0.18 | 0.00001 | UE in Seminoma |
| 193 | Heat shock 70 protein 1-like isoform X1 | 530381921 | 207.0 | 24.3 | H | M | 0.19 | 0.00038 | UE in Seminoma |
| 194 | Nitrilase homolog 1 isoform 3 | 297632348 | 18.7 | 2.0 | L | VL | 0.19 | 0.00095 | UE in Seminoma |
| 195 | T-complex protein 1 subunit eta isoform a | 5453607 | 129.7 | 16.0 | H | L | 0.19 | 0.00010 | UE in Seminoma |
| 196 | Calcium-binding tyrosine phosphorylation-regulated protein isoform a | 24797108 | 63.3 | 9.0 | M | L | 0.20 | 0.00012 | UE in Seminoma |
| 197 | Tricarboxylate transport protein, mitochondrial isoform b | 374717343 | 15.3 | 1.7 | L | VL | 0.20 | 0.00049 | UE in Seminoma |
| 198 | T-complex protein 1 subunit epsilon | 24307939 | 78.7 | 11.3 | M | L | 0.20 | 0.00001 | UE in Seminoma |
| 199 | Tissue alpha-L-fucosidase precursor | 119360348 | 19.0 | 1.7 | L | VL | 0.20 | 0.00008 | UE in Seminoma |
| 200 | GTP-binding nuclear protein Ran | 5453555 | 22.0 | 2.7 | M | VL | 0.20 | 0.00007 | UE in Seminoma |
| 201 | Dipeptidyl peptidase 2 isoform X1 | 530426726 | 17.7 | 1.7 | L | VL | 0.20 | 0.00216 | UE in Seminoma |
| 202 | 3’(2’),5’-Bisphosphate nucleotidase 1 isoform X3 | 530365931 | 19.3 | 2.3 | L | VL | 0.20 | 0.00871 | UE in Seminoma |
| 203 | Lysine-tRNA ligase isoform 1 | 194272210 | 30.0 | 3.0 | M | VL | 0.20 | 0.00462 | UE in Seminoma |
| 204 | Mitochondrial thiamine pyrophosphate carrier isoform X1 | 530412630 | 16.3 | 2.0 | L | VL | 0.21 | 0.00139 | UE in Seminoma |
| 205 | Vesicle-fusing ATPase isoform X1 | 578831007 | 16.7 | 2.3 | L | VL | 0.21 | 0.00045 | UE in Seminoma |
| 206 | FUN14 domain-containing protein 2 | 24371248 | 60.3 | 8.0 | M | L | 0.22 | 0.00023 | UE in Seminoma |
| 207 | Mitochondrial pyruvate carrier 2 | 219521872 | 25.7 | 5.0 | M | VL | 0.22 | 0.00370 | UE in Seminoma |
| 208 | Dynein light chain 1, axonemal isoform 1 | 164607156 | 14.3 | 2.0 | L | VL | 0.23 | 0.00000 | UE in Seminoma |
| 209 | Cytochrome b-c1 complex subunit 2, mitochondrial precursor | 50592988 | 111.0 | 14.0 | H | L | 0.23 | 0.00006 | UE in Seminoma |
| 210 | ADP/ATP translocase 4 | 13775208 | 140.3 | 25.3 | H | M | 0.23 | 0.00180 | UE in Seminoma |
| 211 | 26S protease regulatory subunit 7 isoform 1 | 4506209 | 9.7 | 1.7 | L | VL | 0.23 | 0.00149 | UE in Seminoma |
| 212 | Uncharacterized protein C9orf9 | 33285006 | 45.0 | 6.7 | M | VL | 0.23 | 0.00005 | UE in Seminoma |
| 213 | ADP/ATP translocase 2 | 156071459 | 35.7 | 8.3 | M | L | 0.24 | 0.00081 | UE in Seminoma |
| 214 | Synaptojanin-2-binding protein | 157388993 | 28.3 | 5.0 | M | VL | 0.24 | 0.00057 | UE in Seminoma |
| 215 | Heat shock 70 protein 1A/1B | 167466173 | 54.7 | 8.0 | M | L | 0.24 | 0.00086 | UE in Seminoma |
| 216 | 26S protease regulatory subunit 6B isoform 1 | 5729991 | 16.0 | 2.3 | L | VL | 0.24 | 0.00222 | UE in Seminoma |
| 217 | Mannose-6-phosphate isomerase isoform 1 | 4505235 | 9.0 | 1.3 | L | VL | 0.25 | 0.00220 | UE in Seminoma |
| 218 | Nuclear pore membrane glycoprotein 210 precursor | 27477134 | 23.3 | 3.3 | M | VL | 0.25 | 0.00026 | UE in Seminoma |
| 219 | Arylsulfatase A isoform a precursor | 313569791 | 28.0 | 3.3 | M | VL | 0.25 | 0.00335 | UE in Seminoma |
| 220 | Leucine-rich repeat-containing protein 37A isoform X5 | 530413292 | 165.7 | 27.7 | H | M | 0.26 | 0.00002 | UE in Seminoma |
| 221 | Solute carrier family 2, facilitated glucose transporter member 5 isoform X2 | 578799621 | 31.0 | 7.7 | M | VL | 0.26 | 0.00472 | UE in Seminoma |
| 222 | Protein-glutamine gamma-glutamyltransferase 4 | 156627577 | 232.3 | 44.0 | H | M | 0.26 | 0.00006 | UE in Seminoma |
| 223 | Protein FAM162A | 49355721 | 9.0 | 1.3 | L | VL | 0.26 | 0.00288 | UE in Seminoma |
| 224 | 26S protease regulatory subunit 6A | 21361144 | 22.3 | 4.0 | M | VL | 0.27 | 0.00780 | UE in Seminoma |
| 225 | Myosin regulatory light chain 12B | 15809016 | 11.7 | 2.3 | L | VL | 0.27 | 0.00058 | UE in Seminoma |
| 226 | Hexokinase-1 isoform X2 | 530393498 | 345.3 | 64.0 | H | M | 0.27 | 0.00001 | UE in Seminoma |
| 227 | NADH dehydrogenase (ubiquinone) iron-sulfur protein 7, mitochondrial | 187281616 | 9.7 | 2.0 | L | VL | 0.27 | 0.00101 | UE in Seminoma |
| 228 | Cytochrome b-c1 complex subunit Rieske, mitochondrial | 163644321 | 27.0 | 4.7 | M | VL | 0.27 | 0.00004 | UE in Seminoma |
| 229 | Leucine-rich repeat-containing protein 37B precursor | 53829385 | 176.3 | 40.3 | H | M | 0.27 | 0.00007 | UE in Seminoma |
| 230 | ES1 protein homolog, mitochondrial-like isoform X1 | 578797780 | 35.0 | 5.7 | M | VL | 0.27 | 0.00001 | UE in Seminoma |
| 231 | Lysosomal Pro-X carboxypeptidase isoform 1 preproprotein | 4826940 | 13.3 | 1.7 | L | VL | 0.27 | 0.00077 | UE in Seminoma |
| 232 | Transmembrane protein 190 precursor | 21040263 | 33.3 | 5.7 | M | VL | 0.28 | 0.00004 | UE in Seminoma |
| 233 | UTP-glucose-1-phosphate uridylyltransferase isoform a | 48255966 | 20.3 | 5.0 | M | VL | 0.28 | 0.00603 | UE in Seminoma |
| 234 | 26S protease regulatory subunit 10B | 195539395 | 21.3 | 4.3 | M | VL | 0.29 | 0.00659 | UE in Seminoma |
| 235 | Dynactin subunit 1 isoform 4 | 205277396 | 21.7 | 3.7 | M | VL | 0.29 | 0.00497 | UE in Seminoma |
| 236 | 26S protease regulatory subunit 8 isoform 1 | 24497435 | 17.0 | 3.7 | L | VL | 0.29 | 0.00479 | UE in Seminoma |
| 237 | Ethanolamine-phosphate cytidylyltransferase isoform 6 | 532524977 | 16.0 | 3.0 | L | VL | 0.29 | 0.00018 | UE in Seminoma |
| 238 | 60 heat shock protein, mitochondrial isoform X1 | 530370277 | 125.7 | 16.3 | H | L | 0.30 | 0.00029 | UE in Seminoma |
| 239 | Beta-galactosidase-1-like protein isoform X1 | 530370954 | 35.3 | 4.7 | M | VL | 0.30 | 0.00099 | UE in Seminoma |
| 240 | Adenylate kinase isoenzyme 1 isoform X1 | 530390694 | 45.3 | 7.7 | M | VL | 0.30 | 0.00114 | UE in Seminoma |
| 241 | Dynein light chain Tctex-type 1 | 5730085 | 10.0 | 1.7 | L | VL | 0.30 | 0.00600 | UE in Seminoma |
