Abstract
Allergic rhinitis (AR) is an allergic disease characterized as (immunoglobulin E)-mediated type I hypersensitivity disorder. The interleukin-13 (IL-13) signaling pathway has been implicated in the pathogenesis of AR. In the present study, we investigated the regulatory role and mechanism of long noncoding RNA Linc00632 in IL-13-induced inflammatory cytokine and mucus production in nasal epithelial cells (NECs) from AR patients. We evaluated the expression of Linc00632 in nasal tissues from AR patients and in IL-13-treated NECs. We explored the role of Linc00632 in granulocyte-macrophage colony-stimulating factor (GM-CSF), eotaxin, and MUAC5AC production in IL-13-treated NECs. We searched for the potential target of Linc00632. Downregulation of Linc00632 was identified in nasal tissues of AR patients and in IL-13-treated NECs. Linc00632 inhibited IL-13-induced GM-CSF, eotaxin, and MUAC5AC production. Linc00632 targeted miR-498 and negatively regulated its expression. MiR-498 targeted IL1RN and inhibition of miR-498 suppressed IL-13-induced GM-CSF, eotaxin, and MUC5AC expression. The regulation of IL-13-induced dysfunction of NECs by Linc00632 depended on miR-498. Linc00632 inhibited IL-13-induced GM-CSF, eotaxin, and MUAC5AC production in IL-13-treated NECs by targeting miR-498.
Keywords: Allergic rhinitis, Interleukin-13, Nasal epithelial cells, Linc00632, Granulocyte-macrophage colony-stimulating factor
Introduction
Allergic rhinitis (AR) is a very common allergic disease that affects over 30% of populations over the world [1]. AR generally develops during childhood, and it is the most common chronic allergic disorder in children [2]. AR is characterized as symptoms of sneezing, nasal pruritus, and airflow obstruction caused by immunoglobulin E (IgE)-mediated type I hypersensitivity against inhaled allergens. The type 2 helper T (Th2) cells have been known to drive the mucosal inflammation [3].
The nasal epithelium is the first site of exposure to inhaled antigens. The nasal epithelial cells (NECs) have been shown to play an essential role in the innate immune response to AR. Epithelial cell-secreted cytokines, which included thymic stromal lymphopoietin, interleukin (IL)-25, and IL-33, have been implicated in regulating innate and adaptive immune responses associated with Th2 cytokine-mediated inflammation at nasal mucosal sites [4]. Therefore, understanding the biology and functions of NECs will provide us critical information for developing anti-allergic disease treatments.
IL-13 is a typical Th2 cytokine that has been shown to be the central mediator of physiologic changes induced by allergic inflammation [5]. IL-13 triggers macrophage and eosinophil activation, induces B cells to produce IgE, activates smooth muscle cells, promotes mucus and growth factor production by epithelial cells, promotes eotaxin production to stimulate eosinophil recruitment, and triggers airway fibroblasts activation, proliferation, and migration [6]. IL-13-treated NECs is a commonly used cell model for AR analysis [7]. Preventing and reducing allergic airway responses through targeting IL-13 and its signaling pathway have been attracted more and more attention. For instance, recent studies have indicated that IL-13 can lead to pathologic changes reminiscent of allergic airway diseases, such as eosinophil recruitment, mucus cell metaplasia, subepithelial fibrosis, and smooth muscle hypertrophy [8, 9, 10]. Furthermore, IL-13 has been implicated in mucus hypersecretion and inflammatory mediator release by airway epithelial cells [6]. Based on the fact that IL-13 was so much associated with pathogenesis of AR, the model of IL-13 induction was widely accepted in the research of AR [11], so in this study, we treated NECs with IL-13.
IL1RN gene encodes the protein IL-1 receptor antagonist (IL-1RA). IL-1RA is a member of the IL-1 cytokine family and is a natural inhibitor of the proinflammatory effect of IL-1 [12]. IL-1RA is implicated in several inflammatory diseases including asthma and AR [13, 14]. It has been shown that administration of IL-1RA alleviated nasal congestion, rhinorrhea, and sneezing in AR guinea pig model [15]. Therefore, targeting IL-1 signaling pathway should be a potential therapeutic treatment for AR.
The long noncoding RNAs (lncRNAs) are defined as transcripts longer than 200 nucleotides that are not translated into protein [16]. lncRNAs reside in the nucleus or cytoplasm and interact with nucleic acids or proteins, activate or inhibit signal molecules, decoy for microRNAs (miRNAs), and guide for transcriptional factors. lncRNAs have been shown to play essential biological activities including DNA damage, programmed cell death, development, inflammation, and tumor progression [17]. The lncRNAs have also been implicated in AR [18].
MiRNAs are a class of endogenous, noncoding small RNAs (22 nucleotides) that pair to the 3′ untranslated region (3′-UTR) and regulate gene expression [19]. MiRNAs have been shown to regulate multiple cellular activities including cell proliferation, apoptosis, differentiation, and development. The dysregulation of miRNAs has also been reported in AR, suggesting potential roles of miRNA in AR [20].
In the current study, we evaluated the roles of lncRNA Linc00632 in AR. We showed that lncRNA Linc00632 negatively regulated miR-498 expression and inhibited IL-13-induced inflammatory cytokine and mucus production in NECs from AR patients. Thus, our results suggested that lncRNA Linc00632 and miR-498 could be used as potential targets for the prevention and treatment of AR.
Materials and Methods
Clinical Nasal Mucosal Samples
All patients recruited in the current study were diagnosed based on their medical history, nasal endoscopy, an allergen skin-prick test, and a specific IgE assay. Nasal mucosal samples were obtained from inferior turbinate sections from 30 patients with perennial AR and 30 healthy controls. All patients had not received topical or systemic corticosteroid therapy for 4 weeks prior to study recruitment. Partial inferior turbinectomy was performed for nasal obstruction. This study was approved by the Ethic Committee of the Second Hospital of Hebei Medical University, and written consent for each participant was derived.
NECs Culture and Treatment
Primary NECs were isolated and cultured as described previously [21]. Inferior turbinate NECs were collected by nasal scraping under local anesthesia. Collected NECs were cultured in BEGM medium (Lonza, Walkersville, MD, USA) under submerged conditions. When 80–90% confluence was reached, the NECs were passaged. For certain experiments, the NECs were stimulated with 50 ng/mL IL-13 for 24 h or 10 ng/mL IL-13 for 14 days as described previously [11]. Cell supernatant and pellets were then harvested for analysis.
Plasmid, siRNA, and miRNA Transfection
Linc00632 was cloned into lentiviral vector pSin using following primers: Forward 5′-CTAGAATTCAGTGCGACAGACAGCC-3′, Reverse 5′-CTAGGATCCATTTTTTTTGAGGC AAAG-3′. Linc00632 siRNAs and control siRNA were purchased from Thermo Fisher. miR-498 mimic, inhibitor, and negative control were purchased from RiboBio Co., Ltd (Guangzhou, Guangdong, China). Plasmid, miRNA, and siRNA were transfected into cells using Lipofectamine 2000 following manufacturer's protocol. Then cells were harvested for further treatment or analysis.
