Abstract
ARID1A alterations would compromise mismatch repair pathway and increase the number of tumor-infiltrating lymphocytes and PD-L1 expression in some cancers, which would cooperate with immune checkpoint inhibitors (ICIs) treatment. However, a comprehensive analysis of ARID1A alteration frequency and its predictive value for ICI treatment outcome in cancers has not yet been investigated. Hence, we performed this pan-cancer analysis to evaluate the prevalence and predictive value of ARID1A alterations across >40,000 cases in multiple cancer types. We found a high frequency (6.2%) of ARID1A, which were associated with significantly higher tumor mutation burden level across various cancers. Importantly, patients with ARID1A alterations and advanced cancers had the substantially prolonged overall survival in ICI treatment cohort, suggesting it might be used to predict a survival benefit from ICI therapy across multiple cancer types. Notably, ARID1A alterations were correlated with markedly high immune infiltrates in endometrial, stomach and colon cancer. However, patients with ARID1A-mutant renal clear cell carcinoma had dramatically lower CD8+ T cell infiltrations than those without, indicating the association between ARID1A alterations and immune infiltrates was cancer-dependent. Collectively, our findings highlight the important value of ARID1A alterations as pan-cancer predictive biomarkers for ICI treatment.
Keywords: ARID1A, Immunotherapy, Biomarker, Pan-cancer.
Introduction
Although immune checkpoint inhibitors (ICIs) targeting programmed cell death protein 1 (PD-1), its ligand (PD-L1), or cytotoxic T lymphocyte antigen-4 (CTLA-4) significantly extend overall survival (OS) in patients with diverse types of cancer, it could benefit only a limited subpopulation of patients1. Hence, there is an urgent need to investigate the rational and effective strategies to improve the efficacy of ICIs in patients with cancer. Understanding genomic correlates of response to ICIs could help the development of novel biomarkers and therapies to enhance the clinical response and expand the benefit population2, 3.
The AT-rich interaction domain 1A (ARID1A) is a subunit of the chromatin remodeling complex SWI/SNF, which facilitates access of proteins to DNA. ARID1A is one of the most commonly mutated genes in cancer4, 5. A recent study found that ARID1A loss and impaired ARID1A binding to the mismatch repair (MMR) protein MSH2 similarly reduced MMR and increased mutation frequency and the number of tumor-infiltrating lymphocytes and PD-L1 expression. In mice with ARID1A-deficient ovarian cancer, PD-L1 inhibitor resulted in reduced tumor volume and better survival compared with controls6. Then, several retrospective and exploratory studies reported the relationship between ARID1A alterations and clinical benefit to ICI in some cancers. These findings suggested that ARID1A alteration would not only cooperate with ICI treatment but also have the potential predictive value for ICI therapy. However, a comprehensive analysis of ARID1A alteration frequency and its predictive value for ICI treatment outcome in diverse cancers has not yet been investigated.
In this study, we performed this pan-cancer analysis by using online database to systematically characterize the prevalence and predictive value of ARID1A alterations across multiple cancer types. We found a relatively high frequency (6.2%) of ARID1A alterations and significant predictive value for ICIs treatment in >40,000 patients with cancers. We also investigated the association between ARID1A alterations and tumor mutation burden (TMB) level or immune cell infiltrations. The current evidence suggested that ARID1A alterations would yield promising predictive value for ICI treatment across diverse cancers.