| 242 | Chitinase domain-containing protein 1 isoform X2 | 530395670 | 17.3 | 3.3 | L | VL | 0.31 | 0.00482 | UE in Seminoma |
| 243 | A-kinase anchor protein 4 isoform 1 | 21493037 | 156.3 | 28.0 | H | M | 0.31 | 0.00000 | UE in Seminoma |
| 244 | Hypoxia up-regulated protein 1 isoform X2 | 530397761 | 177.0 | 37.0 | H | M | 0.31 | 0.00002 | UE in Seminoma |
| 245 | Diablo homolog, mitochondrial isoform 1 precursor | 9845297 | 27.3 | 6.7 | M | VL | 0.32 | 0.00223 | UE in Seminoma |
| 246 | Zona pellucida-binding protein 2 isoform 2 precursor | 40556389 | 45.7 | 9.0 | M | L | 0.32 | 0.00009 | UE in Seminoma |
| 247 | Ubiquitin-like modifier-activating enzyme 1 isoform X1 | 530421539 | 75.0 | 11.3 | M | L | 0.32 | 0.00035 | UE in Seminoma |
| 248 | 40S ribosomal protein S15a | 14165469 | 12.7 | 2.3 | L | VL | 0.33 | 0.00231 | UE in Seminoma |
| 249 | Prohibitin isoform 1 | 527498279 | 22.7 | 4.7 | M | VL | 0.33 | 0.00192 | UE in Seminoma |
| 250 | Long-chain-fatty-acid-CoA ligase 6 isoform e | 327412327 | 39.0 | 6.0 | M | VL | 0.33 | 0.00042 | UE in Seminoma |
| 251 | Importin subunit beta-1 isoform 1 | 19923142 | 54.7 | 11.7 | M | L | 0.34 | 0.00045 | UE in Seminoma |
| 252 | Long-chain-fatty-acid-CoA ligase 1 isoform X3 | 530377352 | 170.7 | 45.7 | H | M | 0.34 | 0.00024 | UE in Seminoma |
| 253 | AP-1 complex subunit beta-1 isoform b | 260436860 | 12.0 | 2.3 | L | VL | 0.34 | 0.00257 | UE in Seminoma |
| 254 | Acrosin precursor | 148613878 | 255.7 | 65.3 | H | M | 0.34 | 0.00011 | UE in Seminoma |
| 255 | Glutathione S-transferase omega-2 isoform 2 | 300360567 | 10.3 | 2.0 | L | VL | 0.34 | 0.00400 | UE in Seminoma |
| 256 | Carboxypeptidase D isoform 1 precursor | 22202611 | 36.3 | 7.0 | M | VL | 0.34 | 0.02105 | UE in Seminoma |
| 257 | Phosphate carrier protein, mitochondrial isoform b precursor | 4505775 | 38.3 | 14.0 | M | L | 0.35 | 0.00176 | UE in Seminoma |
| 258 | Heat shock 70 protein 4 | 38327039 | 44.7 | 7.7 | M | VL | 0.35 | 0.00303 | UE in Seminoma |
| 259 | Fatty acid-binding protein, epidermal | 4557581 | 30.7 | 6.0 | M | VL | 0.35 | 0.00328 | UE in Seminoma |
| 260 | Ras-related protein Rab-2A isoform a | 4506365 | 156.7 | 37.3 | H | M | 0.36 | 0.00002 | UE in Seminoma |
| 261 | 3-hydroxyacyl-CoA dehydrogenase type-2 isoform 1 | 4758504 | 43.0 | 9.3 | M | L | 0.36 | 0.00138 | UE in Seminoma |
| 262 | T-complex protein 1 subunit delta isoform a | 38455427 | 108.3 | 25.7 | H | M | 0.36 | 0.00075 | UE in Seminoma |
| 263 | cAMP-dependent protein kinase type II-alpha regulatory subunit isoform X1 | 530372834 | 109.7 | 29.0 | H | M | 0.36 | 0.00000 | UE in Seminoma |
| 264 | Glutamine synthetase isoform X1 | 578800828 | 23.3 | 5.3 | M | VL | 0.36 | 0.01626 | UE in Seminoma |
| 265 | Calmodulin isoform X1 | 578826144 | 75.0 | 20.7 | M | M | 0.37 | 0.00004 | UE in Seminoma |
| 266 | Elongation factor 1-beta | 4503477 | 10.0 | 2.3 | L | VL | 0.37 | 0.00022 | UE in Seminoma |
| 267 | ruvB-like 1 | 4506753 | 99.7 | 20.3 | H | M | 0.37 | 0.00684 | UE in Seminoma |
| 268 | Elongation factor 1-alpha 1 | 4503471 | 155.0 | 51.3 | H | M | 0.38 | 0.00001 | UE in Seminoma |
| 269 | hsc70-interacting protein isoform 1 | 19923193 | 30.3 | 7.3 | M | VL | 0.38 | 0.01230 | UE in Seminoma |
| 270 | Transmembrane protein 126A isoform 1 | 14150017 | 22.0 | 6.0 | M | VL | 0.38 | 0.00020 | UE in Seminoma |
| 271 | 26S proteasome non-ATPase regulatory subunit 2 isoform 1 | 25777602 | 56.0 | 9.0 | M | L | 0.39 | 0.00049 | UE in Seminoma |
| 272 | Arachidonate 15-lipoxygenase B isoform d | 85067501 | 23.7 | 4.3 | M | VL | 0.40 | 0.00236 | UE in Seminoma |
| 273 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex subunit 9, mitochondrial precursor | 6681764 | 9.7 | 2.3 | L | VL | 0.40 | 0.00078 | UE in Seminoma |
| 274 | electron transfer flavoprotein subunit beta isoform 1 | 4503609 | 22.0 | 5.0 | M | VL | 0.41 | 0.00000 | UE in Seminoma |
| 275 | Tubulin alpha-3C/D chain | 156564363 | 242.3 | 60.7 | H | M | 0.41 | 0.00001 | UE in Seminoma |
| 276 | Fumarylacetoacetate hydrolase domain-containing protein 2B | 40786394 | 31.0 | 7.7 | M | VL | 0.41 | 0.00018 | UE in Seminoma |
| 277 | Peroxiredoxin-5, mitochondrial isoform a precursor | 6912238 | 78.0 | 20.7 | M | M | 0.41 | 0.00039 | UE in Seminoma |
| 278 | NADH-ubiquinone oxidoreductase 75 subunit, mitochondrial isoform 4 | 316983156 | 21.0 | 4.0 | M | VL | 0.42 | 0.03074 | UE in Seminoma |
| 279 | Probable C-mannosyltransferase DPY19L2 isoform X1 | 578823598 | 101.0 | 26.0 | H | M | 0.42 | 0.00052 | UE in Seminoma |
| 280 | L-lactate dehydrogenase A-like 6B | 15082234 | 145.0 | 42.7 | H | M | 0.42 | 0.00021 | UE in Seminoma |
| 281 | Acrosin-binding protein precursor | 17999524 | 288.3 | 74.7 | H | M | 0.43 | 0.00003 | UE in Seminoma |
| 282 | Tubulin beta-4B chain | 5174735 | 287.3 | 81.3 | H | L | 0.43 | 0.00004 | UE in Seminoma |
| 283 | Electron transfer flavoprotein subunit alpha, mitochondrial isoform a | 4503607 | 46.0 | 12.0 | M | L | 0.44 | 0.00065 | UE in Seminoma |
| 284 | Serpin B6 isoform d | 425876768 | 79.0 | 33.3 | M | M | 0.46 | 0.00013 | UE in Seminoma |
| 285 | Lysozyme-like protein 4 isoform X2 | 578805633 | 29.3 | 8.0 | M | L | 0.46 | 0.00114 | UE in Seminoma |
| 286 | cAMP-dependent protein kinase type I-alpha regulatory subunit isoform a | 443497964 | 42.0 | 12.0 | M | L | 0.47 | 0.00245 | UE in Seminoma |
| 287 | Vesicle-associated membrane protein-associated protein A isoform 2 | 94721252 | 38.7 | 12.0 | M | L | 0.47 | 0.00220 | UE in Seminoma |
| 288 | Acrosomal protein SP-10 isoform a precursor | 4501879 | 64.3 | 20.0 | M | M | 0.48 | 0.00064 | UE in Seminoma |
| 289 | Carnitine O-acetyltransferase isoform 2 | 383209673 | 41.0 | 11.7 | M | L | 0.48 | 0.00294 | UE in Seminoma |
| 290 | Endoplasmin precursor | 4507677 | 543.7 | 146.0 | H | H | 0.49 | 0.00066 | UE in Seminoma |
| 291 | Izumo sperm-egg fusion protein 4 isoform 1 precursor | 89903025 | 119.3 | 39.0 | H | M | 0.53 | 0.00058 | UE in Seminoma |
