Abstract
Bacterial RNA degradosomes are multienzyme molecular machines that act as hubs for post-transcriptional regulation of gene expression. The ribonuclease activities of these complexes require tight regulation, as they are usually essential for cell survival while potentially destructive. Recent studies have unveiled a wide variety of regulatory mechanisms including autoregulation, post-translational modifications and protein compartmentalization. Recently, the subcellular organization of bacterial RNA degradosomes was found to present similarities with eukaryotic messenger ribonucleoprotein (mRNP) granules, membraneless compartments that are also involved in mRNA and protein storage and/or mRNA degradation.
In this review, we present the current knowledge on the composition and targets of RNA degradosomes, the most recent developments regarding the regulation of these machineries and their similarities with the eukaryotic mRNP granules.
Keywords: RNA degradosome, RNA maturation and degradation, post- transcriptional regulation, mRNP granules, regulation, compartmentalization, membraneless organelles
RNA degradosomes at the center of bacterial post-transcriptional regulation
Post-transcriptional regulation is one of the most important levels of control of gene expression in every kingdom of life [1]. It involves all the mechanisms that affect structure and/or stability of cellular transcripts, including stable RNAs (ribosomal and transfer RNAs, called rRNAs and tRNAs, respectively), as well as messenger RNAs (mRNAs) and regulatory RNAs, designated small non-coding RNAs (ncRNAs) (see Glossary) in bacteria).
Ribonucleases (RNases) are key enzymes in post-transcriptional regulation, involved in RNA maturation and degradation. They act either internally on the RNA molecule as endoribonucleases, or as exoribonucleases by attacking the RNA at its 5’ or 3’-end to initiate degradation. To control such important functions, some RNases act in multi-protein complexes. These complexes are designated exosomes in Eukarya and Archaea and RNA degradosomes in bacteria and chloroplasts [2,3].
The first RNA degradosome was identified in Escherichia coli as a complex bound to RNase E, an endoribonuclease that initiates bulk mRNA degradation [2,4,5]. More recently, RNA degradosomes were found to be more widespread, being present in many different bacteria, including important human pathogens, such as Staphylococcus aureus [6], Pseudomonas aeruginosa [7], Mycobacterium tuberculosis [8] or Helicobacter pylori [9], among others. Since RNases are potentially very destructive enzymes if they degrade RNAs that are important for bacterial survival, their activities must be under tight control. Recent studies have reported diverse regulatory mechanisms including negative-feedback loops, post-translational modifications, bacterial or phagic protein inhibitors, etc., that will be presented in this review. Most interestingly, recent work has revealed that RNA degradosomes can have subcellular localizations that vary between organisms. Even more surprising was the finding that RNA degradosomes form structures similar to the eukaryotic processing bodies (p-bodies, see Glossary) and stress granules, membraneless organelles involved in RNA degradation [10]. These similarities will also be addressed in this review.
RNA degradosomes are widespread and vary in composition
Both Gram-positive and Gram-negative bacteria possess an important repertoire of RNases that varies between species both in number and composition. For instance, the Gram-negative model organism, E. coli, possesses 15 RNases [11], including RNase E. The Gram-positive model, Bacillus subtilis, possesses 20 RNases [12,13] with striking differences such as the absence of RNase E and the presence of RNases J1/J2 and RNase Y.
The number of RNA degradosomes reported in bacteria has been rising over the years. Their composition is variable (Table 1 and references therein), although it cannot be excluded that this is partly due to the various approaches or conditions used to characterize them. Despite this, they are defined by two core components (Fig. 1): at least one RNase (from here on designated dRNase, for “degradosome RNase”) and an RNA helicase of the DEAD-box family. The RNA helicase helps unfolding secondary structures in RNA molecules, allowing the cleavage sites to be accessible for one or more RNases. Often, the complex contains multiple RNases with different enzymatic activities (an endonuclease and an exonuclease), their “combined” activities being compatible with the multiple functions of the degradosome. Several degradosomes include, in addition, a metabolic enzyme (enolase, aconitase or phosphofructokinase) whose role in the complex is not completely clear; it has been proposed that enolase can couple the metabolic status of the cell with RNA degradation in E. coli, as it is important for the degradation of mRNAs encoding central metabolism proteins [14].
Table 1.
Interaction partners of different bacterial RNA degradosomes.
| Main RNase | Organism | Interacting components | Information source | Reference(s) |
|---|---|---|---|---|
| RNase E | Escherichia coli |
RhlBa PNPase Enolase |
In vivo affinity purification, co-purification | [79–81] |
| RNase II Ppk DnaK RNase R |
In vivo affinity purification | [82–85] | ||
| RhlE, CsdA, SrmB, PAPI | In vitro interaction | [61,86] | ||
| Hfq | Structural data | [16] | ||
| Caulobacter crescentus |
RhlB PNPase Aconitase RNase D RhlE Rho S1 ribosomal protein 2-oxoglutarate dehydrogenase E1 component DEAD-box helicase CCNA_01546 Methionine adenosyl transferase Nudix family pyrophosphatase Acetoacetyl-CoA reductase |
In vivo affinity purification | [62,87,88] | |
| Mycobacterium tuberculosis | PNPase RhlE RNase J DnaK, GroEL, Ppk, acetyltransferase, CspA, CspB, Rv29G8c, Rv392Gc |
In vivo affinity purification | [8,89] | |
| Pseudoalteromonas haloplanktis |
RhlB PNPase |
In vivo affinity purification | [90] | |
| Pseudomonas aeruginosa | PNPase DeaD ClpA, DnaK, NADH-quinone oxidoreductase subunit C/D (PA2639), DnaA, ATP synthase F0F1 subunit alpha/beta, FliC, ribosomal proteins, PA3309 |
In vivo affinity purification | [7] | |
| Vibrio cholerae |
RhlB PNPase Enolase |
In vivo affinity purification | [44] | |
| Anabaena spp. | PNPase | In vivo affinity purification, in vitro co-purification | [91] | |
| Synechocystis spp. | PNPase | In vivo affinity purification, in vitro co-purification | [91] | |
| CrhR | Cosedimentation | [92] | ||
| Streptomyces coelicolorb | PNPase | In vivo affinity purification after overexpression | [93] | |
| Pseudomonas syringae | RNase R RhlE |
In vivo affinity purification | [94] | |
| Rhodobacter capsulatus |
DEAD-box RNA helicases Rho |
In vivo affinity purification | [95] | |
| Chloroplasts in Arabidopsis thaliana | RHON1 | In vivo affinity purification | [96] | |
| RNase Y | Bacillus subtilis | PNPase Enolase |
Bacterial two-hybrid assay, in vivo affinity purification after cross-linking and surface plasmon resonance | [23,71,97] |
|
CshA Phosphofructokinase RNase J1 RNase J2 |
Bacterial two-hybrid assay and in vivo affinity purification after cross-linking | [23,71] | ||
| GapA YlbF, YmcA and YaaT |
In vivo affinity purification | [98,99] | ||
| Streptococcus pyogenes | Enolase | In vivo affinity purification after cross-linking | [35] | |
| Staphylococcus aureus |
CshA RNase J1 RNase J2 PNPase Enolase Flotillin |
In vivo affinity purification bacterial and two-hybrid assay | [6,63,100] | |
| Phosphofructokinase RnpA (RNase P) |
Bacterial two-hybrid assay | [6] | ||
| RNase J | Helicobacter pylori |
RhpA Ribosomal proteins |
In vivo affinity purification, in vitro co-purification | [9] |
DEAD-box RNA helicases are highlighted in bold.
The homolog for RNase E of S. coelicolor is called RNase ES.
Figure 1.