Quantitative Real-Time PCR
Total RNA from NECs or nasal mucosal specimens was isolated using Trizol reagent (Thermo Fisher, Waltham, MA, USA) according to the manufacturer's protocol. cDNA was synthesized using 1 µg RNA and SuperScript® III First-Strand Synthesis System (Thermo Fisher, USA). Real-time PCR was performed using QuantiTect SYBR Green PCR Kit (Qiagen, Germantown, MD, USA) on a QuantStudio 3 Real-Time PCR System (Thermo Fisher, USA). miR-498 cDNA was synthesized using specific primer 5′-GTCGTATCCAGTGCA GGGTCCGAGGTATTCGCA CTG GATACGACGAAAAAC -3′. U6 (internal control) cDNA was synthesized using specific primer 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATT CGCACTGGATACGACAAAAAT ATGGAAC-3′. The sequences of primers used for qPCR in the current study were Linc00632 Forward: 5′-CACGCCTGTTATCCC-3′, Reverse: 5′-CAACCTCCGCCTCTT-3′; IL1RN Forward: 5′-AACAGAAAGCAGGACAAG CG-3′, Reverse: 5′-CCTTCGTCAGGCATATTG GT-3′; β-actin Forward: 5′-CGCTCTTCCAG CCTTCCTT-3′, Reverse: 5′-CGTTGTTGGCAT ACAGGTCCT-3′; granulocyte-macrophage colony-stimulating factor (GM-CSF) Forward 5′-ATGTGGCTGCAGAGCCTGCTGC-3′, Reverse: 5′-CTCCCAGCAGTCAAAGGG-3′; eotaxin Forward: 5′-CCCCTTCAGCGACTAGAGAG-3′, Reverse: 5′-TCTTGGGGTCGGCACAGAT-3′; MUC5AC Forward: 5′-TGATCATCCAGCAGGGCT-3′, Reverse: 5′-CCGAG CTCAGAGGACATATGGG-3′; miR-498 Forward: 5′-CTTTCAAGCCAGGGGGC-3′, Reverse: 5′-CAGTG CAGGGTCCGAGGT-3′; and U6 Forward: 5′-CGCAAGGATGACACGCAAATTCG-3′, Reverse 5′-CAGTGCAGGGTCCGAGGT-3′.
Western Blot
Total proteins from NECs were extracted using ReadyPrepTM Protein Extraction Kit (Bio-Rad, Hercules, CA, USA). Protein concentration was measured using Bio-Rad Protein Assay (Bio-Rad, USA). Total 20 µg proteins were loaded onto sodium dodecyl sulfate–polyacrylamide gel electrophoresis gel and transferred to polyvinylidene fluoride membrane. Five percent nonfat milk was used to block the membrane at room temperature for 1 h and then the membranes were incubated with primary antibodies overnight. Next day, after 3 times wash with tris-buffered saline buffer containing 0.1% Tween 20, membranes were incubated with corresponding horse radish peroxidase-conjugated secondary antibodies at room temperature for 1 h. Primary antibodies used in the current study were anti-IL1RN (Thermo Fisher) and anti-β-actin (Sigma, St. Louis, MO, USA). ClarityTM Western ECL Blotting Substrates (Bio-Rad) were used to detect the immunoreactive proteins. The density was quantitated using GS-900TM Calibrated Densitometer (Bio-Rad) and analyzed by using Image Lab (Bio-Rad).
ELISA
Cell supernatant levels GM-CSF and eotaxin were detected using commercial ELISA kits from R&D systems (Minneapolis, MN, USA), and MUC5AC was detected using Human Mucin-5 subtype AC ELISA kit from BluegGene Biotech (Shanghai, China) according to manufacturer's instructions.
3′ UTR Luciferase Reporter Assay
The Linc00632 was amplified by PCR using following primers: Forward: 5′-CTACTCGAG AGTGCGACAGACAGCC-3′, Reverse: 5′-CTAGCGGCCGC ATTTTTTTTGAGGCAAAG-3′ and cloned into psiCHECK2 reporter vector (Promega, Madison, WI, USA) downstream of the luciferase gene. The mutated Linc00632 was generated using QuikChange II Site-Directed Mutagenesis Kit (Aligent, Santa Clara, CA, USA) using following primers: Forward: 5′- TAGACTCATGAGACAGGTGGCCG-3′, Reverse: 5′-CGGCCACCTGTCTCATGAGTCTA-3′. The 3′-UTR of IL1RN containing the putative miR-498-binding site was amplified by PCR using following primers: Forward: 5′-CTACTCGAGTACTGCCCAGGCCTGCC-3′, Reverse: 5′-CTACTCGAGTACTGCCCAGGCCTGCC-3′ and cloned into psiCHECK2 reporter vector downstream of the luciferase gene. The mutated 3′-UTR of IL1RN was generated using QuikChange II Site-Directed Mutagenesis Kit (Aligent, Santa Clara, CA, USA) using following primers: Forward: 5′-GTTTTACAATAAAATCGTCACAA-3′, Reverse: 5′-TTGTGACGATTT TATTGTAAAAC-3′. For luciferase assays, NECs or HEK293 cells were seeded in 24-well plate and then co-transfected with 50 nM miR-498 or miR-NC and 0.2 µg luciferase reporter plasmid using Lipofectamine 2000 (Thermo Fisher). Dual-luciferase reporter system (Promega) was used to measure the luciferase activities according to manufacturer's instructions 48 h post transfection.