Results and discussion
Genetic alterations of ARID1A and its association with TMB level
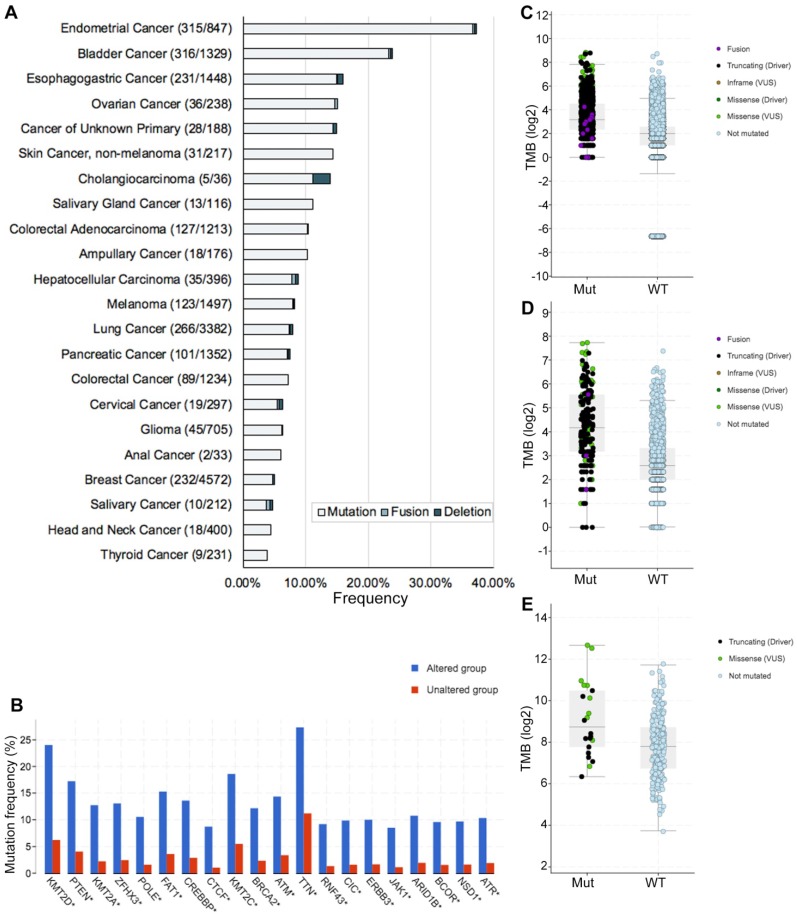
In this study, we defined ARID1A alterations as all kinds of nonsynonymous mutations including missense, frame-shift, splice site, nonstop, nonsense, fusions, deletions and translation start site changes. The frequency of ARID1A alterations in 40167 patients with various cancers was 6.2% (Figure 1A), with patients with endometrial cancer having the highest levels of ARID1A alterations (37.2%). Most of the alterations were missense mutations (39.6%, 979/2471; Supplemental Figure S1). The prevalence and spectrum of ARID1A alterations were slightly different in early-stage (TCGA cohort; Supplemental Figure S1A) versus advanced-stage cancers (MSK-IMPACT cohort; Supplemental Figure S1B). Co-occurring of genetic mutations in cancers with ARID1A alterations were not uncommon and some of them are popular driver genes in cancers (e.g. KMT2D/A/C, PTEN, POLE, etc.) (Figure 1B). Of note, co-occurring landscape of genetic mutations in cancers with ARID1A alterations was totally distinct in early-stage (TCGA cohort; Supplemental Figure S2A) versus advanced-stage cancers (MSK-IMPACT cohort; Supplemental Figure S2B). Considering ARID1A alterations promoting cancer mutability6, 7, we then investigate the difference of TMB level between ARID1A alteration and wild type groups. We selected a subset generated from MSK-IMPACT cohort that ensure the TMB could be comparable8. In the MSK-IMPACT cohort8, 10945 samples were identified and 912 (8.3%) of them had ARID1A alterations. TMB of patients with ARID1A alterations was significantly higher than it in those without (9 vs. 4 mutations/Mb, P < 0.0001; Figure 1C). This was validated in two ICI-treated cohorts (P < 0.0001, P = 0.0012, respectively; Figure 1C and D)3, 9. Notably, cancers with multiple ARID1A alterations had the highest TMB level (Supplemental Figure S2C-E). These results were consistent with a recent study, which also reported that the mutation load was significantly elevated in ARID1A-mutant tumors6. Together, these findings reveal a high prevalence of ARID1A alterations and its close relationship with TMB level across cancer types, suggesting that ARID1A alterations could be considered as biomarkers when conducting ICI treatment.
Figure 1.