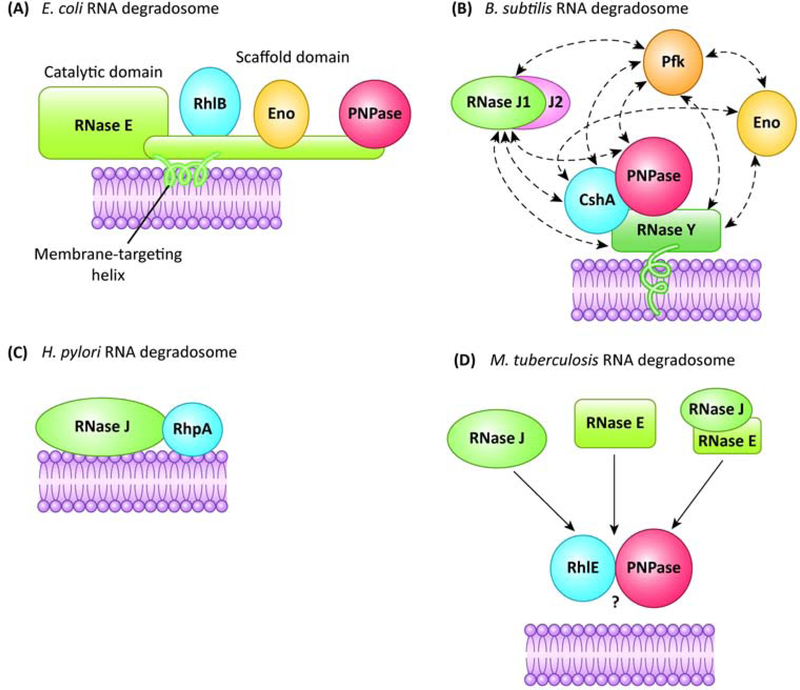
Schematic representation of the core components of the different types of bacterial RNA degradosomes described to date. Solid arrows indicate interactions, dashed arrows possible additional partners. (a) E. coli RNA degradosome, based on RNase E. RNase E (green) possesses a C-terminal domain containing a membrane-targeting helix, as well as interaction sites for the RNA helicase RhlB, enolase (Eno) and polynucleotide phosphorylase (PNPase). Not shown are binding sites for RNA. (b) B. subtilis RNA degradosome, containing RNase Y and also RNases J1 and J2, that potentially interact with other enzymes, namely the RNA helicase CshA, PNPase, enolase and phosphofructokinase (Pfk). (c) H. pylori RNA degradosome, based on RNase J, that interacts with the RNA helicase RhpA. (d) M. tuberculosis possible RNA degradosome(s), containing either RNase J, RNase E or both, that interact with the RNA helicase RhlE and PNPase. The question mark indicates that the membrane targeting has not been explored so far.
The E. coli RNA degradosome is centered on the essential hydrolytic endoribonuclease RNase E (EcoRNase E) [4], an enzyme composed of an N- terminal catalytic domain and a C-terminal unstructured scaffolding region with two RNA-binding sites. This C-terminal domain carries several Short Linear Motifs (SLiMs) that are abundant in proteins carrying intrinsically disordered regions (IDRs), as well as well-defined binding sites for the other components of the complex: the RNA helicase RhlB, the phosphorolytic 3’−5’ exoribonuclease PNPase and the glycolytic enzyme enolase [4,15]. Other studies report additional partners that might correspond to minor constituents (Table 1). In addition, interaction of RNase E with the RNA chaperone Hfq has been proposed to be mediated by RNA [16]. The C-terminus of EcoRNase E contains a short membrane-targeting sequence that interacts with the E. coli inner membrane phospholipids to form a stabilized amphipathic α-helix that acts as a membrane anchor for the whole complex. The anchoring of the RNase E-based degradosome to the membrane is not ubiquitous in bacteria; it is conserved in the gamma-proteobacteria [15] but not in some alpha- proteobacteria that have been recently shown to possess a cytoplasmic RNase E-based RNA degradosome [17,18]. The role of the membrane anchoring of EcoRNase E is still not completely clear but some clues were recently provided and will be discussed below [19–21].
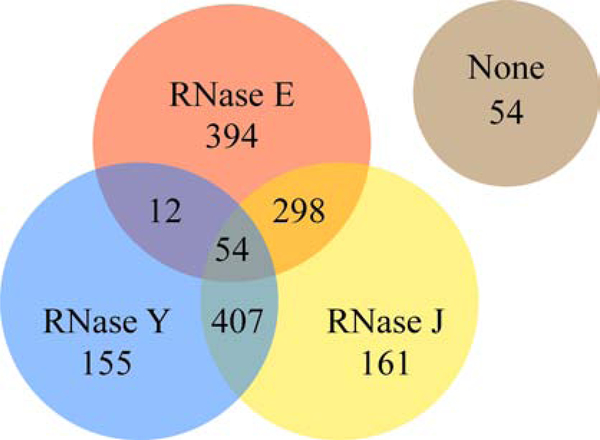
In Gram-positive bacteria, such as the model B. subtilis and the pathogen S. aureus, the existence of an RNA degradosome is less well-established [13]. Like in many bacteria, these organisms lack an RNase E ortholog but contain other RNases that act as functional analogues: the membrane-anchored endoribonuclease RNase Y, and RNases J which possess both endoribonuclease and 5’−3’ exoribonuclease activities [12]. An analysis of the phylogenetic distribution of RNase E, RNase J1/J2 and RNase Y homologs was carried out for this review in a representative set of 1535 bacterial genomes (Fig. 2 and Supp. Table S1). Around 26% of the genomes analyzed encode only RNase E (like E. coli), 11% contain only RNase J and 10% encode only RNase Y, with 19% containing both RNase E and RNase J (like M. tuberculosis) and 27% both RNase J and RNase Y (like B. subtilis or H. pylori). Less than 1% contain both RNase E and RNase Y. Finally, 4% of the analyzed genomes carry all three RNases and 4% none of them. The large majority of the latter mostly belong to the Chlamydiae and Bacteriodetes phylum, 40 out of 54 of these organisms carried RNase G, an enzyme with homology to the catalytic domain of RNase E that might partially compensate for its absence [22]. The few organisms lacking all three RNases and RNase G mainly belong to the Spirochaetes phylum.
Figure 2.
Distribution of the degradosome RNases (RNase E, RNase J and RNase Y) in a representative set of bacterial genomes. Most bacterial species (96.5%) contain at least one of these RNases. Out of the 54 genomes that do not contain any of these RNases (marked as “none” in the Venn diagram), 40 contain RNase G. Analysis was performed on 15S5 representative genomes chosen based on phylogenetic diversity as previously described [101], using the PubSEED database [102]. Details are provided in supplemental Table S1 and the results are also available in the ‘RNAse_2019_Minimal’ subsystem on the public PubSEED server (http://pubseed.theseed.org/SubsysEditor.cgi).
In B. subtilis, interactions between RNase J1/J2, RNase Y, the DEAD-box RNA helicase CshA, the 3’−5’ exoribonuclease PNPase and the glycolytic enzymes phosphofructokinase and enolase have been reported [23] (Fig. 1, Table 1, for a review see [12],). However, the colocalization of the different components and their membrane targeting is not clear (except for RNase Y) [24]. In addition, the whole complex has never been successfully purified from its native organism. Moreover, the interaction data obtained by Bacterial Two Hybrid (BACTH) between RNase Y and RNase J1 are not strong [23] and other authors failed to reproduce it [12]. This has led some authors to consider that these organisms possess a degradosome-like protein network, but that the interactions between its components are transient [12].
Interestingly, three other proteins (YlbF, YmcA and YaaT, collectively called the Y-complex) have been identified as interactors of RNase Y in B. subtilis and have been proposed to act as specificity factors for the cleavage by RNase Y of certain operon mRNAs and riboswitches both in B. subtilis and S. aureus [25].
A final example of the difference in composition of RNA degradosomes is seen in the Gram-negative pathogen H. pylori. Our team demonstrated the existence of a minimal RNA degradosome composed of two partners: RNase J and its sole DEAD-box RNA helicase, RhpA [9]. The simplicity of this degradosome might be related to the reduced genome of H. pylori (1.6 Mb), its restricted colonization niche and the capacity of RNase J to act both as an endo and exoribonuclease.
Thus, the composition of these complexes varies between different bacterial species. Even so, many of their functions and regulation mechanisms are broadly conserved between them, as will be discussed below.
Targets of the RNA degradosomes
RNA degradosomes play a prominent role in the maturation and degradation of many RNA species. In many examples, it has been shown that the endoribonuclease of the complex (be it RNase E, Y or J) can cleave at an internal site of the target RNA molecule, and then, in some cases, the exoribonuclease(s) of the complex, such as PNPase or RNase J, can continue the degradation of one of the resulting RNA molecules [4,12]. Several targets of the dRNases have been identified and validated in vitro [12,26]. Defining, at the global level, the specific RNA targets of dRNases is however more difficult since these enzymes are often essential for normal growth and their global impact results from both direct and indirect effects as they may influence the expression of other pleiotropic regulators.
In E. coli, EcoRNase E is required for the decay of numerous mRNAs [27] and the processing of many sRNAs, although to a lesser extent [26], sometimes in concert with Hfq [28]. It is also required for the maturation of stable RNAs (16S and 5S rRNAs, tRNAs [29] and trans-translation tmRNAs [30]) and is thus important for their respective functions. Though the mechanism by which RNase E targets its substrates is still unclear, an in vivo cleavage map generated using transient inactivation of endonuclease followed by RNA-seq (TIER-seq) in Salmonella enterica, an organism closely related to E. coli, shows that there is a predominant uridine two nucleotides downstream of the RNase E cleavage sites [31]. This suggests an original ruler-and-cut recognition mechanism [31]. Additionally, in E. coli, the membrane anchor of EcoRNase E was recently found to be important for its cleavage specificity by reducing the decay rate of cytosolic ribosome-free transcripts, but had no effect on transcripts encoding inner membrane proteins [20]. In contrast, in another report they found that the membrane anchor reduced the average half-life of transcripts encoding inner membrane proteins [21].