Biotin Pull-Down Assay
The procedure of Biotinylated Micro-RNA pull-down assay was described previously [22]. Briefly, NECs were harvested and lysed in lysis buffer (20 mM Tris-HCl, pH 7.5, 100 mM KCl, 5 mM MgCl2, 0.05% NP-40) containing RNase inhibitor and protease inhibitor cocktail (Sigma, St. Louis, MO, USA) for 30 min on ice. After centrifugation, the supernatant was bound to 3′ biotinylated miR-498 or control RNA (RiboBio Co., Ltd., China) at 4°C for 2 h. The streptavidin magnetic beads were pretreated with lysis buffer containing 0.5 µg/µL RNase-free BSA and 0.2 µg/µL yeast tRNA for 3 h and washed twice with lysis buffer. Then the mixture was bound to streptavidin beads (Thermo Fisher) for 1 h at 4°C. The biotin–miRNA/mRNA complex was eluted with elution buffer (20 mM Tris-HCl pH 7.4, 400 mM KCl, 0.5% NP-40, 5 mM biotin, and 80 U/mL RNase inhibitor) at 42°C for 5 min. Then the miRNA/mRNA complex was extracted with phenol-chloroform and treated with DNase I. After reverse transcription, the quantities of IL1RN mRNA in biotinylated miR-498 and control random RNA pull-down complex were compared by qRT-PCR. For antisense oligomer affinity pull-down assay, 1 µg sense or antisense biotin-labeled DNA oligomers corresponding to Linc00632 were incubated with NEC lysates. One hour after incubation, streptavidin-coupled agarose beads were added to isolate the RNA-complex. The Linc00632 and miR-498 levels were analyzed by qRT-PCR. DNA probes used in the current study were listed below: Linc00632-DNA-1-sense: (biotin-) 5′-CCTGGATCAAATGATACT AAGGGG GACTTG-3′, Linc00632-DNA-1-antisense: (biotin-) 5′-CAAGTCCCCCTTAG TATCATTTGATCCAGG-3′; Linc00632-DNA-2-sense: (biotin-) 5′-CCAATCCCCGTATCA TTAGACTCT CCCAGA-3′, Linc00632-DNA-2-antisense: (biotin-) 5′-TCTGGGAGAGTCT AATGATACGGGGATTGG-3′; Linc00632-DNA-3-sense: (biotin-) 5′-CACTCTGCAGAGC ACGTAGAGTCACCCCGC-3′; Linc00632-DNA-3-antisense: (biotin-) 5′-GCGGGGTGACT CTACGTGCTCTGCAGAGTG-3′.
Statistical Analysis
All data were presented as mean ± SD. Data were analyzed by 2-way analysis of variance or Student t test analysis. The statistical difference was considered as significant when p value is <0.05.
Results
Downregulated Linc00632 Level Was Associated in AR Patients and IL-33-Treated NECs
To investigate the potential role of Linc00632 in AR, first we evaluated the expression levels of Linc00632 in AR patients. As shown in Figure 1a, Linc00632 was significantly downregulated in AR patients when compared to healthy people. We also detected significantly decreased Linc00632 in IL-13-treated NECs when compared to untreated cells (Fig. 1b). These findings strongly indicated that Linc00632 was aberrantly downregulated in AR, suggesting potential roles of Linc00632 in AR.
Fig. 1.
Linc00632 was decreased in nasal mucosal tissues from AR patients and in IL-13-treated NECs. a The expression levels of Linc00632 in mucosal tissues from 30 AR patients and 30 healthy controls were measured by qRT-PCR. b The expression levels of Linc00632 in NECs after IL-13 stimulation (50 ng/mL) for 24 h were measured by qRT-PCR. ** p < 0.01. AR, allergic rhinitis; NECs, nasal epithelial cells; IL-13, interleukin-13.
Linc00632 Inhibited IL-13-Induced GM-CSF, Eotaxin, and MUC5AC Expression in Human NECs
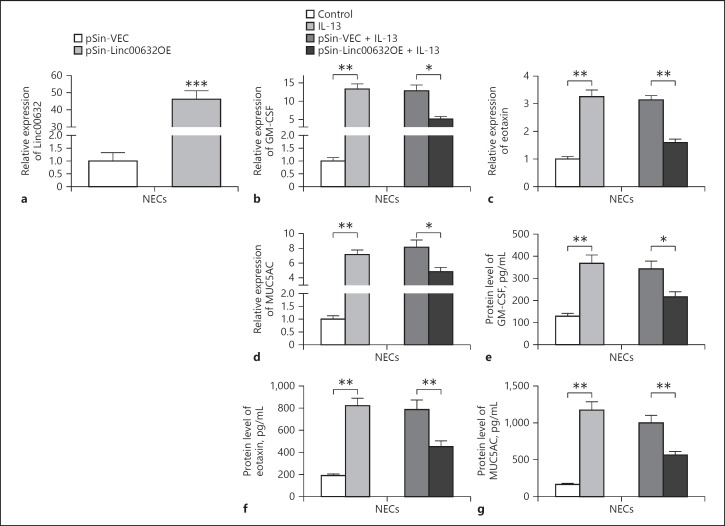
We next explored the potential effect of Linc00632 on AR using the IL-13-treated NECs model and monitored the expression of GM-CSF, eotaxin, and MUC5AC as described previously [11]. To overexpress Linc00632, we transfected NECs with lentiviral plasmid pSin-Linc00632OE, which encoded the linc00632. We detected significantly increased Linc00632 levels in NECs when compared to NECs transfected with control vector (Fig. 2a). The mRNA levels of GM-CSF (Fig. 2b) and eotaxin (Fig. 2c) were significantly increased in NECs treated with 50 ng/mL IL-13 for 24 h. In contrast, GM-CSF and eotaxin mRNA levels were significantly decreased in Linc00632 overexpressing NECs when compared to control transfected cells. Similarly, MUC5AC mRNA level was significantly increased in NECs treated with 10 ng/mL IL-13 for 14 days and overexpression of Linc00632 significantly decreased MUC5AC mRNA level (Fig. 2d). Consistent to the mRNA level, IL-13 treatment significantly increased the protein levels of GM-CSF (Fig. 2e), eotaxin (Fig. 2f), and MUC5A (Fig. 2g), while overexpression of Linc00632 significantly inhibited the IL-13-induced upregulation of these proteins. Collectively, our data indicated that Linc00632 inhibited IL-13-induced GM-CSF, eotaxin, and MUC5AC expression in NECs.
Fig. 2.
Linc00632 inhibited IL-13-induced GM-CSF, eotaxin, and MUC5AC expression in human NECs. a The expression level of Linc00632 in NECs transfected with Linc00632 overexpression plasmid (pSin-Linc00632OE) or empty vector (pSin-VEC) was measured by qRT-PCR. b–d Effects of Linc00632 overexpression on IL-13-induced inflammatory cytokine and mucin mRNA expression in cultured NECs. The mRNA expression levels of GM-CSF, eotaxin, and MUC5AC were measured by qRT-PCR. e–g Effects of Linc00632 overexpression on the synthesis of inflammatory cytokines and MUC5AC protein in IL-13-stimulated NECs. The protein levels of GM-CSF, eotaxin, and MUC5AC were determined by ELISA (50 ng/mL 24 h for GM-CSF and eotaxin detecting; 10 ng/mL 14 days for MUC5AC detecting). The data represent the mean ± SD from 3 independent experiments. * p < 0.05; ** p < 0.01; *** p < 0.001. AR, allergic rhinitis; NECs, nasal epithelial cells; IL-13, interleukin-13; GM-CSF, granulocyte-macrophage colony-stimulating factor.
