Abstract
Autophagy is a major intracellular digestion system that delivers cytoplasmic components for degradation and recycling. In this capacity, autophagy plays an important role in maintaining cellular homeostasis by mediating the degradation of cellular macromolecules and dysfunctional organelles and regeneration of nutrients for cell growth. Autophagy is important in innate immunity, as it is responsible for the clearance of various pathogens. Deficiency of intracellular autophagy can result in exaggerated activation of the inflammasome. The latter is an innate immune complex that senses diverse pathogen-associated or danger-associated molecular patterns and activates the expression of inflammatory cytokines. In autophagy-deficient cells, accumulation of damaged organelles, misfolded proteins, and reactive oxygen species contribute to inflammasome activation. The lung is continuously exposed to pathogens from the environment, rendering it vulnerable to infection. The lung innate immune cells act as a crucial initial barrier against the continuous threat from pathogens. In this review, we will summarize recent findings on the regulation of autophagy and its interaction with innate immunity, focusing on the lung.
Keywords: Autophagy, Inflammasome, Pathogen, Innate immunity and pulmonary inflammation
Autophagy in Regulatory and Signaling Pathways
Autophagy is a conserved intracellular digestion and recycling pathway that is present in all eukaryotic cells [1]. It is a process that involves the engulfment of cytoplasmic components by double membrane-bound compartments, called autophagosomes, and fusion of these autophagosomes with lysosomes. It functions as a waste disposal system removing potential toxic cellular products by targeting aggregate proteins and malfunctioning organelles for degradation and the subsequent release of degraded products for cellular recycling purposes [2]. Autophagy provides substrates for biosynthesis and energy generation which promote cell survival during times of energy and nutrient deprivation [2, 3]. Thus, autophagy is a survival mechanism designed to maintain cellular homeostasis in response to metabolic stress [3]. In addition, autophagy is involved in various aspects of immunity, including the clearance of pathogens, cytokines, and immune signals, suggesting that autophagy also plays important roles in innate [3, 4] and adaptive immunity [5].
Cellular Regulation of Autophagy
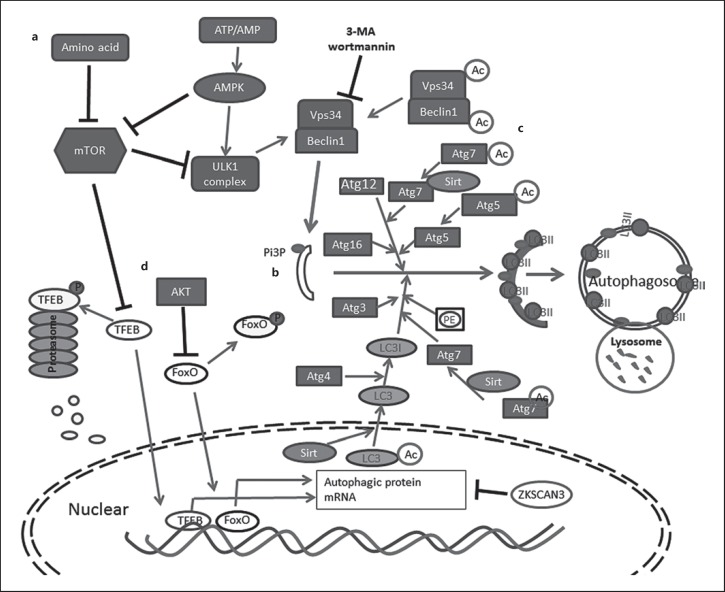
The canonical autophagy pathway can be separated into three major steps: initiation, elongation, and maturation [6]. Each step is regulated by specific autophagy-related (Atg) proteins [6, 7, 8, 9]. In the initiation step, with the activation of the ULK1 kinase complex, a small membranous sac called a phagophore is formed. A second kinase complex called the Vps34 complex is then recruited to the phagophore membrane [6]. Vps34 catalyzes the phosphorylation of cellular phosphatidylinositol to produce phosphatidylinositol 3-phosphate (PI3P), which serves as a platform to recruit more Atg proteins to promote the elongation of autophagic membranes (Fig. 1a) [8]. In the elongation step, two ubiquitin-like conjugation systems, Atg12–Atg5-Atg16L and LC3–PE, are involved. In the first ubiquitin-like system, Atg12 acts as a ubiquitin-like protein and, with the help of Atg7 and Atg10, becomes conjugated to Atg5. The resulting complex subsequently interacts with the coiled-coil protein Atg16L1 to form the Atg12–Atg5-Atg16L complex [10]. In the second ubiquitin-like system, pro-LC3 is cleaved by the cysteine protease Atg4, exposing a C-terminal glycine residue [11]. LC3 is then transiently linked to Atg7 and Atg3, and is eventually conjugated to phosphatidylethanolamine to form the mature form of lipid protein LC3II. The two systems work together to expand phagophore membrane formation that leads to mature autophagosomes (Fig. 1b) [6]. The mature autophagosomes then fuse with lysosomes to create autolysosomes and the acidic proteolytic environment of the autolysosome contributes to the degradation of the inner membrane as well as the luminal contents inside the autophagic vacuoles [6, 9].
Fig. 1.
Simplified autophagy signaling pathway and regulation. a Nutrition deprivation inhibits mTOR, which in turn activates ULK1 complex. Energy deprivation is sensed by AMPK to suppress mTOR and activates ULK1 complex. Activated ULK1 complex recruits Vps34-Beclin-1 complex to produce PI3P and initiates phagosome formation. b Two conjugation systems, Atg12–Atg5-Atg16L and LC3–PE, facilitate phagophore membrane elongation and autophagosome maturation. c NAD-dependent deacetylase Sirt1 catalyzes deacetylation of Vps34, Beclin1, Atg5, Atg7, and LC3 to promote autophagy. d Transcription factors TFEB and FoxO translocate to the nuclear to induce autophagy-related gene expression. Their transcription activity can be inhibited by phosphorylation caused by mTOR or AKT, respectively. Phosphorylated TFEB is digested by proteasome; ZKSCAN3 repress autophagy-related gene transcription.
Autophagosomes can be processed in the absence of some key autophagy components, in a process described as noncanonical autophagy [12, 13]. Noncanonical autophagy can also be processed without the ubiquitin conjugation proteins Atg5 and Atg7 [14, 15], and this type of autophagy has been found to play an important role in mitochondrial digestion during erythroid maturation in vivo [14, 15, 16]. LC3-associated phagocytosis (LAP) is a distinct form of noncanonical autophagy. It engages most of the canonical autophagy components, such as the Class III PI3K complex and ubiquitinylation-like protein-conjugation systems [17]. However, LAP proceeds without the ULK1 complex to form LC3-conjugated single-membraned phagosomes [17].