The targets of RNase Y have been studied in S. aureus [32], B. subtilis [33,34], Streptococcus pyogenes [35] and Clostridium perfringens [36]. These targets are mostly mRNAs, ncRNAs and antisense RNAs (asRNAs). Like with RNase E, the targeting mechanism of RNase Y is unclear, but of only 99 total cleavage sites that were identified in S. aureus, a preference for sites after a guanosine residue was observed [32]. Interestingly, the same cleavage sites were found in the absence of the membrane anchor of RNase Y, but the degradation rate of these targets was not assessed [32].
B. subtilis expresses two paralogous RNase J proteins: RNase J1 and RNase J2. In this organism, RNase J1 has an important effect on the abundance of several mRNAs, ncRNAs and asRNAs [33,37]. RNase J2 forms a complex with RNase J1 and its exoribonuclease activity is less important, having a more structural and/or regulatory role over RNase J1 [38,39]. In H. pylori, depletion of the essential and sole RNase J results in the accumulation of 85% of the mRNAs and 78% of the asRNAs. In this mutant, few sRNAs are affected and no role in the maturation of stable RNAs was observed [40], in agreement with recent findings showing an unusual role of another RNase, RNase III, in H. pylori rRNA maturation [41].
Novel roles of RNA degradosomes have recently been reported. The RNase J2 of Staphylococcus epidermidis was shown to participate in CRISPR-Cas10-mediated antiviral and anti-plasmid protection, and its PNPase in CRISPR RNA (crRNA) maturation [42], suggesting a role for these RNases in concert with other cellular nuclease complexes. RNase E was also found to participate in crRNA maturation in Synechocystis [43]. In addition, the RNA degradosome has been shown to be involved in quality control of tRNAs by clearance of hypomodified tRNA species in Vibrio cholerae [44].
Finally, in some cases the RNA degradosome assembly influences cellular processes other than direct RNA degradation. For instance, it was recently shown that the PNPase-binding site of RNase E is required to maintain a normal polyadenylation state of cellular mRNAs, as PNPase interacts with poly(A) polymerase I (PAP I) and may compete with it for the 3’ end of the RNA molecules [45], showing that RNA degradosomes may have pleiotropic effects at multiple levels.
The vast changes caused by dRNase depletion, their essentiality for normal bacterial growth and for the adaptive response under different conditions, as well as potential deleterious activities, account for a central role of dRNases and the associated RNA degradosomes in bacterial physiology and point to the need for a tight regulation of their activities.
Regulation of the RNA degradosome
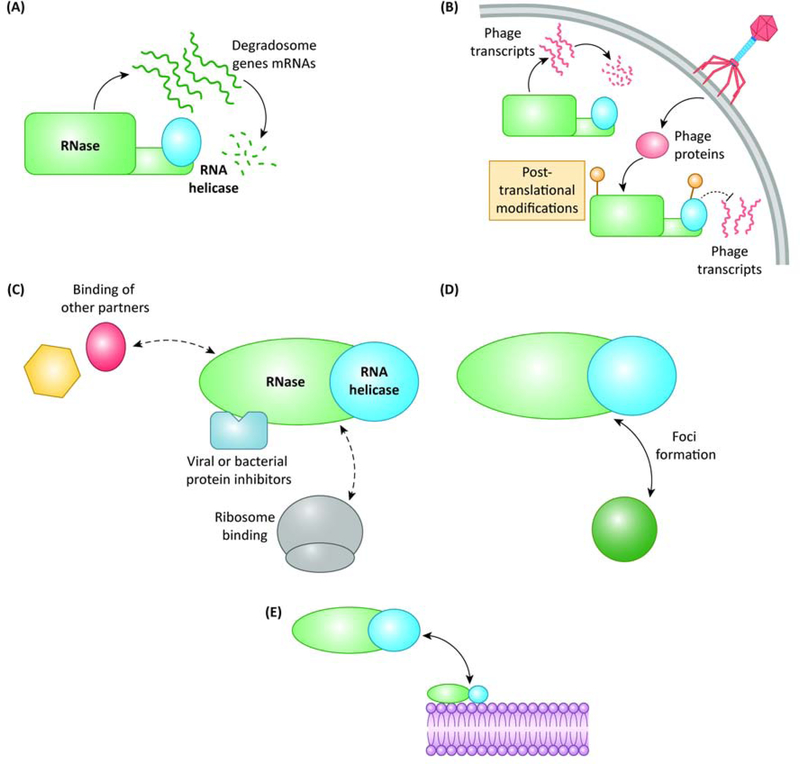
Several mechanisms regulating RNA degradosomes have been reported to date and will be presented below. These processes include autoregulation of their components, post-translational modifications, variations in their components and regulation by spatial localization (Fig. 3, Key Figure). In addition, we will discuss examples in which phages hijack degradosomes for their own benefit.
Figure 3, Key Figure.
Regulation mechanisms of the RNA degradosomes. These mechanisms are not mutually exclusive and may act in concert to adjust the activity of the RNA degradosome. There are five major classes of regulation (a) the components of the RNA degradosome are often autoregulated, and sometimes they also regulate the expression of each other, (b) the partners of the RNA degradosome are susceptible to post-translational modifications, often as a result of a phage infection causing phage RNA to be protected from degradation, (c) different proteins can bind the core RNA degradosome and regulate its activity, including some cellular proteins, ribosomal proteins, as well as dedicated bacterial or viral inhibitors, (d) the central RNases of some degradosomes are compartmentalized within the cell in the form of foci, and (e) the components of the RNA degradosome are often targeted to the bacterial membrane, forming foci or not.
Autoregulation of the RNA degradosome
Negative feedback regulation of the expression of RNases through the control of the stability of their own mRNAs is frequently observed. When EcoRNase E activity exceeds cellular needs, it binds the 5’-untranslated region (5’-UTR) of its own mRNA and cleaves it, a process that results in the destabilization of this messenger [46].
RNase Y has also been reported to cleave within its own mRNA in S. aureus [32]. Interestingly, it also has cleavage sites in the transcripts of other degradosome-related enzymes, such as RNases J1 and J2 and phosphofructokinase [32].
RNases J1 and J2 also regulate each other’s expression in a reciprocal manner in B. subtilis [33] and RNase J1 shows limited autoregulation [47]. In H. pylori, RNase J is strongly up-regulated in the absence of its partner RNA helicase RhpA and has been proposed to be autoregulated through the activity of the degradosome [48].
In a recent report, a marine cyanophage was shown to exploit the Prochlorococcus host RNase E activity for its advantage by preventing its autoregulation [49], constitutively increasing the levels of RNase E that degrade the host’s mRNAs, while phage mRNAs remain protected by asRNAs.
Therefore, autoregulation of the dRNases seems to be a common regulatory mechanism to control and adjust their cellular amounts.
Post-translational modifications
Post-translational modification of proteins can alter either their activity, their capacity to interact with other partners or their stability. In this context, it was shown that, in E. coli, RNase E and RhlB can be phosphorylated by a viral protein kinase upon infection by bacteriophage T7, which helps the phage to successfully infect the cell by stabilizing the mRNAs synthesized by T7 RNA polymerase [50]. Given the high regulatory potential of post-translational modifications, other mechanisms may be uncovered in the future.
Interaction with other protein partners
Several proteins can interact with RNA degradosomes and modify their activities. RraA and RraB are two inhibitors of E. coli RNase E [51]. RraA binds to the RNA-binding regions of its C-terminal domain and hence prevents RNA binding to this domain. It also targets the C-terminus of RhlB, repressing its activity and that of PNPase [52]. In addition, both RraA and RraB can reduce the binding of other degradosome components to RNase E, and RraB causes an increase in the binding of the minor components DnaK and CsdA [51]. The overexpression of these inhibitors impacts several transcripts such as mRNAs (including the one coding for RNase E) and tRNAs [51]. Interestingly, the expression of at least RraA seems to be regulated by the environmental conditions, being overexpressed in stationary phase in E. coli and consequently regulating the activity of the RNA degradosome as a function of the growth conditions [53]. RraA and RraB homologs were found in other organisms, such as Vibrio vulnificus [54], P. aeruginosa [55] and Streptomyces coelicolor [56]. Interestingly, RraAS1, the ortholog for RraA in S. coelicolor, acts by binding the catalytic domain of RNase E [57]. Similarly, the L4 ribosomal protein also modulates the activity of RNase E in E. coli by binding its C-terminal region, stabilizing mRNAs encoding stress-induced proteins [58].
