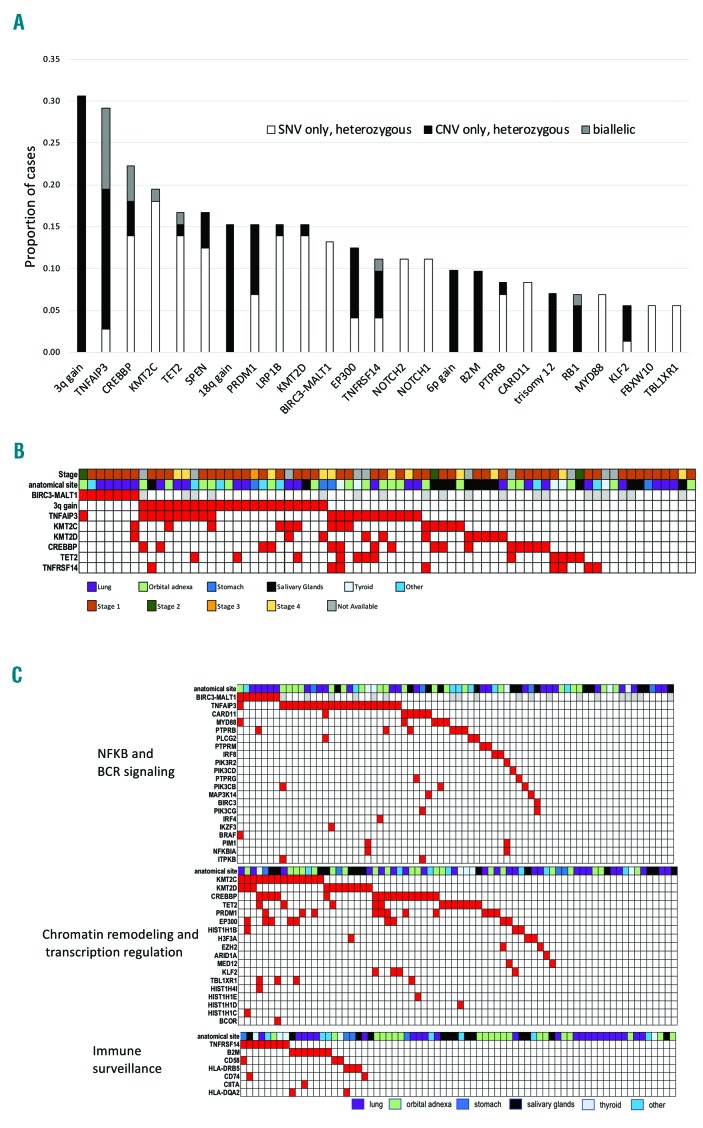
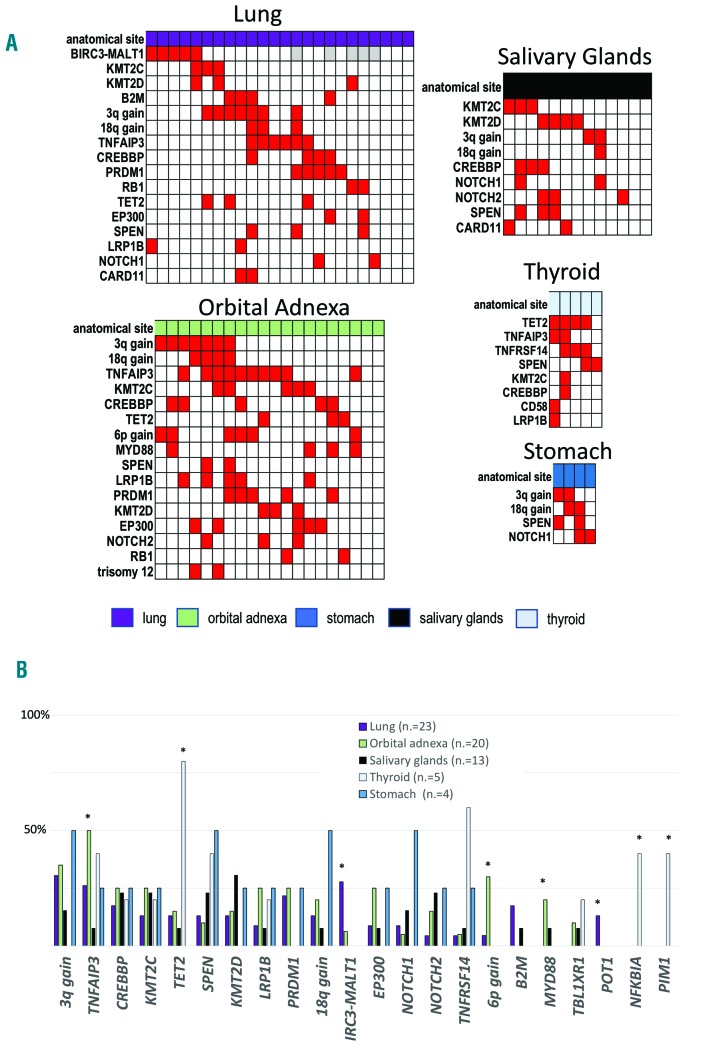
Mucosa-associated lymphoid tissue (MALT) lymphoma is the most common type of marginal zone lymphoma (MZL) representing 5-8% of all B-cell lymphomas and occurs at diverse anatomic sites with site-specific clinical and biological differences.1 Unique among the three MZL (extranodal MZL of MALT type, splenic MZL, and nodal MZL), MALT lymphomas have recurrent chromosomal translocations with the BIRC3-MALT1 being the most frequent event observed in 15-40% of cases, especially those occurring in the lung and stomach.1 We analyzed 72 cases of MALT lymphomas derived from different anatomic sites (lung 32%, ocular adnexa 28%, salivary glands 18%, thyroid 7%, stomach 5%, and other 10%) for DNA copy number variations (CNV) and for the mutational status of a large panel of genes previously reported to be mutated in B-cell lymphomas (Online Supplementary Methods). In addition, for a subset of cases, transcriptional (n=21, 29%) and methylation (n=19, 26%) profiling was evaluated. DNA sequencing achieved at least 100x coverage in > 90% of the target regions (Online Supplementary Table S1 and Online Supplementary Figure S1) with a mean coverage of ~700x. From this analysis, we identified an average of three mutated genes per patient (range: 0-11). Seventy patients (97%) had at least one event (mutations or genomic alteration) and mutations were detected in 90 genes in at least one case (Online Supplementary Table S2). The most frequent recurring lesions were: trisomy 3 (31%), mutation and/or deletion of TNFAIP3 (29%), CREBBP (22%), KMT2C (19%), TET2 and SPEN (17%, each), trisomy 18, mutation and/or deletion of KMT2D, LRP1B and PRDM1 (15%, each), BIRC3-MALT1 translocation and mutation and/or deletion of EP300 (13%, each), mutation and/or deletion of TNFRSF14 and mutations in NOTCH1/NOTCH2 (11%, each), mutation and/or deletion of B2M and 6p gains (10%, each) (Figure 1A and Online Supplementary Figure S2). The presence of trisomy 3, the BIRC3-MALT1 fusion or the inactivation of TNFAIP3, KMT2C, KMT2D, CREBBP, TET2 