Autophagy Regulation
While the autophagy cascade could be affected in all three steps of the pathway, most regulation of autophagy occurs at the initiation step [9, 18]. The initiation complex ULK1 kinase is mainly regulated by mammalian target of rapamycin serine/threonine kinase complex 1 (mTOR) and adenosine monophosphate-activated protein kinase (AMPK). mTOR acts as a cellular nutritional sensor, phosphorylating ULK1, thus inhibiting autophagy initiation in nutrient-sufficient conditions [9, 18]. Under starvation or treatment with mTOR inhibitors such as rapamycin, mTOR activity is inhibited and ULK1 is rapidly dephosphorylated, resulting in activation of the ULK1 kinase (Fig. 1a). AMPK acts as an energy sensor that is activated by a decrease in the ATP/AMP ratio. AMPK activates autophagy initiation through inhibiting mTORC1 and activating the ULK1 complex (Fig. 1a) [18]. Vps34 kinase activity depends on Vps34-Beclin1 interaction as the direct downstream target of ULK, and it is inhibited by kinase inhibitor 3-methyladenine (3-MA) and wortmannin [18]. A recent study showed that the activity of Vps34 is also inhibited by acetyltransferase p300 [19]. Deacetylation of LC3 by Sirt1 affects LC3's distribution between the nuclear and cytoplasmic compartments and modulates the LC3-Atg7 conjugation process [20]. NAD+-dependent deacetylase Sirt1 directly deacetylates other Atg proteins, including Atg5, Atg7 [21], and Beclin1 [22], thus affecting autophagy processing (Fig. 1c).
Autophagy can be regulated at the transcriptional level. Forkhead transcription factors (FoxOs) upregulate autophagy by promoting the transcription of Atg genes, including Atg4, LC3B, and ULK1 [23, 24]. The activity of FoxOs depends on its nuclear localization. It has been shown that AKT/protein kinase B mediates the phosphorylation of FoxOs, leading to nuclear exclusion and inactivation [23, 24] (Fig. 1d). Transcription factor EB (TFEB) is another newly identified master transcription regulator of autophagy [25, 26]. Like FoxOs, the activation of TFEB is also dependent on nuclear translocation of the non-phosphorylated form of TFEB. mTOR and MAPK are extracellular signaling kinases that catalyze TFEB phosphorylation and restrain its nuclear translocation, inhibiting its activity (Fig. 1d) [25, 27, 28]. Our studies show that TFEB activity could also be regulated by proteasome-mediated degradation of phosphorylated TFEB [29, 30]. In contrast to FoxOs and TFEB, the transcriptional factor ZKSCAN3, which controls cell proliferation [31], has recently been identified to act as a repressor to a wide range of Atg genes. Knockdown of ZKSCAN3 has been shown to induce autophagy and lysosome biogenesis (Fig. 1d) [32].
Autophagy and the Elimination of Pulmonary Pathogens
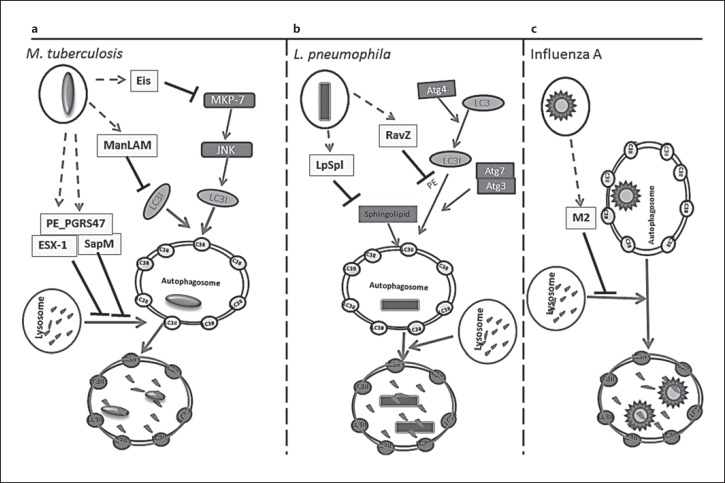
In addition to its role in maintaining cellular homeostasis, autophagy is actively engaged in cellular host defense by reducing the pathogen burden [33]. Mycobacterium tuberculosis bacteriumis a notorious pulmonary pathogen. M. tuberculosis primarily infects alveolar phagocytic cells, in which it resides and multiplies within the host-derived phagosomes [34]. M. tuberculosis developed strategies to escape phagocytotic clearance by preventing phagosome-lysosome fusion [35], disrupting vacuolar H-ATPase recruitment and phagosome acidification [36, 37], and inhibiting PI3P-dependent membrane trafficking [38, 39]. M. tuberculosissuppress the apoptosis of infected macrophages and trigger necrosis that results in the spreading of more bacteria to infect adjacent cells [40, 41]. Multiple studies have shown that autophagy plays an important role in innate defense against M. tuberculosis infection [42, 43, 44, 45]. Stimulation of autophagy by starvation, rapamycin, IFN-γ [4], ATP [46], or lipopolysaccharides (LPS) [44, 47] promoted the transfer of intracellular mycobacteria to lysosomes to be killed. Furthermore, vitamin D, which was used to treat tuberculosis in the preantibiotic era, has been shown to exert anti-M. tuberculosis effects by stimulating autophagy through induction of the antibacterial peptide, cathelicidin [48, 49, 50]. Antituberculosis drugs such as isoniazid and pyrazinamide have been shown to be partly dependent on autophagy activation, because in autophagy-defective cells antibiotic treatment was less effective against mycobacteria [51]. Additionally, autophagy deficiency also indirectly affects M. tuberculosis infection by enhancing macrophage uptake of mycobacteria through upregulation of scavenger receptor expression [52] and inhibiting antigen presentation [5, 47]. Some M. tuberculosis virulence factors facilitate intracellular bacterial survival by autophagy inhibition. M. tuberculosis secrete phosphatase SapM [39] and mycobacterial cell wall glycolipid mannose-capped lipoarabinomannan ManLAM [38], which are major virulence factors that can cause mycobacterial phagosome maturation arrest by interfering with the PI3K/PI3P pathway and suppressing PI3P production. Recent evidence indicates that SapM blocks autophagosome-lysosome fusing by binding with GAPase RAB7 [53]. ManLAM was also found to suppress autophagosome formation [54]. Another mycobacterial secreted protein is Eis, which is an N-acetyltransferase that enhances the survival of mycobacteria in human monocytic cells. It suppresses autophagy by acetylating JNK-specific phosphatase MKP-7 to inhibit JNK-dependent autophagy efflux initiation [55]. The M. tuberculosis ESX-1 secretion system [56] was found to inhibit autophagic flux by blocking autophagosome-lysosome fusion in human dendritic cells [57]. A recent study of M. tuberculosis virulence-related PE_PGR genes revealed that the PE_PGRS47 protein actively suppresses autophagy by blocking mycobacterial phagosome acidification and phagolysosomal fusion. By such inhibition, PE_PGRS47 restricts MHC class II antigen presentation in dendritic cells (Fig. 2a) [58].