| 292 | Heat shock-related 70 protein 2 | 13676857 | 442.0 | 126.0 | H | H | 0.53 | 0.00000 | UE in Seminoma |
| 293 | Elongation factor 2 | 4503483 | 84.3 | 30.0 | H | M | 0.56 | 0.00021 | UE in Seminoma |
| 294 | Clathrin heavy chain 1 isoform X2 | 530411491 | 122.7 | 51.3 | H | M | 0.58 | 0.00014 | UE in Seminoma |
| 295 | Sperm acrosome membrane-associated protein 1 precursor | 13569934 | 176.0 | 83.7 | H | H | 0.60 | 0.00073 | UE in Seminoma |
| 296 | Zona pellucida-binding protein 1 isoform 1 precursor | 229577313 | 278.7 | 110.3 | H | H | 0.61 | 0.00002 | UE in Seminoma |
| 297 | Phosphoglycerate kinase 2 | 31543397 | 204.3 | 79.0 | H | M | 0.65 | 0.00034 | UE in Seminoma |
| 298 | 2,4-Dienoyl-CoA reductase, mitochondrial precursor | 4503301 | 180.7 | 70.0 | H | M | 0.65 | 0.00211 | UE in Seminoma |
| 299 | Aminopeptidase N isoform X1 | 530407092 | 218.0 | 186.3 | H | H | 1.54 | 0.00040 | OE in Seminoma |
| 300 | Calreticulin precursor | 4757900 | 125.0 | 106.7 | H | H | 1.55 | 0.00511 | OE in Seminoma |
| 301 | Dipeptidyl peptidase 4 | 18765694 | 138.0 | 122.0 | H | H | 1.61 | 0.00164 | OE in Seminoma |
| 302 | Plastin-2 isoform X2 | 530402335 | 116.7 | 100.0 | H | H | 1.62 | 0.00036 | OE in Seminoma |
| 303 | Angiotensin-converting enzyme isoform 1 precursor | 4503273 | 141.3 | 124.7 | H | H | 1.62 | 0.01310 | OE in Seminoma |
| 304 | Transitional endoplasmic reticulum ATPase | 6005942 | 145.0 | 97.0 | H | H | 1.75 | 0.00238 | OE in Seminoma |
| 305 | Neprilysin | 116256327 | 85.3 | 59.3 | H | H | 2.01 | 0.00052 | OE in Seminoma |
| 306 | Adipocyte plasma membrane-associated protein | 24308201 | 54.0 | 66.7 | M | M | 2.01 | 0.00011 | OE in Seminoma |
| 307 | Carboxypeptidase Z isoform 1 precursor | 62388877 | 29.3 | 28.0 | M | M | 2.06 | 0.00040 | OE in Seminoma |
| 308 | Annexin A5 | 4502107 | 41.0 | 48.0 | M | M | 2.08 | 0.00049 | OE in Seminoma |
| 309 | Annexin A2 isoform 2 | 50845386 | 46.7 | 57.7 | M | M | 2.09 | 0.00126 | OE in Seminoma |
| 310 | Lysosome-associated membrane glycoprotein 1 precursor | 112380628 | 18.0 | 20.0 | L | M | 2.14 | 0.00275 | OE in Seminoma |
| 311 | Plasma serine protease inhibitor preproprotein | 194018472 | 40.7 | 45.7 | M | M | 2.15 | 0.00208 | OE in Seminoma |
| 312 | Dehydrogenase/reductase SDR family member 7 isoform X1 | 530403978 | 22.3 | 35.7 | M | M | 2.46 | 0.00033 | OE in Seminoma |
| 313 | Cysteine-rich secretory protein 1 isoform 1 precursor | 25121982 | 24.0 | 35.7 | M | M | 2.53 | 0.00587 | OE in Seminoma |
| 314 | Annexin A4 | 4502105 | 22.3 | 31.3 | M | M | 2.56 | 0.01438 | OE in Seminoma |
| 315 | Calnexin precursor | 66933005 | 34.3 | 51.3 | M | M | 2.56 | 0.00094 | OE in Seminoma |
| 316 | Clusterin isoform X1 | 578815184 | 116.7 | 175.3 | H | H | 2.71 | 0.00034 | OE in Seminoma |
| 317 | Gastricsin isoform 1 preproprotein | 4505757 | 18.0 | 29.7 | L | M | 2.76 | 0.00296 | OE in Seminoma |
| 318 | Metalloproteinase inhibitor 1 precursor | 4507509 | 9.3 | 14.7 | L | L | 2.80 | 0.00553 | OE in Seminoma |
| 319 | Lactotransferrin isoform 1 precursor | 54607120 | 702.3 | 996.7 | H | H | 2.87 | 0.00000 | OE in Seminoma |
| 320 | Protein S100-A9 | 4506773 | 23.7 | 45.0 | M | M | 3.35 | 0.00011 | OE in Seminoma |
| 321 | Glyceraldehyde-3-phosphate dehydrogenase, testis-specific | 7657116 | 48.7 | 106.7 | M | H | 3.41 | 0.00000 | OE in Seminoma |
| 322 | Protein disulfide-isomerase precursor | 20070125 | 85.0 | 185.0 | H | H | 3.53 | 0.00012 | OE in Seminoma |
| 323 | Histone H4 | 4504301 | 13.7 | 27.7 | L | M | 3.56 | 0.00040 | OE in Seminoma |
| 324 | Maltase-glucoamylase, intestinal isoform X1 | 578814724 | 16.7 | 33.0 | L | M | 3.58 | 0.00342 | OE in Seminoma |
| 325 | Alpha-actinin-4 | 12025678 | 44.0 | 48.7 | M | M | 3.62 | 0.00001 | OE in Seminoma |
| 326 | Lysozyme C precursor | 4557894 | 7.7 | 16.3 | VL | L | 3.76 | 0.00087 | OE in Seminoma |
| 327 | Alpha-1-antichymotrypsin precursor | 50659080 | 18.7 | 33.7 | L | M | 3.82 | 0.00044 | OE in Seminoma |
| 328 | Protein S100-A8 | 21614544 | 15.0 | 33.3 | L | M | 3.98 | 0.00483 | OE in Seminoma |
| 329 | Thioredoxin-dependent peroxide reductase, mitochondrial isoform b | 32483377 | 3.3 | 8.3 | VL | L | 4.38 | 0.00328 | OE in Seminoma |
| 330 | Semenogelin-2 precursor | 4506885 | 261.3 | 682.7 | H | H | 4.41 | 0.00044 | OE in Seminoma |
| 331 | Prosaposin isoform a preproprotein | 11386147 | 20.0 | 48.0 | M | M | 4.73 | 0.00704 | OE in Seminoma |
| 332 | Olfactomedin-4 precursor | 32313593 | 15.3 | 35.7 | L | M | 4.90 | 0.00039 | OE in Seminoma |
| 333 | Lactadherin isoform a preproprotein | 167830475 | 8.3 | 25.0 | L | M | 4.91 | 0.00144 | OE in Seminoma |
| 334 | Mucin-5B precursor | 301172750 | 22.0 | 65.0 | M | M | 4.97 | 0.00001 | OE in Seminoma |
| 335 | Prolactin-inducible protein precursor | 4505821 | 238.3 | 849.0 | H | H | 4.99 | 0.00045 | OE in Seminoma |
| 336 | Alpha-1-antitrypsin precursor | 189163528 | 13.7 | 34.7 | L | M | 5.01 | 0.00000 | OE in Seminoma |
| 337 | Histone H3.3 | 4885385 | 5.7 | 18.7 | VL | L | 5.70 | 0.00001 | OE in Seminoma |
| 338 | Annexin A11 isoform X1 | 530393508 | 4.0 | 16.0 | VL | L | 6.08 | 0.00010 | OE in Seminoma |
| 339 | Ectonucleotide pyrophosphatase/phosphodiesterase family member 3 | 111160296 | 7.0 | 24.3 | VL | M | 6.24 | 0.00214 | OE in Seminoma |
| 340 | Cathepsin D preproprotein | 4503143 | 3.3 | 11.7 | VL | L | 6.46 | 0.00352 | OE in Seminoma |
| 341 | BPI fold-containing family B member 1 precursor | 40807482 | 4.7 | 15.3 | VL | L | 6.64 | 0.00033 | OE in Seminoma |
| 342 | Fibronectin isoform 1 preproprotein | 47132557 | 112.7 | 505.0 | H | H | 7.15 | 0.00001 | OE in Seminoma |
| 343 | Nucleobindin-2 isoform X1 | 578820554 | 13.0 | 51.3 | L | M | 7.44 | 0.00026 | OE in Seminoma |
| 344 | Semenogelin-1 preproprotein | 4506883 | 94.0 | 422.3 | H | H | 7.78 | 0.00187 | OE in Seminoma |