Genetic alterations of ARID1A and its association with TMB level. A. Prevalence of ARID1A alterations in different cancer types; B. Co-occurring of genetic mutations in cancers with ARID1A alterations; C. The association between TMB and ARID1A mutations in MSK-IMPACT cohort; D. The association between TMB and ARID1A alterations in immune checkpoint inhibitors treatment cohort; E. The association between TMB and ARID1A alterations in patients with microsatellite-stable solid tumors received immune checkpoint inhibitors treatment. TMB, tumor mutation burden; Mut, mutation; WT, wild type.
Predictive and prognostic value of ARID1A alterations
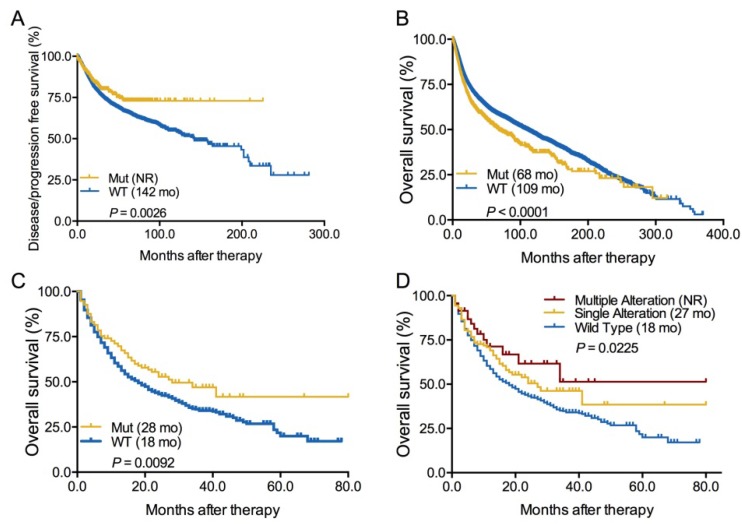
Next, we evaluated the association between ARID1A alterations and clinical outcome in both whole group and ICI-treated cohort. We firstly found that patients with ARID1A alterations showed a significantly longer disease-free survival or progression-free survival [DFS/PFS, not reached vs 142 months, hazard ratio (HR) = 0.74, 95% confidence interval (CI) 0.64-0.91, P = 0.0026; Figure 2A] but markedly shorter OS (68 vs 109 months, HR = 1.30, 95% CI 1.22-1.47, P < 0.0001; Figure 2B) than those without in whole group. In early-stage cancers, ARID1A alterations were also correlated with longer DFS/PFS (P = 0.0005; Supplemental Figure S3A). However, the prognostic value of ARID1A alterations was only found in advanced-stage cancers (P = 0.0094; Supplemental Figure S3C) instead of early-stage cancers (P = 0.2086; Supplemental Figure S3B). In the ICI treatment cohort9, we firstly identified 1661 patients with different cancers receiving ICI therapy and 193 of them with ARID1A alterations (Supplemental Table S1). Interestingly, patients with ARID1A alterations had a substantially prolonged OS of 28 months vs 18 months in the wild-type group (HR = 0.73, 95% CI 0.61-0.93, P = 0.0092; Figure 2C). Subgroup analysis revealed that patients with multiple ARID1A alterations had the longest OS than those with single ARID1A alterations or without (not reached vs. 27 vs. 18 months, P = 0.0225; Figure 2D). Of note, we did not observe the association between ARID1A alterations and OS in patients with microsatellite-stable (MSS) solid tumors (P = 0.4075; Supplemental Figure S4A). Even in patients with multiple ARID1A alterations and MSS solid tumors, we also did not found the close association (P = 0.4899; Supplemental Figure S4B). These findings were consistent with previous publications that ARID1A alterations contributes to impaired MMR and mutator phenotype in cancer, and may cooperate with ICI treatment6.
Figure 2.
Predictive and prognostic value of ARID1A alterations. A. Predictive value of ARID1A alterations in all cancers; B. Prognostic value of ARID1A alterations in all cancers; C. Predictive value of ARID1A alterations in patients received ICI therapy; D. Subgroup analysis the predictive value of ARID1A alterations subtypes in patients received ICI treatment. Mut, mutation; WT, wild type.