Fig. 2.
Strategies used by pulmonary pathogens to avoid host autophagy. a M. tuberculosis has five identified anti-autophagy factors. M. tuberculosis secretion system Esx-1, the secreted phosphatase SapM, and the virulence protein PE_PGRS47 inhibit autophagy by blocking autophagosome lysosome fusing. Eis is an N-acetyltransferase. It acetylates JNK-specific phosphatase MKP-7 to initiate the inhibition of JNK-dependent autophagy. Mannose-capped lipoarabinomannan ManLAM interferes with trafficking proteins in autophagy and also affects LC3 protein expression levels and inhibits accumulation of autophagic vacuoles. b RavZ, the bacterial effector protein of L. pneumophila, inhibits autophagy by hydrolyzing the release of LC3II phosphatidylethanolamine at the carboxyl-terminal glycine residue. The hydrolyzed LC3 cannot be reconjugated by Atg7 and Atg3 to form mature LC3II for autophagosome localization. LpSpl with sphingosine-1 phosphate lyase activity reduces sphingolipid levels, reducing autophagosome formation. c Influenza A's transmembrane protein M2, which arrests autophagosome degradation by blocking autophagosome and lysosome fusion.
Legionella pneumophila is a common pulmonary pathogen infecting human lung alveolar macrophages and causing pneumonia [59]. It evades the immune response by residing in a special vacuole formed from the endoplasmic reticulum membrane and by inhibiting lysosome fusion [60, 61]. Autophagy was shown to be critical for L. pneumophila elimination in an in vitro study showing that knockdown of Atg5 in mouse macrophages enhanced bacterial replication [62]. Furthermore, in vivo studies using the Atg9 mutant Dictyostelium discoideum showed a critical defect in the clearance of L. pneumophila [63]. Legionella developed strategies to counter cellular autophagy elimination. The Legionella I Dot/Icm type IV secretion system secretes RavZ, a cystine protease, and delipidates the LC3, blocking its membrane conjugation [64]. Another effector protein, LpSpl, acts as sphingosine-1 phosphate lyase, decreasing host cell sphingolipid levels to inhibit autophagosome formation (Fig. 2b) [65]. A common pulmonary virus pathogen, Influenza virus A, induces autophagy but blocks the autophagosome-lysosome fusion by the viral Matrix 2 (M2) ion-channel protein [66]; thus, the virus adapts the multifunctional autophagosomes to reproduce the virus components and replicate (Fig. 2c) [66, 67]. Consistent with the role of autophagy in host defense, recent studies have addressed the augmentation of autophagy as a method to enhance the clearance of pathogens including Pseudomonas aeruginosa[68] and Burkholderia cenocepacia [69].
Autophagy and Lung Inflammation
Recent studies have found that autophagy is a negative regulator of inflammation in general, and of NLRP3 inflammasome in particular. The inflammasome is a multiprotein complex responsible for caspase-1 activation. Activation of caspase-1 leads to the release of the active form of potent inflammatory cytokines, including IL-1β and IL-18, by proteolytic cleavage. Macrophages from Atg16L1-deficient mice produced exaggerated quantities of IL-1β and IL-18 in response to LPS [70]. Depletion of other autophagic proteins such as Atg7, LC3B, or Beclin 1, or treatment with autophagy inhibitors wortmannin or 3-methyladenine, enhanced the production of IL-1β and IL-18 by macrophages [70, 71]. These studies indicated that autophagy deficiency is associated with increased inflammasome activity. Furthermore, autophagy deficiency in myeloid-derived cells was shown to cause spontaneous pulmonary inflammation in two independent studies [72, 73]. In both of these studies, mice lacking either Atg5 or Atg7 in myeloid cells spontaneously developed lung inflammation characterized by enhanced recruitment of inflammatory cells into the lung, increased levels of pro-inflammatory cytokines, submucosal thickening, goblet cell metaplasia, and increased collagen content [72, 73]. Following LPS challenge, these autophagy-deficient mice had higher levels of pro-inflammatory cytokines in serum and in bronchoalveolar lavage, severe pulmonary inflammation, as well as increased mortality compared to wild-type mice [72, 73]. In addition, mice lacking Atg5 or Atg7 in myeloid cells were more susceptible to bleomycin and silica challenge [74]. Spontaneous lung inflammation was also found in mice with Atg5 deletion in dendritic cells [75]. Knockout of other autophagy-related genes such as Atg14, Fip200, or Epg5 in myeloid cells led to sterile lung inflammation, thus confirming the essential role of autophagy in lung homeostasis, which is not specific to a particular autophagy-related gene [76].
During active infection, autophagy also functions to prevent extensive inflammation [56, 76, 77]. In vivo studies of M. tuberculosis infection showed that, compared to wild-type, mice with myeloid cell-specific Atg5 knockout had a higher bacterial burden, severe necrotic lung lesions, elevated levels of IL-17 and IL-1α, and higher mortality. These studies suggest that in addition to suppressing M. tuberculosis growth, autophagy in myeloid-derived cells is responsible for controlling damaging inflammation [56, 77]. A recent study showed that the loss of Atg5 in polymorphonuclear cells causes excessive inflammation and predisposes to M. tuberculosis infection. This study suggested that the role of Atg5 in M. tuberculosis inhibition could be at least partially independent of autophagy [78].