Again, viruses have evolved mechanisms to regulate RNase E activity. The Srd protein encoded in the genome of bacteriophage T4 binds the catalytic domain of EcoRNase E, increasing its activity against certain targets and favoring phage growth [59]. Another example is the protein called Dip, encoded by phage ϕKZ, that blocks the RNA-binding sites in the C-terminus of RNase E in P. aeruginosa, inhibiting RNA degradation and processing [7].
In response to changes in the environmental conditions, different proteins can become part of the RNA degradosome. Under cold-shock conditions, the DEAD-box RNA helicase CsdA replaces RhlB on EcoRNase E, probably allowing a more efficient unwinding of RNA structures that are known to be stabilized at lower temperature [60]. In addition, the other E. coli DEAD-box RNA helicases, RhlE and SrmB, also interact with EcoRNase E in vitro, and RhlE can functionally replace RhlB in vitro [61]. Interestingly, in C. crescentus, RhlE is part of the degradosome under cold-shock conditions [62]. The influence of these cold-shock helicases on the targets of the RNA degradosome has not been assessed.
In S. aureus, it was shown by BACTH that RNase Y interacts with flotillin (FloA). FloA is a membrane protein that acts as a scaffold, being more abundant in the so-called detergent-resistant membrane fractions, similar to eukaryotic lipid rafts [63]. The absence of FloA affects the oligomerization state and the activity of RNase Y, upregulating some target sRNAs of RNase Y. This could underlie a potential regulatory mechanism linking membrane properties to RNA degradosome activity.
To sum up, several proteins interact with dRNases and have the potential of modulating their activities. It is also likely that some of these proteins will affect other properties of the degradosome besides their activity, such as their localization, which is emerging as a potential regulatory level.
Multiple and variable subcellular localizations of RNA degradosomes
As stated above, several dRNases have been found to localize at the bacterial membrane. This is the case for EcoRNase E in E. coli (and predicted for almost all its Y-proteobacteria orthologs as they possess a predicted amphipathic helix [15]) and for RNase Y in B. subtilis and S. aureus. EcoRNase E is attached to the membrane through an amphipathic helix [64], whereas RNase Y contains a single pass transmembrane region [65]. In contrast, in Caulobacter crescentus, RNase E (CcRNase E) was recently reported to lack a membrane anchor and hence to be cytoplasmic [17]. In H. pylori, although no transmembrane region is predicted from the sequence of either RNase J or RhpA, and no interaction with other membrane proteins has been found to date, Tejada-Arranz et al. (in prep.) recently showed that both HpRNase J and RhpA are associated with the inner membrane of this organism.
In two recent studies, the consequences of deleting the membrane-targeting sequence of EcoRNase E were analyzed [20,21]. In one of them, the deletion did not result in massive transcriptomic changes but rather in a global slowdown of RNA degradation [20]. Interestingly, some transcripts appeared to be degraded faster, such as those synthesized by T7 RNA polymerase, which is faster than the bacterial RNA polymerase. It was concluded that RNA molecules that are not readily bound and protected by ribosomes become more accessible for a cytoplasmic form of RNase E. In contrast, in the other study it was found that this deletion results in an increase in the average half-life of transcripts encoding inner membrane proteins [21]. In the case of RNase Y of S. aureus, deletion of its transmembrane region does not change its target molecules [32]. However, the deletion strain grows slower as compared to the wild-type strain, which could correspond to a differential degradation rate of some of its targets.
The RNA degradosomes of E. coli, H. pylori and B. subtilis have, in addition, been shown to be associated with ribosomes [9,66]. Whether this is due to a coupling between translation and RNA decay of a subset of transcripts or to a role of some of their components in rRNA maturation remains to be elucidated.
To study the localization and dynamics of the degradosomes, several groups have analyzed the behavior of dRNases fused to fluorescent proteins in live bacteria. EcoRNase E-YFP rapidly diffuses at the inner membrane of E. coli and forms small short-lived foci [19]. The formation of these foci is regulated, since under anaerobic conditions, EcoRNase E adopts a diffuse distribution in the cell in an enolase binding-dependent fashion [67]. In contrast, the CcRNase E-YFP is located in the cytoplasm, where it forms clusters along the central axis of the cell that change with the cell cycle. These clusters colocalize with ribosomal RNA transcription sites [17]. The CcRNase E foci are dynamically assembled and seem to become more abundant in the presence of stresses such as heat shock or ethanol, among others, and improve the bacterial response to stress [18]. They present characteristics of liquid-liquid phase separation (LLPS, Box 1), analogous to certain eukaryotic structures.
Box 1. Liquid-liquid phase separation (LLPS)
Liquid-liquid phase separation is a biophysical process in which different molecules, such as proteins and/or nucleic acids, form a condensate of a liquidlike nature that allows them to be physically apart from the solution in which they are found, such as the cytoplasm [10]. It often results from the presence of a certain concentration of multivalent molecules, mainly proteins or nucleic acids, that contain binding sites for several other molecules, or proteins containing intrinsically disordered regions (IDRs) that can act as a scaffold for the formation of the condensate. This phenomenon is largely responsible for the formation of membraneless subcellular compartments, allowing the organization of certain enzymatic reactions and/or the sequestration of molecules that may be necessary in a freely diffusing state in a different situation. Some examples of structures formed by LLPS in eukaryotic cells are p-bodies, stress granules or germ granules, and they were recently reported for RNA degradosome foci in prokaryotic organisms.
RNA degradosome compartmentalization and analogies with eukaryotic structures
Compartmentalization of metabolic processes is crucial for any cell in order to optimize enzymatic activities and avoid unwanted reactions. While evident in eukaryotes (mitochondria, nucleus, Golgi apparatus, etc.), just a few physically separated compartments are found in bacteria (spores, magnetosomes, etc.) [68].However, independently of these structures, the importance of intracellular compartmentalization is increasingly being recognized in prokaryotes. Emerging structures that are compartmentalized can be associated with membranes or be the so-called membraneless organelles formed by LLPS [10].
Together with their crucial and potentially deleterious function, the RNA degradosomes localization patterns suggest that the activities of these machineries are controlled by compartmentalization. Although many questions remain open about where the active form of the degradosome is localized or how the RNA substrates are discriminated, recent data reveal striking similarities and functional analogies between the bacterial RNA degradosomes and eukaryotic structures.
Compartmentalization of RNA processing structures in eukaryotes
Eukaryotic exosomes are found in both the nucleus and the cytoplasm and contribute to the processing and degradation of almost every class of RNA [69]. In addition to these structures, there have been extensive reports on large microscopic assemblies of RNA and proteins, referred to as messenger ribonucleoprotein (mRNP) granules, such as p-bodies and stress granules [70]. These are membraneless structures that appear in the cytoplasm of eukaryotic cells in response to stress [70]. Under these conditions, many post- transcriptional actors (including RNases and helicases), RNA-binding proteins (RBPs) and RNAs initiate a process of LLPS leading to the formation of concentrated cytoplasmic granules [70]. It has been proposed that these mRNP granules segregate mRNAs from the cytoplasm and regulate their fate, either by storage, decay or eventual reincorporation to the translatable pool [70].
Similarities between RNA degradosome foci and mRNP granules
First, both prokaryotic degradosomes and eukaryotic granules are assembled on components that act as hubs for their assembly and maintenance and are designated scaffolds [4,70]. Like many of the scaffolding proteins found in eukaryotic mRNPs, several bacterial dRNases have IDRs [15,71]. For example, as discussed above for RNase E, its unstructured C-terminus serves as a binding platform for RNA and the degradosome partners. In eukaryotic granules, these IDRs help in the granule assembly process, and thus this may also be the case for bacterial RNA degradosome foci [72].
Another similarity between RNA degradosome foci and eukaryotic granules is that the formation of both is promoted by RNA substrates [18,19,73]. In C. crescentus, CcRNase E requires RNA for foci assembly and RNA cleavage is required for their dissociation [18]. Furthermore, treatment with compounds that lock ribosomes on mRNAs, or dissociate ribosomes and free their mRNAs, respectively decrease or increase foci assembly [18], hinting at a correlation between free RNA and formation of these structures. Similar observations were reported for granules in eukaryotic cells [73,74]. In addition, the membrane-associated EcoRNase E foci depend on transcription and have been proposed to form on transcripts in E. coli cells [19]. The nature of the RNA molecules that are targeted to mRNP granules and RNA degradosomes is still not clear, raising questions about the factors responsible for their specificity. In the case of p-bodies and stress granules, it has been proposed that other co-localizing proteins may be responsible for it, such as proteins that recognize RNA G-quadruplexes or RNA methyltransferases, among others [70]. Such recognition mechanisms could also be an unexplored possibility for RNA degradosomes.