Immune landscape of cancer with ARID1A alterations
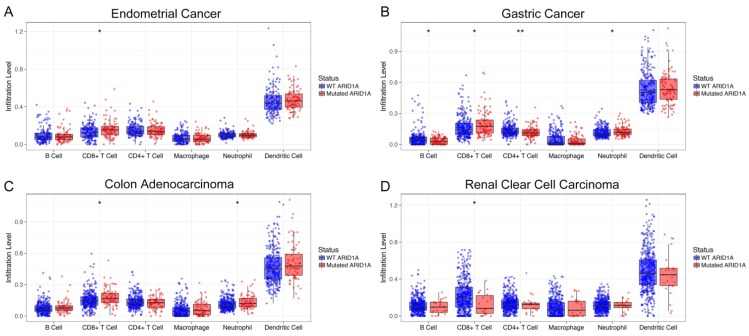
To unravel the potential mechanism of the predictive value of ARID1A alterations for ICI treatment, we then surveyed the relationship between ARID1A alterations and six common immune infiltrates (B cells, CD4+ T cells, CD8+ T cells, macrophages, neutrophils and dendritic cells) across different cancer types10. Interestingly, ARID1A deficiency was correlated with significantly higher six immune infiltrates in most of the cancer types, especially CD8+ T cells, including endometrial cancer (Figure 3A), stomach carcinoma (Figure 3B) and colon adenocarcinoma (Figure 3C). However, patients with ARID1A-mutant renal clear cell carcinoma had dramatically lower CD8+ T cell infiltrations than those without (Figure 3D). Of note, copy number variations (either deletion or amplification) of ARID1A were associated with substantially lower six immune infiltrates in most of the cancer types including endometrial cancer, gastric cancer, colon adenocarcinoma, head and neck cancer, lung squamous cell carcinoma and breast invasive carcinoma (Supplemental Figure S5).
Figure 3.
Immune landscape of cancer with ARID1A alterations. The association between ARID1A alterations and six immune infiltrates in A. Endometrial cancer; B. Stomach carcinoma; C. Colon adenocarcinoma; D. Renal clear cell carcinoma.
Conclusions
To our knowledge, this study firstly reported a high frequency of ARID1A alterations and the predictive significance of ARID1A alterations for ICI treatment in multiple cancer types. We also observed that ARID1A alterations were associated with significantly higher TMB level. Although ARID1A alterations were correlated with significantly inferior OS in total populations, they were associated with significantly prolonged OS in ICI treatment cohort, suggesting it might be used to predict a survival benefit from ICI therapy across multiple cancer types. Notably, patients with ARID1A alterations were correlated with markedly high immune infiltrates in endometrial, stomach and colon cancer but dramatically lower CD8+ T cell infiltrations in ARID1A-mutant renal clear cell carcinoma, indicating the association between ARID1A alterations and immune infiltrates was cancer-dependent. Collectively, our findings highlight the important value of ARID1A alterations as pan-cancer predictive biomarkers for ICI treatment.
Supplementary Material
Supplementary figures and tables.
Acknowledgments
The results here are in whole based upon data generated by the cBioPortal online database.
Funding
This study was supported in part by grants from the National Natural Science Foundation of China (No. 81772467, 81871865, 81874036 and 81972167) and the Backbone Program of Shanghai Pulmonary Hospital (NO. FKGG1802).
Ethics approval and consent to participate
Patient data we used were acquired by publicly available datasets that were collected with patients' informed consent.
Availability of data and materials
All included patients and sequencing data were identified and downloaded from the cBioPortal online database (https://www.cbioportal.org). The immune infiltrates abundances were estimated by using a web server for comprehensive analysis of tumor-infiltrating immune cells, named TIMER (Tumor Immune Estimation Resource, https://cistrome.shinyapps.io/timer/). Kaplan-Meier curves with log-rank tests were used to determine the survival difference.