or TNFRSF14 genes accounted for the genomic changes in 61 of 72 (85%) cases (Figure 1B). There was no association between mutational status and clinical stage. The genes affected in >5% of cases mainly coded for proteins involved in chromatin remodeling and transcription regulation, BCR/NF-κB pathway, immune escape (Figure 1C) and the NOTCH pathway, in agreement with other reports.2–4 Similar to what has been reported in follicular lymphoma and in diffuse large B-cell lymphoma (DLBCL),5 mutations in KMT2C, KMT2D CREBBP and EP300 tended to co-occur, while this was not the case for TET2 mutations. Genetic lesions in MALT lymphomas largely overlapped those reported for SMZL and NMZL6–8 but differed, besides the presence of the BIRC3-MALT1, with a higher frequency of inactivation of TNFAIP3 (29% vs. 14% in SMZL and 9% in NMZL) and CREBBP (22% vs. 8% in SMZL), a lower percentage of mutated cases (P<0.05) for KMT2D (15% vs. 34% in NMZL), KLF2 (6% vs. 21% in SMZL) and PTPRD (3% vs. 14% in NMZL) (Online Supplementary Table S3 and Online Supplementary Figure S3). With the limitation of a distribution of anatomical sites not fully representative of the disease due to the requirement of frozen material, we looked at the occurrence of lesions in the main localizations (Figure 2 and Online Supplementary Table S4). Gains affecting chromosomes 3 and 18, inactivation of chromatin remodeling genes (KMT2C, KMT2D CREBBP, EP300), and mutations of NOTCH1/2 were equally distributed among the different sites. Ocular adnexal cases had the highest frequency of 6p gains (30% vs. 2%; P=0.001, false discovery rate (FDR)=0.07), mutations of MYD88 (20% vs. 2%; P=0.019, FDR=0.33) and TNFAIP3 (50% vs. 21%; P=0.021, FDR=0.33). Thyroid MALT lymphomas had a higher frequency of lesions in TNFRSF14 (60% vs. 7%; P=0.008, FDR=0.04), NFKBIA and PIM1 (40%, each vs. 0%; P=0.004, FDR=0.02) and, particularly, TET2 (80% vs. 12%; P=0.002, FDR=0.02). Lung cases had a higher frequency of POT1 mutations (13% vs. 0%; P=0.03, FDR=0.69) and of BIRC3-MALT1 translocations (27% vs. 6%; P=0.04, FDR=0.69).