Autophagy Regulates Inflammasome Activity
The innate immune system is an important component that acts as an initial barrier to protect against microbial pathogens or damaging agents. Cross-talk between autophagy and the innate immune system balances protection of the host against an exaggerated immune response, while enabling the neutralization of infectious and damaging threats. This is crucial at sites such as the lung, skin, and colon, where the host is continuously exposed to potential hazardous elements, such as inhaled toxins, toxic food products, as well as chemicals and commensal and pathogenic bacteria. The innate immune system is able to recognize and orchestrate a protective inflammatory response against harmful insults. Such responses should be tightly controlled to prevent exaggerated damage to the host.
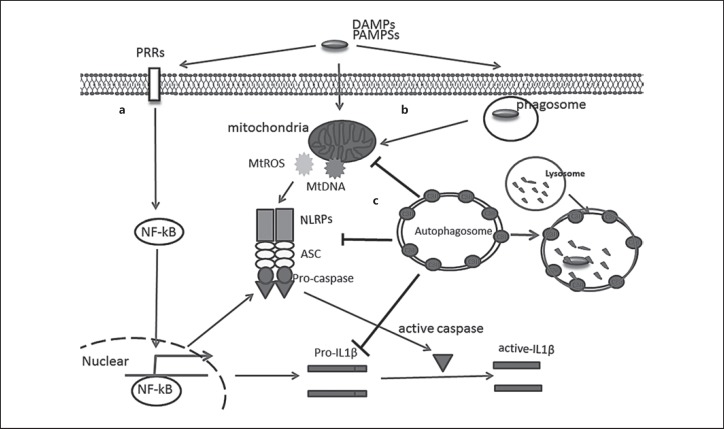
The innate immune system relies on a group of pattern-recognition receptors that include toll-like receptors (TLRs), nucleotide-binding oligomerization domain-like receptors (NLRs), and absent in melanoma (AIM2)-like receptors (ALRs). Both NLRs and ALRs can form a cytoplasmic multiprotein complex called the inflammasome upon sensing a wide variety of ligands. Inflammasome assembly involves the adapter protein apoptosis-associated speck-like protein containing a caspase-recruitment domain (ASC) which recruits caspase-1. Activation of caspase-1 leads to the release of the active form of IL-1β and IL-18 by proteolytic cleavage and can also lead to a form of cell death called pyroptosis [79]. The most widely studied inflammasome is NLRP3. Its activation depends on two steps. In the first step, pathogen-associated molecular patterns (PAMPs) and damage-associated molecular patterns (DAMPs) are recognized by TLRs to activate NF-κB signaling-dependent expression of the inflammasome components and pro-cytokines (Fig. 3a) [47, 80]. In the second step, specific stimuli trigger inflammasome complex assembly and the inflammasome processes the pro-cytokine to generate mature cytokines by active caspases (Fig. 3b) [81, 82]. Many forms of such stimuli have been discovered, such as DAMP/PAMP-induced mitochondria damage [81, 82]. A recent study revealed that newly synthesized mitochondrial DNA may act as an NLRP3 ligand and directly associate with the NLRP3 inflammasome complex, thereby promoting its activity [80]. There have been several mechanisms proposed for how autophagy deficiency can lead to inflammasome activation. Accumulation of damaged mitochondria, leading to the release of reactive oxygen species (ROS) and/or mitochondrial DNA, and failure to degrade misfolded proteins have been proposed as mechanisms for inflammasome activation in autophagy-deficient cells [71, 83]. Autophagy has also been suggested to suppress inflammasome activation by directly digesting inflammasome components such as ubiquitinated ASC (Fig. 3c) [84]. Furthermore, autophagy was found to directly sequester pro-cytokines such as pro-IL-1β for digestion to reduce mature cytokine production (Fig. 3c) [85]. Mice with Atg7 deficiency in myeloid cells developed spontaneous lung inflammation that was mostly mediated by IL-18. Neutralization of IL-18, but not IL-1β or IL-17, attenuated lung inflammation in these mice. In contrast, increased mortality in response to endotoxin was caused by increased IL-1β [62]. In addition to the effect of autophagy on inflammasome-associated cytokines, several studies have suggested an effect of autophagy or autophagic proteins on cytokines that are not associated with inflammasome activation. In mice, Atg5-deficient macrophages produced more pro-inflammatory cytokine IL-1α in an inflammasome-independent way [77]. In Influenza A virus infection, excessive immune responses, including increased neutrophil and macrophage infiltration, contribute to lung injury and pathology more than the effects of viral replication [86].
Fig. 3.
Autophagy in host defense and inflammasome regulation. a PAMPs or DAMPs, recognized by pattern-recognition receptors, result in NF-κB activation. Active NK-κB promotes inflammasome components and cytokine expression. b PAMPs or DAMPs cause mitochondrial damage and the release of mitochondrial ROS and DNA, triggering the assembling of NLRP3, ASC, and Pro-caspase into active inflammasome. Caspase-1 is activated by autocleavage and then cleaves the pro-inflammatory cytokines IL-1β into active cytokines. Bacteria-containing phagosome membrane disruption leads to the release of PAMPs. c Activated autophagosomes can engulf damaged mitochondria, NLRP3, ASC, and Pro-caspase, and target them to lysosomal degradation, reducing the production and secretion of active cytokines.
Conclusion and Future Prospects
Autophagy is an important intracellular recycling system with diverse functions implicated in multiple cellular signaling pathways. Autophagy is regulated at the transcriptional, translational, and posttranslational levels. Phosphorylation and de-phosphorylation on some key proteins in the initiation complexes has been found to be a major mechanism of autophagy regulation [18]. Recent studies revealed that acetylation could modify autophagy proteins and influence the autophagy cascade [21]. Further elucidation of these regulatory mechanisms could provide potential therapeutic targets in diseases in which autophagy modulation is desired.
Since the discovery of autophagy's involvement in the innate immune response, extensive in vitro and in vivo studies have shown that autophagy plays an important role in lung homeostasis and tissue protection [68, 72, 73, 77]. During host infection, autophagy eliminates pathogens by mediating pathogen autolysosomal killing and facilitating antimicrobial antigen presentation [5, 77, 87]. In addition to pathogen elimination, autophagy tames the host inflammatory response by negative regulation of inflammasome activity. Multiple studies have shown that the induction of autophagy can have beneficial effects in combating infections, suggesting that promoting autophagy may be a beneficial strategy to control lung infection [43, 44]. However, some pathogens have evolved adaptive strategies to resist autophagy elimination, potentially limiting the impact of autophagy in immune defense. Some of these anti-autophagy strategies employed by pathogens include anti-autophagy factors such as SapM, ManLAM [54, 88], Eis [55], PE_PGRS47 [58], and ESX-1 from M. tuberculosis [57], RavZ and LpSpl from L. pneumophila [64, 65], and M2 ion-channel protein from Influenza A [67] (Fig. 2). These virulence factors contribute to the drug resistance of those pathogens by enhancing pathogen survival in spite of host autophagy. Thus, developing drugs that inactivate pathogen virulence factors involved in autophagy avoidance may represent the next generation of anti-microbial agents.