Linc00632 Negatively Regulated miR-498
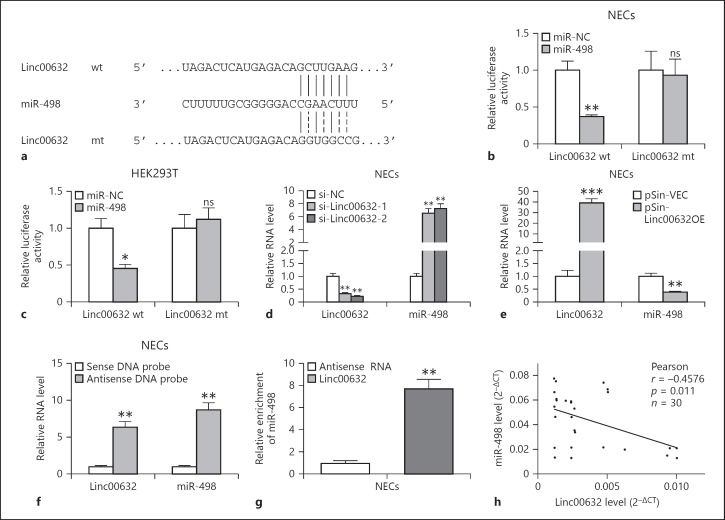
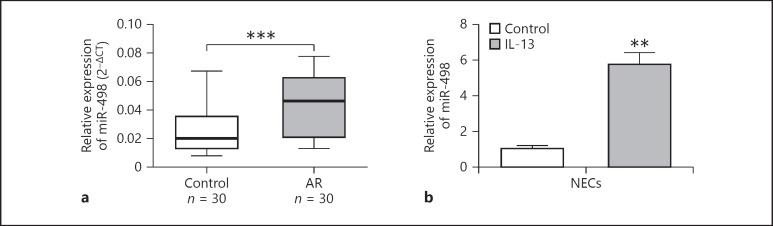
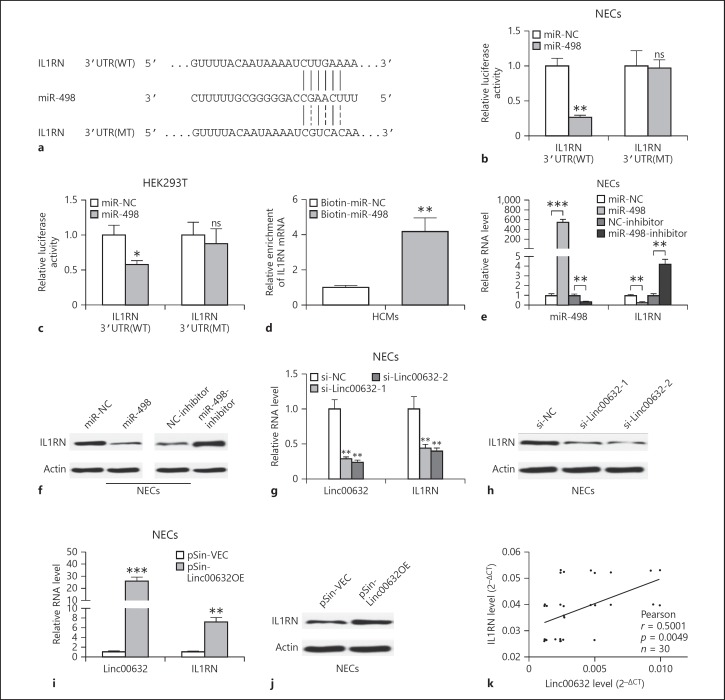
We searched for the potential targets of Linc00632 using online software targetscan (http://www.targetscan.org/vert_71/). We found that miR-498 was predicted to bind to Linc00632 (Fig. 3a). To validate whether miR-498 was a functional target of Linc00632, we utilized the dual-luciferase reporter system. As shown in Figure 3b, the wild-type Linc00632 or mutant was cloned into the luciferase reporter vector and co-transfected with miR-498 or control miRNA into NECs. Luciferase assay results showed that expression of miR-498 significantly decreased the luciferase activity of the reporter gene with wild type but not mutated Linc00632 construct. Similar results were obtained using HEK293 cells (Fig. 3c). To test whether Linc00632 regulated miR-498, we knocked down endogenous Linc00632 by transfecting its specific siRNA or overexpressed Linc00632 in NECs and then monitored the miR-498 level. As shown in Figure 3d, transfecting siRNA against Linc00632 significantly decreased the endogenous level of Linc00632. Knocking down Linc00632 resulted in significantly increased level of miR-498. In contrast, overexpression of Linc00632 by transfecting pSin-Linc00632OE resulted in significantly increased Linc00632 level while significantly decreased the miR-498 level (Fig. 3e). Therefore, our results demonstrated that Linc00632 negatively regulated miR-498 expression. In addition, we demonstrated that biotinylated antisense Linc00632 DNA probe-enriched endogenous Linc00632 was able to pull down miR-498 (Fig. 3f). Similarly, in vitro-synthesized biotinylated Linc00632 was also able to pull down miR-498 successfully (Fig. 3g). These results demonstrated that Linc00632 interacted with miR-498 and negatively regulated miR-498 expression. We performed Pearson correlation analysis to evaluate the expression relationship between Linc00632 and miR-498. We found that Linc00632 expression level was significantly negatively correlated with miR498 expression level (Fig. 3h). In addition, we detected upregulated miR-498 in AR patients (Fig. 4a) and IL-13-treated NECs (Fig. 4b), which was corresponding to the downregulation of Linc00632 in AR patients and IL-13-treated NECs.
Fig. 3.
Linc00632 negatively regulated miR-498 as an RNA sponge. a The prediction for miR-498-binding sites on Linc00632 transcript and schematic of luciferase reporter vector constructs Linc00632 wt and the miR-498-binding site Linc00632 mt one. b, c The luciferase activities in NECs and HEK293T cells co-transfected with miR-498 or miR-NC mimics and luciferase reporters containing Linc00632 wt or Linc00632 mt. Data are presented as the relative ratio of hRluc luciferase activity to hluc+ luciferase activity. d, e The expression levels of Linc00632 and miR-498 in NECs transfected with Linc00632 siRNAs (si-Linc00632–1, si-Linc00632–2) or negative control (si-NC) and Linc00632 overexpression plasmid (pSin-Linc00632OE) or empty vector (pSin-VEC) were measured by qRT-PCR. f Lysates from NECs were incubated with in vitro-synthesized biotin-labeled sense or antisense DNA probes against Linc00632 for biotin pull-down assay, followed by qRT-PCR analysis to examine miR-498 levels. g Lysates from NECs were incubated with in vitro-synthesized biotin-labeled Linc00632 and antisense RNA for biotin pull-down assay, followed by real-time-PCR analysis to examine miR-498 levels. h Correlation analysis between Linc00632 and miR-498 in mucosal tissues from 30 AR patients. Data are representative of 3 or more independent experiments. * p < 0.05; ** p < 0.01; *** p < 0.001. NECs, nasal epithelial cells; ns, not significant.