| 345 | Cytoskeleton-associated protein 4 | 19920317 | 1.3 | 8.0 | VL | L | 8.24 | 0.00084 | OE in Seminoma |
| 346 | Ribonuclease pancreatic precursor | 38201684 | 0.7 | 4.7 | VL | VL | 8.79 | 0.00074 | OE in Seminoma |
| 347 | Transketolase isoform 1 | 205277463 | 3.7 | 15.3 | VL | L | 8.84 | 0.00097 | OE in Seminoma |
| 348 | Neutrophil defensin 1 precursor | 124248516 | 8.7 | 36.0 | L | M | 9.32 | 0.00010 | OE in Seminoma |
| 349 | Neutrophil gelatinase-associated lipocalin precursor | 38455402 | 9.3 | 58.7 | L | M | 9.97 | 0.00000 | OE in Seminoma |
| 350 | Myeloperoxidase precursor | 4557759 | 69.3 | 368.7 | M | H | 10.30 | 0.00000 | OE in Seminoma |
| 351 | Myeloblastin precursor | 71361688 | 6.0 | 34.0 | VL | M | 10.33 | 0.00005 | OE in Seminoma |
| 352 | Catalase | 4557014 | 1.7 | 8.3 | VL | L | 10.41 | 0.00009 | OE in Seminoma |
| 353 | Azurocidin preproprotein | 11342670 | 15.7 | 95.0 | L | H | 11.52 | 0.00000 | OE in Seminoma |
| 354 | Carcinoembryonic antigen-related cell adhesion molecule 1 isoform 1 precursor | 19923195 | 2.0 | 14.0 | VL | L | 12.42 | 0.00321 | OE in Seminoma |
| 355 | Erythrocyte band 7 integral membrane protein isoform a | 38016911 | 9.3 | 58.7 | L | M | 13.46 | 0.00055 | OE in Seminoma |
| 356 | Apolipoprotein B-100 precursor | 105990532 | 1.7 | 12.0 | VL | L | 13.65 | 0.00031 | OE in Seminoma |
| 357 | Cysteine-rich secretory protein 3 isoform 1 precursor | 300244560 | 0.7 | 5.3 | VL | VL | 14.12 | 0.00098 | OE in Seminoma |
| 358 | Mucin-6 isoform X1 | 578840955 | 3.7 | 29.3 | VL | M | 14.30 | 0.00214 | OE in Seminoma |
| 359 | ERO1-like protein alpha precursor | 7657069 | 0.7 | 8.7 | VL | L | 15.45 | 0.00087 | OE in Seminoma |
| 360 | Annexin A3 | 4826643 | 4.0 | 45.7 | VL | M | 17.62 | 0.00001 | OE in Seminoma |
| 361 | Neutrophil elastase preproprotein | 4503549 | 2.3 | 23.0 | VL | M | 17.84 | 0.00006 | OE in Seminoma |
| 362 | Phospholipase B-like 1 precursor | 110227598 | 1.7 | 22.7 | VL | M | 22.14 | 0.00036 | OE in Seminoma |
| 363 | Laminin subunit alpha-5 precursor | 21264602 | 4.3 | 39.0 | VL | M | 22.30 | 0.00613 | OE in Seminoma |
| 364 | Moesin isoform X1 | 530421753 | 0.3 | 4.0 | VL | VL | 25.18 | 0.00003 | OE in Seminoma |
| 365 | Eosinophil cationic protein precursor | 45243507 | 1.0 | 20.3 | VL | M | 27.03 | 0.00057 | OE in Seminoma |
| 366 | Carcinoembryonic antigen-related cell adhesion molecule 6 precursor | 40255013 | 2.0 | 24.3 | VL | M | 27.08 | 0.00170 | OE in Seminoma |
| 367 | Syntenin-1 isoform X1 | 530388518 | 0.3 | 5.7 | VL | VL | 30.17 | 0.00085 | OE in Seminoma |
| 368 | CD63 antigen isoform A | 383872447 | 1.0 | 14.7 | VL | L | 31.84 | 0.00518 | OE in Seminoma |
| 369 | Collagen alpha-1 (XVIII) chain isoform 1 precursor | 110611235 | 1.0 | 27.0 | VL | M | 33.62 | 0.00154 | OE in Seminoma |
| 370 | Laminin subunit gamma-1 precursor | 145309326 | 1.0 | 22.0 | VL | M | 40.70 | 0.00062 | OE in Seminoma |
| 371 | Integrin alpha-M isoform 1 precursor | 224831239 | 5.3 | 176.0 | VL | H | 50.91 | 0.00002 | OE in Seminoma |
| 372 | Laminin subunit beta-2 isoform X1 | 530372442 | 1.3 | 45.0 | VL | M | 61.08 | 0.00000 | OE in Seminoma |
| 373 | Alpha-1-acid glycoprotein 1 precursor | 167857790 | 0.7 | 20.7 | VL | M | 64.68 | 0.00005 | OE in Seminoma |
| 374 | Integrin beta-2 precursor | 188595677 | 2.3 | 124.0 | VL | H | 71.24 | 0.00000 | OE in Seminoma |
| 375 | Carcinoembryonic antigen-related cell adhesion molecule 8 precursor | 21314600 | 0.3 | 18.0 | VL | L | 103.73 | 0.00001 | OE in Seminoma |
| 376 | Cytochrome b-245 heavy chain | 6996021 | 0.3 | 15.0 | VL | L | 112.04 | 0.00002 | OE in Seminoma |
| 377 | Bactericidal permeability-increasing protein precursor | 157276599 | 0.3 | 49.0 | VL | M | 300.57 | 0.00002 | OE in Seminoma |
| 378 | Matrix metalloproteinase-9 preproprotein | 74272287 | 0.0 | 90.0 | ni | H | Seminoma only | 0.00000 | Unique to Seminoma |
| 379 | Leukocyte elastase inhibitor | 13489087 | 0.0 | 23.0 | ni | M | Seminoma only | 0.00006 | Unique to Seminoma |
| 380 | Arachidonate 5-lipoxygenase isoform 2 | 371877525 | 0.0 | 12.7 | ni | L | Seminoma only | 0.00000 | Unique to Seminoma |
| 381 | Prostate and testis expressed protein 4 precursor | 221554530 | 0.0 | 8.0 | ni | L | Seminoma only | 0.00002 | Unique to Seminoma |
| 382 | Chitinase-3-like protein 1 precursor | 144226251 | 0.0 | 10.3 | ni | L | Seminoma only | 0.00003 | Unique to Seminoma |
| 383 | ADP-ribosyl cyclase 2 precursor | 168229159 | 0.0 | 13.7 | ni | L | Seminoma only | 0.00009 | Unique to Seminoma |
| 384 | Peptidoglycan recognition protein 1 precursor | 4827036 | 0.0 | 10.7 | ni | L | Seminoma only | 0.00059 | Unique to Seminoma |
| 385 | Neutrophil collagenase preproprotein | 4505221 | 0.0 | 16.3 | ni | L | Seminoma only | 0.00120 | Unique to Seminoma |
| 386 | Haptoglobin isoform 2 preproprotein | 186910296 | 0.0 | 11.3 | ni | L | Seminoma only | 0.00207 | Unique to Seminoma |
| 387 | Resistin precursor | 301129180 | 0.0 | 6.0 | ni | VL | Seminoma only | 0.00001 | Unique to Seminoma |
| 388 | Matrilin-2 isoform a precursor | 62548860 | 0.0 | 3.0 | ni | VL | Seminoma only | 0.00002 | Unique to Seminoma |
| 389 | Immunoglobulin alpha Fc receptor isoform a precursor | 4503673 | 0.0 | 3.7 | ni | VL | Seminoma only | 0.00018 | Unique to Seminoma |
| 390 | Vascular non-inflammatory molecule 2 isoform X1 | 578813045 | 0.0 | 6.0 | ni | VL | Seminoma only | 0.00025 | Unique to Seminoma |
| 391 | Integrin beta-2 isoform X1 | 578836536 | 0.0 | 6.3 | ni | VL | Seminoma only | 0.00029 | Unique to Seminoma |
| 392 | Flotillin-2 isoform X1 | 530410971 | 0.0 | 6.3 | ni | VL | Seminoma only | 0.00055 | Unique to Seminoma |
| 393 | Cathepsin G preproprotein | 4503149 | 0.0 | 3.3 | ni | VL | Seminoma only | 0.00065 | Unique to Seminoma |
H: high; L: low; M: medium; ni: not identified; NSAF: normalized spectral abundance factor; OE: overexpressed; SC: spectral counts; UE: underexpressed; VL: very low
Figure 1.