Figure 1.
Genetic landscape of mucosal-associated lymphoid tissue (MALT) lymphomas. (A) Frequency of genetic events occurring in at least 5% of cases. Although PRDM1 appeared among the most frequently inactivated genes, most of the deletions also comprised TNFAIP3, making it difficult to ascertain the exact role of this lesion in MALT lymphomas. (B) Heatmap of the genetic landscape of MALT lymphomas. (C) Heatmap of pathways most commonly affected by genetic events in MALT lymphomas. (B and C) Each column represents one case. Red: present; White: absent; Gray: not available.
Figure 2.
Genetic lesions at different anatomic sites. (A) Heatmap of lesions by anatomical sites. Only lesions occurring in at least two cases are shown. Each column represents one case. Red: present; White: absent; Gray: not available. (B) Distribution by anatomical sites for the lesions occurring in >10% of cases from the whole mucosa-associated lymphoid tissue lymphoma series or showing a preferential distribution to one specific site. *P<0.05.
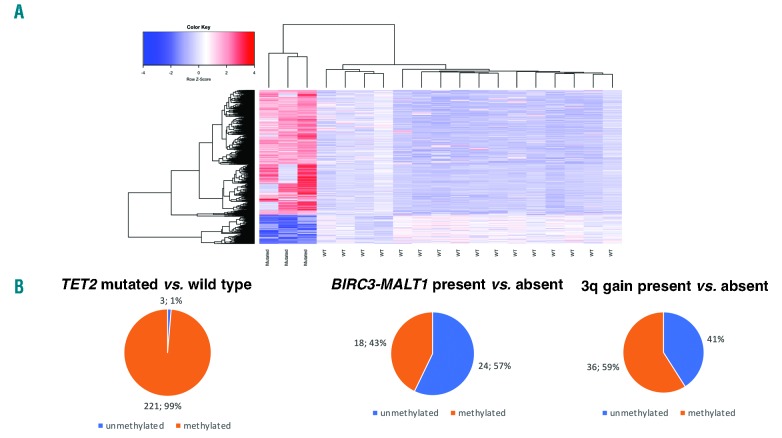
While the pattern of mutations of TET2 and TNFRSF14 was similar to that found in a recent independent series,2 other findings in our study differed, possibly due to differences in the panel design (e.g. GPR4 and CCR62 were not sequenced by us while the opposite was true for PIM1, NFKBIA, and POT1), sample size, and technical aspects (target enrichment modality, next generation sequencing platform, data mining). Since TET2 mutations are associated with changes in the tumor methylation landscape of DLBCL,9 we compared the genome-wide DNA methylation profiles of MALT lymphomas with TET2 mutations (n=3, mutations were validated by Sanger sequencing in 3 of 3 cases tested) versus cases with a wild-type gene (n=16). The presence of TET2 mutations was associated with increased DNA methylation (Figure 3A and Online Supplementary Table S5A). In particular, almost all the differentially methylated probes interrogating promoter regions were more frequently fully or partially methylated in mutated than in wild-type cases (Figure 3B and Online Supplementary Table S5B). The genes with hypermethylated promoters in TET2 mutated cases were mostly enriched for the PRC2-complex (EZH2, EED and SUZ12) targets, genes bearing histone H3 dimethylation at K4 (H3K4me2) and trimethylation at K27 (H3K27me3) in the brain (FDR < 0.001) (Online Supplementary Table S5D). Conversely, other recurrent events such as BIRC3-MALT1 translocation or trisomy 3 showed similar percentages of differentially hyper- or hypomethylated probes between cases with or without the lesions (Figure 3B and Online Supplementary Table S5C). We then compared the transcriptome of cases with (n=3) versus those without (n=16) the BIRC3-MALT1 fusion. We limited the analysis to lung MALT lymphomas due to the enrichment of the translocation in MZL derived from this anatomic site. As a positive control, we first assessed whether signatures previously reported for translocated cases10,11 were enriched in our series of MALT lymphomas bearing the t(11:18), and this was the case (Online Supplementary Table S6 and Online Supplementary Figure S4). Transcripts expressed in cases with the translocation were also enriched in MYC target genes, genes involved in the cell cycle, oxidative phosphorylation and in DNA repair, as well as in DLBCL driver genes (such as MALT1, IRF4, CD89A, BCL2, PI3KCD, BTK, CARD11 and SPIB), and transcripts down-regulated in lymphomas by epigenetic drugs, such as inhibitors of histone deacetylases (HDAC) or inhibitors of the bromodomain and extra-terminal (BET) family of bromodomain-containing proteins (Online Supplementary Table S6 and Online Supplementary Figure S5). Among the genes most up-regulated in this subgroup of MALT lymphomas were members of the TNF-receptor superfamily, TNFSF12-TNFSF13 (TWE-PRIL), TNFRSF17 (BCMA), LTB (lymphotoxin β), and CXCR3. Trisomy 3 was mutually exclusive with BIRC3-MALT1 in MALT lymphomas, but cases with this lesion showed overlapping gene expression signatures with cases harboring the fusion (Online Supplementary Table S4 and Online Supplementary Figure S6). This observation suggests that the two recurrent lesions might have a similar biological role. Ten individual transcripts were significantly up-regulated in cases with the BIRC3-MALT1 fusion or trisomy 3: CCZ1B, CDHR1, EIF3CL, LTB, NLRP2, PARP14, SASH3, SLFN14, TBX21 (coding for the transcription factor T-BET), and TIGIT. TBX21 (T-BET) is known to be expressed in B cells, especially memory B cells, including CD11c+ and CXCR3+ subsets, which have roles in antimicrobial immunity, autoimmunity and aging.12 A variable proportion of MZL, hairy cell leukemia and chronic lymphocytic leukemia cases express T-BET.13,14 Since T-BET expression has been associated with monocytoid morphology in MZL,13,14 we evaluated the cytomorphological features of a subset of MALT lymphomas bearing the fusion or trisomy 3 (n=12) or none of the two lesions (n=7). Monocytoid morphology was observed in all cases bearing one of the two genetic lesions (100%) and in 4 of 7 (57%) without lesions while extensive plasmacytic differentiation was seen only in the absence of trisomy 3 or fusion (Fisher’s exact test, P=0.067). No enrichment of specific gene sets was observed for cases with inactivation of TNFAIP3 or of KMT2C/KMT2D. Besides the frequent recurrent mutations in chromatin remodeling genes in MALT lymphomas, some observations provide a rationale for epigenetic therapy in MZL. The presence of TET2 mutations is associated with a higher response to demethylating agents in acute myeloid leukemia,15 and since demethylating agents can revert the methylator phenotype, these agents might be explored in patients with TET2 mutated MALT lymphomas. As the gene expression signatures associated with the presence of the BIRC3-MALT fusion and with trisomy 3 appeared negatively correlated with the transcriptome changes induced by other epigenetic agents, there might be the potential to study BET inhibitors and HDAC inhibitors in these patients.
Figure 3.
TET2 mutations in mucosa-associated lymphoid tissue lymphomas. (A) DNA methylation heatmap constructed using supervised hierarchical Ward clustering of the 2,208 differentially methylated CpG probes between TET2 mutated and TET2 wild-type marginal zone lymphoma (Limma moderated t-test, false discovery rate <0.05) (Online Supplementary Table S3A). (B) Distribution of probes interrogating promoter regions discriminating fully/partially methylated versus unmethylated states (Fisher’s test, P<0.05) in cases bearing or not harboring a specific genetic lesion.
In conclusion, MALT lymphomas are characterized by frequent lesions affecting chromatin remodeling, BCR/NF-κB and NOTCH pathways. Lymphomas from different anatomic sites exhibit a different spectrum of genetic lesions. TET2 is recurrently inactivated and associated with a specific methylation signature. The observed genetic landscape, methylation and transcriptome data suggest exploration of epigenetic drugs therapy in at least a subgroup of MZL patients.