Finally, both bacterial foci and eukaryotic mRNP granules were found to be dynamic. In C. crescentus, CcRNase E foci rapidly form and dissolve over time, and their numbers are increased under some stresses [18]. In E. coli, the membrane-associated degradosome foci are also dynamic and form by transient clustering of EcoRNase E [19], although no impact of stress exposure on the formation of these structures has been reported yet. Despite the fact that eukaryotic granules are larger than bacterial structures, they rapidly form upon stress and clear after recovery, partly due to their LLPS properties, although not all of them are liquid in nature [18,19,75].
What is the “raison d’être” of RNP granules/foci?
Overall, the function of RNP granules or foci is still poorly understood. As stated above, it was shown that eukaryotic cells that lack the ability to properly form stress granules are more sensitive to stress [76], and the same was seen in C. crescentus [18]. However, it is not clear whether this is due to the fact that stress granules are important to sequester a specific subset of mRNAs or proteins during stress; or if they are required for cell recovery upon stress withdrawal by release of their components, bypassing the need for de novo transcription and nuclear export [77]. P-bodies were originally proposed to be hubs of RNA degradation with accumulation of RNA degradation intermediates in these structures [70]. However, it is not clear whether RNA degradation actually occurs in the p-bodies or elsewhere, and it has been proposed that these have more of a storage role, as their disruption does not affect global RNA decay [78]. Another possible function could be to sequester translation initiation factors and the RNA degradation machinery to reduce their effective concentration in the cytoplasm during stress. All of these possible roles of eukaryotic mRNP granules could potentially also apply to RNA degradosomes, although this has not been explored yet.
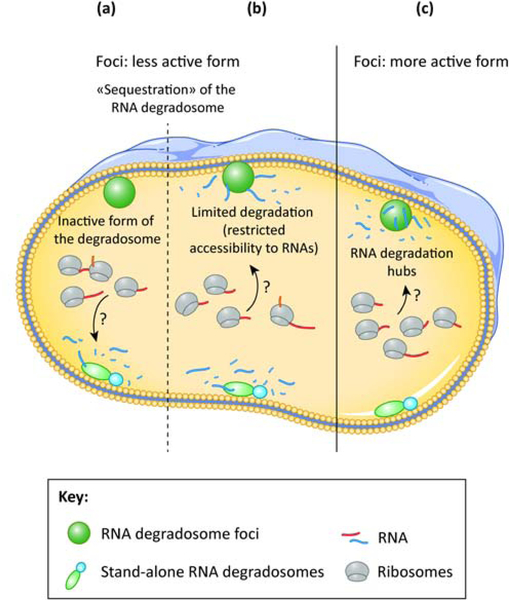
The striking analogy between eukaryotic mRNPs and bacterial foci opens many pathways for future research. Important questions on the mechanisms leading to the formation of RNA degradosome foci, their composition, their regulation and specific roles are still to be answered. As illustrated in Fig. 4, we need to determine whether the foci correspond to RNA degradation hubs or rather inactive clusters and define whether the foci and/or membrane localization leads to sequestration of the degradosome away from some of its targets and thereby limits its activity and whether these structures are important for its target discrimination and subsequent degradation.
Figure 4.
Models for the possible physiological roles of RNA degradosome foci. In (a) and (b), the foci represent a less active form of the RNA degradosome. In (a), the foci-forming degradosomes are catalytically inactive, whereas the complexes outside foci are able to degrade RNA. In (b), the foci-forming degradosomes retain their catalytic activity, but they are sequestered away from some cellular RNAs, limiting their degradation. In (c), the foci-forming degradosomes are the active form, making foci RNA degradation hubs, whereas the complexes outside foci retain comparatively little or no activity. The targeting mechanisms of the RNAs that are degraded by the degradosomes are still not clear.
Concluding remarks
RNA degradosomes are diverse protein machineries that play an essential role in the maintenance of cell homeostasis. Although a better view of the dRNase activities and their important number of cellular RNA targets is emerging, the influence of the composition of the degradosome on the selectivity of the targets is not yet established. The vast changes caused by dRNase depletion, their essentiality for normal bacterial growth and for the adaptive response under different conditions, as well as potential deleterious activities, account for a central role in bacterial physiology and tight regulation of the activities of dRNases and their associated RNA degradosomes. Further research is required to understand more precisely the mechanisms that govern the activity of these machines (see Outstanding Questions) and how they can fine-tune cell physiology depending on factors such as their subcellular localization and stress response. Furthermore, similarities between RNA degradosome foci and eukaryotic mRNP granules suggest a conserved requirement for compartmentalization of these structures, a feature that is most likely central in global post-transcriptional regulation and response to stresses both in prokaryotic and eukaryotic organisms.
Outstanding questions
-
-
What determines the RNA target specificity of RNA degradosomes in bacteria?
-
-
What is the interplay between the different dRNases (RNase E, RNase Y, RNase J) when they are co-expressed in an organism?
-
-
Are there additional mechanisms that control the RNA degradosome activity?
-
-
Do degradosomes play a general role in stress response or survival in bacteria?
-
-
What is the physiological relevance of the membrane localization and foci formation of degradosomes?
-
-
Do degradosomes play different roles as a function of their subcellular localization?
-
-
What is the degradosome foci composition and what are the determinants of their formation?
-
-
Do foci represent an active RNA degradation hub or rather a sequestered form of the degradosome to prevent it from acting when or where it should not?
-
-
How analogous are bacterial RNA degradosomes and eukaryotic p-bodies and stress granules?
Supplementary Material
Highlights
-
-
RNA degradosomes, molecular machines composed of at one RNase and one RNA helicase, are major players in post-transcriptional regulation of gene expression in bacteria.
-
-
RNA degradosomes are very diverse in composition and are tightly regulated by a wealth of mechanisms, including autoregulation, post-translational modifications, protein localization or interaction with different partners.
-
-
Some RNA degradosomes seem to be compartmentalized at the membrane, which could affect their activity.
-
-
Some bacterial RNA degradosomes show similarities with eukaryotic mRNP granules at the level of their physicochemical properties.
Acknowledgements
ATA is part of the Pasteur - Paris University (PPU) International PhD Program. This project has received funding from the European Union’s Horizon 2020 research and innovation program under the Marie Sklodowska-Curie grant agreement No 665807, and from the Institut Carnot Pasteur Microbes & Santé. Figures were generated with images from Servier Medical Art (https://smart.servier.com/), licensed under the Creative Commons Attribution 3.0 Unported License (http://creativecommons.org/license/by/3.0/). We thank the “Fondation pour la Recherche Médicale” for the support to the Institut Pasteur laboratory (grant DBF20161136767 to HDR). The degradosome studies of the Institut Pasteur laboratory are supported by the Pasteur- Weizmann Consortium of “The Roles of Noncoding RNAs in Regulation of Microbial Life Styles and Virulence”. The work of V dC-L was partially funded by the National Institutes of Health (R01 GM70614).
Glossary
- 5’-UTR
5’ untranslated region, part of the mRNA molecule that does not code for a protein.
- asRNA
antisense RNA, RNA molecule transcribed from the opposite strand to that of the mRNA and is therefore perfectly complementary to its corresponding mRNA molecule.
- BACTH
bacterial two hybrid, technique that allows the study of the interaction between two proteins in E. coli.
- CRISPR-Cas
Clustered Regularly Interspaced Short Palindromic Repeats- CRISPR-associated protein, bacterial system that allows the cleavage of foreign DNA by the Cas enzymes in a CRISPR-directed manner.
- IDRs
intrinsically disordered regions, parts of proteins with no stable tertiary structure under physiological conditions in vitro. Some of these regions can bind other proteins, interact with nucleic acids, or serve as scaffold domains. They are also involved in signaling pathways and have important roles in protein regulation.
- Membraneless organelles
cellular structures that allow proteins to become concentrated in sub-compartments without being surrounded by a barrier to diffusion such as a membrane and that are responsible for performing specific cellular functions. In contrast to organelles with a lipid bilayer membrane, membraneless structures are formed through a process known as liquid-liquid phase separation.
- ncRNA
non-coding RNAs, RNA molecules that do not encode a protein and that have a regulatory function.
- Processing bodies (p-bodies)
membraneless organelles characteristic of eukaryotic cells that are composed of proteins and RNA and whose function is not clear, but may serve as storage of different components.
- RNA G-quadruplex
extremely stable RNA secondary structure that is formed by guanine-rich sequences.
- RNA helicase of the DEAD-box family
proteins that unwind secondary structures of RNA molecules and allow for them to be cleaved by an RNase or be acted upon by other RNA-modifying enzymes. This family of helicases contains a DEAD sequence motif that gives it the name.
- Stress granules
stress granules are dense membraneless aggregates characteristic of the cytosol of eukaryotic cells. They are composed of proteins and RNA molecules and usually form in a reversible manner upon cellular stress.
- TIER-seq
transient inactivation of endonuclease followed by RNA-seq, technique allowing the mapping of the 5’-ends of transcripts under normal conditions and after the temporary inactivation of an essential endonuclease.