Statement of Ethics
The authors have no ethical conflicts to disclose.
Disclosure Statement
The authors have no conflicts of interest to declare.
Acknowledgments
We thank Anindita Ravindran, Elmoataz Abdel Fattah, and Li-Yuan Yu-Lee for critical review of the manuscript.
References
- 1.Harnett MM, Pineda MA, Latré de Laté P, Eason RJ, Besteiro S, Harnett W, et al. From Christian de Duve to Yoshinori Ohsumi: more to autophagy than just dining at home. Biomed J. 2017 Feb;40((1)):9–22. doi: 10.1016/j.bj.2016.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Klionsky DJ, Emr SD. Autophagy as a regulated pathway of cellular degradation. Science. 2000 Dec;290((5497)):1717–21. doi: 10.1126/science.290.5497.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011 Nov;147((4)):728–41. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 4.Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell. 2004 Dec;119((6)):753–66. doi: 10.1016/j.cell.2004.11.038. [DOI] [PubMed] [Google Scholar]
- 5.Jagannath C, Lindsey DR, Dhandayuthapani S, Xu Y, Hunter RL, Jr, Eissa NT. Autophagy enhances the efficacy of BCG vaccine by increasing peptide presentation in mouse dendritic cells. Nat Med. 2009 Mar;15((3)):267–76. doi: 10.1038/nm.1928. [DOI] [PubMed] [Google Scholar]
- 6.Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007 Oct;9((10)):1102–9. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- 7.Matsunaga K, Saitoh T, Tabata K, Omori H, Satoh T, Kurotori N, et al. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat Cell Biol. 2009 Apr;11((4)):385–96. doi: 10.1038/ncb1846. [DOI] [PubMed] [Google Scholar]
- 8.He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43((1)):67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feng Y, He D, Yao Z, Klionsky DJ. The machinery of macroautophagy. Cell Res. 2014 Jan;24((1)):24–41. doi: 10.1038/cr.2013.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mizushima N, Noda T, Yoshimori T, Tanaka Y, Ishii T, George MD, et al. A protein conjugation system essential for autophagy. Nature. 1998 Sep;395((6700)):395–8. doi: 10.1038/26506. [DOI] [PubMed] [Google Scholar]
- 11.Ichimura Y, Kirisako T, Takao T, Satomi Y, Shimonishi Y, Ishihara N, et al. A ubiquitin-like system mediates protein lipidation. Nature. 2000 Nov;408((6811)):488–92. doi: 10.1038/35044114. [DOI] [PubMed] [Google Scholar]
- 12.Cheong H, Lindsten T, Wu J, Lu C, Thompson CB. Ammonia-induced autophagy is independent of ULK1/ULK2 kinases. Proc Natl Acad Sci USA. 2011 Jul;108((27)):11121–6. doi: 10.1073/pnas.1107969108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu JH, Horbinski C, Guo F, Watkins S, Uchiyama Y, Chu CT. Regulation of autophagy by extracellular signal-regulated protein kinases during 1-methyl-4-phenylpyridinium-induced cell death. Am J Pathol. 2007 Jan;170((1)):75–86. doi: 10.2353/ajpath.2007.060524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nishida Y, Arakawa S, Fujitani K, Yamaguchi H, Mizuta T, Kanaseki T, et al. Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature. 2009 Oct;461((7264)):654–8. doi: 10.1038/nature08455. [DOI] [PubMed] [Google Scholar]
- 15.Chang TK, Shravage BV, Hayes SD, Powers CM, Simin RT, Wade Harper J, et al. Uba1 functions in Atg7- and Atg3-independent autophagy. Nat Cell Biol. 2013 Sep;15((9)):1067–78. doi: 10.1038/ncb2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Codogno P, Mehrpour M, Proikas-Cezanne T. Canonical and non-canonical autophagy: variations on a common theme of self-eating? Nat Rev Mol Cell Biol. 2011 Dec;13((1)):7–12. doi: 10.1038/nrm3249. [DOI] [PubMed] [Google Scholar]
- 17.Martinez J, Malireddi RK, Lu Q, Cunha LD, Pelletier S, Gingras S, et al. Molecular characterization of LC3-associated phagocytosis reveals distinct roles for Rubicon, NOX2 and autophagy proteins. Nat Cell Biol. 2015 Jul;17((7)):893–906. doi: 10.1038/ncb3192. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Yang Z, Klionsky DJ. Mammalian autophagy: core molecular machinery and signaling regulation. Curr Opin Cell Biol. 2010 Apr;22((2)):124–31. doi: 10.1016/j.ceb.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Su H, Yang F, Wang Q, Shen Q, Huang J, Peng C, Zhang Y, Wan W, Wong CCL, Sun Q, Wang F, Zhou T, Liu W. VPS34 Acetylation Controls Its Lipid Kinase Activity and the Initiation of Canonical and Non-canonical Autophagy. Molecular cell. 2017;67:907–921. doi: 10.1016/j.molcel.2017.07.024. e907. [DOI] [PubMed] [Google Scholar]
- 20.Huang R, Xu Y, Wan W, Shou X, Qian J, You Z, et al. Deacetylation of nuclear LC3 drives autophagy initiation under starvation. Mol Cell. 2015 Feb;57((3)):456–66. doi: 10.1016/j.molcel.2014.12.013. [DOI] [PubMed] [Google Scholar]
- 21.Lee IH, Cao L, Mostoslavsky R, Lombard DB, Liu J, Bruns NE, et al. A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proc Natl Acad Sci USA. 2008 Mar;105((9)):3374–9. doi: 10.1073/pnas.0712145105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun T, Li X, Zhang P, Chen WD, Zhang HL, Li DD, et al. Acetylation of Beclin 1 inhibits autophagosome maturation and promotes tumour growth. Nat Commun. 2015 May;6((1)):7215. doi: 10.1038/ncomms8215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao J, Brault JJ, Schild A, Cao P, Sandri M, Schiaffino S, et al. FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab. 2007 Dec;6((6)):472–83. doi: 10.1016/j.cmet.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 24.Mammucari C, Milan G, Romanello V, Masiero E, Rudolf R, Del Piccolo P, et al. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab. 2007 Dec;6((6)):458–71. doi: 10.1016/j.cmet.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 25.Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, Erdin S, et al. TFEB links autophagy to lysosomal biogenesis. Science. 2011 Jun;332((6036)):1429–33. doi: 10.1126/science.1204592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sardiello M, Palmieri M, di Ronza A, Medina DL, Valenza M, Gennarino VA, et al. A gene network regulating lysosomal biogenesis and function. Science. 2009 Jul;325((5939)):473–7. doi: 10.1126/science.1174447. [DOI] [PubMed] [Google Scholar]