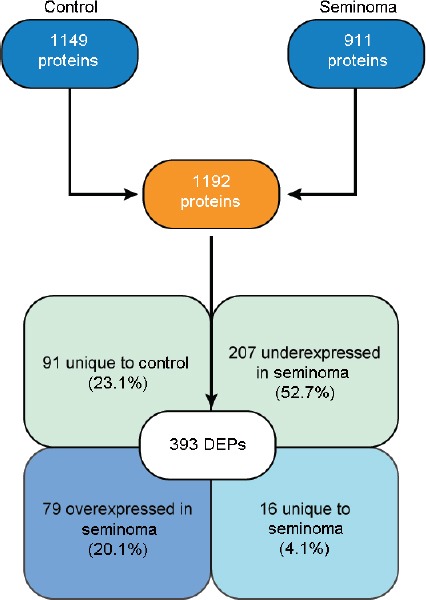
Number of proteins identified by proteomic analysis of spermatozoa samples obtained from fertile men (control) and men with testicular cancer seminoma, and expression profile of the DEPs identified after comparative analysis between the experimental groups. DEPs: differentially expressed proteins.
Selection of proteins for validation
According to the IPA analysis, among the top diseases and bio-functions related to “physiological system development and function,” the category with the highest P value was “reproductive system development and function.” Within this category, we selected seven proteins involved in specific reproductive processes (Table 2): angiotensin-converting enzyme (ACE), acrosin precursor (ACR), T-complex protein 1 subunit gamma (CCT3), sperm surface protein Sp17 (SPA17), sodium/potassium-transporting ATPase subunit alpha-4 (ATP1A4), heat shock-related 70 kDa protein 2 (HSPA2), and proteasome activator complex subunit 4 (PSME4). Some of these proteins were also involved in the top canonical pathways identified in this dataset. While HSPA2 participates in the “protein ubiquitination pathway” and “unfolded protein response,” ACE is related to “phagosome maturation.” Other top five canonical pathways included “mitochondrial dysfunction” and “oxidative phosphorylation.” Among the proteins involved in those pathways were NADH-ubiquinone oxidoreductase 75 kDa subunit (NDUFS1), cytochrome b-c1 complex subunit 2 (UQCRC2), and ATP synthase subunit alpha (ATP5A), which are subunits of the mitochondrial complexes I, III, and V, respectively. These three proteins were also selected for analysis by western blot. The abundance and expression pattern of the ten selected proteins obtained by the proteomic analysis is presented in Table 3.
Table 2.
Specific functions of the differentially expressed proteins related to reproductive system development and function identified by the bioinformatic analysis when comparing the sperm proteome of patients with testicular cancer seminoma with fertile men
| Process | Protein | P |
|---|---|---|
| Binding of sperm | ACE, ACR, CCT3, SPA17 | <0.0001 |
| Fertilization | ACE, ACR, ATP1A4, SPA17 | <0.0001 |
| Cell movement of sperm | ATP1A4 | <0.0001 |
| Spermatogenesis | ACE, ATP1A4, HSPA2, PSME4, SPA17 | 0.0003 |
| Function of sperm | ATP1A4 | 0.0028 |
| Acrosome reaction | ACR | 0.0037 |
| Fertility | ACE, ACR, PSME4 | 0.0067 |
| Morphology of male germ cells | ACR, PSME4 | 0.0089 |
| Morphology of sperm | ACR | 0.0120 |
| Hyperactivation of sperm | ATP1A4 | 0.0133 |
ACE: angiotensin-converting enzyme; ACR: acrosin precursor; ATP1A4: sodium/potassium-transporting ATPase subunit alpha-4; CCT3: T-complex protein 1 subunit gamma; HSPA2: heat shock-related 70 kDa protein 2; PSME4: proteasome activator complex subunit 4; SPA17: sperm surface protein Sp17
Table 3.
Proteomic data of the differentially expressed proteins identified in the spermatozoa samples of fertile men (control) and men with testicular cancer seminoma before cancer therapy, which were selected for validation by western blot
| Protein | Abundance | NSAF ratio | Expression profile | P | |
|---|---|---|---|---|---|
| Control | Seminoma | ||||
| ACE | High | High | 1.62 | Overexpressed in seminoma | 0.0131 |
| ACR | High | Medium | 0.34 | Underexpressed in seminoma | 0.0001 |
| ATP1A4 | Medium | Very low | 0.07 | Underexpressed in seminoma | 0.0001 |
| ATP5A1 | High | Medium | 0.18 | Underexpressed in seminoma | <0.0001 |
| CCT3 | High | Very low | 0.09 | Underexpressed in seminoma | <0.0001 |
| HSPA2 | High | High | 0.53 | Underexpressed in seminoma | <0.0001 |
| NDUFS1 | Medium | Very low | 0.42 | Underexpressed in seminoma | 0.0307 |
| PSME4 | Medium | Very low | 0.13 | Underexpressed in seminoma | 0.0006 |
| SPA17 | Medium | Very low | 0.02 | Underexpressed in seminoma | 0.0001 |
| UQCRC2 | High | Low | 0.23 | Underexpressed in seminoma | 0.0001 |
ACE: angiotensin-converting enzyme; ACR: acrosin; ATP1A4: sodium/potassium-transporting ATPase subunit alpha-4; ATP5A: ATP synthase subunit alpha; CCT3: T-complex protein 1 subunit gamma; HSPA2: heat shock-related 70 kDa protein 2; NDUFS1: NADH-ubiquinone oxidoreductase 75 kDa subunit; NSAF: normalized spectral abundance factor; PSME4: proteasome activator complex subunit 4; SPA17: sperm surface protein Sp17; UQCRC2: cytochrome b-c1 complex subunit 2
Prediction of the upstream regulators
The IPA analysis predicted the activation or inhibition of several proteins, which could be responsible for the altered expression in the sperm proteome of men with seminoma. The rapamycin-insensitive companion of mammalian target of rapamycin (RICTOR) was predicted to be activated, thus leading to the underexpression of NDUFS1, UQCRC2, ATP5A1, and PSME4. Moreover, it was predicted that the underexpression of ATP5A1 and ATP1A4 may involve the activation of the amyloid-beta A4 protein (APP). On the other hand, the inhibition of the heat shock factor protein 2 (HSF2) was predicted to regulate the underexpression of CCT3, as well as six other chaperonins of the T-complex protein-1 (TCP-1) family (CCT2, CCT4, CCT5, CCT6A, CCT7, and CCT8).
Western blot analysis
All proteins selected for western blot analysis were identified. There was an increase in the protein expression of ACE (P = 0.005) and ACR (P = 0.009) in the seminoma group (2.61 ± 0.38 and 2.02 ± 0.26-fold variation to control, respectively) in comparison with the control (1.00 ± 0.25 and 1.00 ± 0.19, respectively) (Figure 2). On the other hand, there was a decrease in the protein levels of ATP1A4 (P = 0.016) and HSPA2 (P = 0.041) in men with seminoma (0.53 ± 0.03 and 0.32 ± 0.11-fold variation to control, respectively) when compared with the control group (1.00 ± 0.25 and 1.00 ± 0.22, respectively) (Figure 2). The protein levels of CCT3, SPA17, and PSME4 were similar between the study groups. There was also a decrease (P = 0.026) in the protein expression levels of UQCRC2 (0.34 ± 0.14-fold variation to control) in the seminoma group relative to the control (1.00 ± 0.14) (Figure 3). No differences were found in the protein levels of NDUFS1 or ATP5A1 between the experimental groups.
Figure 2.