- tmRNA
transfer-messenger RNA, RNA molecule responsible for the rescue of stalled ribosomes by trans-translation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- [1].Felden B, Paillard L (2017) When eukaryotes and prokaryotes look alike: the case of regulatory RNAs. FEMS Microbiol. Rev. 41, 624–639. [DOI] [PubMed] [Google Scholar]
- [2].Aït-Bara S, Carpousis AJ (2015) RNA degradosomes in bacteria and chloroplasts: classification, distribution and evolution of RNase E homologs. Mol. Microbiol. 97, 1021–1135. [DOI] [PubMed] [Google Scholar]
- [3].Mitchell P, Petfalski E, Shevchenko A, Mann M, Tollervey D (1997) The exosome: a conserved eukaryotic RNA processing complex containing multiple 3’−5’ exoribonucleases. Cell 91, 457–66. [DOI] [PubMed] [Google Scholar]
- [4].Bandyra KJ, Bouvier M, Carpousis AJ, Luisi BF (2013) The social fabric of the RNA degradosome. Biochim. Biophys. Acta 1829, 514–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Carpousis AJ, Van Houwe G, Ehretsmann C, Krisch HM (1994) Copurification of E. coli RNAase E and PNPase: evidence for a specific association between two enzymes important in RNA processing and degradation. Cell 76, 889–900. [DOI] [PubMed] [Google Scholar]
- [6].Roux CM, DeMuth JP, Dunman PM (2011) Characterization of components of the Staphylococcus aureus mRNA degradosome holoenzyme-like complex. J. Bacteriol. 193, 5520–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Van den Bossche A, Hardwick SW, Ceyssens P-J, Hendrix H, Voet M, Dendooven T, Bandyra KJ, De Maeyer M, Aertsen A, Noben J- P, Luisi BF, Lavigne R (2016) Structural elucidation of a novel mechanism for the bacteriophage-based inhibition of the RNA degradosome. eLife 5, e16413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Plocinski P, Macios M, Houghton J, Niemiec E, Plocinska R, Brzostek A, Slomka M, Dziadek J, Young D, Dziembowski A (2019) Proteomic and transcriptomic experiments reveal an essential role of RNA degradosome complexes in shaping the transcriptome of Mycobacterium tuberculosis. Nucleic Acids Res. 47, 5892–5905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Redko Y, Aubert S, Stachowicz A, Lenormand P, Namane A, Darfeuille F, Thibonnier M, De Reuse H (2013) A minimal bacterial RNase J-based degradosome is associated with translating ribosomes. Nucleic Acids Res. 41, 288–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Banani SF, Lee HO, Hyman AA, Rosen MK (2017) Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 18, 285–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Deutscher MP (2015) Twenty years of bacterial RNases and RNA processing: how we’ve matured. RNA 21, 597–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Durand S, Condon C (2018) RNases and Helicases in Gram-Positive Bacteria. Microbiol. Spectr. 6, 37–53. [DOI] [PubMed] [Google Scholar]
- [13].Redder P (2018) Molecular and genetic interactions of the RNA degradation machineries in Firmicute bacteria. Wiley Interdiscip. Rev. RNA 9, e1460. [DOI] [PubMed] [Google Scholar]
- [14].Morita T, Kawamoto H, Mizota T, Inada T, Aiba H (2004) Enolase in the RNA degradosome plays a crucial role in the rapid decay of glucose transporter mRNA in the response to phosphosugar stress in Escherichia coli. Mol. Microbiol. 54, 1063–1075. [DOI] [PubMed] [Google Scholar]
- [15].Aït-Bara S, Carpousis AJ, Quentin Y (2015) RNase E in the y- Proteobacteria: conservation of intrinsically disordered noncatalytic region and molecular evolution of microdomains. Mol. Genet. Genomics 290, 847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Bruce HA, Du D, Matak-Vinkovic D, Bandyra KJ, Broadhurst RW, Martin E, Sobott F, Shkumatov A V, Luisi BF (2018) Analysis of the natively unstructured RNA/protein-recognition core in the Escherichia coli RNA degradosome and its interactions with regulatory RNA/Hfq complexes. Nucleic Acids Res. 46, 387–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Bayas CA, Wang J, Lee MK, Schrader JM, Shapiro L, Moerner WE (2018) Spatial organization and dynamics of RNase E and ribosomes in Caulobacter crescentus. Proc. Natl. Acad. Sci. U. S. A. 115, E3712–E3721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Al-Husini N, Tomares DT, Bitar O, Childers WS, Schrader JM (2018) α- Proteobacterial RNA Degradosomes Assemble Liquid-Liquid Phase- Separated RNP Bodies. Mol. Cell 71, 1027–1039.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Strahl H, Turlan C, Khalid S, Bond PJ, Kebalo J-M, Peyron P, Poljak L, Bouvier M, Hamoen L, Luisi BF, Carpousis AJ (2015) Membrane recognition and dynamics of the RNA degradosome. PLoS Genet. 11, e1004961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Hadjeras L, Poljak L, Bouvier M, Morin-Ogier Q, Canal I, Cocaign-Bousquet M, Girbal L, Carpousis AJ (2019) Detachment of the RNA degradosome from the inner membrane of Escherichia coli results in a global slowdown of mRNA degradation, proteolysis of RN ase E and increased turnover of ribosome-free transcripts. Mol. Microbiol. mmi.14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Moffitt JR, Pandey S, Boettiger AN, Wang S, Zhuang X (2016) Spatial organization shapes the turnover of a bacterial transcriptome. eLife 5,. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Ow MC, Perwez T, Kushner SR (2004) RNase G of Escherichia coli exhibits only limited functional overlap with its essential homologue, RNase E. Mol. Microbiol. 49, 607–622. [DOI] [PubMed] [Google Scholar]
- [23].Commichau FM, Rothe FM, Herzberg C, Wagner E, Hellwig D, Lehnik- Habrink M, Hammer E, Völker U, Stülke J (2009) Novel Activities of Glycolytic Enzymes in Bacillus subtilis. Mol. Cell. Proteomics 8, 1350–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Cascante-Estepa N, Gunka K, Stülke J (2016) Localization of Components of the RNA-Degrading Machine in Bacillus subtilis. Front. Microbiol. 07, 1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].DeLoughery A, Lalanne J-B, Losick R, Li G- W (2018) Maturation of polycistronic mRNAs by the endoribonuclease RNase Y and its associated Y-complex in Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 115, E5585–E5594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Mackie GA (2013) RNase E: at the interface of bacterial RNA processing and decay. Nat. Rev. Microbiol. 11, 45–57. [DOI] [PubMed] [Google Scholar]
- [27].Clarke JE, Kime L, Romero AD, McDowall KJ (2014) Direct entry by RNase E is a major pathway for the degradation and processing of RNA in Escherichia coli. Nucleic Acids Res. 42, 11733–11751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Waters SA, McAteer SP, Kudla G, Pang I, Deshpande NP, Amos TG, Leong KW, Wilkins MR, Strugnell R, Gally DL, Tollervey D, Tree JJ (2017) Small RNA interactome of pathogenic E. coli revealed through crosslinking of RNase E. EMBO J. 36, 374–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Kime L, Clarke JE, Romero A D, Grasby JA, McDowall KJ (2014) Adjacent single-stranded regions mediate processing of tRNA precursors by RNase E direct entry. Nucleic Acids Res. 42, 4577–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Lin-Chao S, Wei CL, Lin YT (1999) RNase E is required for the maturation of ssrA RNA and normal ssrA RNA peptide-tagging activity. Proc. Natl. Acad. Sci. U. S. A. 96, 12406–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Chao Y, Li L, Girodat D, Förstner KU, Said N, Corcoran C, Smiga M, Papenfort K, Reinhardt R, Wieden H-J, Luisi BF, Vogel J (2017) In Vivo Cleavage Map Illuminates the Central Role of RNase E in Coding and Non-coding RNA Pathways. Mol. Cell 65, 39–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Khemici V, Prados J, Linder P, Redder P (2015) Decay-Initiating Endoribonucleolytic Cleavage by RNase Y Is Kept under Tight Control via Sequence Preference and Sub-cellular Localisation. PLOS Genet. 11, e1005577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Durand S, Gilet L, Bessières P, Nicolas P, Condon C (2012) Three Essential Ribonucleases—RNase Y, J1, and III—Control the Abundance of a Majority of Bacillus subtilis mRNAs. PLoS Genet. 8, e1002520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Lehnik-Habrink M, Schaffer M, Mäder U, Diethmaier C, Herzberg C, Stülke J (2011) RNA processing in Bacillus subtilis: identification of targets of the essential RNase Y. Mol. Microbiol. 81, 1459–1473. [DOI] [PubMed] [Google Scholar]