Fig. 4.
miR-498 was increased in nasal mucosal tissues from AR patients and in IL-13-treated NECs. a The expression levels of miR-498 in mucosal tissues from 30 AR patients and 30 healthy controls were measured by qRT-PCR. b The expression levels of miR-498 in NECs after IL-13 stimulation (50 ng/mL) for 24 h were measured by qRT-PCR. ** p < 0.01; *** p < 0.001. AR, allergic rhinitis; NECs, nasal epithelial cells; IL-13, interleukin-13.
Inhibition of miR-498 Prevented IL-13-Induced GM-CSF, Eotaxin, and MUC5AC Expression in Human NECs
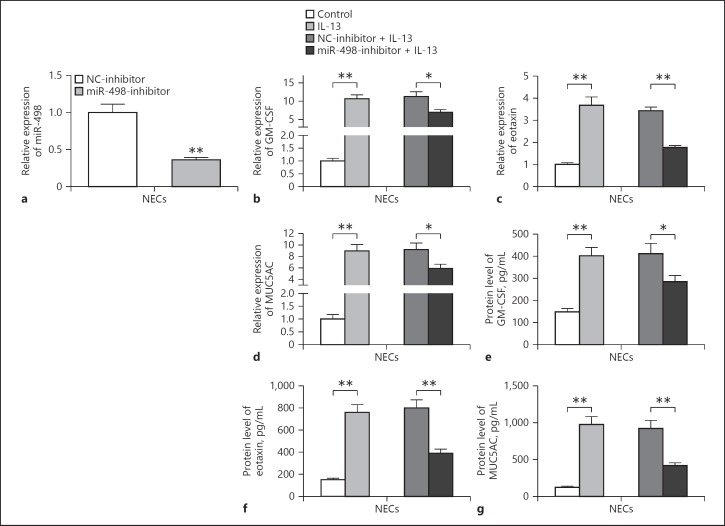
As Linc00632 regulated miR-498 expression, we next evaluated the potential roles of miR-498 in AR using the IL-13-treated NECs model. We transfected NECs with miR-498 inhibitor and found that the miR-498 expression was significantly decreased in NECs (Fig. 5a). The inhibition of miR-498 significantly prevented IL-13-induced mRNA and protein expression of GM-CSF (Fig. 5b, e), eotaxin (Fig. 5c, f), and MU5AC (Fig. 5d, g). In conclusion, our results indicated that miR-498 regulated the expressions of GM-CSF, eotaxin, and MU5AC suggesting the potential roles of miR-498 played in AR.
Fig. 5.
Inhibition of miR-498 suppressed IL-13-induced GM-CSF, eotaxin, and MUC5AC expression. a The knockdown efficiency of miR-498 in NECs transfected with miR-498 inhibitor (miR-498-inhibitor) or negative control inhibitor (NC-inhibitor) was measured by qRT-PCR. b–d Effects of miR-498 inhibition on IL-13-induced inflammatory cytokine and mucin mRNA expression in cultured NECs. The mRNA expression levels of GM-CSF, eotaxin, and MUC5AC were measured by qRT-PCR. e–g Effects of miR-498 inhibition on the synthesis of inflammatory cytokines and MUC5AC protein in IL-13-stimulated NECs. The protein levels of GM-CSF, eotaxin, and MUC5AC were determined by ELISA (50 ng/mL 24 h for GM-CSF and eotaxin detecting; 10 ng/mL 14 days for MUC5AC detecting). The data represent the mean ± SD from 3 independent experiments. * p < 0.05; ** p < 0.01. AR, allergic rhinitis; NECs, nasal epithelial cells; IL-13, interleukin-13; GM-CSF, granulocyte-macrophage colony-stimulating factor.
miR-498 Directly Targeted IL1RN
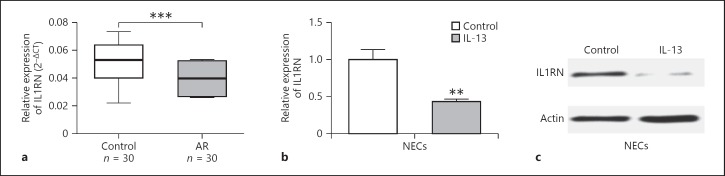
We continued to explore the underlying functional mechanism of miR-498 and used targetscan to predict the target of miR-498. As shown in Figure 6a, miR-498 was predicted to bind to 3′UTR of IL1RN. We also utilized the dual-luciferase reporter system to further confirm the functional interaction between miR-498 and IL1RN 3′UTR. Luciferase assay results showed that expression of miR-498 significantly decreased the luciferase activity of the reporter gene with wild type but not mutated IL1RN 3′UTR construct in both NECs (Fig. 6b) and HEK293T cells (Fig. 6c). In addition, in vitro-synthesized biotinylated miR-498 was also able to pull down IL1RN mRNA successfully (Fig. 6d). These results indicated that miR-498 targeted IL1RN. So we continued to test whether miR-498 regulates IL1RN expression. We overexpressed miR-498 or inhibited miR-498 by transfecting miR-498 mimics or inhibitors. Transfecting miR-498 significantly increased miR-498 level and decreased both IL1RN mRNA and protein levels (Fig. 6e, f). In contrast, transfecting miR-498 inhibitor significantly decreased miR-498 level while increasing IL1RN mRNA and protein levels (Fig. 6e, f). As Linc00632 regulated miR-498 expression, we supposed Linc00632 could regulate IL1RN expression through regulating miR-498. As predicted, knocking down Linc00632 significantly decreased both mRNA level (Fig. 6g) and protein level (Fig. 6h) of IL1RN. In contrast, overexpression of Linc00632 significantly increased IL1RN mRNA and protein levels (Fig. 6i, j). Furthermore, we performed Pearson correlation analysis to evaluate the expression relationship between Linc00632 and IL1RN. We found that Linc00632 expression level was significantly positively correlated with IL1RN expression level (Fig. 6k). Taken together, our data demonstrated that miR-498 targeted IL1RN and negatively regulated IL1RN expression, while Linc00632 positively regulated IL1RN expression. We also detected significantly downregulated IL1RN mRNA in AR patients (Fig. 7a) and significantly downregulated IL1RN mRNA and protein in IL-13-treated NECs (Fig. 7b, c), which were correlated to the upregulation of Linc00632 in AR patients and IL-13-treated NECs. Taken together, our data demonstrated that miR-498 targeted IL1RN and negatively regulated IL1RN expression.