- 27.Martina JA, Chen Y, Gucek M, Puertollano R. MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy. 2012 Jun;8((6)):903–14. doi: 10.4161/auto.19653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Settembre C, Zoncu R, Medina DL, Vetrini F, Erdin S, Erdin S, et al. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J. 2012 Mar;31((5)):1095–108. doi: 10.1038/emboj.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sha Y, Rao L, Settembre C, Ballabio A, Eissa NT. STUB1 regulates TFEB-induced autophagy-lysosome pathway. EMBO J. 2017 Sep;36((17)):2544–52. doi: 10.15252/embj.201796699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rao L, Sha Y, Eissa NT. The E3 ubiquitin ligase STUB1 regulates autophagy and mitochondrial biogenesis by modulating TFEB activity. Mol Cell Oncol. 2017 Sep;4((6)):e1372867. doi: 10.1080/23723556.2017.1372867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang L, Hamilton SR, Sood A, Kuwai T, Ellis L, Sanguino A, et al. The previously undescribed ZKSCAN3 (ZNF306) is a novel “driver” of colorectal cancer progression. Cancer Res. 2008 Jun;68((11)):4321–30. doi: 10.1158/0008-5472.CAN-08-0407. [DOI] [PubMed] [Google Scholar]
- 32.Chauhan S, Goodwin JG, Chauhan S, Manyam G, Wang J, Kamat AM, et al. ZKSCAN3 is a master transcriptional repressor of autophagy. Mol Cell. 2013 Apr;50((1)):16–28. doi: 10.1016/j.molcel.2013.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakagawa I, Amano A, Mizushima N, Yamamoto A, Yamaguchi H, Kamimoto T, et al. Autophagy defends cells against invading group A Streptococcus. Science. 2004 Nov;306((5698)):1037–40. doi: 10.1126/science.1103966. [DOI] [PubMed] [Google Scholar]
- 34.Jordao L, Vieira OV. Tuberculosis: new aspects of an old disease. Int J Cell Biol. 2011;2011:403623. doi: 10.1155/2011/403623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Armstrong JA, Hart PD. Response of cultured macrophages to Mycobacterium tuberculosis, with observations on fusion of lysosomes with phagosomes. J Exp Med. 1971 Sep;134((3 Pt 1)):713–40. doi: 10.1084/jem.134.3.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sturgill-Koszycki S, Schlesinger PH, Chakraborty P, Haddix PL, Collins HL, Fok AK, et al. Lack of acidification in Mycobacterium phagosomes produced by exclusion of the vesicular proton-ATPase. Science. 1994 Feb;263((5147)):678–81. doi: 10.1126/science.8303277. [DOI] [PubMed] [Google Scholar]
- 37.Wong D, Bach H, Sun J, Hmama Z, Av-Gay Y. Mycobacterium tuberculosis protein tyrosine phosphatase (PtpA) excludes host vacuolar-H+-ATPase to inhibit phagosome acidification. Proc Natl Acad Sci USA. 2011 Nov;108((48)):19371–6. doi: 10.1073/pnas.1109201108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fratti RA, Chua J, Vergne I, Deretic V. Mycobacterium tuberculosis glycosylated phosphatidylinositol causes phagosome maturation arrest. Proc Natl Acad Sci USA. 2003 Apr;100((9)):5437–42. doi: 10.1073/pnas.0737613100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vergne I, Chua J, Lee HH, Lucas M, Belisle J, Deretic V. Mechanism of phagolysosome biogenesis block by viable Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 2005 Mar;102((11)):4033–8. doi: 10.1073/pnas.0409716102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gan H, Lee J, Ren F, Chen M, Kornfeld H, Remold HG. Mycobacterium tuberculosis blocks crosslinking of annexin-1 and apoptotic envelope formation on infected macrophages to maintain virulence. Nat Immunol. 2008 Oct;9((10)):1189–97. doi: 10.1038/ni.1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Park JS, Tamayo MH, Gonzalez-Juarrero M, Orme IM, Ordway DJ. Virulent clinical isolates of Mycobacterium tuberculosis grow rapidly and induce cellular necrosis but minimal apoptosis in murine macrophages. J Leukoc Biol. 2006 Jan;79((1)):80–6. doi: 10.1189/jlb.0505250. [DOI] [PubMed] [Google Scholar]
- 42.Lam KK, Zheng X, Forestieri R, Balgi AD, Nodwell M, Vollett S, et al. Nitazoxanide stimulates autophagy and inhibits mTORC1 signaling and intracellular proliferation of Mycobacterium tuberculosis. PLoS Pathog. 2012;8((5)):e1002691. doi: 10.1371/journal.ppat.1002691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Floto RA, Sarkar S, Perlstein EO, Kampmann B, Schreiber SL, Rubinsztein DC. Small molecule enhancers of rapamycin-induced TOR inhibition promote autophagy, reduce toxicity in Huntington's disease models and enhance killing of mycobacteria by macrophages. Autophagy. 2007 Nov-Dec;3((6)):620–2. doi: 10.4161/auto.4898. [DOI] [PubMed] [Google Scholar]
- 44.Xu Y, Fattah EA, Liu XD, Jagannath C, Eissa NT. Harnessing of TLR-mediated autophagy to combat mycobacteria in macrophages. Tuberculosis (Edinb) 2013 Dec;93(Suppl):S33–7. doi: 10.1016/S1472-9792(13)70008-8. [DOI] [PubMed] [Google Scholar]
- 45.Singh SB, Davis AS, Taylor GA, Deretic V. Human IRGM induces autophagy to eliminate intracellular mycobacteria. Science. 2006 Sep;313((5792)):1438–41. doi: 10.1126/science.1129577. [DOI] [PubMed] [Google Scholar]