Authors' contributions
Caicun Zhou had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis. Study concept and design: Tao Jiang, Caicun Zhou. Acquisition, analysis, or interpretation of data: All authors. Drafting of the manuscript: Tao Jiang, Caicun Zhou. Critical revision of the manuscript for important intellectual content: All authors. Statistical analysis: Tao Jiang, Xiaoxia Chen, Chunxia Su. Obtained funding: Chunxia Su, Shengxiang Ren, Caicun Zhou. Administrative, technical, or material support: All authors. Study supervision: Tao Jiang, Caicun Zhou.
Abbreviations
- ARID1A
AT-rich interaction domain 1A
- CI
confidence interval
- CTLA-4
cytotoxic T lymphocyte antigen-4
- HR
hazard ratio
- ICIs
immune checkpoint inhibitors
- MMR
mismatch repair
- MSS
microsatellite-stable
- OS
overall survival
- PD-1
programmed cell death protein 1
- PD-L1
PD-1 ligand
- TMB
tumor mutation burden
References
- 1.Sacher AG, Gandhi L. Biomarkers for the Clinical Use of PD-1/PD-L1 Inhibitors in Non-Small-Cell Lung Cancer: A Review. JAMA Oncol. 2016;2(9):1217–1222. doi: 10.1001/jamaoncol.2016.0639. [DOI] [PubMed] [Google Scholar]
- 2.Keenan TE, Burke KP, Van Allen EM. Genomic correlates of response to immune checkpoint blockade. Nat Med. 2019;25(3):389–402. doi: 10.1038/s41591-019-0382-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miao D, Margolis CA, Vokes NI, Liu D, Taylor-Weiner A, Wankowicz SM, Adeegbe D, Keliher D, Schilling B, Tracy A. et al. Genomic correlates of response to immune checkpoint blockade in microsatellite-stable solid tumors. Nat Genet. 2018;50(9):1271–1281. doi: 10.1038/s41588-018-0200-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu JN, Roberts CW. ARID1A mutations in cancer: another epigenetic tumor suppressor? Cancer Discov. 2013;3(1):35–43. doi: 10.1158/2159-8290.CD-12-0361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karachaliou N, Paulina Bracht JW, Rosell R. ARID1A Gene Driver Mutations in Lung Adenocarcinomas. J Thorac Oncol. 2018;13(12):e255–e257. doi: 10.1016/j.jtho.2018.07.099. [DOI] [PubMed] [Google Scholar]
- 6.Shen J, Ju Z, Zhao W, Wang L, Peng Y, Ge Z, Nagel ZD, Zou J, Wang C, Kapoor P. et al. ARID1A deficiency promotes mutability and potentiates therapeutic antitumor immunity unleashed by immune checkpoint blockade. Nat Med. 2018;24(5):556–562. doi: 10.1038/s41591-018-0012-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mathur R, Alver BH, San Roman AK, Wilson BG, Wang X, Agoston AT, Park PJ, Shivdasani RA, Roberts CW. ARID1A loss impairs enhancer-mediated gene regulation and drives colon cancer in mice. Nat Genet. 2017;49(2):296–302. doi: 10.1038/ng.3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zehir A, Benayed R, Shah RH, Syed A, Middha S, Kim HR, Srinivasan P, Gao J, Chakravarty D, Devlin SM. et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. 2017;23(6):703–713. doi: 10.1038/nm.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Samstein RM, Lee CH, Shoushtari AN, Hellmann MD, Shen R, Janjigian YY, Barron DA, Zehir A, Jordan EJ, Omuro A. et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat Genet. 2019;51(2):202–206. doi: 10.1038/s41588-018-0312-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li T, Fan J, Wang B, Traugh N, Chen Q, Liu JS, Li B, Liu XS. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res. 2017;77(21):e108–e110. doi: 10.1158/0008-5472.CAN-17-0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures and tables.
Data Availability Statement
All included patients and sequencing data were identified and downloaded from the cBioPortal online database (https://www.cbioportal.org). The immune infiltrates abundances were estimated by using a web server for comprehensive analysis of tumor-infiltrating immune cells, named TIMER (Tumor Immune Estimation Resource, https://cistrome.shinyapps.io/timer/). Kaplan-Meier curves with log-rank tests were used to determine the survival difference.