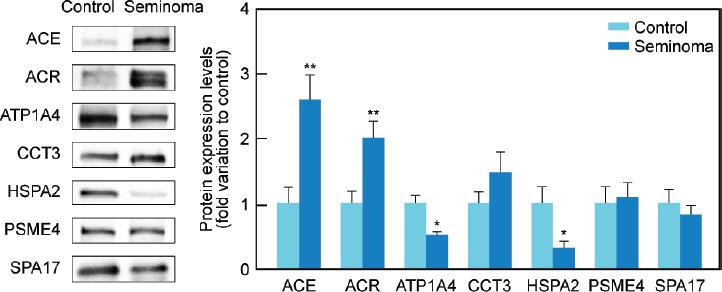
Graphical representation of the expression levels of proteins involved in reproductive functions (ACE, ACR, ATP1A4, CCT3, HSPA2, PSME4, and SPA17) in spermatozoa samples obtained from fertile men (control) and men with testicular cancer seminoma. Results are presented as fold variation to control and expressed as mean±standard error of the mean (n = 15 per group). Statistical significance is indicated as: *P < 0.05, **P < 0.01, seminoma versus control. Representative blots for each protein are also presented. ACE: angiotensin-converting enzyme; ACR: acrosin precursor; ATP1A4: sodium/potassium-transporting ATPase subunit alpha-4; CCT3: T-complex protein 1 subunit gamma; HSPA2: heat shock-related 70 kDa protein 2; PSME4: proteasome activator complex subunit 4; SPA17: sperm surface protein Sp17.
Figure 3.
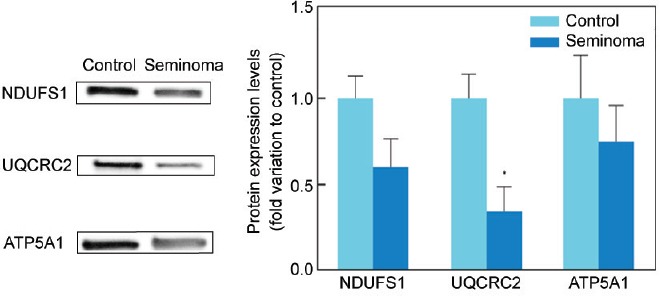
Graphical representation of the protein expression levels of mitochondrial complex subunits NDUFS1, UQCRC2, and ATP5A1 in spermatozoa samples obtained from fertile men (control) and men with testicular cancer seminoma. Results are presented as fold variation to control and expressed as mean ± standard error of the mean (n = 15 per group). Statistical significance is indicated as: *P < 0.05, seminoma versus control. Representative blots for each protein are also presented. NDUFS1: NADH-ubiquinone oxidoreductase 75 kDa subunit; UQCRC2: cytochrome b-c1 complex subunit 2; ATP5A: ATP synthase subunit alpha.
DISCUSSION
The present study is the first attempt to identify alterations in spermatozoa proteome of patients with seminoma before initiating cancer therapy, using fertile donors as control group. Our goal was to evaluate the expression levels of proteins involved in reproductive function from spermatogenesis to sperm function and fertilization. This may provide new insights on the underlying mechanisms responsible for the reduced sperm quality in men with seminoma.
Spermatogenesis consists of a complex process of spermatozoa production that involves several steps of germ cell differentiation. The bioinformatic analysis identified an underexpression of PSME4 in spermatozoa of patients with seminoma. PSME4 plays a role in the morphology of male germ cells; it is particularly important for histone replacement during chromatin remodeling and DNA double-strand break repair.18 It has been reported that mice lacking this protein present impaired spermatogenesis and reduced fertility.19 Thus, the downregulation of this protein may contribute to reduced fertility in men with seminoma. Although we were not able to confirm the underexpression of PSME4 by western blot in our dataset, we observed the underexpression of the molecular chaperone HSPA2 by both proteomics and western blot analysis. Molecular chaperones are essential for normal sperm production and functional transformation. HSPA2 acts as a protein quality control system as it ensures the correct folding/refolding of proteins and activates the degradation of misfolded proteins.20 It has been described that HSPA2 participates in the stability of the microtubules during the meiotic process of germ cell differentiation.21 In fact, animal studies show that knockout mice for Hspa2 exhibit an enormous number of apoptotic germ cells, resulting in infertility.22 Men with abnormal spermatogenesis frequently present a reduced hspA2 mRNA expression.23 Thus, the downregulation of HSPA2 protein in men with seminoma may contribute to the decreased production of normal spermatozoa during spermatogenesis, which is in accordance with the observed reduction in sperm concentration and total sperm count in seminoma group.
The protein ATP1A4 was identified as downregulated in seminoma group by the proteomic analysis, and this result was confirmed by the western blot technique. IPA analysis revealed that ATP1A4 participates in several reproductive processes, including spermatogenesis, function of sperm, cell movement of sperm, hyperactivation, and fertilization. ATP1A4 is the catalytic subunit of the Na+/K+-ATPase membrane protein, which controls the exchange of sodium and potassium ions across the plasma membrane in an ATP-dependent reaction.24 The regulation of ions in spermatozoa is essential for the acquisition of motility and fertilizing ability. ATP1A4 plays a key role in maintaining human sperm motility.25 It has been shown that male mice lacking this subunit are completely sterile and their spermatozoa present not only reduced motility but also impaired hyperactivation and inability to fertilize in vitro.26 These studies highlight the importance of ATP1A4 for male fertility, and the underexpression of this protein in spermatozoa of men with seminoma may explain the decrease in sperm motility and total motile count relative to proven fertile men (control group). The downregulation of ATP1A4 was related to the activation of APP. In fact, this protein has been identified in human spermatozoa and suggested to play an important role in sperm function, especially in signaling events involved in sperm motility.27
Another important process crucial for sperm function is mitochondrial function. It is required for energy production necessary for spermatozoa movement and production of reactive oxygen species (ROS) in physiological amounts to trigger capacitation and regulate hyperactivation and acrosome reaction.28 Mitochondrial function relies on the expression of the mitochondrial complexes I–IV for oxidative phosphorylation (OXPHOS) and complex V for ATP production.29 Our proteomic data showed a downregulation of NDUFS1, UQCRC2, and ATP5A1 in the seminoma group, which are subunits of complex I, III, and V, respectively. The downregulation of these three proteins was predicted to be induced by the activation of RICTOR, which plays a key role in spermatogenesis and sperm maturation signaling pathways.28 The mitochondrial subunits are essential for the proper assembly of the complexes; thus, alterations in their protein expression in spermatozoa are indicative of mitochondrial dysfunction, as reported by the IPA canonical pathways.30 Although the western blot analysis demonstrated a tendency of reduced expression of the three mitochondrial subunits, only the UQCRC2 was decreased in patients with seminoma. Downregulation of UQCRC2 was associated with reduced sperm kinematics, ATP production, and capacitation, which ultimately compromises sperm binding and fertilization.31 In fact, an underexpression of UQCRC2 was observed in infertile men with varicocele.32
The acquisition of sperm fertilizing ability involves a timed triggering of events in the female reproductive system, culminating in sperm–oocyte binding. SPA17 and CCT3 are two sperm proteins involved in this function, which were identified as downregulated in the seminoma group by the proteomic analysis. SPA17 is a mannose-binding protein that binds to zona pellucida carbohydrates during fertilization.33 It also plays an important role in germ cell differentiation during spermatogenesis, as its expression increases from early to late stages.34 CCT3 is one of the subunits of the TCP-1 complex. Although we selected to evaluate the expression levels of this subunit, six other subunits of this complex (CCT2, CCT4, CCT5, CCT6A, CCT7, and CCT8) were also downregulated in men with seminoma. These subunits mediate capacitation-dependent binding of spermatozoa to the zona pellucida.35 Thus, the downregulation of this system may compromise sperm fertilization.36 The downregulation of TCP-1 complex subunits was predicted to be due to HSF2 inhibition. In fact, disruption of hsf2 in mice affected testicular size37 and induced spermatogenic defects.38 When active, HSF2 is likely to induce the upregulation of HSPA2.39 Thus, the predicted inhibition of HSF2 in men with seminoma is in accordance with the downregulation of HSPA2. Although the underexpression of SPA17 and CCT3 was not confirmed by the western blot, the downregulation of HSPA2 in men with seminoma may contribute to the loss of sperm function. In fact, this protein is known to regulate the formation of zona pellucida-binding sites in spermatozoa during spermatogenesis.40 In addition, it regulates fertilization by mediating the function of sperm surface receptors, such as sperm adhesion molecule 1 (SPAM1) and arylsulfatase A (ARSA), during sperm-egg recognition.41 Previous proteomic studies have shown low expression levels of HSPA2 in men with asthenozoospermia42 and primary or secondary infertility.43 Another study also reported a downregulation of HSPA2, ATP1A4, and SPA17 in infertile varicocele patients.32 Our results suggest that the altered expression levels of these proteins in men with seminoma may contribute to the impairment of male fertility.