- 46.Biswas D, Qureshi OS, Lee WY, Croudace JE, Mura M, Lammas DA. ATP-induced autophagy is associated with rapid killing of intracellular mycobacteria within human monocytes/macrophages. BMC Immunol. 2008 Jul;9((1)):35. doi: 10.1186/1471-2172-9-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu Y, Jagannath C, Liu XD, Sharafkhaneh A, Kolodziejska KE, Eissa NT. Toll-like receptor 4 is a sensor for autophagy associated with innate immunity. Immunity. 2007 Jul;27((1)):135–44. doi: 10.1016/j.immuni.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martineau AR, Timms PM, Bothamley GH, Hanifa Y, Islam K, Claxton AP, et al. High-dose vitamin D(3) during intensive-phase antimicrobial treatment of pulmonary tuberculosis: a double-blind randomised controlled trial. Lancet. 2011 Jan;377((9761)):242–50. doi: 10.1016/S0140-6736(10)61889-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu PT, Stenger S, Li H, Wenzel L, Tan BH, Krutzik SR, et al. Toll-like receptor triggering of a vitamin D-mediated human antimicrobial response. Science. 2006 Mar;311((5768)):1770–3. doi: 10.1126/science.1123933. [DOI] [PubMed] [Google Scholar]
- 50.Liu PT, Stenger S, Tang DH, Modlin RL. Cutting edge: vitamin D-mediated human antimicrobial activity against Mycobacterium tuberculosis is dependent on the induction of cathelicidin. J Immunol. 2007 Aug;179((4)):2060–3. doi: 10.4049/jimmunol.179.4.2060. [DOI] [PubMed] [Google Scholar]
- 51.Kim JJ, Lee HM, Shin DM, Kim W, Yuk JM, Jin HS, et al. Host cell autophagy activated by antibiotics is required for their effective antimycobacterial drug action. Cell Host Microbe. 2012 May;11((5)):457–68. doi: 10.1016/j.chom.2012.03.008. [DOI] [PubMed] [Google Scholar]
- 52.Bonilla DL, Bhattacharya A, Sha Y, Xu Y, Xiang Q, Kan A, et al. Autophagy regulates phagocytosis by modulating the expression of scavenger receptors. Immunity. 2013 Sep;39((3)):537–47. doi: 10.1016/j.immuni.2013.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hu D, Wu J, Wang W, Mu M, Zhao R, Xu X, et al. Autophagy regulation revealed by SapM-induced block of autophagosome-lysosome fusion via binding RAB7. Biochem Biophys Res Commun. 2015 May;461((2)):401–7. doi: 10.1016/j.bbrc.2015.04.051. [DOI] [PubMed] [Google Scholar]
- 54.Shui W, Petzold CJ, Redding A, Liu J, Pitcher A, Sheu L, et al. Organelle membrane proteomics reveals differential influence of mycobacterial lipoglycans on macrophage phagosome maturation and autophagosome accumulation. J Proteome Res. 2011 Jan;10((1)):339–48. doi: 10.1021/pr100688h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim KH, An DR, Song J, Yoon JY, Kim HS, Yoon HJ, et al. Mycobacterium tuberculosis Eis protein initiates suppression of host immune responses by acetylation of DUSP16/MKP-7. Proc Natl Acad Sci USA. 2012 May;109((20)):7729–34. doi: 10.1073/pnas.1120251109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Watson RO, Manzanillo PS, Cox JS. Extracellular M. tuberculosis DNA targets bacteria for autophagy by activating the host DNA-sensing pathway. Cell. 2012 Aug;150((4)):803–15. doi: 10.1016/j.cell.2012.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Romagnoli A, Etna MP, Giacomini E, Pardini M, Remoli ME, Corazzari M, et al. ESX-1 dependent impairment of autophagic flux by Mycobacterium tuberculosis in human dendritic cells. Autophagy. 2012 Sep;8((9)):1357–70. doi: 10.4161/auto.20881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Saini NK, Baena A, Ng TW, Venkataswamy MM, Kennedy SC, Kunnath-Velayudhan S, et al. Suppression of autophagy and antigen presentation by Mycobacterium tuberculosis PE_PGRS47. Nat Microbiol. 2016 Aug;1((9)):16133. doi: 10.1038/nmicrobiol.2016.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Joshi AD, Swanson MS. Secrets of a successful pathogen: legionella resistance to progression along the autophagic pathway. Front Microbiol. 2011 Jun;2:138. doi: 10.3389/fmicb.2011.00138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Swanson MS, Isberg RR. Association of Legionella pneumophila with the macrophage endoplasmic reticulum. Infect Immun. 1995 Sep;63((9)):3609–20. doi: 10.1128/iai.63.9.3609-3620.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Amer AO, Swanson MS. Autophagy is an immediate macrophage response to Legionella pneumophila. Cell Microbiol. 2005 Jun;7((6)):765–78. doi: 10.1111/j.1462-5822.2005.00509.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Matsuda F, Fujii J, Yoshida S. Autophagy induced by 2-deoxy-D-glucose suppresses intracellular multiplication of Legionella pneumophila in A/J mouse macrophages. Autophagy. 2009 May;5((4)):484–93. doi: 10.4161/auto.5.4.7760. [DOI] [PubMed] [Google Scholar]
- 63.Tung SM, Unal C, Ley A, Peña C, Tunggal B, Noegel AA, et al. Loss of Dictyostelium ATG9 results in a pleiotropic phenotype affecting growth, development, phagocytosis and clearance and replication of Legionella pneumophila. Cell Microbiol. 2010 Jun;12((6)):765–80. doi: 10.1111/j.1462-5822.2010.01432.x. [DOI] [PubMed] [Google Scholar]