Fig. 6.
miR-498 directly targeted IL1RN in human NECs. a Schematic of predicted wild-type (IL1RN 3′UTR[WT]) or mutated (IL1RN 3′UTR[MT]) miR-498-binding sites in the 3′UTR of IL1RN. b, c Relative luciferase activities of plasmids carrying wide-type (IL1RN 3′UTR[WT]) or mutant (IL1RN 3′UTR[MT]) IL1RN 3′UTR in NECs and HEK293T cells co-transfected with miR-498 mimics or miR-NC. d Detection of IL1RN mRNAs in biotinylated miRNA/target mRNA complex by RT-PCR. The relative level of IL1RN mRNA in the complex pulled down by using biotinylated miR-498 was compared to that of the complex pulled down by using the biotinylated control random RNA. e, f The relative expression levels of IL1RN in NECs transfected with indicated miRNA mimics and miRNA inhibitors or their respective negative controls were detected by qRT-PCR and Western blot. g–j The relative expression levels of IL1RN in NECs transfected with Linc00632 siRNAs (si-Linc00632-1, si-Linc00632-2) or negative control (si-NC) and Linc00632 overexpression plasmid (pSin-Linc00632OE) or empty vector (pSin-VEC) were detected by qRT-PCR and Western blot. k Correlation analysis between Linc00632 and IL1RN in mucosal tissues from 30 AR patients. Data are representative of 3 or more independent experiments. Data are the mean ± SD. * p < 0.05; ** p < 0.01; *** p < 0.001. ns, not significant; IL1RN, interleukin 1 receptor antagonist; NECs, nasal epithelial cells; 3′-UTR, 3′ untranslated region; WT, wild type; miRNA, microRNA.
Fig. 7.
IL1RN was decreased in nasal mucosal tissues from AR patients and in IL-13-treated NECs. a The expression levels of IL1RN in mucosal tissues from 30 AR patients and 30 healthy controls were measured by qRT-PCR. b, c The expression level of IL1RN in NECs after IL-13 stimulation (50 ng/mL) for 24 h was measured by qRT-PCR and Western blot. ** p < 0.01; *** p < 0.001. AR, allergic rhinitis; NECs, nasal epithelial cells; IL-13, interleukin-13; IL1RN, interleukin 1 receptor antagonist.
Linc00632 Mediated IL-13-Induced Dysfunction of NECs via Regulating miR-498/IL1RN Expression
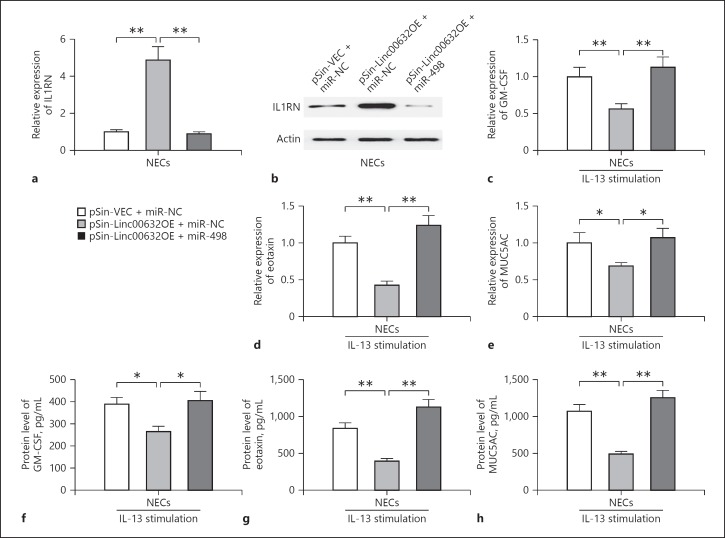
We have demonstrated that Linc00632 inhibited IL-13-induced dysfunction of NECs and positively regulated IL1RN expression, while miR-498 functioned the opposite way. As Linc00632 negatively regulated miR-498, we further explored whether the inhibitory effect of Linc00632 on IL-13-induced dysfunction and positive regulation on IL1RN depended on miR-498. We overexpressed both Linc00632 and miR-498 in NECs. We found that miR-498 significantly inhibited Linc000632-induced IL1RN expression at both mRNA and protein levels (Fig. 8a, b). In addition, miR-498 rescued both mRNA and protein expressions of GM-CSF (Fig. 8c, f), eotaxin (Fig. 8d, g), and MUC5AC (Fig. 8e, h), which were inhibited by Linc00632 in IL-13-treated NECs. Therefore, our results demonstrated that Linc00632 mediated IL-13-induced dysfunction of NECs via regulating miR-498/IL1RN expression.
Fig. 8.
Linc00632 mediated IL-13-induced dysfunction of NECs via regulating miR-498/IL1RN expression. a, b The relative mRNA and protein levels of IL1RN in NECs co-transfected with empty vector and negative control mimics (pSin-VEC + miR-NC), Linc00632 overexpression plasmid and negative control mimics (pSin-Linc00632OE + miR-NC) or Linc00632 overexpression plasmid and miR-498 mimics (pSin-Linc00632OE + miR-498) were detected by qRT-PCR and Western blot. c–e MiR-498 reverted the decreased IL-13-induced inflammatory cytokine and mucin mRNA expression in cultured NECs due to Linc00632 overexpression. The mRNA expression levels of GM-CSF, eotaxin, and MUC5AC were measured by qRT-PCR. f–h miR-498 reverted the decreased synthesis of inflammatory cytokines and MUC5AC protein in IL-13-stimulated NECs due to Linc00632 overexpression. The protein levels of GM-CSF, eotaxin, and MUC5AC were determined by ELISA (50 ng/mL 24 h for GM-CSF and eotaxin detecting; 10 ng/mL 14 days for MUC5AC detecting). The data represent the mean ± SD from 3 independent experiments. * p < 0.05; ** p < 0.01. AR, allergic rhinitis; NECs, nasal epithelial cells; IL-13, interleukin-13; GM-CSF, granulocyte-macrophage colony-stimulating factor.
Discussion
In the current study, we found that the lncRNA Linc00632 was strongly downregulated in nasal mucosal tissues from AR patients, while miR-498 was strongly upregulated. We demonstrated that Linc00632 targeted miR-498 and negatively regulated its expression. In addition, we demonstrated the opposite effects of Linc00632 and miR-498 on IL-13-induced inflammatory cytokines and mucus productions in primary NECs from AR patients. Linc00632 inhibited both mRNA and protein expressions of GM-CSF, eotaxin, and MUC5AC in IL-13-treated NECs. In contrast, miR-498 positively regulated IL-13-induced production of GM-CSF, eotaxin, and MUC5AC as inhibiting miR-498 abolished these effects. Moreover, miR-498 targeted IL1RN and inhibited IL1RN expression. Therefore, we demonstrated the regulatory role of Linc00632/miR-498/IL1RN axis in inflammatory response of IL-13-induced NECs in AR.