The proteomic analysis also identified ACE as overexpressed in the seminoma group, and this result was confirmed by western blot. This protein is a zinc metallopeptidase responsible for the conversion of angiotensin I to angiotensin II.44 The role of ACE in male reproductive function is not completely understood. Studies with ACE-deficient mice reported that these animals produce a normal number of spermatozoa and present normal motility and morphology. However, the spermatozoa were unable to bind and fertilize the egg.45,46 A negative correlation between sperm-bound ACE activity and sperm motility has also been observed.47 The testis-specific isoform of this protein (tACE) is believed to be released from functional spermatozoa during capacitation and acrosome reaction to increase the fertilizing ability.48 In fact, a lower tACE activity was detected in spermatozoa from normozoospermic men relative to those with oligoasthenozoospermia.47 Thus, the overexpression of this protein in spermatozoa from men with seminoma may be responsible for the decrease in sperm motility observed in this group, and possibly explains why some men with seminoma are not able to have children even before the treatment.
Finally, the protein ACR, in its precursor form (proacrosin), was identified as underexpressed in the seminoma group by the proteomic analysis. This protein is activated and converted to its active form during acrosome reaction, playing a role in sperm–oocyte binding.49 In contrast, using western blot, we found a high overexpression of this protein in men with seminoma. Although we cannot clearly infer about the molecular mechanisms, any of the scenarios (underexpression/overexpression) could lead to a defective acrosome reaction and impaired fertilization. The difference on these results may be due to the sensitivity of each technique and to the sample size. Further studies to assess acrosin activity in men with seminoma are needed to clarify the impact of this condition in acrosome reaction.
Overall, our study points toward important alterations in sperm proteins with a key role in male fertility in men with seminoma. As of today, no specific sperm markers have been identified for the clinical diagnosis and monitoring of testicular cancer seminoma development. The expression levels of HSPA2, ATP1A4, UQCRC2, and ACE can be helpful sperm biomarkers when evaluating the fertility status of a man, which may allow the early diagnosis of seminomas in a noninvasive approach. Although there is still a lot to explore in the pathophysiology of male subfertility/infertility in men with seminoma, our results represent a step forward in understanding the molecular mechanisms behind the reduced sperm quality in these patients. Future advances in mass spectrometry and bioinformatics will improve our understanding on human sperm function in healthy and disease conditions.
AUTHOR CONTRIBUTIONS
AA and RS were responsible for the conception and design of the study. TRD was responsible for the acquisition and interpretation of data, as well as writing the first draft. GA helped in samples processing and PNP performed the bioinformatic analysis. All authors read and approved the final manuscript.
COMPETING INTERESTS
All authors declared no competing interests.
ACKNOWLEDGMENTS
The authors would like to thank Dr. Belinda Willard (Director, Proteomics Core Laboratory, Lerner Research Institute, Cleveland Clinic) for her support in the proteomic analysis and Dr. Ralf Henkel and Dr. Saradha Baskaran for their help in reviewing the manuscript. Financial support for this study was provided by the American Center for Reproductive Medicine, Cleveland Clinic, OH, USA. Tania R Dias was supported by the Portuguese Foundation for Science and Technology (FCT, SFRH/BD/109284/2015) and Fulbright Program (E0585639). Sponsors were not involved in the experiments or writing/submitting the paper.
Supplementary Information is linked to the online version of the paper on the Asian Journal of Andrology website.
REFERENCES
- 1.Ruf CG, Isbarn H, Wagner W, Fisch M, Matthies C, et al. Changes in epidemiologic features of testicular germ cell cancer: age at diagnosis and relative frequency of seminoma are constantly and significantly increasing. Urol Oncol. 2014;32:33.e1–6. doi: 10.1016/j.urolonc.2012.12.002. [DOI] [PubMed] [Google Scholar]
- 2.Noone A, Howlader N, Krapcho M, Miller D, Brest A, et al. Bethesda: National Cancer Institute; 1975-2015. [Last accessed on 2019 Jan 7]. SEER Cancer Statistics Review. Available from: https://www.seer.cancer.gov/csr/1975_2015/ [Google Scholar]
- 3.Bahadur G, Ozturk O, Muneer A, Wafa R, Ashraf A, et al. Semen quality before and after gonadotoxic treatment. Hum Reprod. 2005;20:774–81. doi: 10.1093/humrep/deh671. [DOI] [PubMed] [Google Scholar]
- 4.Morrish DW, Venner PM, Siy O, Barron G, Bhardwaj D, et al. Mechanisms of endocrine dysfunction in patients with testicular cancer. J Natl Cancer Inst. 1990;82:412–8. doi: 10.1093/jnci/82.5.412. [DOI] [PubMed] [Google Scholar]
- 5.Ping P, Gu BH, Li P, Huang YR, Li Z. Fertility outcome of patients with testicular tumor: before and after treatment. Asian J Androl. 2014;16:107. doi: 10.4103/1008-682X.122194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huddart R, Norman A, Moynihan C, Horwich A, Parker C, et al. Fertility, gonadal and sexual function in survivors of testicular cancer. Br J Cancer. 2005;93:200. doi: 10.1038/sj.bjc.6602677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meistrich ML. Effects of chemotherapy and radiotherapy on spermatogenesis in humans. Fertil Steril. 2013;100:1180–6. doi: 10.1016/j.fertnstert.2013.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Agarwal A. Semen banking in patients with cancer: 20-year experience. Int J Androl. 2000;23:16–9. doi: 10.1046/j.1365-2605.2000.00005.x. [DOI] [PubMed] [Google Scholar]
- 9.Huyghe E, Matsuda T, Daudin M, Chevreau C, Bachaud JM, et al. Fertility after testicular cancer treatments: results of a large multicenter study. Cancer. 2004;100:732–7. doi: 10.1002/cncr.11950. [DOI] [PubMed] [Google Scholar]
- 10.Magelssen H, Haugen T, Von Düring V, Melve K, Sandstad B, et al. Twenty years experience with semen cryopreservation in testicular cancer patients: who needs it? Eur Urol. 2005;48:779–85. doi: 10.1016/j.eururo.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 11.Vakalopoulos I, Dimou P, Anagnostou I, Zeginiadou T. Impact of cancer and cancer treatment on male fertility. Hormones (Athens, Greece) 2015;14:579–89. doi: 10.14310/horm.2002.1620. [DOI] [PubMed] [Google Scholar]
- 12.Agarwal A, Bertolla RP, Samanta L. Sperm proteomics: potential impact on male infertility treatment. Expert Rev Proteomics. 2016;13:285–96. doi: 10.1586/14789450.2016.1151357. [DOI] [PubMed] [Google Scholar]
- 13.Sharma R, Agarwal A, Mohanty G, Du Plessis SS, Gopalan B, et al. Proteomic analysis of seminal fluid from men exhibiting oxidative stress. Reprod Biol Endocrinol. 2013;11:85. doi: 10.1186/1477-7827-11-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baker MA, Nixon B, Naumovski N, Aitken RJ. Proteomic insights into the maturation and capacitation of mammalian spermatozoa. Syst Biol Reprod Med. 2012;58:211–7. doi: 10.3109/19396368.2011.639844. [DOI] [PubMed] [Google Scholar]
- 15.World Health Organization. WHO Laboratory Manual for the Examination and Processing of Human Semen. 5th ed. Geneva: World Health Organization; 2010. [Google Scholar]
- 16.Agarwal A, Gupta S, Sharma R. Andrological Evaluation of Male Infertility. Cham: Springer; 2016. Cryopreservation of client depositor semen; pp. 113–33. [Google Scholar]