- 64.Choy A, Dancourt J, Mugo B, O'Connor TJ, Isberg RR, Melia TJ, et al. The Legionella effector RavZ inhibits host autophagy through irreversible Atg8 deconjugation. Science. 2012 Nov;338((6110)):1072–6. doi: 10.1126/science.1227026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rolando M, Escoll P, Nora T, Botti J, Boitez V, Bedia C, et al. Legionella pneumophila S1P-lyase targets host sphingolipid metabolism and restrains autophagy. Proc Natl Acad Sci USA. 2016 Feb;113((7)):1901–6. doi: 10.1073/pnas.1522067113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Iwasaki A, Pillai PS. Innate immunity to influenza virus infection. Nat Rev Immunol. 2014 May;14((5)):315–28. doi: 10.1038/nri3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gannagé M, Dormann D, Albrecht R, Dengjel J, Torossi T, Rämer PC, et al. Matrix protein 2 of influenza A virus blocks autophagosome fusion with lysosomes. Cell Host Microbe. 2009 Oct;6((4)):367–80. doi: 10.1016/j.chom.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Junkins RD, Shen A, Rosen K, McCormick C, Lin TJ. Autophagy enhances bacterial clearance during P. aeruginosa lung infection. PLoS One. 2013 Aug;8((8)):e72263. doi: 10.1371/journal.pone.0072263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Abdulrahman BA, Khweek AA, Akhter A, Caution K, Kotrange S, Abdelaziz DH, et al. Autophagy stimulation by rapamycin suppresses lung inflammation and infection by Burkholderia cenocepacia in a model of cystic fibrosis. Autophagy. 2011 Nov;7((11)):1359–70. doi: 10.4161/auto.7.11.17660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Saitoh T, Fujita N, Jang MH, Uematsu S, Yang BG, Satoh T, et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature. 2008 Nov;456((7219)):264–8. doi: 10.1038/nature07383. [DOI] [PubMed] [Google Scholar]
- 71.Nakahira K, Haspel JA, Rathinam VA, Lee SJ, Dolinay T, Lam HC, et al. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat Immunol. 2011 Mar;12((3)):222–30. doi: 10.1038/ni.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Abdel Fattah E, Bhattacharya A, Herron A, Safdar Z, Eissa NT. Critical role for IL-18 in spontaneous lung inflammation caused by autophagy deficiency. J Immunol. 2015 Jun;194((11)):5407–16. doi: 10.4049/jimmunol.1402277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kanayama M, He YW, Shinohara ML. The lung is protected from spontaneous inflammation by autophagy in myeloid cells. J Immunol. 2015 Jun;194((11)):5465–71. doi: 10.4049/jimmunol.1403249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jessop F, Hamilton RF, Rhoderick JF, Shaw PK, Holian A. Autophagy deficiency in macrophages enhances NLRP3 inflammasome activity and chronic lung disease following silica exposure. Toxicol Appl Pharmacol. 2016 Oct;309:101–10. doi: 10.1016/j.taap.2016.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Suzuki Y, Maazi H, Sankaranarayanan I, Lam J, Khoo B, Soroosh P, Barbers RG, James Ou JH, Jung JU, Akbari O. Lack of autophagy induces steroid-resistant airway inflammation. The Journal of allergy and clinical immunology. 2016;137:1382–1389. doi: 10.1016/j.jaci.2015.09.033. e1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lu Q, Yokoyama CC, Williams JW, Baldridge MT, Jin X, DesRochers B, et al. Homeostatic Control of Innate Lung Inflammation by Vici Syndrome Gene Epg5 and Additional Autophagy Genes Promotes Influenza Pathogenesis. Cell Host Microbe. 2016 Jan;19((1)):102–13. doi: 10.1016/j.chom.2015.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Castillo EF, Dekonenko A, Arko-Mensah J, Mandell MA, Dupont N, Jiang S, et al. Autophagy protects against active tuberculosis by suppressing bacterial burden and inflammation. Proc Natl Acad Sci USA. 2012 Nov;109((46)):E3168–76. doi: 10.1073/pnas.1210500109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kimmey JM, Huynh JP, Weiss LA, Park S, Kambal A, Debnath J, et al. Unique role for ATG5 in neutrophil-mediated immunopathology during M. tuberculosis infection. Nature. 2015 Dec;528((7583)):565–9. doi: 10.1038/nature16451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Saitoh T, Akira S. Regulation of inflammasomes by autophagy. J Allergy Clin Immunol. 2016 Jul;138((1)):28–36. doi: 10.1016/j.jaci.2016.05.009. [DOI] [PubMed] [Google Scholar]
- 80.Zhong Z, Liang S, Sanchez-Lopez E, He F, Shalapour S, Lin XJ, et al. New mitochondrial DNA synthesis enables NLRP3 inflammasome activation. Nature. 2018 Aug;560((7717)):198–203. doi: 10.1038/s41586-018-0372-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhou R, Yazdi AS, Menu P, Tschopp J. A role for mitochondria in NLRP3 inflammasome activation. Nature. 2011 Jan;469((7329)):221–5. doi: 10.1038/nature09663. [DOI] [PubMed] [Google Scholar]
- 82.Schroder K, Tschopp J. The inflammasomes. Cell. 2010 Mar;140((6)):821–32. doi: 10.1016/j.cell.2010.01.040. [DOI] [PubMed] [Google Scholar]
- 83.Zhong Z, Umemura A, Sanchez-Lopez E, Liang S, Shalapour S, Wong J, et al. NF-κB Restricts Inflammasome Activation via Elimination of Damaged Mitochondria. Cell. 2016 Feb;164((5)):896–910. doi: 10.1016/j.cell.2015.12.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shi CS, Shenderov K, Huang NN, Kabat J, Abu-Asab M, Fitzgerald KA, et al. Activation of autophagy by inflammatory signals limits IL-1β production by targeting ubiquitinated inflammasomes for destruction. Nat Immunol. 2012 Jan;13((3)):255–63. doi: 10.1038/ni.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Harris J, Hartman M, Roche C, Zeng SG, O'Shea A, Sharp FA, et al. Autophagy controls IL-1beta secretion by targeting pro-IL-1beta for degradation. J Biol Chem. 2011 Mar;286((11)):9587–97. doi: 10.1074/jbc.M110.202911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lupfer C, Thomas PG, Anand PK, Vogel P, Milasta S, Martinez J, et al. Receptor interacting protein kinase 2-mediated mitophagy regulates inflammasome activation during virus infection. Nat Immunol. 2013 May;14((5)):480–8. doi: 10.1038/ni.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Xu Y, Eissa NT. Autophagy in innate and adaptive immunity. Proc Am Thorac Soc. 2010 Feb;7((1)):22–8. doi: 10.1513/pats.200909-103JS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Vergne I, Chua J, Deretic V. Tuberculosis toxin blocking phagosome maturation inhibits a novel Ca2+/calmodulin-PI3K hVPS34 cascade. J Exp Med. 2003 Aug;198((4)):653–9. doi: 10.1084/jem.20030527. [DOI] [PMC free article] [PubMed] [Google Scholar]