Footnotes
Funding: the authors would like to thank the financial support from the Nelia et Amadeo Barletta Foundation (Lausanne, Switzerland), the Oncosuisse (grant OCS-02034-02- 2007), the Helmut Horten Foundation (to FB), the Special Program Molecular Clinical Oncology 5 x 1000 No. 10007, Associazione Italiana per la Ricerca sul Cancro Foundation Milan, Italy (to GG), and the Cancer Center Support Grant - P30CA013696 (to GB).
Information on authorship, contributions, and financial & other disclosures was provided by the authors and is available with the online version of this article at www.haematologica.org.
References
- 1.Zucca E, Bertoni F. The spectrum of MALT lymphoma at different sites: biological and therapeutic relevance. Blood. 2016;127(17):2082–2092. [DOI] [PubMed] [Google Scholar]
- 2.Moody S, Thompson JS, Chuang SS, et al. Novel GPR34 and CCR6 mutation and distinct genetic profiles in MALT lymphomas of different sites. Haematologica. 2018;103(8):1329–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jung H, Yoo HY, Lee SH, et al. The mutational landscape of ocular marginal zone lymphoma identifies frequent alterations in TNFAIP3 followed by mutations in TBL1XR1 and CREBBP. Oncotarget. 2017;8(10):17038–17049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johansson P, Klein-Hitpass L, Grabellus F, et al. Recurrent mutations in NF-kappaB pathway components, KMT2D, and NOTCH1/2 in ocular adnexal MALT-type marginal zone lymphomas. Oncotarget. 2016;7(38):62627–62639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rossi D, Trifonov V, Fangazio M, et al. The coding genome of splenic marginal zone lymphoma: activation of NOTCH2 and other pathways regulating marginal zone development. J Exp Med. 2012;209(9):1537–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Spina V, Khiabanian H, Messina M, et al. The genetics of nodal marginal zone lymphoma. Blood. 2016;128(10):1362–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rinaldi A, Mian M, Chigrinova E, et al. Genome-wide DNA profiling of marginal zone lymphomas identifies subtype-specific lesions with an impact on the clinical outcome. Blood. 2011;117(5):1595–1604. [DOI] [PubMed] [Google Scholar]
- 8.Green MR. Chromatin modifying gene mutations in follicular lymphoma. Blood. 2018;131(6):595–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Asmar F, Punj V, Christensen J, et al. Genome-wide profiling identifies a DNA methylation signature that associates with TET2 mutations in diffuse large B-cell lymphoma. Haematologica. 2013;98(12):1912–1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chng WJ, Remstein ED, Fonseca R, et al. Gene expression profiling of pulmonary mucosa-associated lymphoid tissue (MALT) lymphoma identifies new biological insights with potential diagnostic and therapeutic applications. Blood. 2009;113(3):635–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hamoudi RA, Appert A, Ye H, et al. Differential expression of NF-kappaB target genes in MALT lymphoma with and without chromosome translocation: insights into molecular mechanism. Leukemia. 2010;24(8):1487–1497. [DOI] [PubMed] [Google Scholar]
- 12.Rubtsov AV, Rubtsova K, Fischer A, et al. Toll-like receptor 7 (TLR7)-driven accumulation of a novel CD11c(+) B-cell population is important for the development of autoimmunity. Blood. 2011;118(5):1305–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lohneis P, Wienert S, Klauschen F, Ullrich A, Anagnostopoulos I, Johrens K. Marginal zone lymphomas with monocytoid morphology express T-bet and are associated with a low number of T cells in extranodal locations. Leuk Lymphoma. 2014;55(1):143–148. [DOI] [PubMed] [Google Scholar]
- 14.Dorfman DM, Hwang ES, Shahsafaei A, Glimcher LH. T-bet, a T-cell-associated transcription factor, is expressed in a subset of B-cell lymphoproliferative disorders. Am J Clin Pathol. 2004;122(2):292–297. [DOI] [PubMed] [Google Scholar]
- 15.Falini B, Sportoletti P, Brunetti L, Martelli MP. Perspectives for therapeutic targeting of gene mutations in acute myeloid leukaemia with normal cytogenetics. Br J Haematol. 2015;170(3):305–322. [DOI] [PubMed] [Google Scholar]