- 17.Agarwal A, Ayaz A, Samanta L, Sharma R, Assidi M, et al. Comparative proteomic network signatures in seminal plasma of infertile men as a function of reactive oxygen species. Clin Proteomics. 2015;12:23. doi: 10.1186/s12014-015-9094-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ustrell V, Hoffman L, Pratt G, Rechsteiner M. PA200, a nuclear proteasome activator involved in DNA repair. EMBO J. 2002;21:3516–25. doi: 10.1093/emboj/cdf333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khor B, Bredemeyer AL, Huang CY, Turnbull IR, Evans R, et al. Proteasome activator PA200 is required for normal spermatogenesis. Mol Cell Biol. 2006;26:2999–3007. doi: 10.1128/MCB.26.8.2999-3007.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Radons J. The human HSP70 family of chaperones: where do we stand? Cell Stress Chaperones. 2016;21:379–404. doi: 10.1007/s12192-016-0676-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu M, Shi X, Bi Y, Qi L, Guo X, et al. SHCBP1L, a conserved protein in mammals, is predominantly expressed in male germ cells and maintains spindle stability during meiosis in testis. Mol Hum Reprod. 2014;20:463–75. doi: 10.1093/molehr/gau014. [DOI] [PubMed] [Google Scholar]
- 22.Dix DJ, Allen JW, Collins BW, Mori C, Nakamura N, et al. Targeted gene disruption of Hsp70-2 results in failed meiosis, germ cell apoptosis, and male infertility. Proc Natl Acad Sci U S A. 1996;93:3264–8. doi: 10.1073/pnas.93.8.3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Son WY, Han CT, Hwang SH, Lee JH, Kim SS, et al. Repression of hspA2 messenger RNA in human testes with abnormal spermatogenesis. Fertil Steril. 2000;73:1138–44. doi: 10.1016/s0015-0282(00)00496-9. [DOI] [PubMed] [Google Scholar]
- 24.Hlivko JT, Chakraborty S, Hlivko TJ, Sengupta A, James PF. The human Na, K-ATPase alpha4 isoform is a ouabain-sensitive alpha isoform that is expressed in sperm. Mol Reprod Dev. 2006;73:101–15. doi: 10.1002/mrd.20383. [DOI] [PubMed] [Google Scholar]
- 25.Sanchez G, Nguyen AN, Timmerberg B, Tash JS, Blanco G. The Na, K-ATPase α4 isoform from humans has distinct enzymatic properties and is important for sperm motility. Mol Hum Reprod. 2006;12:565–76. doi: 10.1093/molehr/gal062. [DOI] [PubMed] [Google Scholar]
- 26.Jimenez T, McDermott JP, Sánchez G, Blanco G. Na, K-ATPase α4 isoform is essential for sperm fertility. Proc Natl Acad Sci U S A. 2011;108:644–9. doi: 10.1073/pnas.1016902108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fardilha M, Vieira SI, Barros A, Sousa M, Da Cruz e Silva OA, et al. Differential distribution of Alzheimer's amyloid precursor protein family variants in human sperm. Ann N Y Acad Sci. 2007;1096:196–206. doi: 10.1196/annals.1397.086. [DOI] [PubMed] [Google Scholar]
- 28.de Lamirande E, O’Flaherty C. Sperm activation: role of reactive oxygen species and kinases. Biochim Biophys Acta. 2008;1784:106–15. doi: 10.1016/j.bbapap.2007.08.024. [DOI] [PubMed] [Google Scholar]
- 29.Boekema EJ, Braun HP. Supramolecular structure of the mitochondrial oxidative phosphorylation system. J Biol Chem. 2006;282:1–4. doi: 10.1074/jbc.R600031200. [DOI] [PubMed] [Google Scholar]
- 30.Agarwal A, Sharma R, Samanta L, Durairajanayagam D, Sabanegh E. Proteomic signatures of infertile men with clinical varicocele and their validation studies reveal mitochondrial dysfunction leading to infertility. Asian J Androl. 2016;18:282. doi: 10.4103/1008-682X.170445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shukla KK, Kwon WS, Rahman MS, Park YJ, You YA, et al. Nutlin-3a decreases male fertility via UQCRC2. PLoS One. 2013;8:e76959. doi: 10.1371/journal.pone.0076959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Samanta L, Agarwal A, Swain N, Sharma R, Gopalan B, et al. Proteomic signatures of sperm mitochondria in varicocele: clinical utility as biomarkers of varicocele associated infertility. J Urol. 2018;200:414–22. doi: 10.1016/j.juro.2018.03.009. [DOI] [PubMed] [Google Scholar]
- 33.O’rand MG, Richardson RT, Yamasaki N. Expression of the rabbit sperm protein Sp17 in COS cells and interaction of recombinant Sp17 with the rabbit zona pellucida. Mol Reprod Dev. 1995;40:48–55. doi: 10.1002/mrd.1080400107. [DOI] [PubMed] [Google Scholar]
- 34.Grizzi F, Chiriva-Internati M, Franceschini B, Hermonat PL, Soda G, et al. Immunolocalization of sperm protein 17 in human testis and ejaculated spermatozoa. J Histochem Cytochem. 2003;51:1245–8. doi: 10.1177/002215540305100916. [DOI] [PubMed] [Google Scholar]
- 35.Redgrove KA, Anderson AL, Dun MD, McLaughlin EA, O’Bryan MK, et al. Involvement of multimeric protein complexes in mediating the capacitation-dependent binding of human spermatozoa to homologous zonae pellucidae. Dev Biol. 2011;356:460–74. doi: 10.1016/j.ydbio.2011.05.674. [DOI] [PubMed] [Google Scholar]
- 36.Fraser LR, Dudley K. New insights into the t-complex and control of sperm function. Bioessays. 1999;21:304–12. doi: 10.1002/(SICI)1521-1878(199904)21:4<304::AID-BIES6>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 37.Wang G, Ying Z, Jin X, Tu N, Zhang Y, et al. Essential requirement for both hsf1 and hsf2 transcriptional activity in spermatogenesis and male fertility. Genesis. 2004;38:66–80. doi: 10.1002/gene.20005. [DOI] [PubMed] [Google Scholar]
- 38.Mou L, Wang Y, Li H, Huang Y, Jiang T, et al. A dominant-negative mutation of HSF2 associated with idiopathic azoospermia. Hum Genet. 2013;132:159–65. doi: 10.1007/s00439-012-1234-7. [DOI] [PubMed] [Google Scholar]
- 39.Sarge KD, Park-Sarge OK, Kirby JD, Mayo KE, Morimoto RI. Expression of heat shock factor 2 in mouse testis: potential role as a regulator of heat-shock protein gene expression during spermatogenesis. Biol Reprod. 1994;50:1334–43. doi: 10.1095/biolreprod50.6.1334. [DOI] [PubMed] [Google Scholar]
- 40.Huszar G, Stone K, Dix D, Vigue L. Putative creatine kinase M-isoform in human sperm is identifiedas the 70-kilodalton heat shock protein HspA2. Biol Reprod. 2000;63:925–32. doi: 10.1095/biolreprod63.3.925. [DOI] [PubMed] [Google Scholar]
- 41.Redgrove KA, Nixon B, Baker MA, Hetherington L, Baker G, et al. The molecular chaperone HSPA2 plays a key role in regulating the expression of sperm surface receptors that mediate sperm-egg recognition. PLoS One. 2012;7:e50851. doi: 10.1371/journal.pone.0050851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hashemitabar M, Sabbagh S, Orazizadeh M, Ghadiri A, Bahmanzadeh M. A proteomic analysis on human sperm tail: comparison between normozoospermia and asthenozoospermia. J Assist Reprod Genet. 2015;32:853–63. doi: 10.1007/s10815-015-0465-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Intasqui P, Agarwal A, Sharma R, Samanta L, Bertolla R. Towards the identification of reliable sperm biomarkers for male infertility: a sperm proteomic approach. Andrologia. 2018;50:e12919. doi: 10.1111/and.12919. [DOI] [PubMed] [Google Scholar]
- 44.Rigat B, Hubert C, Alhenc-Gelas F, Cambien F, Corvol P, et al. An insertion/deletion polymorphism in the angiotensin I-converting enzyme gene accounting for half the variance of serum enzyme levels. J Clin Invest. 1990;86:1343–6. doi: 10.1172/JCI114844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krege JH, John SW, Langenbach LL, Hodgin JB, Hagaman JR, et al. Male-female differences in fertility and blood pressure in ACE-deficient mice. Nature. 1995;375:146. doi: 10.1038/375146a0. [DOI] [PubMed] [Google Scholar]
- 46.Hagaman JR, Moyer JS, Bachman ES, Sibony M, Magyar PL, et al. Angiotensin-converting enzyme and male fertility. Proc Natl Acad Sci U S A. 1998;95:2552–7. doi: 10.1073/pnas.95.5.2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shibahara H, Kamata M, Hu J, Nakagawa H, Obara H, et al. Activity of testis angiotensin converting enzyme (ACE) in ejaculated human spermatozoa. Int J Androl. 2001;24:295–9. doi: 10.1046/j.1365-2605.2001.00301.x. [DOI] [PubMed] [Google Scholar]
- 48.Köhn FM, Miska W, Schill WB. Release of angiotensin-converting enzyme (ACE) from human spermatozoa during capacitation and acrosome reaction. J Androl. 1995;16:259–65. [PubMed] [Google Scholar]
- 49.Yamagata K, Murayama K, Okabe M, Toshimori K, Nakanishi T, et al. Acrosin accelerates the dispersal of sperm acrosomal proteins during acrosome reaction. J Biol Chem. 1998;273:10470–4. doi: 10.1074/jbc.273.17.10470. [DOI] [PubMed] [Google Scholar]