- [35].Kang SO, Caparon MG, Cho KH (2010) Virulence gene regulation by CvfA, a putative RNase: the CvfA-enolase complex in Streptococcus pyogenes links nutritional stress, growth-phase control, and virulence gene expression. Infect. Immun. 78, 2754–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Obana N, Nakamura K, Nomura N (2017) Role of RNase Y in Clostridium perfringens mRNA Decay and Processing. J. Bacteriol. 199,. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Mäder U, Zig L, Kretschmer J, Homuth G, Putzer H (2008) mRNA processing by RNases J1 and J2 affects Bacillus subtilis gene expression on a global scale. Mol. Microbiol. 70, 183–196. [DOI] [PubMed] [Google Scholar]
- [38].Mathy N, Hébert A, Mervelet P, Bénard L, Dorléans A, Li de la Sierra- Gallay I, Noirot P, Putzer H, Condon C (2010) Bacillus subtilis ribonucleases J1 and J2 form a complex with altered enzyme behaviour. Mol. Microbiol. 75, 489–498. [DOI] [PubMed] [Google Scholar]
- [39].Linder P, Lemeille S, Redder P, Francois P, Corvaglia A (2014) Transcriptome-Wide Analyses of 5’-Ends in RNase J Mutants of a Gram¬Positive Pathogen Reveal a Role in RNA Maturation, Regulation and Degradation. PLoS Genet. 10, e1004207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Redko Y, Galtier E, Arnion H, Darfeuille F, Sismeiro O, Coppée J-Y, Médigue C, Weiman M, Cruveiller S, De Reuse H (2016) RNase J depletion leads to massive changes in mRNA abundance in Helicobacter pylori. RNA Biol. 13, 243–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Iost I, Chabas S, Darfeuille F (2019) Maturation of atypical ribosomal RNA precursors in Helicobacter pylori. Nucleic Acids Res. 47, 5906–5921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Chou-Zheng L, Hatoum-Aslan A (2019) A type III-A CRISPR-Cas system employs degradosome nucleases to ensure robust immunity. eLife 8,. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Behler J, Sharma K, Reimann V, Wilde A, Urlaub H, Hess WR (2018) The host-encoded RNase E endonuclease as the crRNA maturation enzyme in a CRISPR-Cas subtype III-Bv system. Nat. Microbiol. 3, 367–377. [DOI] [PubMed] [Google Scholar]
- [44].Kimura S, Waldor MK (2019) The RNA degradosome promotes tRNA quality control through clearance of hypomodified tRNA. Proc. Natl. Acad. Sci. U. S. A. 116, 1394–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Mildenhall KB, Wiese N, Chung D, Maples VF, Mohanty BK, Kushner SR (2016) RNase E-based degradosome modulates polyadenylation of mRNAs after Rho-independent transcription terminators in Escherichia coli. Mol. Microbiol. 101, 645–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Schuck A, Diwa A, Belasco JG (2009) RNase E autoregulates its synthesis in Escherichia coli by binding directly to a stem-loop in the rne 5’ untranslated region. Mol. Microbiol. 72, 470–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Jamalli A, Hébert A, Zig L, Putzer H (2014) Control of expression of the RNases J1 and J2 in Bacillus subtilis. J. Bacteriol. 196, 318–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].El Mortaji L, Aubert S, Galtier E, Schmitt C, Anger K, Redko Y, Quentin Y, De Reuse H (2018) The sole DEAD-box RNA helicase of the gastric pathogen Helicobacter pylori is essential for colonization. MBio 9, e02071–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Stazic D, Pekarski I, Kopf M, Lindell D, Steglich C (2016) A Novel Strategy for Exploitation of Host RNase E Activity by a Marine Cyanophage. Genetics 203, 1149–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Marchand I, Nicholson AW, Dreyfus M (2001) Bacteriophage T7 protein kinase phosphorylates RNase E and stabilizes mRNAs synthesized by T7 RNA polymerase. Mol. Microbiol. 42, 767–76. [DOI] [PubMed] [Google Scholar]
- [51].Gao J, Lee K, Zhao M, Qiu J, Zhan X, Saxena A, Moore CJ, Cohen SN, Georgiou G (2006) Differential modulation of E. coli mRNA abundance by inhibitory proteins that alter the composition of the degradosome. Mol. Microbiol. 61, 394–406. [DOI] [PubMed] [Google Scholar]
- [52].Górna MW, Pietras Z, Tsai Y-C, Callaghan AJ, Hernández H, Robinson C V, Luisi BF (2010) The regulatory protein RraA modulates RNA-binding and helicase activities of the E. coli RNA degradosome. RNA 16, 553–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Zhao M, Zhou L, Kawarasaki Y, Georgiou G (2006) Regulation of RraA, a Protein Inhibitor of RNase E-Mediated RNA Decay. J. Bacteriol. 188, 3257–3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Kim D, Kim Y-H, Jang J, Yeom J- H, Jun JW, Hyun S, Lee K (2016) Functional Analysis of Vibrio vulnificus Orthologs of Escherichia coli RraA and RNase E. Curr. Microbiol. 72, 716–722. [DOI] [PubMed] [Google Scholar]
- [55].Tang J, Luo M, Niu S, Zhou H, Cai X, Zhang W, Hu Y, Yin Y, Huang A, Wang D (2010) The Crystal Structure of Hexamer RraA from Pseudomonas Aeruginosa Reveals Six Conserved Protein-Protein Interaction Sites. Protein J. 29, 583–590. [DOI] [PubMed] [Google Scholar]
- [56].Yeom J-H, Go H, Shin E, Kim H-L, Han SH, Moore CJ, Bae J, Lee K (2008) Inhibitory effects of RraA and RraB on RNAse E-related enzymes imply conserved functions in the regulated enzymatic cleavage of RNA. FEMS Microbiol. Lett. 285, 10–15. [DOI] [PubMed] [Google Scholar]
- [57].Seo S, Kim D, Song W, Heo J, Joo M, Lim Y, Yeom J-H, Lee K (2017) RraAS1 inhibits the ribonucleolytic activity of RNase ES by interacting with its catalytic domain in Streptomyces coelicolor. J. Microbiol. 55, 37–43. [DOI] [PubMed] [Google Scholar]
- [58].Singh D, Chang S-J, Lin P-H, Averina OV., Kaberdin VR, Lin-Chao S (2009) Regulation of ribonuclease E activity by the L4 ribosomal protein of Escherichia coli. Proc. Natl. Acad. Sci. 106, 864–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Qi D, Alawneh AM, Yonesaki T, Otsuka Y (2015) Rapid Degradation of Host mRNAs by Stimulation of RNase E Activity by Srd of Bacteriophage T4. Genetics 201, 977–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Prud’homme-Généreux A, Beran RK, Iost I, Ramey CS, Mackie GA, Simons RW (2004) Physical and functional interactions among RNase E, polynucleotide phosphorylase and the cold-shock protein, CsdA: evidence for a ‘cold shock degradosome. Mol. Microbiol. 54, 1409–1421. [DOI] [PubMed] [Google Scholar]
- [61].Khemici V, Toesca I, Poljak L, Vanzo NF, Carpousis AJ (2004) The RNase E of Escherichia coli has at least two binding sites for DEAD-box RNA helicases: functional replacement of RhlB by RhlE. Mol. Microbiol. 54, 1422–30. [DOI] [PubMed] [Google Scholar]
- [62].Aguirre AA, Vicente AM, Hardwick SW, Alvelos DM, Mazzon RR, Luisi BF, Marques MV. (2017) Association of the Cold Shock DEAD-Box RNA Helicase RhlE to the RNA Degradosome in Caulobacter crescentus. J. Bacteriol. 199, e00135–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Koch G, Wermser C, Acosta IC, Kricks L, Stengel ST, Yepes A, Lopez D (2017) Attenuating Staphylococcus aureus Virulence by Targeting Flotillin Protein Scaffold Activity. Cell Chem. Biol. 24, 845–857.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Khemici V, Poljak L, Luisi BF, Carpousis AJ (2008) The RNase E of Escherichia coli is a membrane-binding protein. Mol. Microbiol. 70, 799–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Hunt A, Rawlins JP, Thomaides HB, Errington J (2006) Functional analysis of 11 putative essential genes in Bacillus subtilis. Microbiology 152, 2895–2907. [DOI] [PubMed] [Google Scholar]