IL-13 is produced by Th2 cells and is an important cytokine that contributes to AR [23]. By using the IL-13-deficient mice, Miyahara and colleagues reported that IL-13-deficient mice demonstrated significantly reduced late nasal response to antigen challenge. Binding of IL-13 to its receptor resulted in the activation of transcription factor STAT6 and expression of downstream genes. IL-13 has been shown to induce production of multiple cytokines and factors including GM-CSF, eotaxin, and leukotriene. In addition, IL-13 promoted IgE synthesis, eosinophil recruitment, mucus hyperproduction, subepithelial fibrosis, and airway hyperresponsiveness. Therefore, inhibition of IL-13 signaling pathway could be a potential approach to treat allergic diseases.
GM-CSF and eotaxin are proinflammatory cytokines produced by airway epithelial cells during allergic airway inflammation. We also demonstrated that IL-13 treatment induced GM-CSF and eotaxin production in primary NECs. GM-CSF can stimulate the differentiation of hematopoietic stem cells into granulocytes and monocytes and support eosinophil survival [24]. Eotaxins are a family of CC chemokines that recruit eosinophils, basophils, and mast cells to inflammatory sites. Our results showed that IL-13 treatment also induced MUC5AC production in NECs. MUC5AC is a glycoprotein belonging to the superfamily of mucins. Mucus hypersecretion is a common feature of allergic airway disorder. Therefore, inhibiting proinflammatory cytokines and mucus production could be another potential approach to treat allergic diseases.
lncRNAs are transcribed RNA molecules longer than 200 nucleotides without protein-coding function. lncRNAs have been shown to regulate gene expression and participate in various physiological and pathological processes by basepairing with DNA or RNA in a sequence-specific manner [25]. Interestingly, our findings identified the association of downregulated lncRNA Linc00632 with AR and IL-13-treated NECs. These findings suggested the potential roles of Linc00632 in AR. We further demonstrated that Linc00632 inhibited IL-13-induced GM-CSF, eotaxin, and MUC5AC production, indicating a protective role of Linc00632 in AR.
The target-mimetic, sponge/decoy function of lncRNA on miRNAs has been reported [26]. Until now, there was no description about Linc00632 function as miRNA sponge and the precise role of Linc00632 is not well elucidated. Our current study showed that Linc00632 targeted miR-498 and negatively regulated its expression. Therefore, our study was the first to demonstrate that Linc00632 functions as miRNA sponge and targeted miR-498. miRNAs are the class of small noncoding RNA molecules functioning in RNA silencing and posttranscriptional regulation of gene expression by basepairing with complementary sequences with mRNA. Now the functions of miR-498 that have been reported are mainly in cancer field. For example, miR-498 has been shown to inhibit cell proliferation, migration, and invasion in non-small-cell lung cancer [27]. Liu et al. [28] reported that miR-498 targeted and negatively regulated FOXO3 expression, which resulted in the inhibition of human ovarian cancer cells proliferation. Zhang et al. [29] demonstrated that miR-498 inhibited growth and metastasis of liver cancer by targeting zinc finger E‑box binding homeobox 2. In the current study, we demonstrated that the regulation of IL-13-induced GM-CSF, eotaxin, and MUC5AC production by Linc00632 depended on miR498. This is the first description of miR-498 function in AR. We further identified that miR-498 targeted IL1RN and positively regulated IL-13-induced GM-CSF, eotaxin, and MUC5AC production. IL1RN encoded the protein IL-1RA that functions as IL-1 inhibitor to inhibit the activities of IL-1α and IL-1β and modulates IL-1-related immune and inflammatory responses. IL-1RA binds to IL-1 receptor but does not induce the downstream intracellular response. The binding of ligand to IL-1 receptor results in activation of nuclear factor kappa B signaling and the mitogen-activated protein kinase pathways and induces the expression of IL-1-targeted genes including IL-6, IL-8, MCP-1, and COX-2, which have been implicated in AR pathogenesis. Therefore, targeting IL-1 signaling pathway and its downstream factors could be a promising strategy to treat AR. Zhang et al. [15] demonstrated that direct administration of IL-1RA remarkably relieved AR symptoms in guinea pig. Inhibition of nuclear factor kappa B activation has been also shown to inhibit allergic response in a murine model of AR [30]. Therefore, our data strongly suggested that inhibiting IL-1 signaling pathway and its downstream factors could be used as therapeutic treatment to AR.
Conclusion
We demonstrated that lncRNA Linc00632 was upregulated in AR and IL-13-treated NECs. Linc00632 targeted miR-498 and Linc00632 regulated IL-13-induced inflammatory cytokine and mucus production in a miR-498-dependent manner.
Disclosure Statement
The authors declare that they have no conflicts of interest.
Funding Sources
None.
References
- 1.Scadding GK, Richards DH, Price MJ. Patient and physician perspectives on the impact and management of perennial and seasonal allergic rhinitis. Clin Otolaryngol Allied Sci. 2000 Dec;25((6)):551–7. doi: 10.1046/j.1365-2273.2000.00417.x. [DOI] [PubMed] [Google Scholar]
- 2.Garg N, Silverberg JI. Association between childhood allergic disease, psychological comorbidity, and injury requiring medical attention. Ann Allergy Asthma Immunol. 2014 Jun;112((6)):525–32. doi: 10.1016/j.anai.2014.03.006. [DOI] [PubMed] [Google Scholar]
- 3.Deo SS, Mistry KJ, Kakade AM, Niphadkar PV. Role played by Th2 type cytokines in IgE mediated allergy and asthma. Lung India. 2010 Apr;27((2)):66–71. doi: 10.4103/0970-2113.63609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saenz SA, Taylor BC, Artis D. Welcome to the neighborhood: epithelial cell-derived cytokines license innate and adaptive immune responses at mucosal sites. Immunol Rev. 2008 Dec;226((1)):172–90. doi: 10.1111/j.1600-065X.2008.00713.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Minty A, Chalon P, Derocq JM, Dumont X, Guillemot JC, Kaghad M, et al. Interleukin-13 is a new human lymphokine regulating inflammatory and immune responses. Nature. 1993 Mar;362((6417)):248–50. doi: 10.1038/362248a0. [DOI] [PubMed] [Google Scholar]