- [66].Tsai Y-C, Du D, Domínguez-Malfavón L, Dimastrogiovanni D, Cross J, Callaghan AJ, García-Mena J, Luisi BF (2012) Recognition of the 70S ribosome and polysome by the RNA degradosome in Escherichia coli. Nucleic Acids Res. 40, 10417–10431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Murashko ON, Lin-Chao S (2017) Escherichia coli responds to environmental changes using enolasic degradosomes and stabilized DicF sRNA to alter cellular morphology. Proc. Natl. Acad. Sci. 201703731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Cornejo E, Abreu N, Komeili A (2014) Compartmentalization and organelle formation in bacteria. Curr. Opin. Cell Biol. 26, 132–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Januszyk K, Lima CD (2014) The eukaryotic RNA exosome. Curr. Opin. Struct. Biol. 24, 132–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Guzikowski AR, Chen YS, Zid BM (2019) StressD induced mRNP granules: Form and function of processing bodies and stress granules. Wiley Interdiscip. Rev. RNA 10, e1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Lehnik-Habrink M, Newman J, Rothe FM, Solovyova AS, Rodrigues C, Herzberg C, Commichau FM, Lewis RJ, Stülke J (2011. ) RNase Y in Bacillus subtilis: a Natively disordered protein that is the functional equivalent of RNase E from Escherichia coli. J. Bacteriol. 193, 5431–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Protter DSW, Rao BS, Van Treeck B, Lin Y, Mizoue L, Rosen MK, Parker R (2018) Intrinsically Disordered Regions Can Contribute Promiscuous Interactions to RNP Granule Assembly. Cell Rep. 22, 1401–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Teixeira D, Sheth U, Valencia-Sanchez MA, Brengues M, Parker R (2005) Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA 11, 371–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Kedersha N, Cho MR, Li W, Yacono PW, Chen S, Gilks N, Golan DE, Anderson P (2000) Dynamic shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules. J. Cell Biol. 151, 1257–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Kroschwald S, Munder MC, Maharana S, Franzmann TM, Richter D, Ruer M, Hyman AA, Alberti S (2018) Different Material States of Pub1 Condensates Define Distinct Modes of Stress Adaptation and Recovery. Cell Rep. 23, 3327–3339. [DOI] [PubMed] [Google Scholar]
- [76].Riback JA, Katanski CD, Kear-Scott JL, Pilipenko E V., Rojek AE, Sosnick TR, Drummond DA (2017) Stress-Triggered Phase Separation Is an Adaptive, Evolutionarily Tuned Response. Cell 168, 1028–1040.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Saad S, Cereghetti G, Feng Y, Picotti P, Peter M, Dechant R (2017) Reversible protein aggregation is a protective mechanism to ensure cell cycle restart after stress. Nat. Cell Biol. 19, 1202–1213. [DOI] [PubMed] [Google Scholar]
- [78].Eulalio A, Behm-Ansmant I, Schweizer D, Izaurralde E (2007) P-Body Formation Is a Consequence, Not the Cause, of RNA-Mediated Gene Silencing. Mol. Cell. Biol. 27, 3970–3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Carpousis AJ, Van Houwe G, Ehretsmann C, Krisch HM (1994) Copurification of E. coli RNAase E and PNPase: evidence for a specific association between two enzymes important in RNA processing and degradation. Cell 76, 889–900. [DOI] [PubMed] [Google Scholar]
- [80].Py B, Causton H, Mudd EA, Higgins CF (1994) A protein complex mediating mRNA degradation in Escherichia coli. Mol. Microbiol. 14, 717–29. [DOI] [PubMed] [Google Scholar]
- [81].Py B, Higgins CF, Krisch HM, Carpousis AJ (1996) A DEAD-box RNA helicase in the Escherichia coli RNA degradosome. Nature 381, 169–172. [DOI] [PubMed] [Google Scholar]
- [82].Lu F, Taghbalout A (2014) The Escherichia coli major exoribonuclease RNase II is a component of the RNA degradosome. Biosci. Rep. 34, 879–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Miczak A, Kaberdin VR, Wei CL, Lin-Chao S (1996) Proteins associated with RNase E in a multicomponent ribonucleolytic complex. Proc. Natl. Acad. Sci. U. S. A. 93, 3865–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Blum E, Py B, Carpousis AJ, Higgins CF (1997) Polyphosphate kinase is a component of the Escherichia coli RNA degradosome. Mol. Microbiol. 26, 387–98. [DOI] [PubMed] [Google Scholar]
- [85].Carabetta VJ, Silhavy TJ, Cristea IM (2010) The response regulator SprE (RssB) is required for maintaining poly(A) polymerase I-degradosome association during stationary phase. J. Bacteriol. 192, 3713–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Raynal LC, Carpousis AJ (1999) Poly(A) polymerase I of Escherichia coli: characterization of the catalytic domain, an RNA binding site and regions for the interaction with proteins involved in mRNA degradation. Mol. Microbiol. 32, 765–75. [DOI] [PubMed] [Google Scholar]
- [87].Hardwick SW, Chan VSY, Broadhurst RW, Luisi BF (2011. ) An RNA degradosome assembly in Caulobacter crescentus. Nucleic Acids Res. 39, 1449–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Voss JE, Luisi BF, Hardwick SW (2014) Molecular recognition of RhlB and RNase D in the Caulobacter crescentus RNA degradosome. Nucleic Acids Res. 42, 13294–13305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Kovacs L, Csanadi A, Megyeri K, Kaberdin VR, Miczak A (2005) Mycobacterial RNase E-associated proteins. Microbiol. Immunol. 49, 1003–7. [DOI] [PubMed] [Google Scholar]
- [90].Aït-Bara S, Carpousis AJ (2010) Characterization of the RNA degradosome of Pseudoalteromonas haloplanktis: conservation of the RNase E-RhlB interaction in the gammaproteobacteria. J. Bacteriol. 192, 5413–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Zhang J-Y, Deng X-M, Li F-P, Wang L, Huang Q-Y, Zhang C-C, Chen W-L (2014) RNase E forms a complex with polynucleotide phosphorylase in cyanobacteria via a cyanobacterial-specific nonapeptide in the noncatalytic region. RNA 20, 568–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Rosana ARR, Whitford DS, Fahlman RP, Owttrim GW (2016) Cyanobacterial RNA Helicase CrhR Localizes to the Thylakoid Membrane Region and Cosediments with Degradosome and Polysome Complexes in Synechocystis sp. Strain PCC 6803. J. Bacteriol. 198, 2089–2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Lee K, Cohen SN (2003) A Streptomyces coelicolor functional orthologue of Escherichia coli RNase E shows shuffling of catalytic and PNPase- binding domains. Mol. Microbiol. 48, 349–360. [DOI] [PubMed] [Google Scholar]
- [94].Purusharth RI, Klein F, Sulthana S, Jäger S, Jagannadham MV, Evguenieva-Hackenberg E, Ray MK, Klug G (2005) Exoribonuclease R Interacts with Endoribonuclease E and an RNA Helicase in the Psychrotrophic Bacterium Pseudomonas syringae Lz4W. J. Biol. Chem. 280, 14572–14578. [DOI] [PubMed] [Google Scholar]
- [95].Jäger S, Fuhrmann O, Heck C, Hebermehl M, Schiltz E, Rauhut R, Klug G (2001) An mRNA degrading complex in Rhodobacter capsulatus. Nucleic Acids Res. 29, 4581–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Stoppel R, Manavski N, Schein A, Schuster G, Teubner M, Schmitz- Linneweber C, Meurer J (2012) RHON1 is a novel ribonucleic acid- binding protein that supports RNase E function in the Arabidopsis chloroplast. Nucleic Acids Res. 40, 8593–8606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Newman JA, Hewitt L, Rodrigues C, Solovyova AS, Harwood CR, Lewis RJ (2012) Dissection of the network of interactions that links RNA processing with glycolysis in the Bacillus subtilis degradosome. J. Mol. Biol. 416, 121–136. [DOI] [PubMed] [Google Scholar]
- [98].Gimpel M, Brantl S (2016) Dual-function sRNA encoded peptide SR1P modulates moonlighting activity of B. subtilis GapA. RNA Biol. 13, 916–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].DeLoughery A, Dengler V, Chai Y, Losick R (2016) Biofilm formation by Bacillus subtilis requires an endoribonuclease-containing multisubunit complex that controls mRNA levels for the matrix gene repressor SinR. Mol. Microbiol. 99, 425–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Giraud C, Hausmann S, Lemeille S, Prados J, Redder P, Linder P (2015) The C-terminal region of the RNA helicase CshA is required for the interaction with the degradosome and turnover of bulk RNA in the opportunistic pathogen Staphylococcus aureus. RNA Biol. 12, 658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Niehaus TD, Gerdes S, Hodge-Hanson K, Zhukov A, Cooper AJ, ElBadawi-Sidhu M, Fiehn O, Downs DM, Hanson AD (2015) Genomic and experimental evidence for multiple metabolic functions in the RidA/YjgF/YER057c/UK114 (Rid) protein family. BMC Genomics 16, 382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T, Edwards RA, Gerdes S, Parrello B, Shukla M, Vonstein V, Wattam AR, Xia F, Stevens R (2014) The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res. 42, D206–D214. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.