- 6.Ingram JL, Kraft M. IL-13 in asthma and allergic disease: asthma phenotypes and targeted therapies. J Allergy Clin Immunol. 2012 Oct;130((4)):829–42. doi: 10.1016/j.jaci.2012.06.034. [DOI] [PubMed] [Google Scholar]
- 7.Haenuki Y, Matsushita K, Futatsugi-Yumikura S, Ishii KJ, Kawagoe T, Imoto Y, Fujieda S, Yasuda M, Hisa Y, Akira S, Nakanishi K, Yoshimoto T. A critical role of il-33 in experimental allergic rhinitis. J Allergy Clin Immunol. 2012;130:184–194. doi: 10.1016/j.jaci.2012.02.013. e111. [DOI] [PubMed] [Google Scholar]
- 8.Matsukura S, Stellato C, Georas SN, Casolaro V, Plitt JR, Miura K, et al. Interleukin-13 upregulates eotaxin expression in airway epithelial cells by a STAT6-dependent mechanism. Am J Respir Cell Mol Biol. 2001 Jun;24((6)):755–61. doi: 10.1165/ajrcmb.24.6.4351. [DOI] [PubMed] [Google Scholar]
- 9.Wills-Karp M. Interleukin-13 in asthma pathogenesis. Immunol Rev. 2004 Dec;202((1)):175–90. doi: 10.1111/j.0105-2896.2004.00215.x. [DOI] [PubMed] [Google Scholar]
- 10.Corren J. Role of interleukin-13 in asthma. Curr Allergy Asthma Rep. 2013 Oct;13((5)):415–20. doi: 10.1007/s11882-013-0373-9. [DOI] [PubMed] [Google Scholar]
- 11.Teng Y, Zhang R, Liu C, Zhou L, Wang H, Zhuang W, et al. miR-143 inhibits interleukin-13-induced inflammatory cytokine and mucus production in nasal epithelial cells from allergic rhinitis patients by targeting IL13Rα1. Biochem Biophys Res Commun. 2015 Jan;457((1)):58–64. doi: 10.1016/j.bbrc.2014.12.058. [DOI] [PubMed] [Google Scholar]
- 12.Perrier S, Darakhshan F, Hajduch E. IL-1 receptor antagonist in metabolic diseases: Dr Jekyll or Mr Hyde? FEBS Lett. 2006 Nov;580((27)):6289–94. doi: 10.1016/j.febslet.2006.10.061. [DOI] [PubMed] [Google Scholar]
- 13.Joki-Erkkilä VP, Karjalainen J, Hulkkonen J, Pessi T, Nieminen MM, Aromaa A, et al. Allergic rhinitis and polymorphisms of the interleukin 1 gene complex. Ann Allergy Asthma Immunol. 2003 Sep;91((3)):275–9. doi: 10.1016/S1081-1206(10)63530-2. [DOI] [PubMed] [Google Scholar]
- 14.Gohlke H, Illig T, Bahnweg M, Klopp N, André E, Altmüller J, et al. Association of the interleukin-1 receptor antagonist gene with asthma. Am J Respir Crit Care Med. 2004 Jun;169((11)):1217–23. doi: 10.1164/rccm.200302-281OC. [DOI] [PubMed] [Google Scholar]
- 15.Zhang HQ, Sun Y, Xu F. Therapeutic effects of interleukin-1 receptor antagonist on allergic rhinitis of guinea pig. Acta Pharmacol Sin. 2003 Mar;24((3)):251–5. [PubMed] [Google Scholar]
- 16.Perkel JM. Visiting “noncodarnia”. Biotechniques. 2013 Jun;54((6)):301. doi: 10.2144/000114037. [DOI] [PubMed] [Google Scholar]
- 17.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011 Sep;43((6)):904–14. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ma Z, Teng Y, Liu X, Li J, Mo J, Sha M, et al. Identification and functional profiling of differentially expressed long non-coding rnas in nasal mucosa with allergic rhinitis. Tohoku J Exp Med. 2017 Jun;242((2)):143–50. doi: 10.1620/tjem.242.143. [DOI] [PubMed] [Google Scholar]
- 19.Shukla GC, Singh J, Barik S. Micrornas: Processing, maturation, target recognition and regulatory functions. Mol Cell Pharmacol. 2011;3((3)):83–92. [PMC free article] [PubMed] [Google Scholar]
- 20.Shaoqing Y, Ruxin Z, Guojun L, Zhiqiang Y, Hua H, Shudong Y, et al. Microarray analysis of differentially expressed microRNAs in allergic rhinitis. Am J Rhinol Allergy. 2011 Nov-Dec;25((6)):e242–6. doi: 10.2500/ajra.2011.25.3682. [DOI] [PubMed] [Google Scholar]
- 21.Shi J, Luo Q, Chen F, Chen D, Xu G, Li H. Induction of IL-6 and IL-8 by house dust mite allergen Der p1 in cultured human nasal epithelial cells is associated with PAR/PI3K/NFkappaB signaling. ORL J Otorhinolaryngol Relat Spec. 2010;72((5)):256–65. doi: 10.1159/000312687. [DOI] [PubMed] [Google Scholar]
- 22.Yamamoto K, Ito S, Hanafusa H, Shimizu K, Ouchida M. Uncovering direct targets of mir-19a involved in lung cancer progression. PLoS One. 2015 Sep;10((9)):e0137887. doi: 10.1371/journal.pone.0137887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miyahara S, Miyahara N, Matsubara S, Takeda K, Koya T, Gelfand EW. IL-13 is essential to the late-phase response in allergic rhinitis. J Allergy Clin Immunol. 2006 Nov;118((5)):1110–6. doi: 10.1016/j.jaci.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 24.Daffern PJ, Jagels MA, Saad JJ, Fischer W, Hugli TE. Upper airway epithelial cells support eosinophil survival in vitro through production of GM-CSF and prostaglandin E2: regulation by glucocorticoids and TNF-alpha. Allergy Asthma Proc. 1999 Jul-Aug;20((4)):243–53. doi: 10.2500/108854199778339008. [DOI] [PubMed] [Google Scholar]
- 25.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009 Mar;10((3)):155–9. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 26.Paraskevopoulou MD, Hatzigeorgiou AG. Analyzing mirna-lncrna interactions. Methods Mol Biol. 2016;1402:271–86. doi: 10.1007/978-1-4939-3378-5_21. [DOI] [PubMed] [Google Scholar]
- 27.Gao N, Wang FX, Wang G, Zhao QS. Targeting the HMGA2 oncogene by miR-498 inhibits non-small cell lung cancer biological behaviors. Eur Rev Med Pharmacol Sci. 2018 Mar;22((6)):1693–9. doi: 10.26355/eurrev_201803_14582. [DOI] [PubMed] [Google Scholar]
- 28.Liu R, Liu F, Li L, Sun M, Chen K. MiR-498 regulated FOXO3 expression and inhibited the proliferation of human ovarian cancer cells. Biomed Pharmacother. 2015 May;72:52–7. doi: 10.1016/j.biopha.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 29.Zhang X, Xu X, Ge G, Zang X, Shao M, Zou S, et al. miR‑498 inhibits the growth and metastasis of liver cancer by targeting ZEB2. Oncol Rep. 2019 Mar;41((3)):1638–48. doi: 10.3892/or.2018.6948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wee JH, Zhang YL, Rhee CS, Kim DY. Inhibition of allergic response by intranasal selective nf-kappab decoy oligodeoxynucleotides in a murine model of allergic rhinitis. Allergy Asthma Immunol Res. 2017 Jan;9((1)):61–9. doi: 10.4168/aair.2017.9.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]